ABSTRACT
SigB is the main stress gene regulator in Listeria monocytogenes affecting the expression of more than 150 genes and thus contributing to multiple-stress resistance. Despite its clear role in most stresses, its role in oxidative stress is uncertain, as results accompanying the loss of sigB range from hyperresistance to hypersensitivity. Previously, these differences have been attributed to strain variation. In this study, we show conclusively that unlike for all other stresses, loss of sigB results in hyperresistance to H2O2 (more than 8 log CFU ml−1 compared to the wild type) in aerobically grown stationary-phase cultures of L. monocytogenes strains 10403S and EGD-e. Furthermore, growth at 30°C resulted in higher resistance to oxidative stress than that at 37°C. Oxidative stress resistance seemed to be higher with higher levels of oxygen. Under anaerobic conditions, the loss of SigB in 10403S did not affect survival against H2O2, while in EGD-e, it resulted in a sensitive phenotype. During exponential phase, minor differences occurred, and this result was expected due to the absence of sigB transcription. Catalase tests were performed under all conditions, and stronger catalase results corresponded well with a higher survival rate, underpinning the important role of catalase in this phenotype. Furthermore, we assessed the catalase activity in protein lysates, which corresponded with the catalase tests and survival. In addition, reverse transcription-PCR (RT-PCR) showed no differences in transcription between the wild type and the ΔsigB mutant in various oxidative stress genes. Further investigation of the molecular mechanism behind this phenotype and its possible consequences for the overall phenotype of L. monocytogenes are under way.
IMPORTANCE SigB is the most important stress gene regulator in L. monocytogenes and other Gram-positive bacteria. Its increased expression during stationary phase results in resistance to multiple stresses. However, despite its important role in general stress resistance, its expression is detrimental for the cell in the presence of oxidative stress, as it promotes hypersensitivity against hydrogen peroxide. This peculiar phenotype is an important element of the physiology of L. monocytogenes, and it might help us explain the behavior of this organism in environments where oxidative stress is present.
INTRODUCTION
Listeria monocytogenes is a Gram-positive bacterium that causes listeriosis, a serious and potentially lethal foodborne illness (1). Despite its low incidence, listeriosis has a high mortality rate (30%), making it the most deadly foodborne disease in the United Kingdom and the United States, as it claims more lives than any other foodborne pathogen (1, 2). One of the key attributes that makes L. monocytogenes such a successful pathogen is its ability to survive and persist in a wide range of harsh environments both outside and within the human host (3). One of the most important stresses L. monocytogenes has to withstand, in order to survive and cause disease, is oxidative stress. Oxidative stress can occur in the environment where metal or nonmetal redox catalysts are present, during disinfection with oxidative disinfectants, and during processing of foods with ozone or plasma. Furthermore, during the intracellular stage of L. monocytogenes infection, the bacterium encounters oxidative stress within the phagolysosome during phagocytosis.
The alternative sigma factor σB plays an important role in the stress responses of several Gram-positive bacteria (4). In L. monocytogenes, SigB regulates the expression of more than 150 genes (5), contributing to resistance to multiple stresses, including oxidative stress, acid, heat, salt, and bile acids (6–9). Recent evidence shows that the induction of SigB in L. monocytogenes occurs in the early exponential phase of growth, eventually reaching maximum levels at early stationary phase (10).
Highlighting its role in stress resistance, the deletion of sigB in L. monocytogenes leads to sensitivity against various stresses, which, in most cases, can be explained at the molecular level (6–9). An exception to this has been observed with oxidative stress in Bacillus cereus (11), where it has been reported that the ΔsigB mutant is more resistant than the wild type (WT). In the case of L. monocytogenes, there is no consensus about the role of SigB in oxidative stress, with various studies showing that the deletion of sigB leads to sensitivity to oxidative stress (7, 9), while others report increased resistance (12). This discrepancy has previously been attributed to strain variability (12). In this study, we investigate possible reasons for previous discrepancies and the role of SigB in oxidative stress under various environmental conditions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
WT L. monocytogenes strains 10403S and EGD-e (both belong to serotype 1/2a) and their isogenic ΔsigB mutants were used throughout this study. EGD-e, 10403S, and ΔsigB mutants were all constructed during previous works (13, 14) and have been used extensively in work on the role of SigB in L. monocytogenes (7, 11). Stock cultures were stored at −80°C in 7% (vol/vol) dimethyl sulfoxide (DMSO) (Sigma-Aldrich, Dorset, United Kingdom). Prior to experiments, stock cultures were streaked onto brain heart infusion (BHI) agar (LAB M, Lancashire, United Kingdom) and incubated at 37°C or 30°C overnight. A single colony from this medium was transferred to 3 ml of sterile BHI broth (LAB M) and incubated overnight at 37°C or 30°C with shaking (160 rpm). Subsequently, a portion of these overnight cultures served as the inoculum (1% [vol/vol]) to prepare the cultures that were used in the experiments. These cultures were prepared in 250-ml conical flasks containing 20 ml of the same medium as the one used for the inoculum and incubated overnight at 37°C or 30°C with shaking (130 rpm). For anaerobic growth, cultures were grown in 20 ml of BHI in Sterilin Quickstart Universal polystyrene containers (Thermo Scientific, Loughborough, United Kingdom) and placed in an anaerobic cabinet at 37°C with a gas atmosphere maintained at a 80:10:10 N2:CO2:H2 ratio.
Survival in the presence of hydrogen peroxide.
Stationary-phase cells were grown for approximately 18 h, while mid-exponential-phase cells were grown for 4 to 5 h until an optical density at 600 nm (OD600) of ∼0.75 was reached on a spectrophotometer (Spectronic 200; Thermo Fisher Scientific, Loughborough, United Kingdom) and subsequently were challenged with H2O2 (Sigma-Aldrich, Gillingham, United Kingdom). To allow comparisons, the survival of anaerobically grown cells had to be performed at a point of growth similar to that of the aerobically grown cells. Therefore, prior to the experiments, OD600 measurements were taken for 24 h to construct aerobic and anaerobic growth curves (data not shown). In these experiments, it was determined that the 21-h point of the anaerobic growth curves corresponded to the 18-h point in the aerobic ones. Subsequently, cells grown anaerobically were challenged at stationary phase following ∼21 h of growth.
A 30% solution of H2O2 (Sigma-Aldrich, Dorset, United Kingdom) was added to the flasks at various concentrations for each condition and type of cells, but only one representative set of results is presented here. Depending on the H2O2 resistance of each strain, final concentrations of 0.4%, 0.43%, 1%, 3%, and 4.5% (vol/vol) are presented here, and samples were taken at 0 min and every 20 min. Prior to assessment of H2O2 survival, preliminary experiments were performed to define the concentrations of H2O2 that should be used with each of the strains and conditions to avoid rapid death or complete survival that would allow comparison between the sigB mutants and their corresponding WT strains. During the experiments, cultures were kept at the temperature at which they were grown overnight (30°C and 37°C). Prior to and after the addition of H2O2, samples were taken, and serial dilutions were prepared in maximum recovery diluent (MRD; Oxoid, United Kingdom) and spread on BHI agar plates that were incubated at 37°C for 2 days. Subsequently, CFU were enumerated to assess the concentration of cells in the cultures at each time point (every 20 min). All experiments were performed at least in triplicate, and the average and standard deviation were calculated.
Disk diffusion assay.
Cells of the EGD-e WT, 10403S WT, and their corresponding ΔsigB mutants were grown overnight in Mueller-Hinton broth at 30°C or 37°C with shaking (160 rpm). Subsequently, overnight cultures were diluted to an OD600 of 0.2, and 100 μl was spread onto Mueller-Hinton agar (MHA; Oxoid, Basingstoke, United Kingdom). Then, 10 μl of 30% (vol/vol) H2O2 was pipetted onto Whatman 3MM paper disks (0.7-cm diameter), and these disks were placed on top of the agar and incubated for 18 h at the same temperature as the overnight culture (30°C or 37°C). The zones of inhibition (in millimeters) were taken as a measure of H2O2 sensitivity. The zones of inhibition were measured in three dimensions, and the mean values and standard deviations were calculated. All experiments were performed on six independent biological replicates, and statistical analysis was performed as described below.
Catalase test.
Overnight cultures were grown as described above, and subsequently, 5 ml was transferred into Sterilin Quickstart Universal polystyrene containers and 100 μl of 30% H2O2 was added to the aerobic and anaerobic cultures, respectively. The oxygen released in the form of bubbles was visually monitored and photographed after a 5-min period as an indication of catalase activity.
Catalase activity in protein extracts.
Measurement of the catalase activity in protein extracts was performed as described previously (15), with modifications to assess the intracellular activity of this enzyme. In short, following the removal of the growth medium, proteins were extracted using a sonication-based method, as described previously (16), using 20 ml stationary-phase cultures grown for 18 h. The concentrations of protein extracts were determined by the RC DC protein assay kit (Bio-Rad). Extracts were normalized to 0.5 mg ml−1, and 25 μl of each was added to 1 ml of an aqueous solution of 1% H2O2 contained in a quartz cuvette already placed in a UV spectrophotometer set up to record absorbance at 240 nm (λ at which H2O2 absorbs). Subsequently, measurements were taken every 10 s, and the reduction in the intensity of signal represented the H2O2 degradation due to catalase. The above-mentioned concentrations of protein and H2O2 used were defined in preliminary experiments, which aimed to avoid any rapid formation of bubbles in the cuvette causing erratic changes in the absorbance that would make any measurement impossible.
DO measurements.
The concentration of dissolved oxygen (DO) present in bacterial cultures was assessed using an optical sensor (InLab OptiOx) attached to a SevenExcellence S900 benchtop instrument (Mettler Toledo, Columbus, OH, USA). The sensor was calibrated daily using a two-point calibration. The instrument was blanked using tablets, and atmospheric saturation was achieved by placing the sensor in the air. Triplicate measurements of DO were taken from both aerobic and anaerobic cultures grown under the same conditions described for the survival assays.
Transcriptional analysis of genes contributing to oxidative stress.
Transcriptional analysis was performed in the 10403S WT and the corresponding sigB mutant. The transcription of genes responsible for resistance against oxidative stress was quantified as previously described by Karatzas et al. (17) following real-time reverse transcription-PCR (RT-PCR). The efficiencies of the primer pairs (shown in Table 1) were all close to 2, and these values were used for efficiency correction in the quantification step. In all cases, aerobic cultures were grown for ∼18 h in BHI, and samples were taken for RNA isolation. RNA was isolated with the use of the RNeasy midi kit (Qiagen, Manchester, United Kingdom). RNA quality was assessed with the use of a 2100 Bioanalyzer (Agilent, Cheshire, United Kingdom), and in all samples used, the RNA integrity number (RIN) was between 8 and 10. Subsequently, RNA was converted to cDNA with the use of random primers and the SuperScript III reverse transcriptase kit (Invitrogen, Thermo Fisher Scientific, Paisley, United Kingdom). Relative expression was calculated as the ratio between the expression of each of the target genes (kat, lmo0367, lmo1604, and tpx) and the expression of the 16S rRNA gene, which served as the reference gene in each cDNA sample. Calculations were carried out according to the advanced relative quantification settings of the LightCycler 480 software program, with PCR efficiency correction performed as described previously (17). The relative expression of each gene was calculated by a comparison of its expression relative to that of the 16S rRNA gene.
TABLE 1.
Primers used in this study
| Primera | Sequence (5′ to 3′) | Efficiency |
|---|---|---|
| katF | CGCACGGGAAATTTGTTACT | 2.05 |
| katR | GGTCAGGCTTCAAGGAATGA | |
| lmo0367F | TCATGTGCGCCTAGCCAAAG | 2.10 |
| lmo0367R | CCAATTGGGACGGTGTATTC | |
| lmo1604F | AGGCACACAAGCTCCAAGAT | 1.93 |
| lmo1604R | CGCAGCAAGAGGGTAGTTTA | |
| tpxF | GCGTTGTTCCTTCCATTGAT | 1.96 |
| tpxR | ATCGCGGTGGTCAGATAAAG | |
| 16SF | TGGGGAGCAAACAGGATTAG | 2.27 |
| 16SR | TAAGGTTCTTCGCGTTGCTT |
Primers were designed based on the L. monocytogenes EGD-e published sequences from the ListiList website (http://genolist.pasteur.fr/ListiList/) except katF, katR, 16SF, and 16SR, which were designed based on the Listeria monocytogenes 10403S cont5.3 whole-genome shotgun sequence (http://www.ncbi.nlm.nih.gov/genome/159?genome_assembly_id=159669). All primers designed on EGD-e background were confirmed to contain no mismatches with the 10403S background using Standard Nucleotide Blast.
Caco-2 proliferation assays.
The gentamicin protection assay was performed with the strains, as described previously (5), with minor modifications. Two days before the invasion assays were performed, 1.5 × 105 Caco-2 human colon adenocarcinoma cells (European Collection of Cell Cultures no. 86010202) were seeded in 24-well plates in Dulbecco's modified Eagle's medium containing 2 mM glutamine, 1% (wt/vol) nonessential amino acids, and 20% (vol/vol) fetal bovine serum supplemented with 100 U ml−1 penicillin-streptomycin (Sigma). Thirty minutes before coincubation, the medium in each well was replaced with prewarmed fresh medium without antibiotics. The OD600 values of stationary-phase bacterial cultures grown in BHI broth overnight at 37°C were determined, all cultures were washed twice with sterile phosphate-buffered saline (PBS), and the concentrations were adjusted to obtain similar OD600 values. We previously confirmed that there was a good correlation between OD600 and the number of cells for 10403S, as assessed by comparing the numbers of CFU and OD600 values. Coincubation was performed with approximately 2 × 107 CFU (multiplicity of infection [MOI], 50) of stationary-phase bacteria of the 10403S strain for 45 min at 37°C. Subsequently, Caco-2 cells were washed twice with PBS and suspended in Dulbecco's modified Eagle's medium containing 150 mg liter−1 gentamicin. After 45 min of incubation at 37°C, cells were washed twice with sterile PBS and lysed with 2 ml of Triton X-100 (1% [vol/vol]) in PBS. Following incubation for 5 min at 37°C, cell lysates were serially diluted and spread on BHI agar to determine the number of intracellular bacteria. Subsequently, to assess intracellular proliferation, intracellular bacteria were determined in a similar way every 2 h until 12 h postinvasion. This experiment was repeated 8 times, and for each time point, statistical analysis was performed using a paired t test. P values of >0.05 were considered statistically significant.
Statistical analysis.
In all cases, experiments were run at least in triplicate (unless stated), and the results were assessed with a paired Student t test. At a P value of <0.05, results were deemed statistically significant.
RESULTS
Survival in the presence of hydrogen peroxide.
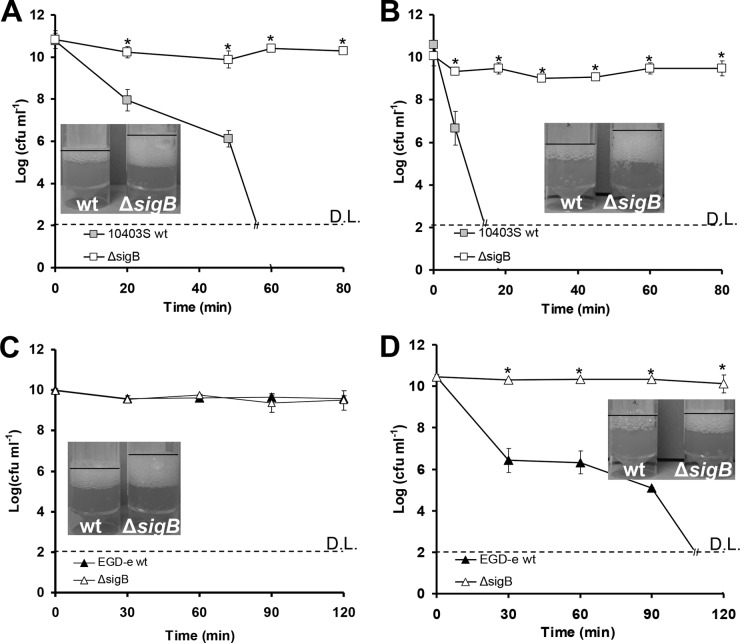
In both 10403S and EGD-e strains, the ΔsigB mutant was significantly more resistant to H2O2 than the WT at stationary phase when cultured under aerobic conditions (P < 0.05). When these cells of WT 10403S were challenged with 3% H2O2 at 30°C (Fig. 1A) or 37°C (Fig. 1B), they showed between 7- and 8-log reduction, while the numbers of the ΔsigB mutant remained almost unaffected (less than 1-log reduction). Similarly, the EGD-e ΔsigB mutant grown at 30°C (Fig. 1C) or 37°C (Fig. 1D) was not affected by 3% H2O2, demonstrating major resistance against H2O2. Notably, the EGD-e WT was not affected at 30°C (Fig. 1C), but it showed a >8-log reduction in CFU at 37°C when exposed to 3% H2O2 (Fig. 1D). To obtain a measurable effect for EGD-e at 30°C, we increased the concentration of H2O2 from 3% to 4.5%. Under these conditions, a reduction of more than 5 logs was achieved for the WT, while the sigB mutant remained unaffected (Fig. 2). From these data, we concluded that WT cells of both strains grown at 30°C were more resistant than those grown at 37°C. Furthermore, EGD-e seemed to be more resistant to H2O2 stress than 10403S.
FIG 1.
Cells of 10403S WT (gray boxes) and its isogenic ΔsigB mutant (white boxes) grown aerobically until stationary phase and challenged with 3% H2O2 at 30°C (A) or 37°C (B). Cells of EGD-e WT (black triangles) and its isogenic ΔsigB mutant (white triangles) grown until stationary phase and challenged with 3% H2O2 at 30°C (C) or 37°C (D). D.L. denotes the detection limit, and the asterisks denote a difference with statistical significance (P < 0.05). Catalase tests were performed in triplicate (one representative experiment is shown) with the addition of 100 μl of 30% H2O2 solution in 5-ml samples of the cultures. Cultures for catalase tests were grown under conditions similar to those used in challenge tests. The error bars represent the standard deviations.
FIG 2.
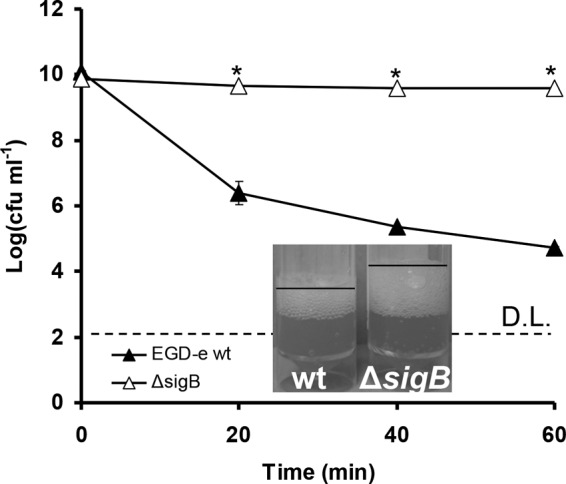
Cells of EGD-e WT (black triangles) and its isogenic ΔsigB mutant (white triangles) grown aerobically until stationary phase and challenged with 4.5% H2O2 at 30°C. D.L. denotes the detection limit, and the asterisks denote a difference with statistical significance (P < 0.05). Catalase tests were performed in triplicate (one representative experiment is shown) with the addition of 100 μl of 30% H2O2 solution in a 5-ml sample of the cultures. Cultures for catalase tests were grown under conditions similar to those used in challenge tests. The error bars represent the standard deviations.
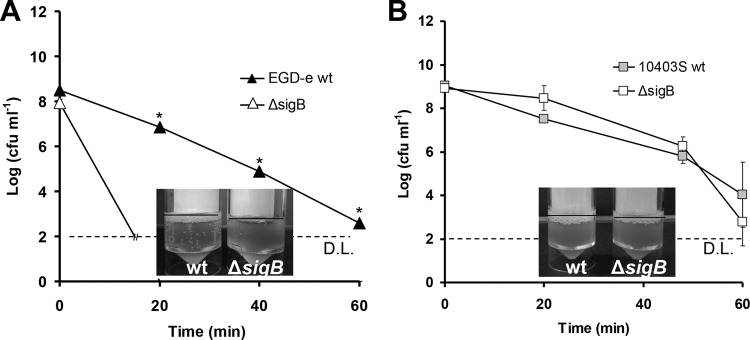
Despite the hyperresistance of the sigB mutant compared to the WT under aerobic conditions at stationary phase, the results obtained under anaerobic conditions were different. When cells of EGD-e grown anaerobically to stationary phase at 37°C were challenged with 0.4% H2O2, no reduction in CFU occurred in both the WT and its isogenic ΔsigB mutant within 60 min, while when they were challenged with 3% H2O2, CFU were reduced to below the detection limit within the first 20 min (data not shown). However, when the same cells were challenged with 1% H2O2, the WT was significantly more resistant than the ΔsigB mutant (P < 0.05; Fig. 3A). There was a statistically significant (P < 0.05) reduction of at least 6 logs for the EGD-e ΔsigB mutant within 20 min, while such a reduction occurred in the WT only after 60 min. However, under anaerobic conditions and in stationary phase for 10403S, there seemed to be no statistically significant difference (P > 0.05) between the WT and ΔsigB mutant at 0.4% H2O2 (Fig. 3B), while when 1% and 3% H2O2 were used, the numbers of both WT and the ΔsigB mutant were reduced to below the detection limit within the first 20 min (data not shown). Under anaerobic conditions, EGD-e was also significantly more resistant than 10403S since, as mentioned above, 0.4% H2O2 resulted in no reduction of the numbers for EGD-e (data not shown), while it caused a 5-log reduction for 10403S (P < 0.05; Fig. 3B).
FIG 3.
Cells of EGD-e (A) and 10403S (B) (WT and ΔsigB mutant) grown anaerobically (static in Sterilin Quickstart Universal polystyrene containers in an anaerobic chamber) at 37°C overnight and then challenged with 1% (A) and 0.4% (B) H2O2, respectively. D.L. denotes the detection limit, and asterisks denote a difference with statistical significance (P < 0.05). Catalase tests were performed in triplicate (one representative experiment is shown), with the addition of 100 μl of 30% H2O2 solution in a 5-ml sample of the cultures. Cultures for catalase tests were grown under conditions similar to those used in challenge tests. The error bars represent the standard deviations.
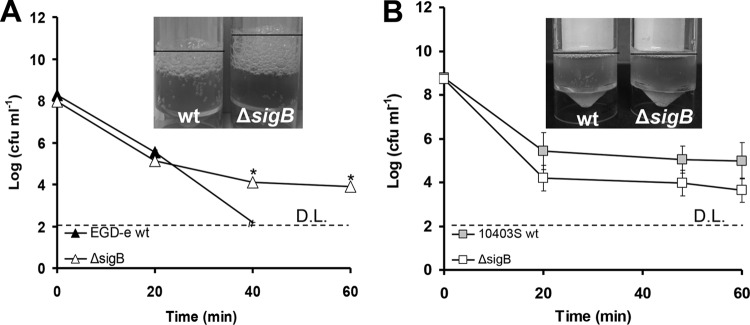
Subsequently, we investigated the effect of growth phase on resistance against H2O2. When cells of WT EGD-e grown aerobically at 37°C to mid-exponential phase were challenged with 0.4% H2O2, CFU remained stable for both the WT and the ΔsigB mutant within 60 min, while when they were challenged with 3% H2O2, the numbers of cells were reduced to below the detection limit within 20 min (data not shown). When these cells were challenged with 1% H2O2, the WT EGD-e was more sensitive than the ΔsigB mutant (Fig. 4A). However, this sensitivity was not as pronounced as in stationary phase, with the log reduction being similar for the first 20 min of the challenge. Interestingly enough, there was no significant difference between the ΔsigB mutant and WT 10403S at mid-exponential phase of growth at 37°C against 0.43% H2O2 (P > 0.05; Fig. 4B). Furthermore, when 1% and 3% H2O2 were used, the CFU of both the WT 10403S and ΔsigB mutant were reduced to below the detection limit within the first 20 min.
FIG 4.
Cells of EGD-e (A) and 10403S (B) (WT and ΔsigB mutant) grown aerobically at 37°C until the mid-exponential phase of growth and then challenged with 1% (A) and 0.43% (B) H2O2. D.L. denotes the detection limit, and the asterisks denote a difference with statistical significance (P < 0.05). Catalase tests were performed in triplicate (one representative experiment is shown), with the addition of 100 μl of 30% H2O2 solution in 5 ml of culture. Cultures for catalase tests were grown under conditions similar to those used in challenge tests. The error bars represent the standard deviations.
Disk diffusion assay.
Disk diffusion assays were performed with 10403S and EGD-e at both temperatures (30°C and 37°C). A statistically significant difference (P < 0.05) was found only with 10403S at 30°C, where the ΔsigB mutant showed a smaller inhibition zone than that of its isogenic WT (Table 2). No significant difference was observed with 10403S at 37°C and with EGD-e at both temperatures.
TABLE 2.
Zones of inhibition diameters for disk diffusion assays with 30% H2O2
| Temp (°C) | Zone of inhibition diam (mm) |
P | |
|---|---|---|---|
| WT | ΔsigB mutant | ||
| 10403S | |||
| 30 | 57 | 48 | 0.03a |
| 37 | 49 | 51 | 0.25 |
| EGD-e | |||
| 30 | 36 | 35 | 0.52 |
| 37 | 38 | 37 | 0.05 |
Denotes statistical significance over 6 replicates using paired t test.
Catalase test.
A catalase test was performed under all the conditions examined in this study. Under aerobic conditions and stationary phase, the ΔsigB mutants produced a more vigorous catalase reaction than their isogenic WT strains (Fig. 1 and 2). Under anaerobic conditions and stationary phase, the WT EGD-e showed stronger catalase activity than the ΔsigB mutant, which coincides with the higher survival rate of the WT over the ΔsigB mutant (Fig. 3A). However, under the same conditions, the WT 10403S did not show a major difference compared to the ΔsigB mutant (Fig. 3B). At mid-exponential phase under aerobic conditions, the ΔsigB mutant had a stronger catalase test result than the WT in EGD-e (Fig. 4A), while in 10403S, the WT and ΔsigB mutant showed no difference in catalase activity (Fig. 4B).
Furthermore, under aerobic conditions, stationary-phase cells produced a more vigorous catalase activity test result than those grown anaerobically. In addition, under aerobic conditions, mid-exponential-phase cells showed weaker catalase activity than those at stationary phase, while EGD-e produced stronger catalase activity than 10403S under all conditions.
Catalase activity in protein extracts.
These experiments were performed with stationary-phase cells of 10403S at both temperatures (30°C and 37°C) to assess intracellular catalase activity. In lysates obtained from 10403S cells grown at 37°C under aerobic conditions, the ΔsigB mutant was able to degrade H2O2 faster than its isogenic WT (Fig. 5). The results were similar with cells obtained at 30°C (data not shown). This suggests a stronger catalase reaction for the ΔsigB mutant at stationary phase.
FIG 5.
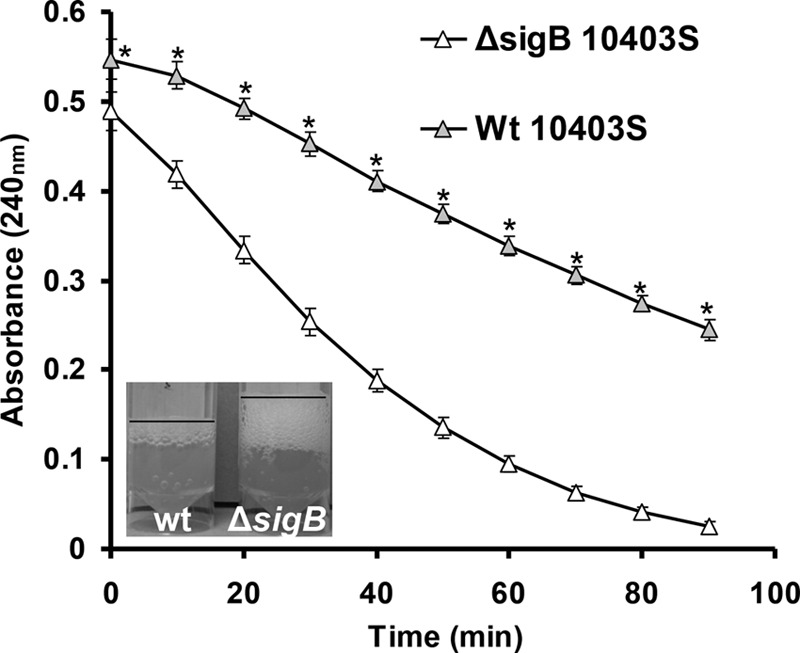
Absorbance at 240 nm (corresponding to concentration of H2O2) indicating the catalase activity of cell lysates of 10403S WT (gray triangles) and its isogenic ΔsigB mutant (white triangles). A more rapid reduction of the absorbance indicates more-rapid H2O2 degradation and therefore higher catalase activity. Cells used for lysate preparation were from the stationary phase and grown at 37°C. Each point is an average of the results from 3 biological and 3 technical replicates, while the error bars represent the standard deviations. The asterisks denote a difference with statistical significance (P < 0.05).
DO measurements.
The DO was measured in bacterial cultures grown aerobically and anaerobically (Table 3). No statistically significant differences were found between the levels of DO in the WT and ΔsigB mutant (P > 0.05) under any of the conditions studied.
TABLE 3.
Dissolved oxygen in Listeria monocytogenes cultures
| Strain | Dissolved oxygen (mean ± SD) (mg/liter) |
||
|---|---|---|---|
| Aerobic growth |
Anaerobic growth | ||
| 30°C | 37°C | ||
| EGD-e | |||
| WT | 7.25 ± 0.07 | 6.53 ± 0.23 | 0.07 ± 0.04 |
| ΔsigB mutant | 7.25 ± 0.08 | 6.57 ± 0.24 | 0.07 ± 0.03 |
| 10403S | |||
| WT | 7.00 ± 0.09 | 6.41 ± 0.12 | 0.06 ± 0.03 |
| ΔsigB mutant | 7.10 ± 0.05 | 6.50 ± 0.22 | 0.21 ± 0.19 |
Transcriptional analysis of genes contributing to oxidative stress.
No statistically significant difference was observed between the ΔsigB mutant and WT in the transcription of any of the genes analyzed. The ΔsigB mutant transcript levels for kat, lmo0367, lmo1604, and tpx were 49.40%, 46.18%, 47.67%, and 45.05% of those for the WT, respectively, but in all cases, the P value was >0.05.
Caco-2 proliferation assays.
The intracellular proliferation of the 10403S WT and ΔsigB mutant was assessed for 12 h (Fig. 6). Except for the first 2 h, where a significant difference was found (P < 0.05), the bacterial proliferation was similar in the WT and ΔsigB mutant (P > 0.05). However, the statistically significant difference (P < 0.05) in the first 2 h is due to the previously well-documented lower invasion of the ΔsigB mutant. Introducing a 1.2-h delay in the WT curve to compensate for the lower initial numbers of the ΔsigB mutant results in similar proliferation curves for the two strains (Fig. 6, inset).
FIG 6.
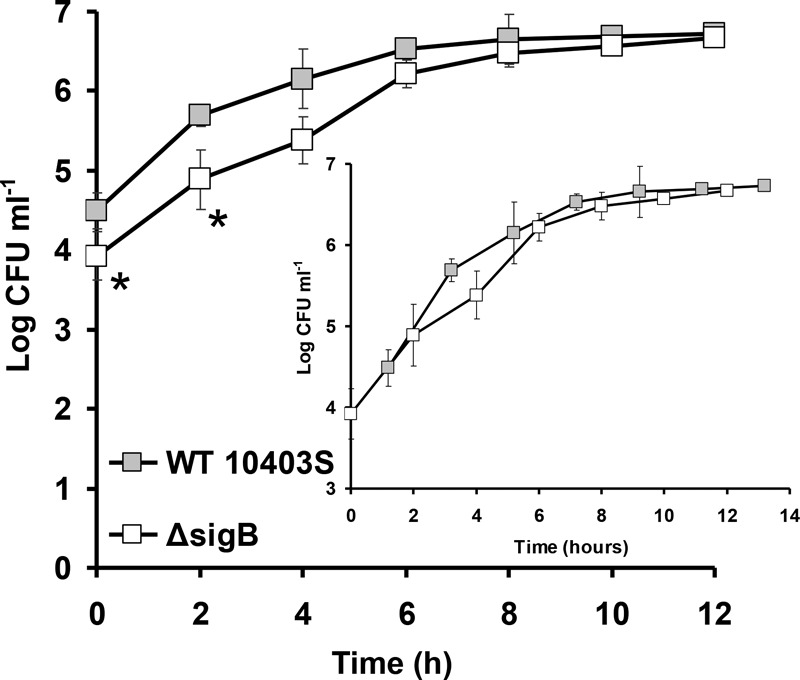
Proliferation assays of cells of L. monocytogenes 10403S WT and its isogenic ΔsigB mutant in Caco-2 epithelial cells. The asterisks denote a difference with statistical significance (P < 0.05). The inset shows both proliferation curves of 10403S WT and the ΔsigB mutant overlaid with WT curve delayed by 1.2 h. The error bars represent the standard deviations.
DISCUSSION
In the present study, we investigate the role of L. monocytogenes SigB in resistance against hydrogen peroxide-mediated oxidative stress. SigB is the central regulator of stress genes in Gram-positive bacteria, such as L. monocytogenes. It is well known that SigB plays an important role against various stresses, such as acid, heat, salt, bile acids, etc. (6–8), as well as oxidative stress (7, 9). Interestingly, despite the above-mentioned well-established role for SigB as an important factor for stress resistance, we show conclusively in two reference strains of L. monocytogenes (EGD-e and 10403S) that SigB confers a survival disadvantage during oxidative stress in stationary-phase cells grown aerobically (Fig. 1 and 2). The loss of SigB exerted such a dramatic effect that the ΔsigB mutant was more resistant to H2O2 by at least 7 logs compared to the WT. In general, stationary-phase cells were more resistant to H2O2 at 30°C than at 37°C (Fig. 1 and 2), which was also apparent with the disk diffusion results (Table 2). However, the hyperresistant phenotype of the ΔsigB mutant was not so pronounced in disk diffusion assays, as it was not observed in EGD-e but only slightly in 10403S at 30°C and not at 37°C. This is most probably due to the disk diffusion assays assessing inhibition of growth, rather than survival after growth, which is mainly assessed in this study.
Overall, our results show a similar behavior between L. monocytogenes and B. cereus, where the presence of sigB also increased sensitivity against oxidative stress (11). In contrast with the mentioned study, where only mid-exponential-phase cells were used, we assessed exponential- and stationary-phase cells. The role of SigB in L. monocytogenes under oxidative stress is controversial, as there are discrepancies in the results of different workers. Our results are partly in agreement with those of Moorhead and Dykes (12), where the ΔsigB mutant of a serotype 4c strain L. monocytogenes was more resistant to oxidative stress than its isogenic WT. However, no other quantitative data apart from this information were presented in this paper to allow comparisons. It was also noted that L61, a serotype 1/2a strain, did not show the above-mentioned phenotype, as the ΔsigB mutant and WT showed similar resistance to oxidative stress. As a result, the authors suggest that this phenotype might be serotype dependent, occurring in serotype 4c strains. However, this does not seem to be the case, since we observed this phenotype in a serotype 1/2a strain (10403S). On the contrary, Oliver et al. (18) showed that SigB plays a limited role in the resistance of L. monocytogenes F2365 against oxidative stress (18), despite the fact that this strain does not utilize an active SigB-dependent response pathway, and therefore, the influence of SigB to its stress resistance is expected to be limited anyway. Most interestingly, SigB has been shown to contribute to resistance against oxidative stress in other strains, including the one we used (10403S) in the current study (7, 9). The above-mentioned discrepancies might be due to the authors (7) using cumene hydroperoxide instead of the H2O2 used in our case, or due to the different length of time cultures were allowed to grow prior to experiments, as we used 18-h cultures instead of 12-h cultures. Cultures grown for a shorter length of time might have a behavior similar to that of exponential-phase cultures, where WT cells might survive better than the ΔsigB mutant cells. We are currently investigating the point during growth that signifies the development of the hyperresistant phenotype. However, the most possible explanation for the discrepancies might be different levels of oxygen tension in the cultures, although strain variability should not be excluded. Specifically, in most of the above-mentioned cases, small culture volumes were used prior to experiments (5 ml of BHI). The limited headspace in this setup might not have allowed sufficient levels of oxygen to dissolve in the medium.
As we demonstrate here, oxygen tension and strain variability influence greatly the H2O2 resistance phenotype. Under all the conditions studied (aerobic at both temperatures and anaerobic), the WT and sigB mutant presented similar levels of oxygen (Table 3), suggesting that the differences in H2O2 resistance between the WT and ΔsigB mutant could not be attributed to different oxygen levels. When grown under anaerobic conditions, and consequently with low levels of oxygen, both EGD-e and 10403S strains were considerably more sensitive than at aerobic growth. However, under anaerobic conditions, the absence of sigB in EGD-e resulted in an H2O2-sensitive phenotype (Fig. 3A). This is in contrast to what has been observed in B. cereus, where the H2O2-resistant phenotype of the ΔsigB mutant was retained following growth under anaerobic conditions, albeit to a lesser extent (11). However, this might be attributed to the different stage of growth affecting sigB expression, as experiments under anaerobic conditions in B. cereus were performed with mid-exponential-phase cells, while we used stationary-phase cells. As observed in B. cereus, the H2O2 resistance was lower under anaerobic than under aerobic conditions (11), as a 3-fold-lower concentration of H2O2 achieved a higher log reduction for EGD-e anaerobically grown cells (Fig. 3A) than those grown aerobically (Fig. 1D). This is because H2O2 is a by-product of aerobic growth, and cellular responses against it (e.g., catalase activity) are expected to appear under aerobic conditions (11). In contrast, under anaerobic conditions, no H2O2 is expected to be formed, and therefore, this might result in the absence of responses to H2O2.
As mentioned earlier, another factor affecting sigB expression is the stage of growth. Cells of EGD-e WT grown aerobically to mid-exponential phase (Fig. 4A) were highly sensitive to low concentrations (1%) of H2O2, surpassing in sensitivity even those grown anaerobically (Fig. 3A). Furthermore, as expected, the role of SigB seemed to be limited at mid-exponential phase (10), as there was only a minor difference between the ΔsigB mutant and WT EGD-e over the first 20 min (Fig. 4A). Following 40 min of H2O2 challenge, the ΔsigB mutant appeared slightly more resistant than the WT, which is consistent with the catalase activity tests, confirming that SigB confers a survival disadvantage for the cells grown aerobically, even if its influence is minimal. However, no statistically significant results (P > 0.05) were obtained with 10403S cells grown aerobically until the exponential phase (Fig. 4B), while the WT appeared slightly more resistant than the ΔsigB mutant to 0.43% H2O2.
In general, we found that the survival results could be explained solely by the catalase tests, with cells producing more vigorous bubbling being more resistant to oxidative stress. At stationary phase under aerobic conditions, in all cases, the ΔsigB mutant showed stronger catalase activity upon addition of 30% H2O2 than the WT (Fig. 1 and 2). To assess the intracellular catalase activity (H2O2 degradation in protein extracts; Fig. 5) we found that it corresponded well with the overall catalase activity of live cells. This confirmed beyond a doubt that mutations in sigB result in enhanced catalase activity and oxidative stress resistance under aerobic conditions, which coincides with what has been described in B. cereus (11).
To identify possible molecular mechanisms behind the higher catalase activity and the hyperresistance of the ΔsigB mutant to oxidative stress, we looked at the transcription of specific genes that protect against oxidative stress and H2O2. Interestingly enough, despite the difference between the catalase activities of the WT and ΔsigB mutant, we found no difference in the transcription of the kat (lmo2785) gene encoding catalase between the ΔsigB mutant and the WT. Since, to our knowledge, L. monocytogenes possesses only one catalase gene, these data suggest that differential expression of the catalase gene might occur posttranscriptionally; alternatively, differential transcription might occur earlier during growth. In addition, we investigated the transcription of thiol peroxidase (tpx or lmo1583), 2-cys peroxiredoxin (lmo1604), and a hypothetical iron peroxidase (lmo0367) that could potentially confer protection against oxidative stress. Interestingly, Lmo1604 has been found to be negatively regulated by σB but in the presence of salt (16). However, similarly to kat, all these genes showed similar transcription in the WT and ΔsigB mutant.
Since oxidative stress is one of the most important stresses occurring within the phagolysosome, it could be expected that this hyperresistance of the ΔsigB mutant to oxidative stress might translate into higher intracellular survival. However, this is not the case, as we show here (Fig. 6), and as was demonstrated previously in other studies (5, 19, 20). We saw a lower level of invasion for the ΔsigB mutant, which has been documented by other workers (5) and occurs due to lower expression of internalins inlA and inlB (21), but not major differences during the proliferation stage. However, the explanation for this lies in the fact that following invasion, L. monocytogenes does not utilize the SigB intracellularly, as was shown previously (22), and as a result, ΔsigB mutants do not show any difference compared to the WT in proliferation assays (20, 22) or in systemic spread in the guinea pig model (19). Therefore, SigB expression occurs in the gut at the extracellular stage of infection, as it is important for internalin expression, but it is subsequently abolished during the intracellular stage. It could be said that L. monocytogenes tries to mimic the behavior of the ΔsigB mutant intracellularly. A logical explanation for this behavior is that the abolishment of SigB expression intracellularly could lead to higher resistance toward oxidative stress, which occurs in the phagolysosome. However, there is no satisfactory explanation on how L. monocytogenes is able to prevent the upregulation of SigB intracellularly, since the phagolysosome is a particularly stressful environment. We currently investigate this further as well as the environmental factors affecting this behavior.
This study illustrates that although sigB is important for general stress resistance, an important deviation to this pattern occurs in the case of oxidative stress under aerobic conditions at stationary phase. In addition, we demonstrate that oxygen tension and growth phase can affect significantly this phenotype in L. monocytogenes. An attempt to identify the molecular mechanism behind this phenotype was unsuccessful, and future work will focus in this direction.
ACKNOWLEDGMENTS
We thank all members of the FMSU and colleagues in the Department of Food and Nutritional Sciences, University of Reading, Reading, United Kingdom, and the Bacterial Stress Response Group and colleagues in microbiology for helpful discussions. The ΔsigB mutant strains used in this study were generously supplied by Kathryn Boor (Cornell University) and Cormac Gahan (University College Cork).
This work was supported by a Marie Curie European Reintegration Grant (grant ERG 265154), a Science Foundation Ireland Starting Investigator Research Grant (SIRG; grant 09/SIRG/B1570) awarded to K. A. G. Karatzas, by an SFI Research Frontiers Programme grant to C. P. O'Byrne (grant 11/RFP.1/GEN/3267), and the University of Reading.
REFERENCES
- 1.Mook P, Grant KA, Little CL, Kafatos G, Gillespie IA. 2010. Emergence of pregnancy-related listeriosis amongst ethnic minorities in England and Wales. Euro Surveill 15:pii=19610 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19610. [DOI] [PubMed] [Google Scholar]
- 2.FDA/USDA. 2003. Docket no. 1999N-1168. 68. U.S. Department of Agriculture, Washington, DC. [Google Scholar]
- 3.O'Byrne CP, Karatzas KAG. 2008. Chapter 5. The role of sigma B (SigB) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence, p 115–140. In Laskin AI, Sariaslani S, Gadd M (ed), Advances in applied microbiology, vol 65 Academic Press, Cambridge, MA. [DOI] [PubMed] [Google Scholar]
- 4.van Schaik W, Abee T. 2005. The role of σB in the stress response of Gram-positive bacteria–targets for food preservation and safety. Curr Opin Biotechnol 16:218–224. doi: 10.1016/j.copbio.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Abram F, Starr E, Karatzas KAG, Matlawska-Wasowska K, Boyd A, Wiedmann M, Boor KJ, Connally D, O'Byrne CP. 2008. Identification of components of the SigB regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl Environ Microbiol 74:6848–6858. doi: 10.1128/AEM.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sue D, Fink D, Wiedmann M, Boor KJ. 2004. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150:3843–3855. doi: 10.1099/mic.0.27257-0. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira A, O'Byrne CP, Boor KJ. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl Environ Microbiol 67:4454–4457. doi: 10.1128/AEM.67.10.4454-4457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira A, Sue D, O'Byrne CP, Boor KJ. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl Environ Microbiol 69:2692–2698. doi: 10.1128/AEM.69.5.2692-2698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver HF, Orsi RH, Wiedmann M, Boor KJ. 2010. L. monocytogenes σB has a small core regulon and a conserved role in virulence but makes differential contributions to stress tolerance across a diverse collection of strains. Appl Environ Microbiol 76:4216–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Utratna M, Cosgrave E, Baustian C, Ceredig RH, O'Byrne CP. 2014. Effects of growth phase and temperature on σB activity within a L. monocytogenes population: evidence for RsbV-independent activation of σB at refrigeration temperatures. Biomed Res Int 2014:641647. doi: 10.1155/2014/641647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Schaik W, Zwietering MH, de Vos WM, Abee T. 2005. Deletion of the sigB gene in Bacillus cereus ATCC 14579 leads to hydrogen peroxide hyperresistance. Appl Environ Microbiol 71:6427–6430. doi: 10.1128/AEM.71.10.6427-6430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moorhead SM, Dykes GA. 2003. The role of the sigB gene in the general stress response of Listeria monocytogenes varies between a strain of serotype 1/2a and a strain of serotype 4c. Curr Microbiol 46:0461–0466. doi: 10.1007/s00284-002-3867-6. [DOI] [PubMed] [Google Scholar]
- 13.Begley M, Sleator RD, Gahan CG, Hill C. 2005. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect Immun 73:894–904. doi: 10.1128/IAI.73.2.894-904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiedmann M, Arvik TJ, Hurley RJ, Boor KJ. 1998. General stress transcription factor sigmaB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol 180:3650–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Schellhorn HE. 2007. Rapid kinetic microassay for catalase activity. J Biomol Tech 18:185–187. [PMC free article] [PubMed] [Google Scholar]
- 16.Abram F, Su W-L, Wiedmann M, Boor KJ, Coote P, Botting C, Karatzas KAG, O'Byrne CP. 2008. Proteomic analyses of a Listeria monocytogenes mutant lacking sigB identify new components of the σΒ regulon and highlight a role for σΒ in the utilization of glycerol. Appl Environ Microbiol 74:594–604. doi: 10.1128/AEM.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karatzas K-AG, Brennan O, Heavin S, Morrissey J, O'Byrne CP. 2010. Intracellular accumulation of high levels of γ-aminobutyrate by L. monocytogenes 10403S in response to low pH: uncoupling of γ-aminobutyrate synthesis from efflux in a chemically defined medium. Appl Environ Microbiol 76:3529–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver HF, Orsi RH, Wiedmann M, Boor KJ. 2013. σΒ plays a limited role in the ability of L. monocytogenes strain F2365 to survive oxidative and acid stress and in its virulence characteristics. J Food Prot 76:2079–2086. [DOI] [PubMed] [Google Scholar]
- 19.Garner MR, Njaa BL, Wiedmann M, Boor KJ. 2006. σΒ contributes to L. monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect Immun 74:876–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H, Boor KJ, Marquis H. 2004. Listeria monocytogenes σΒ contributes to invasion of human intestinal epithelial cells. Infect Immun 72:7374–7378. doi: 10.1128/IAI.72.12.7374-7378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H, Marquis H, Boor KJ. 2005. SigmaB contributes to L. monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151:3215–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. 2006. Intracellular gene expression profile of L. monocytogenes. Infect Immun 74:1323–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]