ABSTRACT
Fairy circles (FCs) are barren circular patches of soil surrounded by grass species. Their origin is poorly understood. FCs feature in both the gravel plains and the dune fields of the Namib Desert. While a substantial number of hypotheses to explain the origin and/or maintenance of fairy circles have been presented, none are completely consistent with either their properties or their distribution. In this study, we investigated the hypothesis that FC formation in dunes and gravel plains is due to microbial phytopathogenesis. Surface soils from five gravel plain and five dune FCs, together with control soil samples, were analyzed using high-throughput sequencing of bacterial/archaeal (16S rRNA gene) and fungal (internal transcribed spacer [ITS] region) phylogenetic markers. Our analyses showed that gravel plain and dune FC microbial communities are phylogenetically distinct and that FC communities differ from those of adjacent vegetated soils. Furthermore, various soil physicochemical properties, particularly the pH, the Ca, P, Na, and SO4 contents, the soil particle size, and the percentage of carbon, significantly influenced the compositions of dune and gravel plain FC microbial communities, but none were found to segregate FC and vegetated soil communities. Nevertheless, 9 bacterial, 1 archaeal, and 57 fungal phylotypes were identified as FC specific, since they were present within the gravel plain and dune FC soils only, not in the vegetated soils. Some of these FC-specific phylotypes were assigned to taxa known to harbor phytopathogenic microorganisms. This suggests that these FC-specific microbial taxa may be involved in the formation and/or maintenance of Namib Desert FCs.
IMPORTANCE Fairy circles (FCs) are mysterious barren circular patches of soil found within a grass matrix in the dune fields and gravel plains of the Namib Desert. Various hypotheses attempting to explain this phenomenon have been proposed. To date, however, none have been successful in fully explaining the etiology of FCs, particularly since gravel plain FCs have been largely ignored. In this study, we investigated the hypothesis that microorganisms could be involved in the FC phenomenon through phytopathogenesis. We show that the microbial communities in FC and control vegetated soil samples were significantly different. Furthermore, we detected 67 FC-specific microbial phylotypes, i.e., phylotypes present solely in both gravel plain and dune FC soils, some of which were closely related to known phytopathogens. Our results, therefore, demonstrate that microorganisms may play a role in the formation and/or maintenance of Namib Desert FCs, possibly via phytopathogenic activities.
INTRODUCTION
Fairy circles (FCs) are circular patches of soil completely devoid of vegetation within a matrix of grass, typically Stipagrostis spp. (1, 2). Common in the Namib Desert, and recently observed in the Australian outback (3), FCs are limited to arid areas that receive between 50 and 100 mm precipitation per annum. FCs are nonpermanent; they typically appear, enlarge, and disappear, with an estimated life span ranging from 22 to 60 years (4, 5).
Although FCs were first reported in the 1970s (6), there is currently no scientific consensus on the mechanisms behind their formation and maintenance (2, 7–9). Fauna-based hypotheses suggest that FCs result from the harvesting activities of termites, ants, or rodents (1, 5, 10–12). The hypothesis of spatial self-organization of plants argues that FCs are the product of extreme aridity (13, 14). During times of drought, competition between plants for nutrients and water resources leads to the formation of barren patches (14), which in turn act as water traps, utilized by the peripheral grasses through extensive root systems and soil-water diffusion (13). Other published hypotheses implicate localized radioactivity hot spots (2), natural gas seepages (8), and the release of allelopathic compounds by dead Euphorbia damarana plants (5, 15). However, all previous studies testing these hypotheses have focused solely on FCs located in Namib Desert dunes (2, 4, 8, 10, 14, 15), whereas FCs also occur, albeit more sparsely, in Namib Desert gravel plains (16). The latter distribution contradicts several of the extant hypotheses. For example, well-defined spatial patterning is not a characteristic of the gravel plain FCs, and E. damarana is not endemic to these regions. A recent hypothesis (16) has suggested the involvement of microorganisms in the FC phenomenon, because two fungal terminal restriction fragments (T-RFs) were detected within gravel plain FCs. However, due to the nature of the fingerprinting technique used, these fragments could not be assigned taxonomic identities (16). The fact that FCs are clear circular patterns with a distinct life cycle (2, 4) is compatible with the presence of a microbial phytotoxic compound that inhibits grass/plant growth (17). Although contested by the findings of recent in situ soil transfer experiments for five dune FCs (7), the phytotoxicity of FC soils has been shown in greenhouse experiments (2), an observation consistent with the presence of an active, possibly microbial plant-inhibitory compound.
To further evaluate the involvement of microorganisms in FCs, we therefore investigated the edaphic microbial communities from both dune and gravel plain FCs, together with control communities from adjacent vegetated soils. This is the first study comparing dune FCs with gravel plain FCs. We used Illumina MiSeq high-throughput sequencing of the 16S rRNA gene and the internal transcribed spacer (ITS) region to target the bacterial, archaeal, and fungal members of each set of soil samples. Our aims were (i) to assess differences in microbial community structures between Namib Desert dune and gravel plain FC soils and control vegetated soils and (ii) to identify microorganisms present solely within dune and gravel plain FC soils. The latter would constitute potential candidates for involvement in the formation and/or maintenance of Namib Desert FCs.
MATERIALS AND METHODS
Sample collection.
Namib Desert gravel plain (23°33′0″S, 15°2′0″E) and dune (24°32′47″S, 15°19′47″E) soil samples were collected in April 2014. At each site, three pseudoreplicates (∼400 g) of surface soils (depth, 0 to 5 cm) were collected aseptically from the center of each of five individual FCs and from adjacent vegetated matrix sites (at a distance of twice the radius from the FC center) (n = 60) (Fig. 1). For each set of soil samples, the three pseudoreplicates were combined in equal amounts (~100 g), for a total of of 20 samples. The samples were transported and were stored at −20°C until further analyses.
FIG 1.
Photographs of Namib Desert dune (A) and gravel plain (B) fairy circles (from J.-B. Ramond). Three pseudoreplicates were taken from inside the fairy circle (red circles) and three from the control vegetated matrix (green circles).
Soil physicochemical analyses.
For each soil sample, the three pseudoreplicates were combined in equal amounts (∼50 g) and were sieved (2 mm) prior to analysis. The soil pH, conductivity, and sodium (Na), potassium (K), calcium (Ca), magnesium (Mg), chloride (Cl), sulfate (SO4), phosphorus (P), ammonium (NH4), and nitrate (NO3) contents were analyzed by Bemlab (Pty) Ltd. (Strand, Western Cape, South Africa) using standard protocols. The percentage of carbon was determined by use of the Walkley-Black method (18), and the particle size by use of the hydrometer method (19), at the Department of Plant Production and Soil Science, University of Pretoria.
Metagenomic DNA extraction and MiSeq amplicon sequencing.
Metagenomic DNA was extracted from all samples using the PowerSoil DNA isolation kit (Mo Bio Laboratories, CA, USA) according to the manufacturer's instructions. DNA concentrations were determined using a Qubit 3.0 fluorometer (Thermo Fisher Scientific, USA). The V4 variable region of the bacterial/archaeal 16S rRNA gene was amplified by PCR using the 515F (5′-GTGYCAGCMGCCGCGGTAA-3′)/806R (5′-GGACTACNVGGGTWTCTAAT-3′) primer set (where N = A, T, C, or G and Y = C or T) (20), and the fungal ITS1 gene region was amplified by PCR using the ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′)/ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) primer set (21). Sequencing was performed by a commercial service provider (MR DNA, Shallowater, TX, USA) on an Illumina MiSeq platform according to the manufacturer's guidelines.
MiSeq sequencing analyses.
Sequence data analysis was performed using the QIIME (version 1.8.0) platform (22). Low-quality sequences were removed using default parameters; during sequence demultiplexing, reads were truncated if individual bases had a Phred score of <20 for 6 consecutive reads for the ITS region data set or for 3 consecutive reads for the 16S rRNA gene data set. Chimeric sequences were removed using the usearch61 method. Sequences were clustered into operational taxonomic units (OTUs) using the UCLUST-based open-reference OTU clustering pipeline with a 97% sequence identity cutoff (22) against the Greengenes database (23) for the 16S rRNA gene and against the UNITE database (24) for the ITS region. Bacterial or archaeal taxonomy was assigned to representative 16S rRNA gene sequences using the UCLUST method and the Greengenes reference database. Fungal taxonomy was assigned to ITS sequences using the RDP classifier (25) and the UNITE database.
Statistical analyses.
Statistical analyses were performed using the R statistical environment (26) with PRIMER software (version 6; Primer-E Ltd., Ivybridge, United Kingdom). Soil physicochemical data were analyzed by a draftsman plot (27) to assess the skewness of the data, after which the K and Cl contents and the percentage of silt were log(x + 1) transformed. Following data normalization, a Euclidean distance matrix was generated. Principal-component analysis (PCA) plots were constructed to assess the relationships among samples, and a permutational analysis of variance (PERMANOVA) test was used to identify significant differences between zones (i.e., dune FC, dune control, gravel plain FC, and gravel plain control).
Samples were randomly subsampled to 14,000 and 34,000 reads for the 16S rRNA gene and ITS region data sets, respectively. Singletons were removed prior to subsequent analyses. Measurements of α-diversity (total number of OTUs; Shannon and Simpson indices), β-diversity (Whittaker index), and γ-diversity were performed using the vegan package in R (28). Differences in diversity between zones were assessed by analysis of variance (ANOVA) and Tukey's honestly significant difference (HSD) post hoc test. Biological data sets were square root transformed prior to community structure analysis. Nonmetric multidimensional scaling (NMDS) ordination plots were generated from 16S rRNA gene and ITS region data sets using Bray-Curtis dissimilarity matrices, and PERMANOVA was performed to identify significant differences between the community structures of each zone. Abiotic drivers of microbial community structure were assessed by redundancy analysis (RDA) using the vegan package in R (26, 28). Plots were constructed using the ggplot2 package in R (26, 29).
Accession number(s).
All sequence data generated as part of this study have been deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information under accession number SRP069846.
RESULTS
Properties of Namib Desert soils.
The physicochemical properties of the soils of the four zones studied (FC and vegetated control soils from both dunes and gravel plains) are summarized in Table 1. A principal-component analysis (PCA) separated the samples by site (dune versus gravel plain) along PC1, explaining 47.4% of the variation observed (Fig. 2, dashed line). This difference was shown to be statistically significant using PERMANOVA (pseudo-F > 29.25; P < 0.001). Overall, the physicochemical properties differed significantly between gravel plain and dune soils (e.g., in Na [P < 0.0001], Ca [P < 0.0001], and SO4 [P < 0.0001] contents and in the percentage of carbon [P < 0.005]). Dune soils had a larger particle size than gravel plain soils (sand, 91.5% ± 2.5% versus 97.5% ± 1.6%; silt, 5.3% ± 1.5% versus 2.5% ± 1.6% [P < 0.05]; clay, 3.3% ± 2.0% versus 0.0% [below the detection limit] [P < 0.05]). However, at both sites, the soil physicochemistries of the different zones sampled (FC versus control soils) could not be statistically distinguished (pseudo-F < 0.31; P > 0.05) (Fig. 2).
TABLE 1.
Physicochemical properties of the soils of the dune and gravel plain fairy circle centers and their respective vegetated controls
| Property | Value for soila |
|||
|---|---|---|---|---|
| Dune |
Gravel plain |
|||
| Control | FC center | Control | FC center | |
| pH | 7.0 (±0.2) | 7.2 (±0.2) | 6.9 (±0.1) | 7.0 (±0.2) |
| Conductivity (mS/m) | 8.62 (±0.49) | 9.22 (±0.91) | 8.50 (±1.04) | 9.59 (±1.71) |
| Concn (mg/liter) of: | ||||
| P | 0.30 (± 0.11) | 0.19 (±0.02) | 0.63 (±0.19) | 0.40 (±0.06) |
| Na | 1.38 (±0.29) | 1.34 (±0.18) | 4.31 (±0.80) | 5.70 (±1.48) |
| K | 3.71 (±0.57) | 5.63 (±5.41) | 7.25 (±5.60) | 4.91 (±1.01) |
| Ca | 12.72 (±0.82) | 14.13 (±1.07) | 9.27 (±1.24) | 9.78 (±1.26) |
| Mg | 1.96 (±0.28) | 2.12 (±0.13) | 2.20 (±0.47) | 2.28 (±0.16) |
| Cl | 6.16 (±0.62) | 13.32 (±15.74) | 15.97 (±17.12) | 10.24 (±3.46) |
| SO4 | 1.86 (±0.38) | 1.57 (±0.18) | 3.96 (±1.02) | 4.09 (±0.70) |
| NH4 | 0.32 (±0.02) | 0.35 (±0.04) | 0.37 (±0.02) | 0.36 (±0.03) |
| NO3 | 0.36 (±0.26) | 0.21 (±0.11) | ND | 0.52 (±0.63) |
| % of: | ||||
| Carbon | 0.03 (±0.02) | 0.03 (±0.02) | 0.08 (±0.01) | 0.05 (±0.01) |
| Clay | ND | ND | 3.53 (±2.24) | 3.02 (±1.73) |
| Silt | 2.09 (±1.32) | 2.97 (±1.72) | 5.74 (±1.11) | 4.79 (±1.68) |
| Sand | 97.91 (±1.32) | 97.03 (±1.72) | 90.73 (±2.61) | 92.19 (±2.22) |
There were five samples for each type of soil. Values are means (± standard deviations). ND, not determined (i.e., below the detection limit).
FIG 2.

Principal-component analysis of physicochemical data from fairy circle and control soils based on a Euclidean distance matrix. Samples are indicated by colored symbols as follows: red squares, dune FC center; blue circles, dune control; green triangles, gravel plain FC center; purple diamonds, gravel plain control. The dashed line indicates the significant difference observed between the physicochemical properties of gravel plain and dune soils (P < 0.001).
Structures of microbial communities in Namib Desert soils.
Illumina MiSeq sequencing generated a total of 537,488 raw reads for the 16S rRNA gene amplicon data set and 1,322,502 raw reads for the ITS region. Sequences were randomly subsampled to 14,000 and 34,000 reads per sample for the 16S rRNA gene and ITS region data sets, respectively (see Fig. SA1 in the supplemental material). After subsampling, averages of 1,081 (±87) bacterial, 35 (±4) archaeal, and 538 (±203) fungal OTUs were assigned per sample based on 97% identity cutoff values (Table 2).
TABLE 2.
α-, β-, and γ-diversities of bacterial, archaeal, and fungal communities in each of the four microenvironments studied
| Community | Microenvironmenta | α-Diversityb |
β-Diversity (Whittaker index) | γ-Diversity | ||
|---|---|---|---|---|---|---|
| No. of species | Shannon index | Simpson index | ||||
| Bacteria | Dune control | 1,153.8 (±69.8) | 5.90 (±0.07) | 0.992 (±0.001) | 1.521 | 1,755 |
| Dune FC center | 1,018.6 (±104.6) | 5.79 (±0.19) | 0.988 (±0.006) | 1.683 | 1,714 | |
| GP control | 1,046.4 (±45.4) | 5.72 (±0.05) | 0.992 (±0.001) | 1.662 | 1,756 | |
| GP FC center | 1,103.2 (±35.3) | 5.84 (±0.14) | 0.991 (±0.005) | 1.678 | 1,834 | |
| Archaea | Dune control | 34.2 (±2.7) | 2.07 (±0.26) | 0.767 (±0.066) | 1.433 | 49 |
| Dune FC center | 35.6 (±4.5) | 2.14 (±0.22) | 0.777 (±0.079) | 1.404 | 50 | |
| GP control | 32.8 (±2.2) | 2.39 (±0.11) | 0.865 (±0.017) | 1.524 | 50 | |
| GP FC center | 36.4 (±3.4) | 2.24 (±0.30) | 0.825 (±0.065) | 1.484 | 54 | |
| Fungi | Dune control | 553.0 (±88.0) | 2.88 (±0.46) | 0.842 (±0.077) | 1.951 | 1,079 |
| Dune FC center | 366.4 (±59.0) | 2.61 (±0.42) | 0.832 (±0.070) | 4.539 | 1,663 | |
| GP control | 586.6 (±80.6) | 3.00 (±0.11) | 0.870 (±0.0152) | 3.074 | 1,803 | |
| GP FC center | 613.0 (±100.8) | 3.20 (±0.32) | 0.888 (±0.042) | 2.845 | 1,744 | |
GP, gravel plain.
Values are means (± standard deviations) of 5 measurements.
Gravel plain bacterial, archaeal, and fungal communities were distinct from their dune counterparts on NMDS plots (Fig. 3). PERMANOVA tests confirmed that the communities were significantly different from each other (for the bacterial communities, pseudo-F was >6.06 and P was <0.001; for the archaeal communities, pseudo-F was >10.37 and P was <0.001; for the fungal communities, pseudo-F was >6.16 and P was <0.001). Furthermore, within the sites sampled, we found that the edaphic bacterial and archaeal communities in dune FC soil differed significantly from those in control vegetated soil by PERMANOVA (for bacterial communities, pseudo-F was >2.98 and P was <0.05; for archaeal communities, pseudo-F was >2.59 and P was <0.05). In contrast, the gravel plain FC and control communities were not significantly different (P > 0.05). The fungal communities were all found to be zone specific, i.e., they differed significantly between the dune FC, dune control, gravel plain FC, and gravel plain control soils (pseudo-F > 2.33; P < 0.05) (Fig. 3).
FIG 3.
Two-dimensional nonmetric multidimensional scaling (NMDS) plots of bacterial (a), archaeal (b), and fungal (c) community structures based on Bray-Curtis similarity matrices of square root-transformed relative abundances of 16S rRNA gene (bacteria and archaea) and ITS region (fungi) OTUs. Communities are indicated by colored symbols as follows: red squares, dune FC center; blue circles, dune control; green triangles, gravel plain FC center; purple diamonds, gravel plain control. Each area bounded by samples from the same zone is shaded in the corresponding color to indicate clustering.
Environmental drivers of soil community composition.
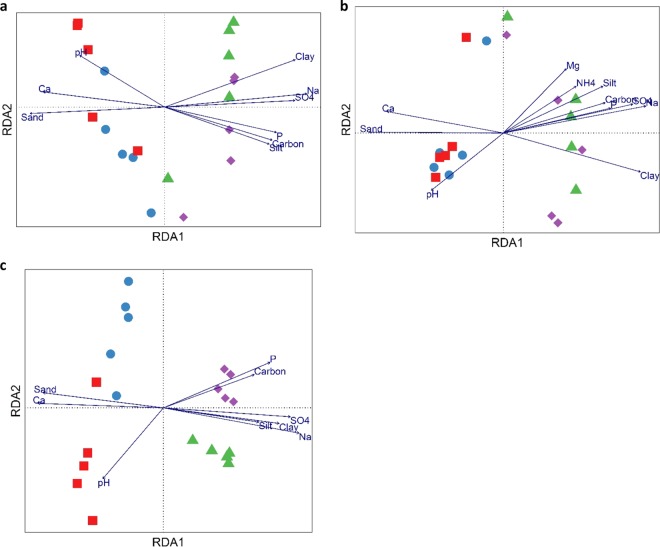
Redundancy analysis (RDA) biplots were used to assess physicochemical factors that influenced microbial community structures (Fig. 4). Overall, the combination of variables explained 61.7%, 80.6%, and 55.6% of the bacterial (Fig. 4a), archaeal (Fig. 4b), and fungal (Fig. 4c) community structure variances, respectively. All communities were clearly separated along the first axis according to their sampling site and were significantly influenced by the same nine physicochemical variables (with the exception of archaea, which were also influenced by two additional properties [Mg and NH4 contents; P, < 0.05 for both]). The percentage of sand, the pH, and the Ca content (P, <0.001, <0.05, and <0.001, respectively) were found to be determining factors in shaping the dune microbial communities, whereas the gravel plain communities were significantly influenced by P, Na, and S concentrations as well as by the percentages of carbon, silt, and clay (P, <0.005, <0.001, <0.001, <0.05, <0.005, and <0.05, respectively). However, we found no soil parameters that significantly influenced FC and control microbial communities (Fig. 4), suggesting that other, unmeasured factors, either biotic or abiotic, may be involved.
FIG 4.
Redundancy analysis (RDA) plots showing the influence of soil physicochemical properties on bacterial (a), archaeal (b), and fungal (c) communities. Vectors indicate significant (P < 0.05) correlations between soil physicochemical properties and microbial community composition. Communities are indicated by colored symbols as follows: red squares, dune FC center; blue circles, dune control; green triangles, gravel plain FC center; purple diamonds, gravel plain control.
Bacterial, archaeal, and fungal richness and diversity.
Analysis of the 16S rRNA gene and ITS region amplicon data sets showed that Namib Desert edaphic bacterial communities were more diverse than the fungal and archaeal communities (Table 2), with a significantly higher number of species (F was >654.5 and P was <0.001 by ANOVA), Shannon index (F > 788.4; P < 0.0001), and Simpson index (F > 56.1; P < 0.001).
Bacterial and archaeal communities showed consistent levels of species richness across the two zones of the two sites studied (i.e., gravel plain and dune FC and control soils) (P, >0.05 by ANOVA), whereas fungal communities had significantly higher numbers of species in the gravel plains than in the dunes (F was >7.74 and P was <0.05 by ANOVA). Both bacterial and archaeal communities also presented high levels of evenness in all of the zones sampled, with no significant differences in α-diversities between the zones sampled (P, >0.05 by ANOVA). The Whittaker index, measuring phylogenetic heterogeneity within zones (β-diversity), ranged from 1.52 to 1.69 for bacterial communities and from 1.40 to 1.53 for archaeal communities. Large numbers of bacterial and archaeal OTUs were also shared by all four zones (59.2% and 58.3%, respectively) (Table 2; Fig. 5a and b).
FIG 5.
Venn diagrams showing the distributions of bacterial (a), archaeal (b), and fungal (c) OTUs within the soils studied. Overlap indicates the occurrence of an OTU in more than one microenvironment. The number of OTUs present solely in both the dune and the gravel plain FC centers is highlighted in red.
In contrast, fungal communities were highly variable in diversity, with significantly different species richness estimates across the four different zones studied (F was >7.15 and P was <0.005 by ANOVA). The dune FC soils had a significantly lower number of fungal OTUs (366 ± 59 OTUs) than all the other zones sampled (553 ± 88 OTUs for the dune control [P, <0.05 by Tukey's post hoc test], 613 ± 101 OTUs for the gravel plain FCs [P < 0.01], and 587 ± 81 OTUs for the gravel plain control [P < 0.005]). Fungal communities had large numbers of zone-specific OTUs (Table 2; Fig. 5c), suggesting that differential assembly processes occur for fungal communities in each of the Namib Desert soil environments studied. This is further emphasized by the fact that Whittaker indices of β-diversity, which ranged from 1.95 to 4.54, were significantly different for all zones studied (F was >17.14 and P was <0.001 by ANOVA).
Globally, Actinobacteria (25 to 38%), Proteobacteria (16 to 33%), and Crenarchaeota (10 to 25%) were the most abundant bacterial and archaeal phyla in all zones (see Fig. SA2a in the supplemental material), while moderately abundant taxa included Acidobacteria (2 to 5%), Bacteroidetes (3 to 20%), Chloroflexi (2 to 4%), Gemmatimonadetes (1 to 2.5%), Verrucomicrobia (0.9 to 2.8%), and Firmicutes (<0.1 to 2.8%). As expected, and based on the diversity indices (Table 2), the compositions of fungal communities were highly variable between samples; each of the four zones studied was dominated by a unique fungal taxon (the class Dothideomycetes for gravel plain control soils [46.2%] and for gravel plain fairy center soils [27.5%], the class Agaricostilbomycetes for dune control soils [23.2%], and the phylum Chytridiomycota for dune fairy center soils [43.6%]) (see Fig. SA2b in the supplemental material). Globally, fungi of the phylum Chytridiomycota were significantly more abundant in dune soils than in gravel plain soils (P, <0.05 by Student's t test), while Dothideomycetes displayed the opposite trend (P < 0.01).
Fairy circle-specific phylotypes.
We identified 9 bacterial, 1 archaeal, and 57 fungal phylotypes that were FC specific; i.e., their phylotypic signals were found only within the soils of dune and the gravel plain FCs (Fig. 5). Their phylogenetic assignments are given in Table 3. The FC-specific archaeal OTU was related to the ammonia-oxidizing genus “Candidatus Nitrososphaera” of the phylum Thaumarchaeota. The bacterial OTUs were assigned to Cyanobacteria (4/9), Firmicutes (2/9), Proteobacteria (2/9), and Actinobacteria (1/9). Although 33 of the 57 (∼58%) FC-specific fungal OTUs could not be classified below the kingdom level, 15 were classified as Ascomycota, 1 as Basidiomycota, and 8 as Chytridiomycota. Five of the eight Chytridiomycota OTUs belonged to the genus Rhizophlyctis, and seven of the Ascomycota OTUs belonged to the class Dothideomycetes. Some of the OTUs assigned to Ascomycota belonged to the Periconia, Phoma, and Curvularia genera, and a single OTU was identified as Aspergillus flavus (Table 3).
TABLE 3.
FC-specific OTUs identifieda
| Kingdom | Phylum | Class | Order | Family | Genus | Species |
|---|---|---|---|---|---|---|
| Archaea | Thaumarchaeota | Nitrososphaeria | Nitrososphaerales | Nitrososphaeraceae | “Candidatus Nitrososphaera” | |
| Bacteria | Actinobacteria | Nitriliruptoria | Nitriliruptorales | Nitriliruptoraceae | ||
| Cyanobacteria | Oscillatoriophycideae | Chroococcales | Xenococcaceae | |||
| Unclassified | ||||||
| Unclassified | ||||||
| Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus | ||
| Paenibacillaceae | Brevibacillus | |||||
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | |||
| Gammaproteobacteria | ||||||
| Fungi | Ascomycota | Dothideomycetes | Pleosporales | Incertae sedis | Periconia | |
| Incertae sedis | Phoma | |||||
| Montagnulaceae | Unidentified | |||||
| Pleosporaceae | Curvularia | |||||
| Curvularia | ||||||
| Curvularia | ||||||
| Sporormiaceae | Unclassified | |||||
| Eurotiomycetes | Eurotiales | Trichocomaceae | Aspergillus | Aspergillus flavus | ||
| Onygenales | Unidentified | |||||
| Incertae sedis | Incertae sedis | Incertae sedis | Calcarisporiella | |||
| Unclassified | ||||||
| Sordariomycetes | Hypocreales | Incertae_sedis_3 | Stachybotrys | Stachybotrys microspora | ||
| Sordariales | Chaetomiaceae | Unclassified | ||||
| Xylariales | Unclassified | |||||
| Xylariales | Unclassified | |||||
| Basidiomycota | Agaricomycetes | Agaricales | Entolomataceae | Clitopilus | ||
| Chytridiomycota | Chytridiomycetes | Unclassified | ||||
| Rhizophlyctidales | Rhizophlyctidaceae | Rhizophlyctis | ||||
| Rhizophlyctis | Rhizophlyctis rosea | |||||
| Rhizophlyctis | Rhizophlyctis rosea | |||||
| Rhizophlyctis | Rhizophlyctis rosea | |||||
| Rhizophlyctis | Rhizophlyctis rosea | |||||
| Unclassified | ||||||
| Unclassified |
FC-specific OTUs were observed in both dune and gravel plain fairy circles. OTUs are classified to the lowest level possible using a high (>80%) level of sequence similarity.
DISCUSSION
Microorganisms play important roles in desert environment bioprocesses, including biogeochemical cycling of carbon and nitrogen and bioweathering of exposed bedrock (30, 31). In addition, they can interact positively (e.g., in mycorrhizae [32]) or negatively (e.g., as phytopathogens [33]) with plants. We thus hypothesize that some microbial groups found in FCs may interact negatively with plant species through phytopathogenic effects and thereby play a role in the formation and/or maintenance of these enigmatic features of the Namib Desert. A corollary to this hypothesis is that FC-specific taxa should be present in the soils of both the gravel plain and dune FCs and absent from the surrounding vegetated soils (2, 16).
In agreement with previous desert phylogenetic surveys, we identified Actinobacteria as the most abundant phylum in our soil samples (30). We also found relatively high abundances of Proteobacteria, Acidobacteria, Bacteroidetes, and Chloroflexi in all the Namib Desert soil zones studied, which is broadly consistent with the findings of other Namib Desert surveys (30, 34, 35). We report a consistently high relative abundance of a limited number of archaeal phylotypes, as observed in Sonoran Desert soils (36). However, a high abundance of Thaumarchaeota OTUs (recently reclassified from Crenarchaeota [37]) was detected in Namib Desert soils, while the crenarchaean Thermoproteus taxon dominated the Sonoran Desert samples (35).
Fungal diversity has not been surveyed comprehensively in desert environments worldwide (30). In the Namib Desert, fungi in mycorrhizae (38) and lichens (39) have been shown to have the potential to participate in local biogeochemical cycling, specifically by decomposing surface litter (40). Although we were able to classify only approximately 50% of the fungal OTUs (due to the underrepresentation of environmental sequences in the UNITE database [41]), this is the first study to use next-generation high-throughput sequencing to assess fungal communities within the Namib Desert. We observed that Ascomycota, Basidiomycota, and Chytridiomycota were detected ubiquitously in these desert soils. Dominant fungal genera and orders, identified as Aspergillus, Chaetomium, Pleosporales, and Stachybotrys, are well-known desert colonists (42).
Microbial community assembly in Namib Desert soils.
In hyperoligotrophic desert environments, soil physicochemical properties, including water content (43), soil pH (44), carbon content (45), nitrate concentration (46), micronutrients (47), and particle size (44), have been shown to significantly shape microbial community structures. In a comparative terminal restriction fragment length polymorphism (T-RFLP) fingerprinting analysis (48), Namib Desert dune and gravel plain bacterial communities were shown to be significantly different from one another, and microbial community structures were significantly governed by soil pH, Na content, and percentages of silt and sand. Using high-throughput sequencing, we demonstrate that the bacterial, archaeal, and fungal communities of Namib Desert dune fields and gravel plains are significantly different. We found that the percentage of sand, the pH, and the Ca content significantly shaped dune microbial (bacterial, archaeal, and fungal) communities, whereas soil P, Na, and S contents and the percentages of clay, silt, and carbon shaped the gravel plain communities. Overall, these findings support the concept that deterministic processes play important roles in shaping Namib Desert soil microbial communities, with microbial communities at each site adapting to local physicochemical conditions (49).
None of the soil physicochemical properties measured were significantly different for FC and control soils in either the dunes or the gravel plain, which is expected given the close proximity of each FC and control sample. Consequently, the soil properties measured could not explain the differences observed between microbial communities in FC and vegetated control soils. This suggests the potential involvement of other factors, such as other edaphic properties (e.g., Fe, Zn, and/or Al content [48]), the presence of a toxin (50), or unknown stochastic processes (51).
The four zones sampled in Namib Desert dunes and gravel plains shared large numbers of bacterial and archaeal OTUs (59.2% and 58.3%, respectively). This large set of cosmopolitan edaphic bacterial and archaeal phylotypes is indicative of a Namib Desert core community that is not subject to dispersal limitation. This indicates that these microbial communities may also be influenced by stochastic (random) events, such as the transport of viable cells over large distances by wind dispersal, rather than deterministic processes, such as differences in soil physicochemical properties.
In contrast, fungal communities were “zone specific”; each zone contained a high number of unique OTUs. Fungal communities also showed a clear and significant distinction between the dune and gravel plain sites. In addition, fungal communities in FC and control vegetated soils were significantly different in composition, a pattern that was not as clearly observed with the distribution of bacterial and archaeal OTUs in the zones sampled. Because there is no significant difference between the physicochemical properties of FC and control soils, we conclude that FC-specific fungal communities are selected through habitat filtration (vegetated soil versus barren soil) (16) or an environmental disturbance (such as the release of a toxin [50]). It is known that the assembly of edaphic bacterial and archaeal communities may differ from that of fungal communities (52, 53).
Could edaphic microbial communities be involved in the fairy circle phenomenon?
While many hypotheses have been proposed in attempts to explain the origin of Namib Desert fairy circles (2, 4, 8, 10, 11, 13–15), none take into account the presence of fairy circles in both the gravel plain and dune environments and the differences between them. Considering that almost all FC studies have focused exclusively on dune FCs, we compared microbial communities in dune and gravel plain FCs.
Our hypothesis that microorganisms may play a role in FC etiology is based on the fact that both bacteria and fungi can have pathogenic or phytotoxic activities (33, 54). Furthermore, microorganisms have been implicated in the formation of various FC-like phenomena, such as the dollar spot (17), fairy rings (55), and circular patches in intertidal mats (56). Additionally, plant pathogens have been shown to be viable and able to infect plants in desert environments (e.g., the fungi Alternaria solani, Stemphylium spp., and Botrytis cinerea, as well as a novel virus of the Potyvirus genus in Albuca rautanenii found in the Namib Desert) (33, 57).
We identified 10 and 57 OTUs from the 16S rRNA gene and ITS region data sets, respectively, that were exclusively and consistently present within dune and gravel FC soils. Of these FC-specific OTUs, many were assigned to phylogenetic groups harboring known phytotoxic or phytopathogenic microorganisms, such as the fungal genera Periconia, Curvularia, and Aspergillus, the fungal order Pleosporales, the fungal family Chaetomiaceae, and the bacterial class Gammaproteobacteria (58–63). While these results do not directly implicate microorganisms in the creation and/or maintenance of Namib Desert FCs, they remain consistent with the hypothesis that microorganisms play a role in the phenomenon.
Conclusions.
Here we report the first phylogenetic study to compare fairy circles from the Namib Desert gravel plains and dune fields. Overall, the Namib Desert edaphic bacterial communities were more diverse than the archaeal and fungal communities. Bacterial and archaeal communities showed consistent diversity across both soil types. The high proportion of shared community members between soil types suggests that stochastic processes are important in their assembly (51). In contrast, fungal communities were more variable overall, with unique sets of fungal phylotypes detected in each zone sampled. This suggests that deterministic niche adaptation is the primary mechanism in the assembly of fungal communities (49). Finally, we have identified microorganisms that are present solely in FC soils and that may play a role in the fairy circle phenomenon. Many of the OTUs found only in FCs are related to known phytopathogen groups (the fungal genera Periconia, Curvularia, and Aspergillus, the fungal order Pleosporales and family Chaetomiaceae, and the bacterial class Gammaproteobacteria [58–63]), suggesting that they may play a role in this phenomenon. To further investigate this hypothesis, the FC-specific phylotypes should be isolated and used in controlled greenhouse experiments to identify patterns related to phytopathology (64). The use of function-based approaches, such as metaproteomics and/or metagenomics (65, 66), could also assist in the identification of phytotoxic proteins or toxins, as well as the enzymes or pathways involved in their biosynthesis.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the South African National Research Foundation (NRF) (grant 90312) and the University of Pretoria for supporting this research. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
This work, including the efforts of Jean-Baptiste Ramond and the bursaries of Andries J. van der Walt and Riegardt M. Johnson, was funded by the National Research Foundation (NRF) (90312).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00844-16.
REFERENCES
- 1.Grube S. 2002. The fairy circles of Kaokoland (Northwest Namibia)—is the harvester termite Hodotermes mossambicus the prime causal factor in circle formation? Basic Appl Ecol 3:367–370. doi: 10.1078/1439-1791-00138. [DOI] [Google Scholar]
- 2.Van Rooyen M, Theron G, Van Rooyen N, Jankowitz W, Matthews W. 2004. Mysterious circles in the Namib Desert: review of hypotheses on their origin. J Arid Environ 57:467–485. doi: 10.1016/S0140-1963(03)00111-3. [DOI] [Google Scholar]
- 3.Getzin S, Yizhaq H, Bell B, Erickson TE, Postle AC, Katra I, Tzuk O, Zelnik YR, Wiegand K, Wiegand T, Meron E. 2016. Discovery of fairy circles in Australia supports self-organization theory. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1522130113:201522130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tschinkel WR. 2012. The life cycle and life span of Namibian fairy circles. PLoS One 7:e38056. doi: 10.1371/journal.pone.0038056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albrecht CF, Joubert JJ, De Rycke PH. 2001. Origin of the enigmatic, circular, barren patches (‘Fairy Rings') of the pro-Namib. S Afr J Sci 97:23–27. [Google Scholar]
- 6.Eicker A, Theron G, Grobbelaar N. 1982. Mikrobiologiese studie van ‘kaal kolle' in die Giribesvlakte van Kaokoland, SWA-Namibie. S Afr J Bot 1:69–74. [Google Scholar]
- 7.Tschinkel WR. 2015. Experiments testing the causes of Namibian fairy circles. PLoS One 10:e0140099. doi: 10.1371/journal.pone.0140099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naude Y, Van Rooyen MW, Rohwer ER. 2011. Evidence for a geochemical origin of the mysterious circles in the Pro-Namib desert. J Arid Environ 75:446–456. doi: 10.1016/j.jaridenv.2010.12.018. [DOI] [Google Scholar]
- 9.Zelnik YR, Meron E, Bel G. 2015. Gradual regime shifts in fairy circles. Proc Natl Acad Sci U S A 112:12327–12331. doi: 10.1073/pnas.1504289112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juergens N. 2013. The biological underpinnings of Namib Desert fairy circles. Science 339:1618–1621. doi: 10.1126/science.1222999. [DOI] [PubMed] [Google Scholar]
- 11.Picker MD, Ross-Gillespie V, Vlieghe K, Moll E. 2012. Ants and the enigmatic Namibian fairy circles—cause and effect? Ecol Entomol 37:33–42. doi: 10.1111/j.1365-2311.2011.01332.x. [DOI] [Google Scholar]
- 12.Vlieghe K, Picker M, Ross-Gillespie V, Erni B. 2015. Herbivory by subterranean termite colonies and the development of fairy circles in SW Namibia. Ecol Entomol 40:42–49. doi: 10.1111/een.12157. [DOI] [Google Scholar]
- 13.Getzin S, Wiegand K, Wiegand T, Yizhaq H, von Hardenberg J, Meron E. 2015. Adopting a spatially explicit perspective to study the mysterious fairy circles of Namibia. Ecography 38:1–11. doi: 10.1111/ecog.00911. [DOI] [Google Scholar]
- 14.Cramer MD, Barger NN. 2013. Are Namibian “fairy circles” the consequence of self-organizing spatial vegetation patterning? PLoS One 8:e70876. doi: 10.1371/journal.pone.0070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer J, Senejoux F, Heyman H, Meyer N, Meyer M. 2015. The occurrence of triterpenoids from Euphorbia gummifera inside the fairy circles of Garub in the southern Namibian pro-desert. S Afr J Bot 98:10–15. doi: 10.1016/j.sajb.2015.01.019. [DOI] [Google Scholar]
- 16.Ramond JB, Pienaar A, Armstrong A, Seely M, Cowan DA. 2014. Niche-partitioning of edaphic microbial communities in the Namib Desert gravel plain Fairy Circles. PLoS One 9:e109539. doi: 10.1371/journal.pone.0109539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh B, Ikeda SS, Boland GJ. 1999. Biology and management of dollar spot (Sclerotinia homoeocarpa); an important disease of turfgrass. Hort Sci 34:13–21. [Google Scholar]
- 18.Walkley A. 1935. An examination of methods for determining organic carbon and nitrogen in soils. J Agric Sci 25:598–609. doi: 10.1017/S0021859600019687. [DOI] [Google Scholar]
- 19.Bouyoucos GJ. 1962. Hydrometer method improved for making particle size analyses of soils. Agron J 54:464–465. doi: 10.2134/agronj1962.00021962005400050028x. [DOI] [Google Scholar]
- 20.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A 82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White TJ, Bruns T, Lee SJWT, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY. [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H. 2013. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ihaka R, Gentleman R. 1996. R: a language for data analysis and graphics. J Comput Graph Stat 5:299–314. doi: 10.1080/10618600.1996.10474713. [DOI] [Google Scholar]
- 27.Härdle WK, Simar L. 2003. Applied multivariate statistical analysis, vol 2 Springer, Berlin, Germany. [Google Scholar]
- 28.Oksanen J, Kindt R, Legendre P, O'Hara B, Stevens MHH, Oksanen MJ, Suggests M. 2009. Vegan: community ecology package, version 2.3-3 J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 29.Ginestet C. 2011. ggplot2: elegant graphics for data analysis. J R Stat Soc A 174:245–246. doi: 10.1111/j.1467-985X.2010.00676_9.x. [DOI] [Google Scholar]
- 30.Makhalanyane TP, Valverde A, Gunnigle E, Frossard A, Ramond J-B, Cowan DA. 2015. Microbial ecology of hot desert edaphic systems. FEMS Microbiol Rev 39:203–221. doi: 10.1093/femsre/fuu011. [DOI] [PubMed] [Google Scholar]
- 31.Belnap J. 2003. The world at your feet: desert biological soil crusts. Front Ecol Environ 1:181–189. doi: 10.1890/1540-9295(2003)001[0181:TWAYFD]2.0.CO;2. [DOI] [Google Scholar]
- 32.Remy W, Taylor TN, Hass H, Kerp H. 1994. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci U S A 91:11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rotem J. 1981. Fungal diseases of potato and tomato in the Negev Desert. Plant Dis 65:315–318. doi: 10.1094/PD-65-315. [DOI] [Google Scholar]
- 34.Makhalanyane TP, Valverde A, Lacap DC, Pointing SB, Tuffin MI, Cowan DA. 2013. Evidence of species recruitment and development of hot desert hypolithic communities. Environ Microbiol Rep 5:219–224. doi: 10.1111/1758-2229.12003. [DOI] [PubMed] [Google Scholar]
- 35.Ronca S, Ramond JB, Jones BE, Seely M, Cowan DA. 2015. Namib Desert dune/interdune transects exhibit habitat-specific edaphic bacterial communities. Front Microbiol 6:845. doi: 10.3389/fmicb.2015.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrew DR, Fitak RR, Munguia-Vega A, Racolta A, Martinson VG, Dontsova K. 2012. Abiotic factors shape microbial diversity in Sonoran Desert soils. Appl Environ Microbiol 78:7527–7537. doi: 10.1128/AEM.01459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. 2008. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 38.Jacobson KM. 1997. Moisture and substrate stability determine VA-mycorrhizal fungal community distribution and structure in an arid grassland. J Arid Environ 35:59–75. doi: 10.1006/jare.1995.0140. [DOI] [Google Scholar]
- 39.Lange O, Meyer A, Zellner H, Heber U. 1994. Photosynthesis and water relations of lichen soil crusts: field measurements in the coastal fog zone of the Namib Desert. Funct Ecol 8:253–264. doi: 10.2307/2389909. [DOI] [Google Scholar]
- 40.Jacobson K, van Diepeningen A, Evans S, Fritts R, Gemmel P, Marsho C, Seely M, Wenndt A, Yang X, Jacobson P. 2015. Non-rainfall moisture activates fungal decomposition of surface litter in the Namib sand sea. PLoS One 10:e0126977. doi: 10.1371/journal.pone.0126977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kõljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E. 2005. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol 166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- 42.Sterflinger K, Tesei D, Zakharova K. 2012. Fungi in hot and cold deserts with particular reference to microcolonial fungi. Fungal Ecol 5:453–462. doi: 10.1016/j.funeco.2011.12.007. [DOI] [Google Scholar]
- 43.Clark JS, Campbell JH, Grizzle H, Acosta-Martìnez V, Zak JC. 2009. Soil microbial community response to drought and precipitation variability in the Chihuahuan Desert. Microb Ecol 57:248–260. doi: 10.1007/s00248-008-9475-7. [DOI] [PubMed] [Google Scholar]
- 44.Lauber CL, Strickland MS, Bradford MA, Fierer N. 2008. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40:2407–2415. doi: 10.1016/j.soilbio.2008.05.021. [DOI] [Google Scholar]
- 45.Kuramae EE, Yergeau E, Wong LC, Pijl AS, van Veen JA, Kowalchuk GA. 2012. Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol Ecol 79:12–24. doi: 10.1111/j.1574-6941.2011.01192.x. [DOI] [PubMed] [Google Scholar]
- 46.Ben-David EA, Zaady E, Sher Y, Nejidat A. 2011. Assessment of the spatial distribution of soil microbial communities in patchy arid and semi-arid landscapes of the Negev Desert using combined PLFA and DGGE analyses. FEMS Microbiol Ecol 76:492–503. doi: 10.1111/j.1574-6941.2011.01075.x. [DOI] [PubMed] [Google Scholar]
- 47.Preem J-K, Truu J, Truu M, Mander Ü, Oopkaup K, Lõhmus K, Helmisaari H-S, Uri V, Zobel M. 2012. Bacterial community structure and its relationship to soil physico-chemical characteristics in alder stands with different management histories. Ecol Eng 49:10–17. doi: 10.1016/j.ecoleng.2012.08.034. [DOI] [Google Scholar]
- 48.Gombeer S, Ramond JB, Eckardt FD, Seely M, Cowan DA. 2015. The influence of surface soil physicochemistry on the edaphic bacterial communities in contrasting terrain types of the Central Namib Desert. Geobiology 13:494–505. doi: 10.1111/gbi.12144. [DOI] [PubMed] [Google Scholar]
- 49.Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. 2010. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4:337–345. doi: 10.1038/ismej.2009.122. [DOI] [PubMed] [Google Scholar]
- 50.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 51.Hubbell SP. 2001. The unified neutral theory of biodiversity and biogeography. Monographs in Population Biology 32. Princeton University Press, Princeton, NJ. [Google Scholar]
- 52.Chase JM, Myers JA. 2011. Disentangling the importance of ecological niches from stochastic processes across scales. Philos Trans R Soc Lond B Biol Sci 366:2351–2363. doi: 10.1098/rstb.2011.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt S, Nemergut DR, Darcy J, Lynch R. 2014. Do bacterial and fungal communities assemble differently during primary succession? Mol Ecol 23:254–258. doi: 10.1111/mec.12589. [DOI] [PubMed] [Google Scholar]
- 54.Starr M. 1959. Bacteria as plant pathogens. Annu Rev Microbiol 13:211–238. doi: 10.1146/annurev.mi.13.100159.001235. [DOI] [PubMed] [Google Scholar]
- 55.Dickinson CH. 1979. Fairy rings in Norfolk. Bull Br Mycol Soc 13:91–94. doi: 10.1016/S0007-1528(79)80005-X. [DOI] [Google Scholar]
- 56.Carreira C, Staal M, Falkoski D, de Vries RP, Middelboe M, Brussaard CP. 2015. Disruption of photoautotrophic intertidal mats by filamentous fungi. Environ Microbiol 17:2910–2921. doi: 10.1111/1462-2920.12835. [DOI] [PubMed] [Google Scholar]
- 57.Zablocki O, Rybicki E, Cowan D. 2014. First report of a potyvirus infecting Albuca rautanenii in the Namib Desert. Plant Dis 98:1749. doi: 10.1094/PDIS-07-14-0737-PDN. [DOI] [PubMed] [Google Scholar]
- 58.Falloon R. 1976. Curvularia trifolii as a high-temperature turfgrass pathogen. N Z J Agric Res 19:243–248. doi: 10.1080/00288233.1976.10426773. [DOI] [Google Scholar]
- 59.Finlay RD. 2006. The fungi in soil, p 107–146. In van Elsas JD, Jansson JK, Trevors JT (ed), Modern soil microbiology, 2nd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 60.Hedayati M, Pasqualotto A, Warn P, Bowyer P, Denning D. 2007. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology 153:1677–1692. doi: 10.1099/mic.0.2007/007641-0. [DOI] [PubMed] [Google Scholar]
- 61.Violi HA, Menge JA, Beaver RJ. 2007. Chaetomium elatum as a root-colonizing fungus in avocado: is it a mutualist, cheater, commensalistic associate, or pathogen? Am J Bot 94:690–700. doi: 10.3732/ajb.94.4.690. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Crous PW, Schoch CL, Hyde KD. 2012. Pleosporales. Fungal Divers 53:1–221. doi: 10.1007/s13225-011-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Daugherty S, Brinkac L, Beanan MJ, Haft DH, Nelson WC, Davidsen T, Zafar N, Zhou L, Liu J, Yuan Q, Khouri H, Fedorova N, Tran B, Russell D, Berry K, Utterback T, Van Aken SE, Feldblyum TV, D'Ascenzo M, Deng WL, Ramos AR, Alfano JR, Cartinhour S, Chatterjee AK, Delaney TP, Lazarowitz SG, Martin GB, Schneider DJ, Tang X, Bender CL, White O, Fraser CM, Collmer A. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A 100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuck S, Weinhold A, Baldwin IT. 2014. Isolating fungal pathogens from a dynamic disease outbreak in a native plant population to establish plant-pathogen bioassays for the ecological model plant Nicotiana attenuata. PLoS One 9:e102915. doi: 10.1371/journal.pone.0102915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilmes P, Bond PL. 2006. Metaproteomics: studying functional gene expression in microbial ecosystems. Trends Microbiol 14:92–97. doi: 10.1016/j.tim.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 66.Handelsman J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.