ABSTRACT
Pseudomonas aeruginosa causes serious intractable infections in humans and animals. Bacteriophage (phage) therapy has been applied to treat P. aeruginosa infections, and phages belonging to the PB1-like virus genus in the Myoviridae family have been used as therapeutic phages. To achieve safer and more effective phage therapy, the use of preadapted phages is proposed. To understand in detail such phage preadaptation, the short-term antagonistic evolution of bacteria and phages should be studied. In this study, the short-term antagonistic evolution of bacteria and PB1-like phage was examined by studying phage-resistant clones of P. aeruginosa strain PAO1 and mutant PB1-like phages that had recovered their infectivity. First, phage KPP22 was isolated and characterized; it was classified as belonging to the PB1-like virus genus in the Myoviridae family. Subsequently, three KPP22-resistant PAO1 clones and three KPP22 mutant phages capable of infecting these clones were isolated in three sets of in vitro experiments. It was shown that the bacterial resistance to phage KPP22 was caused by significant decreases in phage adsorption and that the improved infectivity of KPP22 mutant phages was caused by significant increases in phage adsorption. The KPP22-resistant PAO1 clones and the KPP22 mutant phages were then analyzed genetically. All three KPP22-resistant PAO1 clones, which were deficient for the O5 antigen, had a common nonsense mutation in the wzy gene. All the KPP22 mutant phage genomes showed the same four missense mutations in the open reading frames orf060, orf065, and orf086. The information obtained in this study should be useful for further development of safe and efficient phage therapy.
IMPORTANCE Pseudomonas aeruginosa causes serious intractable infections in humans and animals; bacteriophage (phage) therapy has been utilized to treat P. aeruginosa infections, and phages that belong to the PB1-like virus genus in the family Myoviridae have been used as therapeutic phages. The preadapted phage is trained in advance through the antagonistic evolution of bacteria and phage and is proposed to be used to achieve safer and more effective phage therapy. In this study, to understand the phage preadaptation, the in vitro short-term antagonistic evolution was studied using P. aeruginosa strain PAO1 and the newly isolated PB1-like phage KPP22. Phage KPP22 was characterized, and the molecular framework regarding the phage preadaptation of KPP22 was elucidated. The importance of study of antagonistic evolution of bacteria and phage in phage therapy is discussed.
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative bacterium that causes opportunistic infections. Some P. aeruginosa strains cause intractable chronic infections because of their ability to form biofilms and their drug resistance, in addition to their virulence factors (1–3). Bacteriophage (phage) therapy is the therapeutic use of phages to treat infections caused by pathogenic bacteria. It has a long history of successful clinical use in Eastern societies (4). Phage therapy is useful as an alternative therapeutic measure to chemotherapy for treating intractable P. aeruginosa infections in humans and animals and has begun to be clinically examined in Western countries (5–8).
Among the phages infecting P. aeruginosa, those belonging to the PB1-like virus genus in the Myoviridae family have been isolated worldwide and have relatively minor genetic variation (9–11). The sizes of the heads and tails of the phages are ca. 75 nm in diameter and ca. 140 nm in length, respectively (9). The genome sizes of the phages range from 64 to 67 kbp (9, 11). Phages belonging to the PB1-like virus genus in the Myoviridae family are typically included in the conventional phage mixture used against P. aeruginosa infections (12, 13). A member of this group of viruses has exhibited therapeutic effectiveness and safety in a mouse model of P. aeruginosa keratitis (14). Thus, phages belonging to the PB1-like virus genus are considered to be one of the important groups of therapeutic phages against P. aeruginosa infections.
In the antagonistic evolution of bacteria and phages, bacteria acquire various mechanisms to become resistant to phage infection, and phages evolve to overcome such bacterial immunity (15, 16). The emergence of phage-resistant bacteria is the consequence of such antagonistic evolution of bacteria and phages. At present, the emergence of phage-resistant bacteria appears to be a minor issue for phage therapy because other phages capable of infecting phage-resistant bacteria can be isolated in advance (4, 17).
To achieve safer and more effective phage therapy, the use of a preadapted phage, which is a phage that is evolutionarily trained in advance in in vitro antagonistic evolution, has been proposed (18, 19, 20). A phage preadapted to a clinically isolated P. aeruginosa strain has been shown not only to improve therapeutic efficacy but also to reduce the chance of emergence of phage-resistant bacterial strains (20). However, although the understanding of PB1-like phage preadaptation in detail requires information regarding the molecular framework of the short-term antagonistic evolution of P. aeruginosa and PB1-like phage, such a molecular framework has not been studied genetically, to our knowledge.
In this study, we analyzed a newly isolated phage, KPP22, belonging to the PB1-like virus genus in the Myoviridae family. KPP22-resistant clones of P. aeruginosa strain PAO1 and mutant phages capable of infecting KPP22-resistant PAO1 clones were isolated in in vitro culture and studied at the genetic level.
MATERIALS AND METHODS
Bacteria, phages, culture, culture media, and reagents.
The P. aeruginosa strains used in this study, which had been isolated from the patients in Kochi, Japan, and have been described previously, are listed in Table 1 (14, 21). Phage KPP22 was isolated using strain PA33 as the host by a method described elsewhere (22). Strain PAO1, which was kindly donated by Tetsuya Matsumoto (Department of Microbiology, Tokyo Medical University), was used for phage amplification, for mutant phage isolation, and as an indicator host for phage concentration measurement, unless otherwise stated. The bacteria and phage were cultured overnight at 37°C in Luria-Bertani (LB) broth (Miller; Sigma-Aldrich, St. Louis, MO). The plaque assay was performed using the double-layer agar method, with LB-based medium containing 1.5% agar and LB-based medium containing 0.5% agar used for the lower and upper layers, respectively. All reagents were purchased from Nacalai Tesque (Kyoto, Japan), Wako Pure Chemical Industries (Osaka, Japan), or Sigma-Aldrich, unless otherwise stated.
TABLE 1.
Lytic activity of wild-type phage KPP22, KPP22 mutant phages, and phage KPP12 against P. aeruginosa strains
| P. aeruginosa straina | Lytic activity of tested phageb |
||||
|---|---|---|---|---|---|
| Wild-type phage KPP22 | Mutant phage: |
Phage KPP12c | |||
| KPP22M1 | KPP22M2 | KPP22M3 | |||
| PA1 | I | I | I | I | I |
| PA2 | I | I | I | I | I |
| PA3 | I | I | I | I | − |
| PA4 | I | I | I | I | − |
| PA5 | I | I | I | I | O |
| PA6 | I | I | I | I | I |
| PA7 | I | I | I | I | I |
| PA8 | I | I | I | I | I |
| PA9 | I | I | I | I | O |
| PA10 | I | I | I | I | I |
| PA11 | I | I | I | I | − |
| PA12 | − | − | − | − | O |
| PA13 | I | I | I | I | I |
| PA14 | I | I | I | I | − |
| PA15 | I | I | I | I | − |
| PA16 | − | O | O | O | − |
| PA17 | I | I | I | I | I |
| PA18 | I | I | I | I | I |
| PA19 | I | I | I | I | I |
| PA20 | I | I | I | I | − |
| PA21 | I | I | I | I | I |
| PA22 | − | − | − | − | O |
| PA23 | I | I | I | I | − |
| PA24 | I | I | I | I | I |
| PA25 | I | I | I | I | − |
| PA26 | I | I | I | I | I |
| PA27 | I | I | I | I | − |
| PA28 | I | I | I | I | − |
| PA29 | I | I | I | I | − |
| PA30 | I | I | I | I | I |
| PA31 | I | I | I | I | I |
| PA32 | I | I | I | I | I |
| PA33 | I | I | I | I | I |
| PA34 | I | I | I | I | I |
| PA35 | I | I | I | I | O |
| D4 | I | I | I | I | I |
| S10 | I | I | I | I | O |
| PAO1 | I | I | I | I | I |
Phage purification.
The phages were cultured with bacteria in 250 ml of culture medium. After bacterial lysis, the phage supernatant was collected by centrifugation (10,000 × g, 10 min, 4°C). Polyethylene glycol 6000 and NaCl were added to final concentrations of 10% and 0.5 M, respectively, and the mixture was left overnight at 4°C. After centrifugation (10,000 × g, 40 min, 4°C), the phage pellet was dissolved in TM buffer (10 mM Tris-HCl [pH 7.2], 5 mM MgCl2) containing 100 μg ml−1 of both DNase I and RNase A. After incubation at 37°C for 30 min, the phage suspension was purified by cesium chloride density gradient ultracentrifugation (100,000 × g, 1 h, 4°C), as described elsewhere (14). The phage band was collected and dialyzed against AAS (0.1 M ammonium acetate, 10 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, pH 7.2) for 1 h at 4°C.
Electron microscopy.
Phages, negatively stained with 2% uranyl acetate at pH 4.0, were observed with a JEM-1400Plus transmission electron microscope (JEOL Ltd., Tokyo, Japan) at 80 kV.
Phage genome sequencing.
The phage genomic DNA was extracted from the purified phage sample using a previously described method (14). A single-end library was constructed from the phage DNA and was sequenced using a 454 GS Junior pyrosequencer (Roche, Indianapolis, IN). The sequence reads were assembled into one scaffold using GS Assembler software. The homopolymers were resequenced with a BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions, using an ABI PRISM 3100-Avant genetic analyzer (Applied Biosystems). The open reading frames (ORFs) were determined by considering ribosomal binding sites, based on the automated annotation data obtained from MiGAP (http://www.migap.org/).
The phage genome DNA sequences were compared using BLASTn at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/). The genome was analyzed using In Silico Molecular Cloning Genomic Edition (In Silico Biology, Inc., Yokohama, Japan) and GenomeMatcher (http://www.ige.tohoku.ac.jp/joho/gmProject/gmhomeJP.html) (23).
A maximum-likelihood phylogeny was inferred on the basis of the core genome alignment with PhyML, as implemented in REALPHY (24). Each ORF was analyzed by InterProScan 5 (http://www.ebi.ac.uk/Tools/pfa/iprscan5/), Conserved Domain search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and BLASTp (http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome). Some ORFs were also analyzed by HHpred (http://toolkit.tuebingen.mpg.de/hhpred).
Virion protein analysis.
The N-terminal amino acids of the virion protein were analyzed as described elsewhere (14). Briefly, the virion proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were transferred onto a polyvinylidene fluoride membrane and analyzed by the Edman degradation method using a PPSQ-31A/33A protein sequencer (Shimadzu, Kyoto, Japan). The virion proteins were also analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described elsewhere (22). Briefly, after digestion of the purified phage with trypsin (Trypsin Gold, mass spectrometry grade; Promega, Fitchburg, WI), the peptide sample was subjected to LC-MS/MS analysis using a Direct nanoLC/MALDI fraction system (KYA Technologies, Tokyo, Japan) for liquid chromatography and an AB SCIEX TOF/TOF 5800 system (AB Sciex, Framingham, MA) for matrix-assisted laser desorption ionization (MALDI)–MS and MS/MS analyses. An in-house database was constructed from putative protein data for phage KPP22 for this study. The MS/MS data were analyzed by the Paragon algorithm using ProteinPilot 3.0 (AB Sciex) (25).
Isolation of phage-resistant bacterial clones and mutant phages.
To isolate the phage-resistant bacterial clone, bacteria and phages were incubated on a double-layer agar plate for 24 h. A phage-resistant bacterial colony, which appeared on the top agar of a confluent lysed sample, was isolated. The bacterial colony was purified at least three times; at the final purification, phage resistance was determined via a streak test, as described below. The experiments were done in triplicate, and three phage-resistant bacteria were isolated.
Next, to isolate mutant phages with large-plaque formation, phages were cultured on a double-layer agar plate containing a phage-resistant bacterial clone. After 24 h of incubation, a large and clear plaque was isolated and purified at least three times using the phage-resistant bacterial clone. The experiments were done in triplicate, and three mutant phages were isolated.
Measurement of host spectrum, infectivity, and adsorption efficiency of phage.
The phage host spectrum was determined by examining phage lytic activity against each bacterial strain using a streak test. A loopful of the phage suspension (1010 PFU ml−1) was streaked onto a double-layer agar plate containing a P. aeruginosa strain. After incubation overnight, the bacteriolytic patterns, i.e., lysis with plaques, lysis without plaques, and no lysis, were recorded. Based on the bacteriolytic pattern, the lysis type, i.e., lysis from within, lysis from without, or no lysis, respectively, was determined.
Next, phage infectivity was examined using efficiency of plating (EOP). The EOP was measured as the ratio of PFU of phage plated on a particular bacterial strain to the PFU of phage plated on standard strain PAO1. Moreover, the phage adsorption efficacy was also measured. A 2.7-ml aliquot of a bacterial suspension in logarithmic growth phase (ca. 7.4 × 108 cells ml−1) was supplemented with 0.3 ml of phage suspension (ca. 1.0 × 104 PFU ml−1). The mixture was then incubated for 20 min at 37°C. After pelleting of the bacterial cells by centrifugation (7,000 × g, 2 min) and filtration through a 0.45-μm membrane filter, the concentration of the unbound phages in the supernatant was measured using P. aeruginosa strain PAO1 as an indicator host. Statistical analysis was performed with Student's t test using GraphPad Prism 6.0 (GraphPad Software; La Jolla, CA).
Bacterial genome sequences and mutation analysis.
The bacteria were cultured overnight, and the genomic DNAs were purified using a NucleoSpin tissue kit (Macherey-Nagel, Düren, Germany). A 150-bp paired-end library was prepared and analyzed using Illumina HiSeq4000 (see Table S1 in the supplemental material). The resequencing analysis was performed using the P. aeruginosa strain PAO1 genome as a reference (GenBank accession no. NC_002516), and the mutation sites were identified using the CLC Genomic Workbench (Qiagen, Venlo, Netherlands) (26). Comparing the mutation sites, those common to all bacteria were identified. For the confirmation of the common mutations, the genomic DNA was amplified with primers PAO1_wzy_F (5′-CGTCTGGGGACGGAAATTTTCAATAATCC-3′) and PAO1_wzy_R (5′-CATAGAGTTTTTCCTAAAGACATCTTGAGTGG-3′) and was then sequenced with a BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions using an ABI PRISM 3100-Avant genetic analyzer (Applied Biosystems).
The product of the gene containing the common mutation was analyzed by Conserved Domain search at the NCBI. The transmembrane domain structure of the protein was predicted using TOPCONS (http://topcons.net/) (27).
Analysis of LPS.
Total lipopolysaccharide (LPS) was prepared from the bacteria using the hot aqueous phenol extraction method, essentially as described elsewhere (28). First, the LPS together with a molecular standard (Precision Plus Protein Dual Color Standards; Bio-Rad, Hercules, CA) was separated by SDS-PAGE. The LPS was then fluorescently stained using a Pro-Q Emerald 300 lipopolysaccharide gel stain kit (Molecular Probes, Eugene, OR). Next, Western blotting of the LPS to detect the O5 antigen was performed using a method described elsewhere (29). Briefly, the LPS, together with a molecular standard (Precision Plus Protein Dual Color Standards; Bio-Rad), was separated by SDS-PAGE and transferred onto a nitrocellulose membrane, which was then blocked with 3% bovine serum albumin in phosphate-buffered saline. Mouse anti-P. aeruginosa antigenic serotype O5 (MF15-4) IgM antibody (MédiMabs, Montreal, Quebec, Canada) and alkaline phosphatase-conjugated goat anti-mouse IgM IgG antibody (Bethyl Laboratories, Montgomery, TX) were used as the primary and secondary antibodies, respectively (30). Detection was performed using 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (BCIP-NBT solution kit; Nacalai Tesque).
Accession numbers.
The complete genomic sequences of phage KPP22 and the mutant phages KPP22M1, KPP22M2, and KPP22M3 were deposited in DDBJ/EMBL/GenBank under accession no. LC105987, LC105988, LC105989, and LC105990, respectively. The partial genome sequences of PAO1, PAO1R1, PAO1R2, and PAO1R3 were deposited in DDBJ/EMBL/GenBank under accession no. LC110405, LC110406, LC110407, and LC110408, respectively. The sequencing data for PAO1, PAO1R1, PAO1R2, and PAO1R3 were deposited in the DDBJ Sequence Read Archive (DRA) database under accession no. DRA004286, DRA004287, DRA004288, and DRA004289, respectively. The virion protein sequence was deposited in the UniProt Knowledgebase under accession no. A0A0U5AF03.
RESULTS AND DISCUSSION
Characteristics of the newly isolated PB1-like phage KPP22.
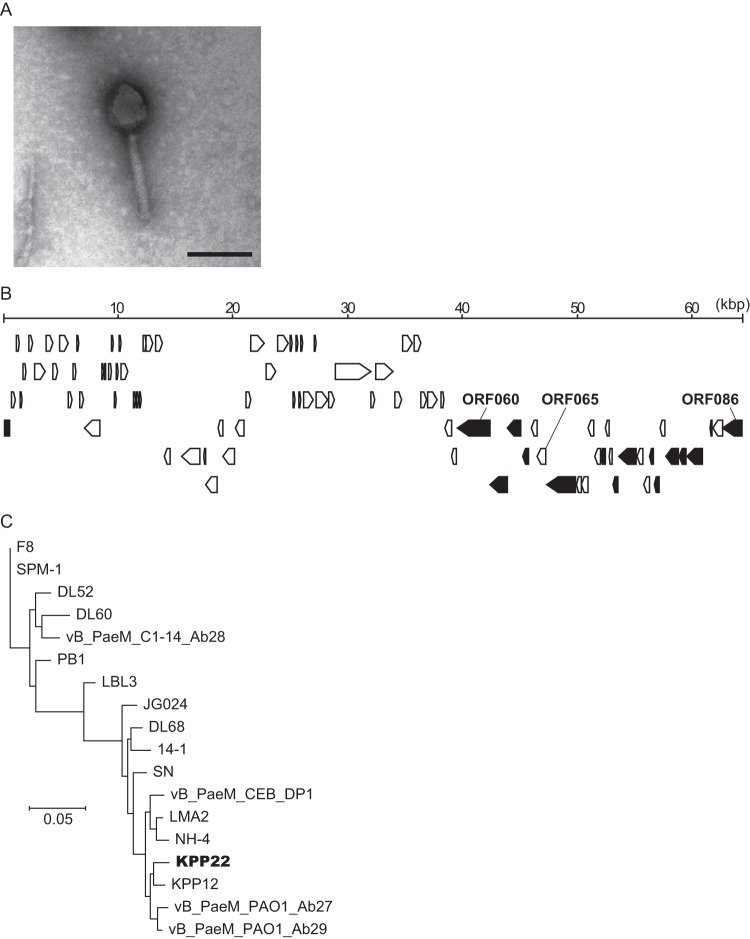
Phage KPP22 was isolated from a water sample collected from an agricultural wastewater drain in Kochi City, Kochi, Japan. First, transmission electron microscopy showed that phage KPP22 had an icosahedral head with a contractile tail and tail fibers (Fig. 1A). Based on this morphology, phage KPP22 was assigned to the family Myoviridae. Next, the whole genome of phage KPP22 was sequenced. It consisted of 64,415 bp, of which the G+C content was 55.6%. Eighty-six ORFs were predicted in the phage KPP22 genome (see Table S2 in the supplemental material). BLASTn analysis of the public database indicated that the DNA sequence of phage KPP22 showed the highest sequence similarity to those of P. aeruginosa phages belonging to the PB1-like virus genus (see Table S3 in the supplemental material). This, together with the observed morphology, meant that phage KPP22 was considered to belong to the PB1-like virus genus in the Myoviridae family.
FIG 1.
Characteristics of phage KPP22. (A) Electron micrograph. The bar indicates 100 nm. The head diameter and the tail length were 66.7 ± 5.6 nm and 123.2 ± 7.7 nm (mean ± standard deviation; n = 20), respectively. (B) Genome map. Arrows indicate ORFs. Black arrows indicate ORFs coding for the virion proteins, which were identified by LC-MS/MS analysis (see Table S4 in the supplemental material). (C) Phylogeny inference for the PB1-like viruses based on the whole genomes. The phylogram was inferred by the maximum-likelihood method using REALPHY (24). The genome data for PB1-like viruses, which were obtained from DDBJ/EMBL/GenBank, include phages DL52, DL60, DL68, F8, KPP12, JG024, LBL3, LMA2, NH-4, PB1, SN, SPM-1, vB_PaeM_PAO1_Ab29, vB_PaeM_CEB_DP1, vB_PaeM_PAO1_Ab27, vB_PaeM_C1-14_Ab28, and 14-1 (GenBank accession no. KR054028, KR054030, KR054033, DQ163917, AB560486, GU815091, FM201281, FM201282, JN254800, EU716414, FM887021, KF981875, LN610588, KR869157, LN610579, LN610589, and FM897211, respectively).
Shotgun proteomic analysis of the phage KPP22 virion proteins identified 15 ORFs as encoding virion proteins (see Table S4 in the supplemental material). These virion protein genes were clustered in the genomic region from orf060 to orf086 (Fig. 1B). The N-terminal protein sequencing of the major virion protein confirmed that it was ORF081 (see Fig. S1 in the supplemental material).
Next, the maximum-likelihood phylogeny was inferred based on genome sequences of PB1-like phages (Fig. 1C). Phage KPP22 was inferred to be most closely related to phage KPP12, which had been isolated from the agricultural wastewater sample at Kochi in Japan and has shown therapeutic effects in experimental P. aeruginosa keratitis (14). Because phage KPP22 was isolated in the same region of Japan after the discovery of phage KPP12, phage KPP22 is highly likely to be related to phage KPP12.
The lytic activity (i.e., lysis from within, lysis from without, or no lysis) of phage KPP22 against clinically isolated P. aeruginosa strains that included various serotypes was compared with that of phage KPP12 (16, 21, 31). Phage KPP22 showed lysis from within on 92.1% (35 of 38) of the tested clinically isolated strains, while phage KPP12 showed lysis from within and lysis from without on 68.4% (26 of 38) of the tested clinically isolated strains (Table 1). Thus, phage KPP22 can infect more P. aeruginosa strains than phage KPP12.
Moreover, phage KPP22 showed lytic activity against the strains that are not sensitive to phage KPP12, and vice versa. Using these phages (KPP12 and KPP22), the clinically isolated P. aeruginosa strains were considered to be lysed (Table 1). The P. aeruginosa strains PA12 and PA22, which are sensitive to phage KPP12, were not sensitive to phage KPP22. Although it is not known why these strains are sensitive to phage KPP12 but not to phage KPP22, which is another possible line of inquiry in the field of phage research, the use of these phages can overcome the issue of the phage host range.
Isolation of KPP22-resistant clones of P. aeruginosa strain PAO1 and KPP22 mutant phages with improved infectivity.
Each KPP22-resistant mutant P. aeruginosa clone, including PAO1R1, PAO1R2, and PAO1R3, was isolated from cultures of P. aeruginosa strain PAO1 with phage KPP22 in in vitro independent experiments (Table 2). Subsequently, the mutant phages, including KPP22M1, KPP22M2, and KPP22M3, that formed plaques on the double-layer agar plate containing the KPP22-resistant PAO1 clones PAO1R1, PAO1R2, and PAO1R3 were independently isolated in in vitro experiments (Table 2).
TABLE 2.
KPP22-resistant clones of P. aeruginosa strain PAO1 and mutant phages capable of infecting them
| Expt no. | Isolated KPP22-resistant PAO1 clone | Isolated mutant phage with large-plaque formation in response to the KPP22-resistant PAO1 clone |
|---|---|---|
| 1 | PAO1R1 | KPP22M1 |
| 2 | PAO1R2 | KPP22M2 |
| 3 | PAO1R3 | KPP22M3 |
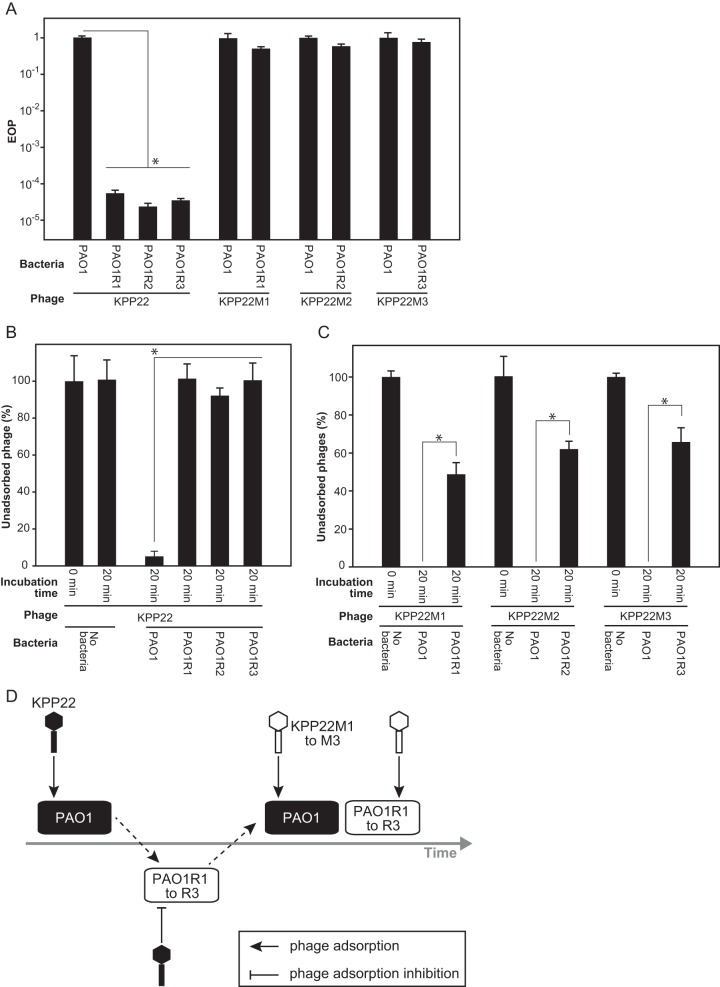
The infectivity level of the phages can be examined by comparison of their EOPs. The EOPs of phage KPP22 and of KPP22 mutant phages were measured. First, the EOP of phage KPP22 on each KPP22-resistant PAO1 clone was compared with that on strain PAO1 (Fig. 2A). The EOPs of phage KPP22 on the KPP22-resistant PAO1 clones were significantly lower (ca. 10−4 to 10−5 times) than that on strain PAO1. In contrast, the EOPs of the KPP22 mutant phages on the KPP22-resistant PAO1 clones were not significantly different from those on strain PAO1.
FIG 2.
Infectivity and adsorption of phage KPP22 and mutant phages to strain PAO1 and to KPP22-resistant PAO1 clones. The KPP22-resistant PAO1 clones were PAO1R1, PAO1R2, and PAO1R3. The KPP22 mutant phages were phages KPP22M1, KPP22M2, and KPP22M3. Asterisks indicate significance (t test; P < 0.05). (A) Infectivities of phage KPP22 and mutant phages. (B) Efficiencies of adsorption of phage KPP22 to strain PAO1 and KPP22-resistant PAO1 clones. (C) Efficiencies of adsorption of mutant phages to strain PAO1 and KPP22-resistant PAO1 clones. (D) Diagram of short-term coevolution of strain PAO1 and phage KPP22.
The host ranges of the KPP22 mutant phages were also compared with each other and with that of phage KPP22. The KPP22 mutant phages KPP22M1, KPP22M2, and KPP22M3 all showed the same host range, which was similar to that of phage KPP22, although a small difference was observed on strain PA16 (Table 1). The KPP22 mutant phages showed lysis-from-without activity against strain PA16, while phage KPP22 showed no lytic activity.
Some mutant phages increase their infectivity by improving their adsorption efficiency (18, 32–34). The adsorption efficiencies of the KPP22 mutant phages were compared with that of phage KPP22 (Fig. 2B and C). Phage KPP22 barely adsorbed to the KPP22-resistant PAO1 clones, although it could efficiently adsorb to strain PAO1. However, the KPP22 mutant phages adsorbed to the KPP22-resistant PAO1 clones. The difference in the infectivity of phage KPP22 and the KPP22 mutant phages was considered to result from the differences in their adsorption efficiency. This short-term coevolutionary arms race between phage KPP22 and strain PAO1 is outlined in Fig. 2D.
Analysis of KPP22-resistant clones of P. aeruginosa strain PAO1.
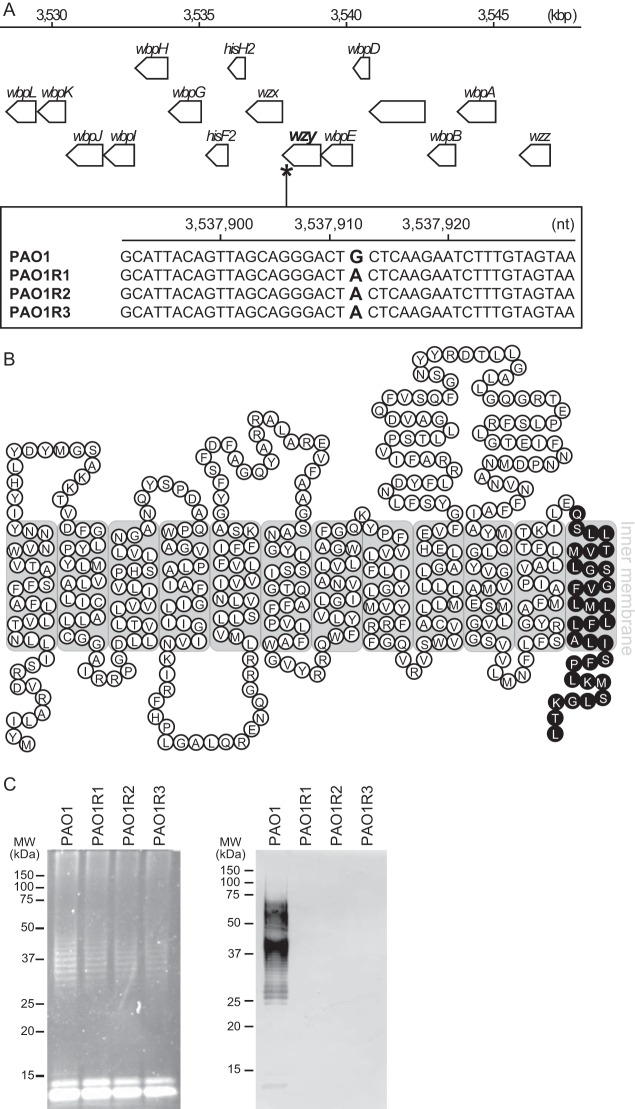
The whole DNAs of strain PAO1 and the KPP22-resistant PAO1 clones were analyzed via next-generation sequencing, and the sequence data were aligned to the PAO1 reference genome (see Table S1 in the supplemental material). The common mutation site shared by the KPP22-resistant PAO1 clones was searched. A common nucleotide substitution from guanine (in our PAO1 genome sequence) to adenine (in the KPP22-resistant PAO1 clone genome sequences) alone was found at nucleotide 3537912 in the tag PA3154 locus (from nucleotide 3537808 to 3539124) of the PAO1 reference genome (Fig. 3A) (26). No other common mutation was found. Genome sequencing using cycle sequencing technology also confirmed this single-nucleotide substitution in the KPP22-resistant PAO1 clones. The locus tag PA3154 in the strain PAO1 genome is known to be the wzy gene. The nucleotide substitution found in the KPP22-resistant PAO1 clones is a nonsense mutation, leading to a C-terminally truncated form of Wzy. Bioinformatic analysis of the Wzy protein suggested a 12-membrane-spanning structure, but no protein domain was predicted (Fig. 3B).
FIG 3.
Analysis of strain PAO1 and KPP22-resistant PAO1 clones PAO1R1, PAO1R2, and PAO1R3. (A) Common mutation site in the genomes of KPP22-resistant PAO1 clones. A common nucleotide substitution was observed at bp 3537912 bp of P. aeruginosa strain PAO1, which is indicated by an asterisk on the genome map at the top. In the bottom box, the site of the nonsense mutation is shown, located in the wzy gene. (B) Diagram of Wzy in P. aeruginosa strain PAO1. Each circle containing a letter indicates an amino acid. Black circles are deficient amino acid sequences relative to the complete Wzy amino acid sequence, demonstrating the deficient Wzy in the KPP22-resistant PAO1 clones. (C) Analysis of LPSs. The LPSs were separated by SDS-PAGE and fluorescently stained (left), and Western blotting detecting the O5 antigen was done (right). Molecular size standards are shown on the left of each gel.
Strain PAO1 is classified as antigenic type O5 according to the International Antigenic Typing Scheme (IATS) classification (35, 36). The O5 antigen in strain PAO1 is produced via a Wzx/Wzy-dependent pathway that includes the sequential activities of such inner membrane proteins as Wzx, Wzy, Wzz, and WaaL (37). The Wzy polymerizes the glycan of the O-specific antigen (38). We hypothesized that such the nonsense mutation affected the O5 antigen production. To test the hypothesis, the LPSs of strain PAO1 and the KPP22-resistant PAO1 clones were analyzed by fluorescent staining and by Western blotting to detect O5 antigen, after separation by SDS-PAGE (Fig. 3C). The results showed that while the patterns of the total LPSs of all the bacteria were very similar, the O5 antigen was not detected on the KPP22-resistant PAO1 clones. Thus, the nonsense mutation seemed to have significantly reduced the activity of Wzy in the KPP22-resistant PAO1 clones, and the O5 antigen did not appear to be produced.
Analysis of KPP22 mutant phages.
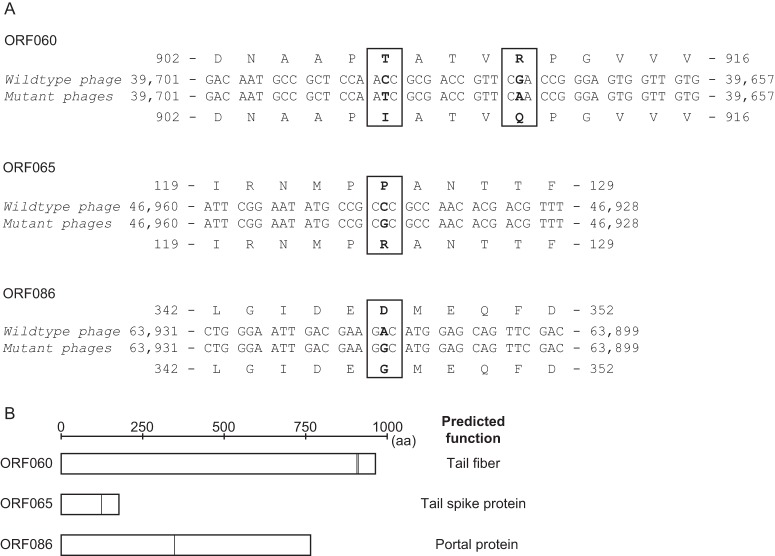
The genomes of the mutant phages KPP22M1, KPP22M2, and KPP22M3 were sequenced and compared with that of phage KPP22. The three mutant phages showed the same four nucleotide substitutions at the same locations on their genomes, at nucleotides 39673, 39685, 46944, and 63915 of the phage KPP22 genome (Fig. 4A). No other mutation sites were found. All four mutations in orf060, orf065, and orf086, which occurred at the second position in the triplet codons, were considered to lead to amino acid substitutions (i.e., missense mutations). The same mutations found in the mutant phage genomes were not considered to be coincident. These mutant phages with the same mutation were likely to have been selectively isolated from the same origin of the mutated phage, assuming that such preadapted phage could have been naturally suspended in the original phage stock (in a minute proportion) before the selection by the KPP22-resistant PAO1 clones.
FIG 4.
Genetic analysis of mutant phages KPP22M1, KPP22M2, and KPP22M3. (A) Mutation sites found in the mutant phages. The same mutations are found at the same locations, which were in orf060, orf065, and orf086 of the mutant phage genomes. All the mutations are missense mutations that lead to amino acid substitutions. (B) ORF060, ORF065, and ORF086. The primary structure of each protein is described as a rectangle with a scale, and the amino acid substitutions in the mutant phages are indicated by vertical lines within the rectangles. The predicted functions are indicated on the right (see Table S5 in the supplemental material).
Moreover, the functions of these ORFs were assessed. ORF060 was predicted by BLASTp to be a tail fiber protein and by HHpred to be a short and long tail fiber protein (Fig. 4B; see Table S5 in the supplemental material). ORF065 was predicted by HHpred to be a tail spike protein, and ORF086 was predicted by HHpred to be a portal protein (Fig. 4B; see Table S5 in the supplemental material). Among these ORFs, orf060 and orf086 were identified in this study as genes that encode virion proteins, as described above (Fig. 1B; see Table S4 in the supplemental material).
Furthermore, the protein sequences of phage KPP22, i.e., ORF060, ORF065, and ORF086, were compared by BLASTp with their homologous protein sequences of phage KPP12, i.e., ORF43, ORF38, and ORF17, respectively (see Fig. S2 in the supplemental material). The sequence identity between ORF060 of phage KPP22 and ORF43 of phage KPP12 (a putative long tail fiber) was 94%, whereas the sequence identities of ORF065 and ORF086 (putative spike and portal proteins, respectively) of phage KPP22 were 100% and 99%, respectively (query coverage of all BLASTp results, 100%). Among these ORFs, ORF060 of phage KPP22 had the lowest protein sequence identity to the homologous ORF of phage KPP12, ORF043, and a dissimilarity was observed particularly in the C-terminal region of the protein. Mutations in the distal portion of tail fibers have been shown to be involved in improvements in adsorption in other phages (33, 39), which supports that ORF060 can be a tail fiber protein.
Phage T4 undergoes specific binding to the host cell surface, which is mediated by the reversible binding of long tail fibers to the LPS or OmpC and the irreversible binding of short tail fibers/spikes to the LPS (40, 41). The long tail fibers/spikes exhibit an increase in the dwell time at the surface of the host bacterium to search for the receptor molecules (41, 42). ORF060 and ORF065 were putative long tail fiber and tail spike proteins, respectively. If this adsorption model of phage T4 applies to phage KPP22, these putative tail proteins, like ORF060 and ORF065, are likely to be highly associated with phage host specificity and adsorption. However, virion proteins other than the tail protein also seem to be important for phage adsorption. For example, the whisker protein of phage T4, which is the neck appendage protein and retracts the tail fibers, is also considered to be associated with T4 adsorption (43). In phage KPP22, ORF086, which was assumed to be a portal protein, may be associated with phage adsorption. Further investigation is required to understand the adsorption mechanism of phage KPP22 in detail.
O5 antigen as one of the receptor molecules to phage KPP22 and KPP22 mutant phages.
The LPS and/or outer membrane proteins of bacteria can be the receptor molecules for phages (39). The PB1-like phages infecting P. aeruginosa have been shown to adsorb to the LPS (44, 45). The LPS is generally divided into two forms of O surface antigens in P. aeruginosa: common polysaccharide antigen (formerly termed the A band) and O-specific antigen (formerly termed the B band) (38). O-specific antigen, which is a repeat of three to five distinct sugars, acts as a receptor molecule for phage adsorption in some cases (38). The O5 antigen is an important receptor molecule for phage KPP22 as described above, and it is also one of the important receptor molecules for KPP22 mutant phages. Observing the adsorption efficiency of the KPP22 mutant phages (Fig. 2B), the rates of adsorption to strain PAO1 and to KPP22-resistant PAO1 clones were almost 100% and ca. 30 to 60% in 20 min, respectively. This suggested that the O5 antigen assisted even the adsorption of KPP22 mutant phages to strain PAO1. Thus, the O5 antigen is one of the important receptor molecules for phage KPP22 and KPP22 mutant phages.
Moreover, some phages adsorb to both the typical LPS and LPS that is deficient in O antigens (39). Phage KPP22 and KPP22 mutant phages showed lytic activity against 92.1% (35/38) and 94.7% (36/38) of the tested P. aeruginosa strains, respectively (Table 1). Not all the tested P. aeruginosa strains had the O5 antigen, according to screening by Western blotting (see Fig. S3 in the supplemental material). Thus, these phages were thought to adsorb to not only the O5 antigens but also the other receptor molecules.
Considering the results above in the analysis of antagonistic evolution, phage KPP22 seemed to increase the affinity to certain receptors on the P. aeruginosa strains with several mutations in its genome, along with the host bacterial phenotypic change.
Study of antagonistic evolution of bacteria and phage in phage therapy.
Because phage therapy failed once in the past and the present phage therapy is a second trial, the framework for phage therapy should be carefully designed based on research and developments in modern science. In addition to the understating of the phage preadaptation, the antagonistic evolution of phage and bacteria is still considered to be an important research topic to develop safe and efficient phage therapy in the future. First, the application of a phage cocktail is considered to be the solution for phage host specificity in phage therapy (46). In this study, the use of phages KPP12 and KPP22 lysed all the clinically isolated P. aeruginosa strains. However, after years of use of the phage mixture including these phages, we may face difficulties in treating bacterial infections because of the emergence of multiphage-resistant bacterial clones, as was the case in the history of chemotherapy (12, 47).
Moreover, the bacterial virulence can be changed through the antagonistic evolution in phage therapy. It has been shown that phage-resistant bacteria lose their virulence during phage therapy (48). In contrast, the virulence of the bacteria may be increased with exposure to phage, according to this study. For example, deletion of the O5 antigen and exposure to phage have been shown to increase the virulence of strain PAO1 (49, 50). Loss of the O5 antigen in strain PAO1 has been shown to increase bacterial cell adhesion to polystyrene (51), which may increase the biofilm formation on the plastic surface. Further studies should be conducted to investigate the antagonistic evolution of bacteria and phage in the use of a phage cocktail for a long period, including risk assessment of the emergence of multiphage-resistant bacteria and changes in their virulence.
Efforts have been made recently in the research and development of phage therapy using preadapted phage to increase therapeutic efficiency and avoid any potential risks (19, 20, 47). In this study, phage KPP22 showed increased infectivity to P. aeruginosa strains through the antagonistic evolution with KPP22-resistant PAO1 clones without changing its phage host range. The accumulation of more knowledge regarding adapted phages together with target bacteria may allow the future development of tailor-made phage therapy, in which the adapted phage that is adequate for the targeted bacteria is selected from the collection of therapeutic phages. We believe that our study will assist in the understanding of phage preadaptation and will contribute to the development of such safe and efficient phage therapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ryu Shigehisa, Kochi University, and Ryuichiro Hirayama, Azabu University, for experimental help. We thank the Science Research Center, Kochi, Japan, for experimental support.
This study was supported by a Grant-in-Aid for Research Activity Start-up (22890129) and Grants-in-Aid for Young Scientists (24791025 and 15K19095). This study was also supported by Kochi System Glycobiology Center, Kochi, Japan, and the Center of Biomembrane Functions Controlling Biological Systems, Kochi, Japan.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00090-16.
REFERENCES
- 1.Lister P, Wolter D, Hanson N. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breidenstein EB, de la Fuente-Núñez C, Hancock RE. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Taylor PK, Yeung AT, Hancock RE. 2014. Antibiotic resistance in Pseudomonas aeruginosa biofilms: towards the development of novel anti-biofilm therapies. J Biotechnol 191:121–130. doi: 10.1016/j.jbiotec.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Matsuzaki S, Rashel M, Uchiyama J, Sakurai S, Ujihara T, Kuroda M, Ikeuchi M, Tani T, Fujieda M, Wakiguchi H, Imai S. 2005. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J Infect Chemother 11:211–219. doi: 10.1007/s10156-005-0408-9. [DOI] [PubMed] [Google Scholar]
- 5.Marza JA, Soothill JS, Boydell P, Collyns TA. 2006. Multiplication of therapeutically administered bacteriophages in Pseudomonas aeruginosa infected patients. Burns 32:644–646. doi: 10.1016/j.burns.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Wright A, Hawkins CH, Anggård EE, Harper DR. 2009. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol 34:349–357. doi: 10.1111/j.1749-4486.2009.01973.x. [DOI] [PubMed] [Google Scholar]
- 7.Hawkins C, Harper D, Burch D, Anggård E, Soothill J. 2010. Topical treatment of Pseudomonas aeruginosa otitis of dogs with a bacteriophage mixture: a before/after clinical trial. Vet Microbiol 146:309–313. doi: 10.1016/j.vetmic.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Rose T, Verbeken G, Vos DD, Merabishvili M, Vaneechoutte M, Lavigne R, Jennes S, Zizi M, Pirnay JP. 2014. Experimental phage therapy of burn wound infection: difficult first steps. Int J Burns Trauma 4:66–73. [PMC free article] [PubMed] [Google Scholar]
- 9.Ceyssens PJ, Miroshnikov K, Mattheus W, Krylov V, Robben J, Noben JP, Vanderschraeghe S, Sykilinda N, Kropinski AM, Volckaert G, Mesyanzhinov V, Lavigne R. 2009. Comparative analysis of the widespread and conserved PB1-like viruses infecting Pseudomonas aeruginosa. Environ Microbiol 11:2874–2883. doi: 10.1111/j.1462-2920.2009.02030.x. [DOI] [PubMed] [Google Scholar]
- 10.Ackermann H. 2011. Bacteriophage taxonomy. Microbiology Aust 32:90–94. [Google Scholar]
- 11.Pires DP, Vilas Boas D, Sillankorva S, Azeredo J. 2015. Phage therapy: a step forward in the treatment of Pseudomonas aeruginosa infections. J Virol 89:7449–7456. doi: 10.1128/JVI.00385-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krylov V, Shaburova O, Krylov S, Pleteneva E. 2013. A genetic approach to the development of new therapeutic phages to fight Pseudomonas aeruginosa in wound infections. Viruses 5:15–53. doi: 10.3390/v5010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krylov V, Pleteneva E, Shaburova O, Bourkaltseva M, Krylov S, Chesnokova E, Polygach O. 2014. Common preconditions for safe phage therapy of Pseudomonas aeruginosa infections. Adv Microbiol 4:766–773. doi: 10.4236/aim.2014.412084. [DOI] [Google Scholar]
- 14.Fukuda K, Ishida W, Uchiyama J, Rashel M, Kato S, Morita T, Muraoka A, Sumi T, Matsuzaki S, Daibata M, Fukushima A. 2012. Pseudomonas aeruginosa keratitis in mice: effects of topical bacteriophage KPP12 administration. PLoS One 7:e47742. doi: 10.1371/journal.pone.0047742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 16.Scanlan PD, Buckling A, Hall AR. 2015. Experimental evolution and bacterial resistance: (co)evolutionary costs and trade-offs as opportunities in phage therapy research. Bacteriophage 5:e1050153. doi: 10.1080/21597081.2015.1050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keen EC. 2012. Phage therapy: concept to cure. Front Microbiol 3:238. doi: 10.3389/fmicb.2012.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uchiyama J, Takemura I, Satoh M, Kato S, Ujihara T, Akechi K, Matsuzaki S, Daibata M. 2011. Improved adsorption of an Enterococcus faecalis bacteriophage φEF24C with a spontaneous point mutation. PLoS One 6:e26648. doi: 10.1371/journal.pone.0026648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betts A, Vasse M, Kaltz O, Hochberg ME. 2013. Back to the future: evolving bacteriophages to increase their effectiveness against the pathogen Pseudomonas aeruginosa PAO1. Evol Appl 6:1054–1063. doi: 10.1111/eva.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friman VP, Soanes-Brown D, Sierocinski P, Molin S, Johansen HK, Merabishvili M, Pirnay JP, De Vos D, Buckling A. 2016. Pre-adapting parasitic phages to a pathogen leads to increased pathogen clearance and lowered resistance evolution with Pseudomonas aeruginosa cystic fibrosis bacterial isolates. J Evol Biol 29:188–198. doi: 10.1111/jeb.12774. [DOI] [PubMed] [Google Scholar]
- 21.Uchiyama J, Rashel M, Takemura I, Kato S, Ujihara T, Muraoka A, Matsuzaki S, Daibata M. 2012. Genetic characterization of Pseudomonas aeruginosa bacteriophage KPP10. Arch Virol 157:733–738. doi: 10.1007/s00705-011-1210-x. [DOI] [PubMed] [Google Scholar]
- 22.Miyata R, Yamaguchi K, Uchiyama J, Shigehisa R, Takemura-Uchiyama I, Kato S, Ujihara T, Sakaguchi Y, Daibata M, Matsuzaki S. 2014. Characterization of a novel Pseudomonas aeruginosa bacteriophage, KPP25, of the family Podoviridae. Virus Res 189:43–46. doi: 10.1016/j.virusres.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 23.Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. 2008. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. 2014. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol 31:1077–1088. doi: 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. 2007. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics 6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 27.Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A. 2015. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43:W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis MR, Goldberg JB. 2012. Purification and visualization of lipopolysaccharide from Gram-negative bacteria by hot aqueous-phenol extraction. J Vis Exp 63:e3916. doi: 10.3791/3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Islam ST, Huszczynski SM, Nugent T, Gold AC, Lam JS. 2013. Conserved-residue mutations in Wzy affect O-antigen polymerization and Wzz-mediated chain-length regulation in Pseudomonas aeruginosa PAO1. Sci Rep 3:3441. doi: 10.1038/srep03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam JS, MacDonald LA, Lam MY, Duchesne LG, Southam GG. 1987. Production and characterization of monoclonal antibodies against serotype strains of Pseudomonas aeruginosa. Infect Immun 55:1051–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Homma JY. 1982. Designation of the thirteen O-group antigens of Pseudomonas aeruginosa; an amendment for the tentative proposal in 1976. Jpn J Exp Med 52:317–320. [PubMed] [Google Scholar]
- 32.Daniels L, Wais A. 1998. Virulence in phage populations infecting Halobacterium cutirubrum. FEMS Microbiol Ecol 25:129–134. doi: 10.1111/j.1574-6941.1998.tb00466.x. [DOI] [Google Scholar]
- 33.Le S, He X, Tan Y, Huang G, Zhang L, Lux R, Shi W, Hu F. 2013. Mapping the tail fiber as the receptor binding protein responsible for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PLoS One 8:e68562. doi: 10.1371/journal.pone.0068562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keen EC. 2014. Tradeoffs in bacteriophage life histories. Bacteriophage 4:e28365. doi: 10.4161/bact.28365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burrows LL, Charter DF, Lam JS. 1996. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol Microbiol 22:481–495. doi: 10.1046/j.1365-2958.1996.1351503.x. [DOI] [PubMed] [Google Scholar]
- 36.Knirel Y, Vinogradov EV, Kocharova NA, Paramonov NA, Kochetkov NK, Dmitriev BA, Stanislavsky ES, Lányi B. 1988. The structure of O-specific polysaccharides and serological classification of Pseudomonas aeruginosa (a review). Acta Microbiol Hung 35:3–24. [PubMed] [Google Scholar]
- 37.Islam ST, Lam JS. 2014. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can J Microbiol 60:697–716. doi: 10.1139/cjm-2014-0595. [DOI] [PubMed] [Google Scholar]
- 38.Lam JS, Taylor VL, Islam ST, Hao Y, Kocíncová D. 2011. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol 2:118. doi: 10.3389/fmicb.2011.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heller K, Braun V. 1979. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. J Bacteriol 139:32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. 2010. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol 59:145–155. [PubMed] [Google Scholar]
- 41.Henning U, Hashemolhosseini S. 1994. Receptor recognition by T-even-type coliphages, p 291–298. In Karam JD. (ed), Molecular biology of bacteriophage T4. ASM Press, Washington, DC. [Google Scholar]
- 42.Chatterjee S, Rothenberg E. 2012. Interaction of bacteriophage λ with its E. coli receptor, LamB. Viruses 4:3162–3178. doi: 10.3390/v4113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conley MP, Wood WB. 1975. Bacteriophage T4 whiskers: a rudimentary environment-sensing device. Proc Natl Acad Sci U S A 72:3701–3705. doi: 10.1073/pnas.72.9.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jarrell KF, Kropinski AM. 1981. Isolation and characterization of a bacteriophage specific for the lipopolysaccharide of rough derivatives of Pseudomonas aeruginosa strain PAO. J Virol 38:529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garbe J, Wesche A, Bunk B, Kazmierczak M, Selezska K, Rohde C, Sikorski J, Rohde M, Jahn D, Schobert M. 2010. Characterization of JG024, a Pseudomonas aeruginosa PB1-like broad host range phage under simulated infection conditions. BMC Microbiol 10:301. doi: 10.1186/1471-2180-10-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan BK, Abedon ST, Loc-Carrillo C. 2013. Phage cocktails and the future of phage therapy. Future Microbiol 8:769–783. doi: 10.2217/fmb.13.47. [DOI] [PubMed] [Google Scholar]
- 47.Ormälä AM, Jalasvuori M. 2013. Phage therapy: should bacterial resistance to phages be a concern, even in the long run? Bacteriophage 3:e24219. doi: 10.4161/bact.24219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.León M, Bastías R. 2015. Virulence reduction in bacteriophage resistant bacteria. Front Microbiol 6:343. doi: 10.3389/fmicb.2015.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Augustin DK, Song Y, Baek MS, Sawa Y, Singh G, Taylor B, Rubio-Mills A, Flanagan JL, Wiener-Kronish JP, Lynch SV. 2007. Presence or absence of lipopolysaccharide O antigens affects type III secretion by Pseudomonas aeruginosa. J Bacteriol 189:2203–2209. doi: 10.1128/JB.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hosseinidoust Z, Tufenkji N, van de Ven TG. 2013. Predation in homogeneous and heterogeneous phage environments affects virulence determinants of Pseudomonas aeruginosa. Appl Environ Microbiol 79:2862–2871. doi: 10.1128/AEM.03817-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makin SA, Beveridge TJ. 1996. The influence of A-band and B-band lipopolysaccharide on the surface characteristics and adhesion of Pseudomonas aeruginosa to surfaces. Microbiology 142:299–307. doi: 10.1099/13500872-142-2-299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.