ABSTRACT
A wealth of biochemical and molecular data have been reported regarding ethanol toxicity in the yeast Saccharomyces cerevisiae. However, direct physical data on the effects of ethanol stress on yeast cells are almost nonexistent. This lack of information can now be addressed by using atomic force microscopy (AFM) technology. In this report, we show that the stiffness of glucose-grown yeast cells challenged with 9% (vol/vol) ethanol for 5 h was dramatically reduced, as shown by a 5-fold drop of Young's modulus. Quite unexpectedly, a mutant deficient in the Msn2/Msn4 transcription factor, which is known to mediate the ethanol stress response, exhibited a low level of stiffness similar to that of ethanol-treated wild-type cells. Reciprocally, the stiffness of yeast cells overexpressing MSN2 was about 35% higher than that of the wild type but was nevertheless reduced 3- to 4-fold upon exposure to ethanol. Based on these and other data presented herein, we postulated that the effect of ethanol on cell stiffness may not be mediated through Msn2/Msn4, even though this transcription factor appears to be a determinant in the nanomechanical properties of the cell wall. On the other hand, we found that as with ethanol, the treatment of yeast with the antifungal amphotericin B caused a significant reduction of cell wall stiffness. Since both this drug and ethanol are known to alter, albeit by different means, the fluidity and structure of the plasma membrane, these data led to the proposition that the cell membrane contributes to the biophysical properties of yeast cells.
IMPORTANCE Ethanol is the main product of yeast fermentation but is also a toxic compound for this process. Understanding the mechanism of this toxicity is of great importance for industrial applications. While most research has focused on genomic studies of ethanol tolerance, we investigated the effects of ethanol at the biophysical level and found that ethanol causes a strong reduction of the cell wall rigidity (or stiffness). We ascribed this effect to the action of ethanol perturbing the cell membrane integrity and hence proposed that the cell membrane contributes to the cell wall nanomechanical properties.
INTRODUCTION
The yeast Saccharomyces cerevisiae is a remarkable ethanol producer that is also very sensitive to its main fermentative product. At low to moderate concentrations (5 to 7%), ethanol mainly affects the growth rate, and at higher concentrations (>10%), it can strongly impair cell integrity, eventually leading to cell death with features of apoptosis (1). These inhibitory and toxic effects are ascribed to the fact that ethanol alters cell membrane fluidity and dissipates the transmembrane electrochemical potential, thereby creating permeability to ionic species and causing leakage of metabolites (2). Recent works using lipidomic methodologies confirmed the relationship between the composition of lipids, notably ergosterol and unsaturated fatty acids, and ethanol tolerance (3, 4). Moreover, as it diffuses freely into cells, ethanol at high concentrations may directly perturb and denature intracellular proteins (reviewed in references 5 and 2). The production of ethanol as an alternative fuel energy from renewable carbon resources by microbial cell factories is a great industrial concern nowadays. For this to become economically attractive, a major challenge is to increase the tolerance of yeast to ethanol, which requires an understanding of the mechanisms of its toxicity.
The remarkable advances in genomic technologies over the last 15 years have raised the possibility of investigating ethanol toxicity on a global (genomic-proteomic-metabolomic) scale. DNA microarrays were used to explore the transcriptomic responses of yeast exposed to ethanol stress (6–9). These works revealed impressive transcriptomic changes which implicate a broad range of functional categories, including protein biosynthesis, metabolism of amino acids, nucleotides, lipids, and sterols, ion homeostasis, the cell cycle, and membrane and cell wall organization (for a review, see reference 5). On the other hand, the genetic basis of ethanol resistance was investigated using transposon mutagenesis and single-gene-knockout (SGKO) mutant collections that were challenged with different concentrations of ethanol (10–13). This was followed by applying genetic/genomic methods to map genomic regions related to ethanol tolerance. This powerful approach, which relies on crossing two parents, one inferior and the other superior for a trait of interest, followed by whole-genome sequencing of a large set of recombination segregants, allowed identification of potential genetic loci linked to high ethanol tolerance and, thereby, determination of causative genes. In particular, MKT1, SWS2, and AJP1 (14) were isolated by this approach, as well as the VPS16, VPS28, and VPS70 genes, which encode components of the vacuole protein sorting system (15, 16). However, the causative genes identified are apparently dependent on several criteria, including the origin of the parental strains, the culture conditions, the size of the segregant sample, and the performance of the algorithm used to analyze the data. Altogether, and regardless of how tolerance to ethanol was defined (2), these genome-scale studies underscored the genetic intricacy of ethanol tolerance and the complexity of the yeast response to this compound at the molecular level. However, these studies did not bring us any clue about the physical effects that ethanol can have on yeast cells, although some transcriptomic and metabolomic data may suggest important modifications of cellular membranes (3, 4, 17–20) and cell wall organization (12) of yeast cells exposed to high levels of ethanol. Thus, obtaining biophysical data on yeast cells exposed to high ethanol concentrations may provide complementary information on how cells can cope with this toxic compound, which may be relevant for further strategies to improve the tolerance of yeast toward ethanol.
These biophysical data can now be obtained using atomic force microscopy (AFM), which has emerged as a powerful technology for probing microbial cells at the nanoscale level and measuring the nanomechanical properties in the natural environment of the microbes (21, 22). It is therefore the best tool for visualizing and quantifying effects of ethanol stress at the single-cell level. This question was previously approached by Canetta et al. (23). However, they used an air-drying immobilization technique to fix ethanol-treated yeast cells on a glass slide. As discussed in a previous work (24), the immobilization technique must keep the cells alive under conditions as close as possible to those of the natural environment. Thus, the immobilization technique has to be “neutral” to avoid interfering with the biological condition under study. As these conditions were apparently not respected in the work of Canetta et al. (23, 25), we revisited ethanol stress by using our innovative immobilization method, which is based on trapping single yeast cells in microchambers produced by microstructured polydimethylsiloxane (PDMS) stamps (26). These microchambers can easily be loaded by convective/capillary deposition of yeast cells. This versatile method was validated by AFM nanomechanical measurements of yeast cells and germinated Aspergillus conidia in growth media (27). In addition, we investigated the role of the transcription factor Msn2/Msn4 in this AFM analysis of ethanol stress. The reason for this was that a large part of the transcriptomic response to ethanol stress is mediated through the general stress-responsive factor encoded by MSN2 and its orthologue MSN4 (7, 29, 30). This response is explained by the ethanol-induced translocation of Msn2 from the cytosol to the nucleus and the consecutive transcriptional activation of the binding of Msn2 to the stress response elements (STRE) present in the promoters of stress-related genes (31). The potential function of YAP1 in this ethanol effect was also evaluated because this gene was inferred to be implicated in ethanol resistance, as indicated by its upregulation in an ethanol-tolerant S. cerevisiae strain obtained by enforced evolutionary adaptation (30).
In this study, we showed that ethanol stress provoked a dramatic reduction of cell wall rigidity (stiffness) that was accompanied by neither a change in the polysaccharide composition nor a change in the thickness of the cell wall. We furthermore found that this biophysical response to ethanol does not occur in cells lacking Msn2/Msn4, although the stiffness of these cells is already significantly reduced relative to that of wild-type cells, pointing to a role of this transcription factor in the control of the nanomechanical properties of the cell wall. Evidence that the effects of ethanol on cell stiffness implicate the cellular membrane was obtained by the finding that amphotericin B (AmB), which, like ethanol, alters the membrane fluidity (32), also caused a dramatic reduction of cell wall rigidity. Taking our data together, we propose that the nanomechanical properties of yeast cells are dependent on some ethanol-sensitive molecular components that build the interconnection between the plasma membrane and the cell wall and that these components are under the transcriptional control of Msn2/Msn4.
MATERIALS AND METHODS
Yeast strains, culture media, and viability assay.
Unless otherwise stated, the Saccharomyces cerevisiae “wild-type” strain was BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), and the isogenic BY yap1Δ, BY rlm1Δ, BY crz1Δ, BY msn2Δ, and BY msn4Δ mutants were from the EUROSCARF collection (Institute of Microbiology of the University of Frankfurt, Germany). The double mutant BY msn2Δ msn4Δ was obtained by crossing the haploid mutant α msn2Δ::KanMX4 with the a msn4Δ::KanMX4 strain, followed by sporulation and selection on G418-containing yeast extract-peptone-dextrose (YPD) agar medium, and the mutant was confirmed by PCR analysis using primers upstream of MSN2 and MSN4 and inside KanMX4, as described previously (33). The BYMSN2up strain, which constitutively expresses MSN2, was constructed as follows. The 2,115-bp MSN2 open reading frame (ORF) was amplified by PCR from the genomic DNA of the BY4741 strain with the primers MSN2_BamH1_S and MSN2_HindIII_AS (see Table S1 in the supplemental material). This PCR fragment was inserted into the BamHI/HindIII sites of the YCplac33-PGK/CYC1 plasmid, which was created with the PGK/CYC1 cassette from pYPGE2 (34) in the centromeric YCplac33 backbone (35). This plasmid, YCplac33-MSN2, was then used as the template to amplify the PGK1-MSN2-CYC1 cassette by using the primers YIplac211_PGK_S and YIplac211_CYC1_AS (see Table S1). The PCR fragment was subcloned into the YIplac211 integrative URA3 plasmid (35) by using an In-Fusion HD cloning kit (Clontech) after digestion by HindIII and EcoRI. The YIplac211-PGK-MSN2-CYC1 plasmid was then linearized by cutting with NcoI in the URA3 marker and used to transform the double mutant BY msn2Δ msn4Δ to obtain integration. Correct integration at the URA3 genomic locus of the PGK-MSN2-CYC1 cassette was confirmed by PCR using the above-mentioned MSN2_HindIII_AS and MSN2_BamH1_S primers (see Table S1). The haploid, prototrophic S. cerevisiae strain CEN.PK-113.7D was also used in this study to compare its resistance to ethanol with that of strain BY4741. The S. cerevisiae strains used in this study were cultivated in rich YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 2% [wt/vol] glucose) at 30°C with agitation (200 rpm). When the optical density at 600 nm (OD600) reached 1 unit, half of the culture was treated with 9% (vol/vol) ethanol for 5 h. Cell viability was estimated using methylene blue following a previously described procedure (36) or by flow cytometry according to the procedure described by Petitjean et al. (37), taking cell samples before and after 1 or 5 h of treatment with 9% (vol/vol) ethanol.
Assays of sensitivity to ethanol, cell wall drugs, and amphotericin B.
Sensitivities to ethanol and cell wall drugs were determined using exponential-phase cells on YPD agar, which were collected by centrifugation at an OD600 of about 2 units, washed with 1 M sorbitol, and resuspended in the same solution at an OD600 of 5 units (around 5 × 107 cells/ml−1). Aliquots of 3 μl at dilutions of 10 to 104 were placed on YPD plates containing ethanol, calcofluor white (Sigma-Aldrich), or Congo red (Sigma-Aldrich) at the concentrations indicated in the corresponding figures. The latter drugs were prepared at 5 mg ml−1 and filter sterilized through 0.2-μm filters (Minisart; Sartorius) prior to their use. The plates were incubated at 30°C, and colonies were scored after 48 h. Amphotericin B (AmB) sensitivity was determined essentially as described previously (32). Briefly, serial dilutions from a stock solution of 2 mg ml−1 AmB (Sigma-Aldrich) in dimethyl sulfoxide (DMSO) were added to 5-ml yeast cultures in YPD at an OD600 of 1.0 (about 1.4 × 107 cells ml−1). The MIC was estimated as the concentration of AmB that resulted in no measurable growth after 24 h at 30°C. Under these growth conditions, the MIC was evaluated to be 4 μg ml−1. For AFM experiments, 5 ml of yeast culture in YPD medium at an OD600 of 1.0 was treated with 8 μg ml−1 of AmB for 1 h at 30°C with shaking (200 rpm).
Yeast transformation and β-galactosidase assays of lacZ gene fusion constructs.
Wild-type and mutant cells were transformed by the standard lithium acetate (LiAc) method (38) with the following plasmids. (i) p1366 is a reporter plasmid for Rlm1 activity that contains the promoter of YIL117c fused to lacZ and has been described previously (39). (ii) pJL1 is a reporter plasmid for the Msn2/Msn4 transcription factor through the STRE. This plasmid contains the promoter of GSY2 fused to lacZ and has been described previously (40). (iii) pSG2 is a reporter for Crz1 transcription factor activity. This plasmid was constructed by inserting 6 CDRE elements (binding sites for Crz1) into the CYC1 promoter upstream of the lacZ gene in plasmid pLG669ZS (41) in a way similar to that described for pFKS2(CDRE)6-lacZ (42) (see Table S1 in the supplemental material for primers used to construct pSG2). The use of this plasmid for activation of Crz1 by calcium was validated by the lack of a response in a crz1Δ mutant transformed with this plasmid to calcium cations (not shown). The transformants were grown for 3 days at 30°C on SD medium (synthetic dextrose medium containing 0.17% [wt/vol] yeast nitrogen base without amino acids, 0.5% [wt/vol] ammonium sulfate, and 2% [wt/vol] glucose) complemented with auxotrophic amino acids (methionine, leucine, histidine, lysine, and tryptophan) and adenine, added at a 0.1% (wt/vol) final concentration. Clones arising from this medium were picked up and further cultivated in SD liquid selective medium overnight, and then cells were harvested by gentle centrifugation, resuspended to an OD600 of 0.5 to 0.8 unit in liquid YPD medium, and grown for 1 h at 30°C. Ethanol stress treatment was conducted exactly as described for the AFM experiments. Cells were collected and assayed for β-galactosidase enzyme activity as described by Rose et al. (43). The activity was expressed as nanomoles of p-nitrophenylgalactoside per minute and per milligram of protein. Concentrations of proteins in the cell extracts were measured using Bradford reagent, with bovine serum albumin as a standard (44).
Gene expression analysis by RT-qPCR.
Three independent cultures of wild-type strain BY4741 were inoculated at an OD600 of 0.05 into 250-ml shake flasks containing 100 ml YPD at 30°C and shaken in a rotary shaker set at 200 rpm. At an OD600 of 0.5, they were collected by centrifugation at 1,000 × g for 10 min, and the cell pellet was resuspended to the same OD in YPD in the absence or presence of 9% (vol/vol) ethanol. After 5 h of incubation at 30°C, the cells were collected by centrifugation as described above, and the tubes containing the cell pellet were immediately frozen in liquid nitrogen and stored at −80°C. Total RNA extraction was performed as described previously (45). Quantification was performed at 260 nm (1 OD260 unit = 40 μg RNA/ml), and purity was analyzed by determining the 260 nm/280 nm ratio (purities ranged from 1.9 to 2.1). The integrity of RNA was also verified by the sharpness and intensity of bands corresponding to 26S and 18S rRNAs as a parameter for agarose gel electrophoresis. For cDNA synthesis, 1 μg of total RNA was used in a 40-μl reverse transcription (RT) reaction mixture with the ImProm-II reverse transcription system (Promega) following the manufacturer's instructions. Primer design, nucleotide sequences, and PCR optimization were performed as previously described (45). Quantitative real-time PCR (qPCR) was conducted in an ABI Prism 7300 machine (Applied Biosystems, Foster City, CA), using a SYBR green PCR master mix kit (Applied Biosystems) under previously described conditions (45). Possible contamination by genomic DNA was evaluated. The quantification cycle (Cq) values were given automatically for independent amplifications. Raw Cq values for all the samples were then plotted in Microsoft Excel 2007 worksheets to create a suitable input file for geNorm that complied with the user's guide. Reference genes TDH3, RDN18, and FPR2 were chosen on the basis of expression stability and M values to normalize test gene values. RT-qPCR assays and analysis followed the MIQE guidelines (46), and the reliability of these parameters has been reported elsewhere (45–47).
Biophysical analysis of the cell surface by AFM.
Cultures of yeast cells (5 ml at an OD600 of 1.0 in YPD), treated or not with ethanol or AmB, were quickly collected by centrifugation, washed once with 1 ml acetate buffer (18 mM sodium acetate, 1 mM CaCl2, 1 mM MnCl2, pH 5.2), and resuspended in 5 ml of the same buffer. They were then immediately immobilized in microchambers made by PDMS stamps and prepared as described previously (26). Briefly, freshly oxygen-activated microstructured PDMS microchambers were covered by a total of 100 μl of cell suspension deposited into the microstructures by convective/capillary assembly. AFM images were recorded in contact mode with MLCT AUWH cantilevers (nominal spring constant of 0.01 N/m; Bruker). The cantilever spring constants were determined prior to each experiment by using the thermal noise method (48). The applied force was kept lower than 0.5 nN. To determine the nanomechanical properties of cells, force curves were recorded in force volume mode with an applied force of 0.5 nN. Data were processed using JPK data processing software (JPK Instruments, Berlin, Germany). For nanomechanical data, the Hertz model, which gives the force (F) as a function of the indentation (δ) and Young's modulus (E), was used to extract Young's modulus values according to equation 1, as follows:
| (1) |
where α is the tip opening angle (35°) and ν the Poisson ratio (arbitrarily assumed to be 0.5). The reported Young's modulus value is the value obtained at the maximal height of the Gaussian curve ± the σ value (corresponding to the midheight width) of this Gaussian distribution function.
The roughness was calculated according to equation 2, as follows:
| (2) |
The images used to measure roughness contained n ordered, equally spaced points, and xi is the vertical distance from the mean plane to the ith data point. For both imaging and force spectroscopy, we used AFM Nanowizard III (JPK Instruments, Berlin, Germany). Data processing was realized using JPK data processing software as well as OpenFovea software (49).
Isolation and determination of the biochemical composition of yeast cell walls.
Preparation of purified cell walls and acid treatment of the cell walls to release sugar monomers from β-glucan and mannans were performed as described previously (50). Quantification of chitin, β-1,3-glucan, and β-1,6-glucan was performed by use of a combined enzymatic/chemical method recently developed in the laboratory (51). Released monosaccharides (N-acetylglucosamine from chitin, glucose from β-glucan, and mannose from mannans) were determined by high-performance anionic exchange chromatography coupled to pulsed amperometric detection (HPAEC-PAD) as described previously (52).
TEM and fluorescence microscopy analyses.
For transmission electron microscopy (TEM) analysis, yeast cells (about 20 OD600 units; treated or not for 5 h with 9% ethanol) were harvested by centrifugation and fixed with 1× FIX solution (2% glutaraldehyde, 100 mM Sörensen buffer, pH 7.4). The suspension was then collected by centrifugation (2,000 × g, 5 min), and the pellet was incubated in 2% agarose (low melting point). This mixture was then washed once with 200 mM Sörensen buffer, pH 7.4, resuspended in 1% OsO4 containing 250 mM glucose in 100 mM Sörensen buffer, pH 7.4, and incubated for 1 h at room temperature. The fixed cells were dehydrated with increasing concentrations of ethanol (30 to 70% [vol/vol]) and transferred to 70% ethanol for 1 min at 37°C, followed twice by incubation in acetone for 2 min at 37°C. The cells were then infiltrated for 18 h in a 3:1 mixture of acetone-LR White. This mixture was replaced by LR White for 12 h, and the cells were resuspended in fresh LR White, which was polymerized for 24 h at 60°C. Sections (70 to 80 nm thick) were cut using a Leica Ultracut UCT ultramicrotome, mounted on single-slot grids, and poststained with 2% uranyl acetate in water and lead citrate. The ultrathin sections were observed with a Hitachi TEM HT7700 electron microscope at an acceleration voltage of 80 kV. For each condition, 30 cells from 2 independent cultures were examined.
Labeling of yeast cells with 1-[4-(trimethylamine)]-6-phenyl-1,3,5-hexatriene (TMA-DPH; Life Technologies) was carried out according to the protocol described by Chazotte (53). Briefly, 1 ml of yeast culture in YPD that was treated or not for 5 h with 9% ethanol was collected, washed once with phosphate-buffered saline (PBS), pH 7.4 (Euromedex), and immersed in the labeling solution (1 μM TMA-DPH in water). After a 10-min incubation in the dark, the labeling suspension was centrifuged, the cells were rinsed once with 100 μl of PBS, and then 10 μl of the cell suspension was mounted on a coverslip for imaging. A confocal laser scanning microscope equipped with a 405-nm UV laser (Leica SP2-AOBS) was used to visualize TMA-DPH labeling of the cells.
Statistical analyses.
All results were statistically analyzed using two-sample Student's t test (analysis of variance [ANOVA]) with Statgraphics Centurion XVI.II software, with a significance cutoff of 0.01. Significant differences are denoted by asterisks in the corresponding figures.
RESULTS
The nanomechanical properties of yeast cells are significantly altered upon ethanol treatment.
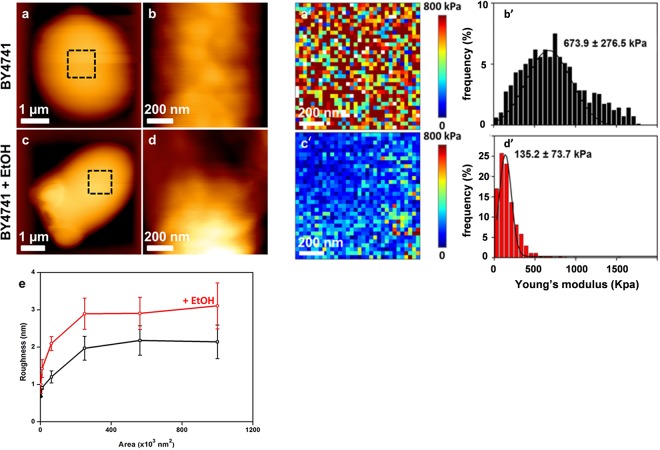
The scientific literature is not consistent with respect to the concentration of ethanol used for investigations of toxic effects on yeast cells or with respect to the duration of this treatment. In this study, we employed 9% (vol/vol) ethanol because this concentration roughly represents the median titer during traditional industrial fermentation (54), and we exposed yeast cells for 5 h, which is long enough to observe morphological and biophysical consequences caused by this alcoholic compound. Following this experimental design, untreated and ethanol-treated yeast cells were embedded in PDMS-fabricated microchambers for AFM analysis. Typical deflection images obtained by contact mode for a wild-type yeast cell trapped in a microchamber before and after ethanol treatment are shown in Fig. 1. At low resolution, the surface morphology appeared to be smooth and not altered by ethanol treatment. However, high-resolution deflection images taken at different areas on the surface revealed that this treatment caused a 50% increase of the cell roughness (Fig. 1e). To obtain quantitative values for cell surface changes caused by ethanol stress, we employed AFM in force spectroscopy mode, in which the cantilever and the tip successively approach and are retracted from the surface, and collected a large number of force curves (n = 65,536) across a defined (usually away from any bud scar) region of the cell. Using appropriate software (OpenFovea [49]), these force curves were converted into forces versus indentation to generate elasticity maps (Fig. 1a′ and c′). They were also fitted with the Hertz model to provide the distribution of values for Young's modulus, which is a quantitative expression of cell stiffness (Fig. 1b′ and d′). Qualitatively, the elasticity maps of the untreated cells were relatively heterogeneous, which was reflected by a large Gaussian distribution of Young's modulus values, with a maximal value of 674 kPa and a σ value of 276 kPa. In contrast, treatment of yeast cells for 5 h with 9% ethanol resulted in an apparently homogeneous elasticity map, consistent with a narrow Gaussian curve for the Young's modulus values, with a maximal value that was nonetheless 5-fold lower than that for the untreated wild-type cells (Fig. 1b′ and d′; Table 1). To verify that this ethanol effect on cell wall stiffness was independent of the strain background, we repeated the same experiments with strain CEN.PK113-7D, another laboratory strain most often employed in physiological studies (55). We found that the stiffness of this strain was also reduced, albeit only 3-fold, upon exposure to ethanol (see Fig. S1 in the supplemental material). This lower effect of ethanol was in line with our finding that the CEN.PK strain background is more ethanol tolerant than BY4741 (data not shown). It is worth noting that such a low stiffness of ethanol-treated cells is comparable to that measured for a double crh1Δ crh2Δ mutant defective in transglycosylases that catalyze the cross-linkages between chitin and β-glucan (24).
FIG 1.
AFM imaging of wild-type cells treated with ethanol. High-resolution AFM images (z range = 100 nm) of exponentially growing BY4741 cells on YPD that were not treated (a and b) or were treated for 5 h with 9% ethanol (EtOH) (c and d). (e) Levels of roughness measured at different areas on the cell surface for untreated (black line) and ethanol-treated (red line) BY4741 cells. Elasticity maps (z range = 800 kPa) were recorded for an untreated cell (a′) and a cell treated with 9% ethanol (c′). Young's modulus values (from 1,024 force curves) corresponding to the elasticity maps are shown for an untreated cell (b′) and an ethanol-treated cell (d′).
TABLE 1.
Ethanol effects on Young's modulus and on roughness of wild-type and mutant yeast cellsa
| Yeast strain | Young's modulus (kPa) |
Roughness (nm) |
||||
|---|---|---|---|---|---|---|
| No ethanol | Ethanol treatment | Ratio (with/without ethanol) | No ethanol | Ethanol treatment | Ratio (with/without ethanol) | |
| BY4741 | 673.9 ± 276.5 | 135.2 ± 73.7 | 0.20 | 2.05 ± 0.35 | 3.2 ± 0.5 | 1.56 |
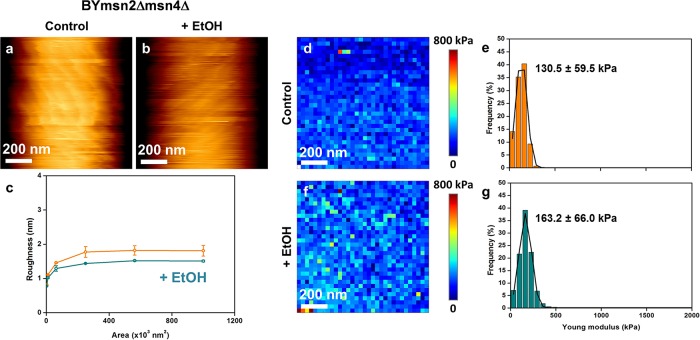
| BY msn2Δ msn4Δ | 130.5 ± 59.5 | 163.2 ± 66 | 1.25 | 1.85 ± 0.35* | 1.55 ± 0.10* | 0.85 |
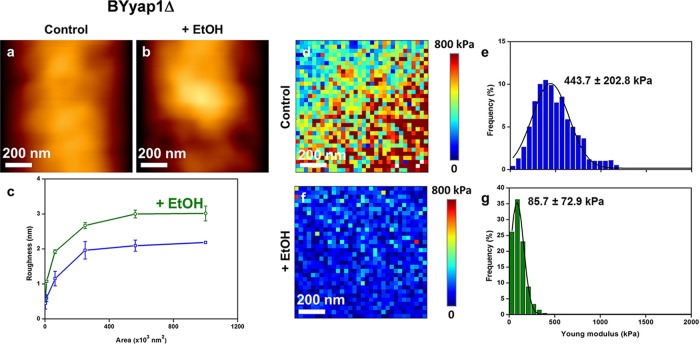
| BY yap1Δ | 443.7 ± 202.8 | 85.7 ± 72.9 | 0.19 | 2.2 ± 0.1 | 3.05 ± 0.3 | 1.38 |
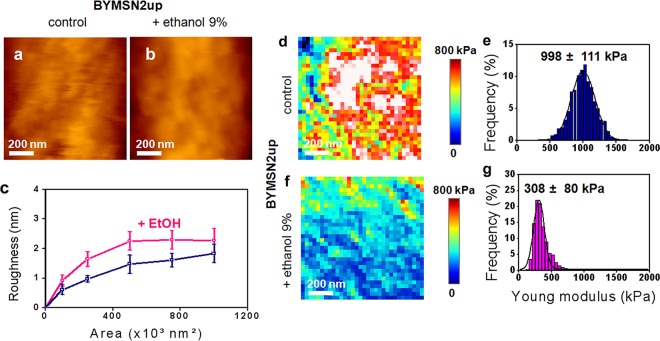
| BYMSN2up | 998 ± 111 | 308 ± 80 | 0.31 | 1.7 ± 0.3* | 2.2 ± 0.3* | 1.39 |
Ethanol was added to a 9% final concentration to yeast cells cultivated in YPD medium at an OD600 of around 1. After 5 h of incubation, the cells were collected and analyzed by AFM as described in Materials and Methods to determine the biophysical properties. Young's modulus (maximal value of the Gaussian distribution ± σ, expressed as the standard deviation) was determined for 15 independent cells (5 cells from 3 biological replicates). Differences between mutant and wild-type cells were analyzed statistically using two-sample Student's t test (ANOVA) with Statgraphics Centurion XVI.II software. *, P < 0.01 (significant).
Ethanol stress alters neither biochemical composition nor cell wall thickness but causes a reduction of cell membrane thickness.
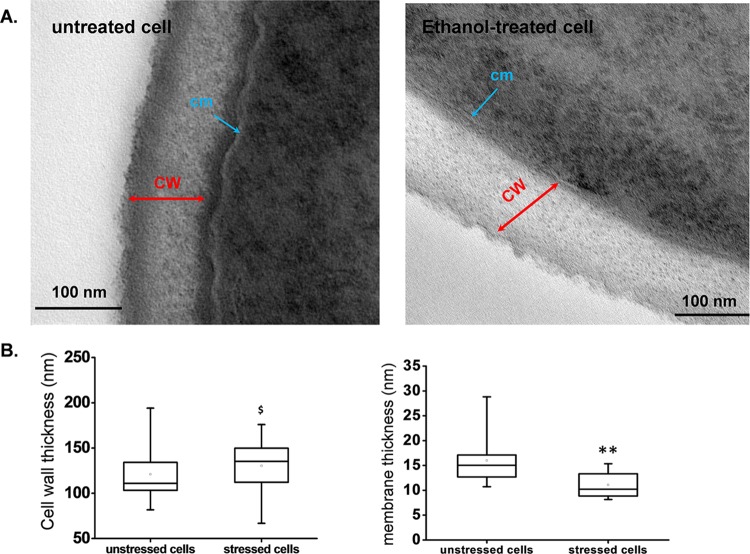
The above results together with our previous works (24, 56) led us to examine whether the effect of ethanol on cell stiffness was due to a change in the polysaccharide composition and/or in the cross-linkages between these cell wall components. To answer the first question, we determined the cell wall composition by using a combined enzymatic and chemical method that allowed us to quantify the four components, namely, β-1,3-glucan, β-1,6-glucan, chitin, and mannan, of isolated walls (51). As reported in Table 2, the cell wall composition of yeast cultivated on glucose was not significantly altered after a 5-h treatment with 9% (vol/vol) ethanol. In addition, we evaluated the cell wall thickness by TEM. Figure 2A shows representative TEM images of untreated and ethanol-treated cells, each magnified to show the cell wall. This allowed us to distinctly visualize the outer, electron-dense layer (mainly composed of mannoproteins) and the inner, less electron-dense layer (mainly constituted of β-glucan) (57, 58). According to the recommendation of Backhaus et al. (59), we measured the cell wall thickness at several points on independent mother cells far away from the growing buds, as the latter clearly had an impact on the cell thickness (for instance, see Fig. S2 in the supplemental material). From the collected values, we determined median values of 115 nm for untreated cells and 130 nm for ethanol-treated cells. However, the difference is not significant due to the relatively high dispersion of the values over the median in the box plot diagram (Fig. 2B). This result is thus consistent with the failure of ethanol to change the cell wall composition. Moreover, the cell wall thickness was similar to that reported by others using the same method (59, 60) but slightly (10%) lower than that estimated by AFM (61).
TABLE 2.
Cell wall compositions of wild-type, msn2Δ msn4Δ, and yap1Δ yeast strains upon exposure to 9% (vol/vol) ethanol for 5 ha
| Polysaccharide | % of total cell wall polysaccharides (mean ± SD)d |
|||||||
|---|---|---|---|---|---|---|---|---|
| BY4741 (wild type) |
BY msn2Δ msn4Δ |
BY yap1Δ |
BYMSN2up |
|||||
| Control | With ethanol | Control | With ethanol | Control | With ethanol | Control | With ethanol | |
| β-(1,3)-Glucan | 36.8 ± 5.0 | 37.7 ± 2.0 | 42 ± 6.0 | 38.5 ± 2.5 | ND | ND | 37.2 ± 5.5 | 36.7 ± 5.0 |
| β-(1,6)-Glucan | 20 ± 1.0 | 21 ± 2.0 | 23 ± 2.0 | 24 ± 2.0 | ND | ND | 22.8 ± 4.0 | 26.3 ± 5.0 |
| β-Glucanb | 57 ± 1.0 | 59 ± 4.0 | 65 ± 5.0# | 63.5 ± 4.5# | 58 ± 6.0c# | 60 ± 5.0 | 57.4. ± 5.5 | 63.0 ± 5.0 |
| Chitin | 4.9 ± 0.2 | 4.9 ± 0.2 | 4.2 ± 0.2 | 3.5 ± 0.2 | 3.0 ± 0.5 | 3.2 ± 0.3 | 3.8 ± 0.3 | 3.5 ± 0.32 |
| Mannan | 37.4 ± 3.3 | 36 ± 3.1 | 32 ± 1.0# | 33 ± 2.0# | 39 ± 3.0# | 36 ± 2.0 | 38.9 ± 4.0 | 36.5 ± 5.0 |
| Glucan/mannan ratio | 1.52 | 1.63 | 2.1 | 1.95 | 1.49 | 1.67 | 1.52 | 1.72 |
Cell wall compositions were determined for exponentially growing yeast cells in YPD, treated or not for 5 h with 9% ethanol as described in Materials and Methods. Values are means and SD for three independent biological samples that were technically repeated two times.
Sum of β-(1,3)- and β-(1,6)-glucan.
Determined by a chemical method.
#, P < 0.05 (statistical difference between wild-type and mutant yeast cells as calculated by Student's t test); ND, not determined.
FIG 2.
Estimation of the thicknesses of the cell wall and cell membrane for untreated and ethanol-treated yeast cells from transmission electron micrographs. (A) Representative TEM images of untreated and ethanol-treated cells obtained as described in the legend to Fig. 1. (B) Cell wall and cell membrane thicknesses, represented by box plots obtained for the analysis of 30 cells each at 2 different points. The lower and upper boundaries of each box represent the 25th and 75th percentiles, respectively. The line within each box corresponds to the median, and the cross to the mean distribution of the values. The results were statistically analyzed using two-sample Student's t test (ANOVA) with Statgraphics Centurion XVI.II software. $, P > 0.05 (not significant); **, P < 0.001 (significant). Abbreviations: CW, cell wall; cm, cell membrane.
The cell membrane was also clearly visible on TEM images of untreated cells but was less discernible in ethanol-treated cells (Fig. 2A; also see Fig. S2 in the supplemental material), probably because the membrane was damaged by this treatment, as emphasized in previous works (62–65). We therefore tentatively estimated the thickness of the cell membrane by taking measurements of at least 5 different points per cell and made these measurements for 20 (each) untreated and ethanol-treated cells. A membrane thickness of about 15 nm was determined for the unstressed cells, in accordance with a previous report of a three-dimensional analysis of cell structure (66). However, the cell membrane of ethanol-treated cells was 35% thinner than that of untreated cells (Fig. 2B). To further investigate whether cell membranes were damaged by ethanol treatment, we used TMA-DPH, a fluorescent membrane probe that has the unique property of labeling the outer leaflet of a membrane bilayer due to its charged head group (53). Accordingly, the fluorophore localized exclusively to the plasma membrane in the untreated cells, perfectly delimiting their shape. In contrast, in ethanol-treated cells, the TMA-DPH probe did not mark solely the plasma membrane, as other internal structures were also labeled, likely because the probe was able to diffuse inside the cell (see Fig. S3). In addition, we also used annexin V coupled to a fluorescent dye. This marker is commonly used to assess membrane integrity during apoptosis because it binds to phosphatidylserine exposed on the external face of the membrane (67, 68). We found that the number of fluorescent cells detected by fluorescence-activated cell sorting (FACS) rose from 3% for untreated cells to 35% for yeast cells treated with ethanol for 5 h, which is actually in frame with the loss of cell viability (see Fig. S4). Taken together, these results are in accordance with the well-recognized actions of ethanol, i.e., perturbing the plasma membrane fluidity (63) and modifying the phospholipid and ergosterol content (17, 69, 70), thereby compromising the selectively permeable nature of the cell membrane.
The effect of ethanol on cell stiffness does not involve cell wall remodeling.
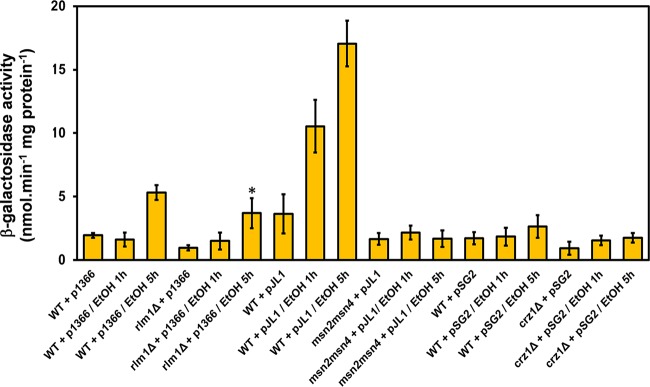
In a previous work, we provided evidence that cross-links between the polysaccharides of the cell wall contribute to the nanomechanical properties of yeast cells (71). This molecular architecture that ensures cell wall integrity is in great part under the control of the PKC1/SLT2-dependent cell wall integrity (CWI) pathway, and Rlm1 is the transcription factor responsible for the bulk of the CWI transcription program (72). Therefore, we asked whether the effect of ethanol stress on the nanomechanical properties involves the CWI pathway. To address this question directly, we used a transcriptional readout that is specifically activated by Rlm1. This readout consists of measuring the β-galactosidase activity in yeast cells transformed with the bacterial lacZ gene fused to the promoter for the Rlm1-dependent activation of YIL117c (73). As shown in Fig. 3, the 2.5-fold increase of β-galactosidase activity of this readout after 5 h of exposure of yeast cells to 9% ethanol was not statistically different from the activation measured in an rlm1Δ mutant. This result indicates that ethanol stress does not activate the CWI pathway, although it was reported that the Slt2/Mpk1 kinase is phosphorylated in response to ethanol stress (13). However, it has been shown that phosphorylation of Slt2/Mpk1 does not always imply its activation (74). On the other hand, it has been reported that the transcriptional response of yeast to ethanol stress is largely dependent on the transcription factor encoded by MSN2 and MSN4 (5, 29). To confirm this statement, we used the transcription readout GSY2-lacZ (75), which contains 2 STRE in the GSY2 promoter to which Msn2/Msn4 binds. The results shown in Fig. 3 support this statement, as the 5- and 7-fold increases of β-galactosidase activity measured after 1 and 5 h of treatment with 9% ethanol, respectively, were totally abolished in an msn2Δ msn4Δ mutant. We also assessed the Ca2+ signaling cascade that relies on the Crz1 transcription factor (76) and which was identified to be part of the cell wall compensatory mechanism (42, 77). However, the CDRE-lacZ transcriptional readout (76) was totally unresponsive to ethanol stress (Fig. 3). Overall, these results confirmed that the transcriptional response induced by ethanol stress belongs to the Msn2/Msn4-dependent general stress response. This transcriptional activation can be explained mainly by the ethanol-induced translocation of the transcription factor Msn2 into the nucleus (13, 31). Furthermore, these results complemented the transcriptional data of Lewis et al. (78) showing that the transcriptomic response of oak-soil yeast to 5% ethanol was seriously compromised upon deletion of the MSN2 gene. However, despite a lack of ethanol effects on the transcriptional readouts specific to CWI, RT-qPCR was performed on essential CWI-regulated cell wall genes in wild-type cells exposed to 9% ethanol. The results showed significant transcriptional activation (ranging from 3- to 10-fold) of FKS1 (coding for the major β-1,3-glucan synthase [79]), GAS1 (coding for a β-1,3-glucanosyltransferase [80]), MNN9 and MNN10 (encoding Golgi-located mannosyltransferase enzymes [81]), YLR194c (renamed NCW2, for new cell wall protein [http://www.yeastgenome.org/cgi-bin/locus.fpl?locus=YLR194c#S000151886]), and RLM1 upon exposure of yeast cells to 9% ethanol (see Fig. S4 in the supplemental material). Upregulation of these genes by this ethanol stress could be explained by the presence of cis STRE in their promoters (as analyzed by Yeastract [http://www.yeastract.com/]). These data are indirectly supported by the fact that CTT1, a well-established Msn2/Msn4 stress-responsive gene, was also upregulated under these conditions (82).
FIG 3.
Transcriptional readouts depending on the Rlm1, Msn2/Msn4, and Crz1 transcription factors in response to ethanol stress. Wild-type BY4741 cells transformed with an empty plasmid or with plasmid p1366 (pYIL117c-LacZ; transcriptional readout of Rlm1 transcriptional activity), pJL1 [pGSY2(6×STRE)-LacZ; transcriptional readout of Msn2/Msn4 transcriptional activity], or pSG2 (6×CDRE-lacZ; transcriptional readout of Crz1 transcriptional activity) were cultivated in selective YN glucose liquid medium supplemented with auxotrophic requirements until an OD600 of 1. The culture was then transferred to YPD medium for 1 h, and to half of the culture ethanol was added to a 9% (vol/vol) final concentration. Samples were collected at 1 and 5 h at 30°C for measurement of the β-galactosidase activity. Values shown are the means for three independent experiments, with standard deviations represented by error bars.
The effect of ethanol on the nanomechanical properties of the cell wall is not directly dependent on Msn2/Msn4, albeit this property is under the control of this transcription factor.
Based on the fact that ethanol stress is mediated through Msn2/Msn4 (83), we used a mutant defective in this transcription factor as well as a strain that constitutively expressed MSN2 under the control of the strong PGK1 promoter to directly evaluate the role of Msn2/Msn4 in the ethanol effect on cell wall biophysics. Quite unexpectedly, we found that the cell wall stiffness of an msn2 msn4 mutant was very similar to that of a wild-type strain treated for 5 h with 9% ethanol (compare the data in Fig. 1d′ with those in Fig. 4e and Table 1). Moreover, the low cell stiffness of this mutant was not reduced further after ethanol treatment (Fig. 4g), suggesting that the maximal effect on cell elasticity was reached upon deletion of MSN2/MSN4. It is also interesting that in contrast to what was observed in wild-type cells, ethanol treatment caused a 15% decrease of the roughness of the msn2Δ msn4Δ mutant (Fig. 4c). Despite this dramatic effect of MSN2/MSN4 deletion on cell wall stiffness, the cell wall composition of this mutant was barely affected, showing only a slightly higher (25%) β-glucan/mannan ratio than that of the wild type (Table 2). In addition, further treatment of this mutant with ethanol had no effect on its cell wall composition. We then investigated the effects of overexpressing MSN2 on the biophysical properties of yeast cells. We first verified that the Msn2 transcription factor was overproduced in BYMSN2up by showing that this strain was slightly more viable than the wild type after ethanol treatment, consistent with an earlier report (83), and that it was the sole strain to accumulate trehalose during growth on glucose, due to an Msn2/Msn4-dependent transcriptional activation of the trehalose synthetic genes (75) (see Fig. S5 in the supplemental material). We noticed that high and constitutive expression of MSN2 had no effect on the cell wall composition, which was not modified after ethanol stress (Table 2). The results of AFM analyses of BYMSN2up are reported in Fig. 5 and Table 1. We found that the average stiffness of the cells was 35% higher than that of the wild-type cells. However, despite this higher stiffness, the median value of Young's modulus for this mutant dropped about 3.5-fold after ethanol treatment. Figure 5 also shows that the roughness of BYMSN2up cells was statistically lower (P < 0.05) than that of the wild-type cells and increased by 50% upon exposure to ethanol, as in the wild-type strain. We also checked the consequences of deleting YAP1 on these biophysical properties of the cells, since it was recently suggested that this gene may also be implicated in the cell wall integrity pathway (45) and in the response to ethanol stress (5). As shown in Fig. 6, a mutant defective in YAP1 exhibited a roughness similar to that of the wild type before and after ethanol treatment. In addition, the value for Young's modulus for this mutant, while being 35% lower than that for the wild type, was still reduced 5-fold after treatment with 9% ethanol.
FIG 4.
AFM imaging and elasticity mapping of the BY msn2Δ msn4Δ mutant before and after ethanol treatment. The same procedure as that described in the legend to Fig. 1 was used, except that high-resolution AFM images (z range = 100 nm) were obtained for untreated (a) and ethanol-treated (b) msn2Δ msn4Δ mutant cells. (c) Levels of roughness. The elasticity maps (z range = 800 kPa) (d and f) and distributions of Young's modulus values (e and g) were obtained from 1,024 force versus indentation curves for control and 5-h ethanol-treated cells.
FIG 5.
AFM imaging and elasticity mapping of the BYMSN2up mutant before and after ethanol treatment. The same procedure as that described in the legend to Fig. 1 was used, except that high-resolution AFM images (z range = 100 nm) were obtained for untreated (a) and ethanol-treated (b) mutant cells that expressed MSN2 under the control of the strong PGK1 promoter. (c) Levels of roughness. The elasticity maps (z range = 800 kPa) (d and f) and distributions of Young's modulus values (e and g) were obtained from 1,024 force versus indentation curves for control and 5-h ethanol-treated mutant cells.
FIG 6.
AFM imaging and elasticity mapping of the BY yap1Δ mutant before and after ethanol treatment. The same procedure as that described in the legend to Fig. 1 was used, except that high-resolution AFM images (z range = 100 nm) were obtained for the untreated (a) and ethanol-treated (b) yap1Δ mutant. (c) Levels of roughness. The elasticity maps (z range = 800 kPa) (d and f) and distributions of Young's modulus values (e and g) were obtained from 1,024 force versus indentation curves for control and 5-h ethanol-treated mutant cells.
Complementary to these biophysical data, we evaluated the growth phenotype of these mutant strains on YPD agar plates in the presence of ethanol (0, 5, 9, and 12%) or the cell wall-interfering drugs calcofluor white and Congo red (0 to 100 μg/ml). While the yap1Δ mutant exhibited higher sensitivities to ethanol and to cell wall drugs, the deletion of MSN2/MSN4 or the high expression of MSN2 had little effect on the sensitivity to these compounds, and both relevant strains showed similar reductions of ethanol resistance (see Fig. S6 in the supplemental material). These results indicated that any alteration of the nanomechanical properties of the cell wall is not necessarily a consequence of cell wall defects. Taken together, these results show that the biophysical properties of yeast cells as identified by wall stiffness are to a great extent under the control of the Msn2/Msn4 general stress response pathway. However, the effects of ethanol on the nanomechanical properties of the cells are mediated neither by this transcription factor nor by Yap1.
The cell membrane contributes to the nanomechanical properties of cells.
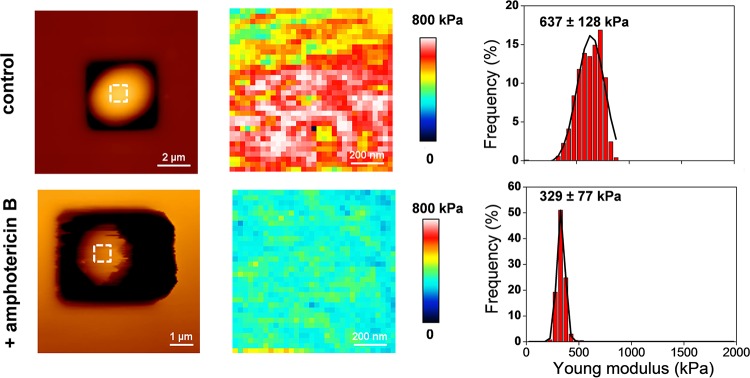
The data reported above suggest that the reduction of cell stiffness in response to ethanol stress can be due to an alteration of the plasma membrane. To support this idea, we carried out an AFM analysis of yeast cells treated with the antifungal amphotericin B (AmB). We chose this drug because it is well reported to alter proper functioning of yeast cell membranes (84, 85) by direct binding to ergosterol (32). Figure 7 shows that treatment of yeast cells with AmB at 2 times its MIC (i.e., 8 μg ml−1) for only 1 h (short enough to not cause any drop in cell viability [data not shown]) resulted in a 2-fold decrease of the median Young's modulus value, confirming that cell wall stiffness is in part dependent on the integrity of the cell membrane.
FIG 7.
AFM imaging and elasticity mapping of the BY4741 wild-type strain before and after treatment with amphotericin B. The same procedure as that described in the legend to Fig. 1 was used, except that the cells were treated with 8 μg ml−1 of amphotericin B for 1 h and the AFM images for untreated and treated cells were obtained at a low resolution (z range = 500 nm). The elasticity maps (z range = 800 kPa) and distributions of Young's modulus values were obtained from 1,024 force versus indentation curves for untreated and amphotericin B-treated cells.
DISCUSSION
At the molecular level, ethanol stress triggers a remarkable remodeling of the yeast transcriptome (6, 7, 78), proteome (54), and metabolome (86), but the consequences on the physical properties of the cells are unknown. Here we addressed this question by using AFM, a technology that is increasingly being used in biology to investigate nanomechanical properties of microbial cells (87, 88). The main result of this study was to show that the stiffness of yeast cells was considerably reduced upon exposure to 9% (vol/vol) ethanol (or, conversely, the elasticity of the cell was dramatically increased), and this reduced stiffness was not accompanied by any significant change in the cell wall polysaccharide composition. It is also worth noting that despite a higher elasticity, the cell size and cell shape of ethanol-treated yeasts were similar to those of untreated yeast cells, suggesting that these parameters are likely dependent on cell wall composition. Our results thus differ from the previous work of Canetta et al. (23), who actually reported shrinkage of cells after incubation for more than 1 h with ethanol at a 10% or higher concentration. That we did not observe such a shrinkage in our work can most likely be ascribed to the difference in the methods of immobilizing cells for AFM analysis, as our method kept the cells alive, whereas yeast cells were fixed and dried on a glass slide in the work of Canetta et al. (23).
In a previous work, we showed that the stiffness of yeast cells is affected by cross-links between cell wall components, not by their actual polysaccharide content (24). Consistent with this finding, we found that the reduction of cell stiffness caused by ethanol was not accompanied by any change in the composition of the cell wall, suggesting that ethanol alters this physical property by activating the cell wall remodeling mechanism. However, published transcriptomic data sets for ethanol-stressed cells did not highlight an upregulation of genes encoding cell wall remodeling enzymes (2, 5), which is confirmed by our finding that ethanol stress does not activate the CWI pathway. Moreover, we did not obtain any conclusive data on an in vitro effect of ethanol on the activity of exo- and endoglucanases as assayed on purified yeast cell walls by use of laminarin as the substrate according to the analytical method described by Mrsa et al. (89; M. Schiavone and J. M. François, unpublished data). Another possible explanation for the alteration of the nanomechanical properties could be a disorganization of the F-actin cytoskeleton, as this was reported to be caused by a high ethanol concentration (90). This explanation actually does not hold either, because treatment of yeast cells with latrunculin A, a toxin molecule known to disrupt the actin cytoskeleton (91), did not modify Young's modulus (F. Pillet, M. Schiavone, E. Dague, and J. M. François, unpublished data).
Ethanol stress is known to cause a rapid translocation of Msn2 from the cytosol to the nucleus, where it binds to STRE and thereby promotes transcriptional activation of STRE-responsive genes (30, 31). We confirmed this mechanism by showing a strong upregulation of the GSY2-lacZ gene fusion as a readout of Msn2 transcriptional activation in the presence of 9% ethanol. Therefore, we investigated whether the reduced stiffness of yeast cells in response to this ethanol stress is mediated through Msn2/Msn4 by using a mutant defective in MSN2/MSN4 as well as a strain that overexpressed the MSN2 gene. Our results pointed to an unexpected role of Msn2/Msn4 in controlling the mechanical properties of the cell wall, since a mutant defective in this transcription factor harbored a low cell wall stiffness comparable to that of the wild type after ethanol treatment, while a strain overproducing Msn2 showed a 35% higher stiffness than that of the wild type. We found these data difficult to immediately reconcile with a role of Msn2/Msn4 in mediating the effect of ethanol on cell stiffness. Indeed, owing to the fact that ethanol activates the Msn2/Msn4 transcriptional response, one might expect that cell stiffness might not be altered upon the loss of MSN2/MSN4. Alternatively, if the effect of ethanol is to activate the transcription of an inhibitor of the cell wall organization in an Msn2/Msn4-dependent manner, then a yeast strain that overexpresses the MSN2 gene should have a lower cell stiffness than the wild type, which is not the case either. Overall, we therefore favor a model in which ethanol may act directly by inhibiting some putative Mns2/Msn4 transcriptionally regulated cell integrity components that contribute to the rigidity of the yeast cell wall.
What could be the nature of these putative ethanol-sensitive Msn2/Msn4-regulated cell integrity components? It is worth recalling the well-established effects of ethanol in perturbing membrane structure and fluidity (reviewed in reference 3). Consistently, we found that the fluorescent TMA-DPH probe, which specifically labels only the plasma membrane in a wild-type cell, diffused inside yeast cells treated with ethanol. In addition, we reported that the cell membrane thickness of ethanol-treated yeast cells was about 35% thinner than that of untreated cells. These data are in accordance with studies on model membrane systems which reported a 30% reduction of the membrane thickness at high concentrations (>6%) of ethanol. It is proposed that this important reduction of membrane thickness is due to a transition from a bilayer to an interdigitated phase that takes place when the fatty acyl chains cross the bilayer midplane and that ethanol shields the fatty acid methyl groups from the aqueous phase (64). As a consequence, this ethanol-induced membrane reduction may have dramatic effects on membrane-associated protein conformation and function (reviewed in reference 3). Based on this model, proper delivery of glycosylphosphatidylinositol (GPI)-linked proteins to the membrane via the secretory pathway (and subsequently their linkage to cell wall polysaccharides) can be impaired when the membrane is disorganized due to the presence of an elevated concentration of ethanol. The in silico analysis carried out by the Klis group (92) underscored at least 20 GPI-linked proteins distributed among four different families, namely, the Gas1, Yap3, Sps2, and Plb1 families (92), and a fifth family containing different proteins with a predicted membrane localization, including Exg2 (a β-1,3-exoglucanase enzyme) (93) and Kre1 (a glycoprotein that serves as a killer K1 receptor) (94). In addition, the genes encoding these proteins possesses one to several STRE in their promoters (checked by use of Yeastract), suggesting that they can potentially be regulated transcriptionally by Msn2/Msn4, and hence this may account for the similar effects of ethanol and the lack of Msn2/Msn4 on cell wall stiffness. However, no transcriptomic data are yet available in public databases (i.e., Gene Expression Omnibus at http://www.ncbi.nlm.nih.gov/gds) to support that these GPI-linked protein-encoding genes are under the control of Msn2/Msn4. Another potential target of ethanol is the plasma membrane protein Hsp12, which was shown to be implicated in cell wall elasticity (95) and which is transcriptionally regulated by Msn2/Msn4 (96).
This study on the physical effects of ethanol on yeast cells opens up the more general view that the plasma membrane contributes to the nanomechanical properties, namely, stiffness and roughness, of the cell wall. This suggestion is supported by the finding that cell wall stiffness is dramatically reduced upon treatment of yeast with amphotericin B, a polyene antifungal agent that binds to ergosterol and causes a loss of membrane structure (32). However, the reduction of stiffness caused by this drug is lower than that after ethanol stress, indicating some difference in the mechanism. It is possible that in contrast to the effects of ethanol, this effect is in part counterbalanced by the activation of the CWI pathway, since it was reported by Straede et al. (97) that AmB as well as other plasma membrane-interfering compounds, such as chlorpromazine and nystatin, caused an activation of this pathway. However, and in accordance with our model, it can be expected that any stretching of the plasma membrane caused, for instance, by an increase of the turgor pressure or by some organic solvents (98) might also alter the stiffness of the cell.
Conclusions.
In this work, we investigated the physical effects of ethanol on yeast by employing the AFM methodology. This approach allowed us to find that ethanol at a concentration reached under industrial fermentation conditions provokes a dramatic reduction of the cell wall stiffness and an alteration of the cell surface (increase of the roughness). We found that this effect is not mediated by the Msn2/Msn4-dependent general stress response pathway, although the nanomechanical properties of the cell wall, namely, stiffness and roughness, are in part under the control of this pathway. Taking into account the well-established effect of ethanol of destroying the membrane organization, these data led us to propose that the plasma membrane contributes to the nanomechanical properties of the cell wall and that the loss of cell wall stiffness in response to ethanol stress is due to a disorganization of some cell membrane integrity components, most likely GPI-anchored membrane proteins that make the interconnection between the cell wall and the cell membrane and which are under the transcriptional control of Msn2/Msn4.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Isabelle Fourquaux (Center of Electronic Microscopy, Faculty of Medicine, Federal University of Toulouse) for technical assistance with TEM preparations. We thank members of the lab teams of M.D.M., E.D., and J.M.F. for technical and scientific support throughout this work.
This work was supported by a grant from the Young Scientist Program of the ANR (Agence Nationale de la Recherche) (project ANR-11-JSV5-001-01 to E.D.) and by a grant from the Region Midi Pyrénées (project 10051296 to J.M.F.). C.F.D. was supported by a grant from the Direction Générale de l'Armement (DGA), and M.S. was supported by a CIFRE grant from ANRT and Lallemand Inc. during Ph.D. studies. C.E. was supported by grants from the CAPES-FACEPE/PNPD program (grants 681/2011 and APQ-0201-2.02/10).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01213-16.
REFERENCES
- 1.Kitagaki H, Araki Y, Funato K, Shimoi H. 2007. Ethanol-induced death in yeast exhibits features of apoptosis mediated by mitochondrial fission pathway. FEBS Lett 581:2935–2942. doi: 10.1016/j.febslet.2007.05.048. [DOI] [PubMed] [Google Scholar]
- 2.Stanley D, Bandara A, Fraser S, Chambers PJ, Stanley GA. 2010. The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J Appl Microbiol 109:13–24. doi: 10.1111/j.1365-2672.2009.04657.x. [DOI] [PubMed] [Google Scholar]
- 3.Henderson CM, Block DE. 2014. Examining the role of membrane lipid composition in determining the ethanol tolerance of Saccharomyces cerevisiae. Appl Environ Microbiol 80:2966–2972. doi: 10.1128/AEM.04151-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao X, Zheng D, Liu T, Wang P, Zhao W, Zhu M, Jiang X, Zhao Y, Wu X. 2012. A novel strategy to construct yeast Saccharomyces cerevisiae strains for very high gravity fermentation. PLoS One 7:e31235. doi: 10.1371/journal.pone.0031235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma M, Liu ZL. 2010. Mechanisms of ethanol tolerance in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 87:829–845. doi: 10.1007/s00253-010-2594-3. [DOI] [PubMed] [Google Scholar]
- 6.Alexandre H, Ansanay-Galeote V, Dequin S, Blondin B. 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett 498:98–103. doi: 10.1016/S0014-5793(01)02503-0. [DOI] [PubMed] [Google Scholar]
- 7.Chandler M, Stanley G, Rogers P, Chambers P. 2004. A genomic approach to defining the ethanol stress response in the yeast Saccharomyces cerevisiae. Ann Microbiol 54:427–454. [Google Scholar]
- 8.Dinh TN, Nagahisa K, Yoshikawa K, Hirasawa T, Furusawa C, Shimizu H. 2009. Analysis of adaptation to high ethanol concentration in Saccharomyces cerevisiae using DNA microarray. Bioprocess Biosyst Eng 32:681–688. doi: 10.1007/s00449-008-0292-7. [DOI] [PubMed] [Google Scholar]
- 9.Gasch AP, Eisen MB. 2002. Exploring the conditional coregulation of yeast gene expression through fuzzy k-means clustering. Genome Biol 3:RESEARCH0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita K, Matsuyama A, Kobayashi Y, Iwahashi H. 2006. The genome-wide screening of yeast deletion mutants to identify the genes required for tolerance to ethanol and other alcohols. FEMS Yeast Res 6:744–750. doi: 10.1111/j.1567-1364.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi T, Shimoi H, Ito K. 2001. Identification of genes required for growth under ethanol stress using transposon mutagenesis in Saccharomyces cerevisiae. Mol Genet Genomics 265:1112–1119. doi: 10.1007/s004380100510. [DOI] [PubMed] [Google Scholar]
- 12.Teixeira MC, Raposo LR, Mira NP, Lourenco AB, Sa-Correia I. 2009. Genome-wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol. Appl Environ Microbiol 75:5761–5772. doi: 10.1128/AEM.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Voorst F, Houghton-Larsen J, Jonson L, Kielland-Brandt MC, Brandt A. 2006. Genome-wide identification of genes required for growth of Saccharomyces cerevisiae under ethanol stress. Yeast 23:351–359. doi: 10.1002/yea.1359. [DOI] [PubMed] [Google Scholar]
- 14.Swinnen S, Schaerlaekens K, Pais T, Claesen J, Hubmann G, Yang Y, Demeke M, Foulquie-Moreno MR, Goovaerts A, Souvereyns K, Clement L, Dumortier F, Thevelein JM. 2012. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res 22:975–984. doi: 10.1101/gr.131698.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duitama J, Sanchez-Rodriguez A, Goovaerts A, Pulido-Tamayo S, Hubmann G, Foulquie-Moreno MR, Thevelein JM, Verstrepen KJ, Marchal K. 2014. Improved linkage analysis of quantitative trait loci using bulk segregants unveils a novel determinant of high ethanol tolerance in yeast. BMC Genomics 15:207. doi: 10.1186/1471-2164-15-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu XH, Wang MH, Tan T, Li JR, Yang H, Leach L, Zhang RM, Luo ZW. 2007. Genetic dissection of ethanol tolerance in the budding yeast Saccharomyces cerevisiae. Genetics 175:1479–1487. doi: 10.1534/genetics.106.065292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexandre H, Rousseaux I, Charpentier C. 1994. Relationship between ethanol tolerance, lipid composition and plasma membrane fluidity in Saccharomyces cerevisiae and Kloeckera apiculata. FEMS Microbiol Lett 124:17–22. doi: 10.1111/j.1574-6968.1994.tb07255.x. [DOI] [PubMed] [Google Scholar]
- 18.Thomas DS, Hossack JA, Rose AH. 1978. Plasma-membrane lipid composition and ethanol tolerance in Saccharomyces cerevisiae. Arch Microbiol 117:239–245. doi: 10.1007/BF00738541. [DOI] [PubMed] [Google Scholar]
- 19.Walker-Caprioglio HM, Casey WM, Parks LW. 1990. Saccharomyces cerevisiae membrane sterol modifications in response to growth in the presence of ethanol. Appl Environ Microbiol 56:2853–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You KM, Rosenfield CL, Knipple DC. 2003. Ethanol tolerance in the yeast Saccharomyces cerevisiae is dependent on cellular oleic acid content. Appl Environ Microbiol 69:1499–1503. doi: 10.1128/AEM.69.3.1499-1503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dague E, Gilbert Y, Verbelen C, Andre G, Alsteens D, Dufrene YF. 2007. Towards a nanoscale view of fungal surfaces. Yeast 24:229–237. doi: 10.1002/yea.1445. [DOI] [PubMed] [Google Scholar]
- 22.Dufrene YF. 2010. Atomic force microscopy of fungal cell walls: an update. Yeast 27:465–471. doi: 10.1002/yea.1773. [DOI] [PubMed] [Google Scholar]
- 23.Canetta E, Adya AK, Walker GM. 2006. Atomic force microscopic study of the effects of ethanol on yeast cell surface morphology. FEMS Microbiol Lett 255:308–315. doi: 10.1111/j.1574-6968.2005.00089.x. [DOI] [PubMed] [Google Scholar]
- 24.Dague E, Bitar R, Ranchon H, Durand F, Yken HM, Francois JM. 2010. An atomic force microscopy analysis of yeast mutants defective in cell wall architecture. Yeast 27:673–684. doi: 10.1002/yea.1801. [DOI] [PubMed] [Google Scholar]
- 25.Canetta E, Walker GM, Adya AK. 2006. Correlating yeast cell stress physiology to changes in the cell surface morphology: atomic force microscopic studies. Sci World J 6:777–780. doi: 10.1100/tsw.2006.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Formosa C, Pillet F, Schiavone M, Duval RE, Ressier L, Dague E. 2015. Generation of living cell arrays for atomic force microscopy studies. Nat Protoc 10:199–204. doi: 10.1038/nprot.2015.004. [DOI] [PubMed] [Google Scholar]
- 27.Dague E, Jauvert E, Laplatine L, Viallet B, Thibault C, Ressier L. 2011. Assembly of live micro-organisms on microstructured PDMS stamps by convective/capillary deposition for AFM bio-experiments. Nanotechnology 22:395102. doi: 10.1088/0957-4484/22/39/395102. [DOI] [PubMed] [Google Scholar]
- 28.Reference deleted.
- 29.Ding J, Huang X, Zhang L, Zhao N, Yang D, Zhang K. 2009. Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol 85:253–263. doi: 10.1007/s00253-009-2223-1. [DOI] [PubMed] [Google Scholar]
- 30.Ma M, Liu LZ. 2010. Quantitative transcription dynamic analysis reveals candidate genes and key regulators for ethanol tolerance in Saccharomyces cerevisiae. BMC Microbiol 10:169. doi: 10.1186/1471-2180-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev 12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray KC, Palacios DS, Dailey I, Endo MM, Uno BE, Wilcock BC, Burke MD. 2012. Amphotericin primarily kills yeast by simply binding ergosterol. Proc Natl Acad Sci U S A 109:2234–2239. doi: 10.1073/pnas.1117280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wach A, Brachat A, Pohlmann R, Philippsen P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 34.Brunelli JP, Pall ML. 1993. A series of yeast/Escherichia coli lambda expression vectors designed for directional cloning of cDNAs and cre/lox-mediated plasmid excision. Yeast 9:1309–1318. doi: 10.1002/yea.320091204. [DOI] [PubMed] [Google Scholar]
- 35.Gietz RD, Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 36.Cot M, Loret MO, Francois J, Benbadis L. 2007. Physiological behaviour of Saccharomyces cerevisiae in aerated fed-batch fermentation for high level production of bioethanol. FEMS Yeast Res 7:22–32. doi: 10.1111/j.1567-1364.2006.00152.x. [DOI] [PubMed] [Google Scholar]
- 37.Petitjean M, Teste MA, Francois JM, Parrou JL. 2015. Yeast tolerance to various stresses relies on the trehalose-6P synthase (Tps1) protein, not on trehalose. J Biol Chem 290:16177–16190. doi: 10.1074/jbc.M115.653899. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Gietz RD, Schiestl RH. 2007. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:35–37. doi: 10.1038/nprot.2007.14. [DOI] [PubMed] [Google Scholar]
- 39.Jung US, Levin DE. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol Microbiol 34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- 40.Parrou JL, Enjalbert B, Francois J. 1999. STRE- and cAMP-independent transcriptional induction of Saccharomyces cerevisiae GSY2 encoding glycogen synthase during diauxic growth on glucose. Yeast 15:1471–1484. [DOI] [PubMed] [Google Scholar]
- 41.Guarente L, Ptashne M. 1981. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagorce A, Hauser NC, Labourdette D, Rodriguez C, Martin-Yken H, Arroyo J, Hoheisel JD, Francois J. 2003. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J Biol Chem 278:20345–20357. doi: 10.1074/jbc.M211604200. [DOI] [PubMed] [Google Scholar]
- 43.Rose M, Casadaban MJ, Botstein D. 1981. Yeast genes fused to beta-galactosidase in Escherichia coli can be expressed normally in yeast. Proc Natl Acad Sci U S A 78:2460–2464. doi: 10.1073/pnas.78.4.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 45.Elsztein C, de Lucena RM, de Morais MA Jr. 2011. The resistance of the yeast Saccharomyces cerevisiae to the biocide polyhexamethylene biguanide: involvement of cell wall integrity pathway and emerging role for YAP1. BMC Mol Biol 12:38. doi: 10.1186/1471-2199-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 47.Teste MA, Duquenne M, Francois JM, Parrou JL. 2009. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol 10:99. doi: 10.1186/1471-2199-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hutter JL, Bechhoefer J. 1993. Calibration of atomic force microscope tips. Rev Sci Instrum 64:1868–1873. doi: 10.1063/1.1143970. [DOI] [Google Scholar]
- 49.Roduit C, Saha B, Alonso-Sarduy L, Volterra A, Dietler G, Kasas S. 2012. OpenFovea: open-source AFM data processing software. Nat Methods 9:774–775. doi: 10.1038/nmeth.2112. [DOI] [PubMed] [Google Scholar]
- 50.Francois JM. 2006. A simple method for quantitative determination of polysaccharides in fungal cell walls. Nat Protoc 1:2995–3000. [DOI] [PubMed] [Google Scholar]
- 51.Schiavone M, Vax A, Formosa C, Martin-Yken H, Dague E, Francois JM. 2014. A combined chemical and enzymatic method to determine quantitatively the polysaccharide components in the cell wall of yeasts. FEMS Yeast Res 14:933–947. doi: 10.1111/1567-1364.12182. [DOI] [PubMed] [Google Scholar]
- 52.Dallies N, Francois J, Paquet V. 1998. A new method for quantitative determination of polysaccharides in the yeast cell wall. Application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast 14:1297–1306. [DOI] [PubMed] [Google Scholar]
- 53.Chazotte B. 2011. Labeling the plasma membrane with TMA-DPH. Cold Spring Harb Protoc 2011:pdb.prot5622. doi: 10.1101/pdb.prot5622. [DOI] [PubMed] [Google Scholar]
- 54.Cheng JS, Qiao B, Yuan YJ. 2008. Comparative proteome analysis of robust Saccharomyces cerevisiae insights into industrial continuous and batch fermentation. Appl Microbiol Biotechnol 81:327–338. doi: 10.1007/s00253-008-1733-6. [DOI] [PubMed] [Google Scholar]
- 55.van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, Gancedo C, Giuseppin ML, Heijnen JJ, Hoare M, Lange HC, Madden EA, Niederberger P, Nielsen J, Parrou JL, Petit T, Porro D, Reuss M, van Riel N, Rizzi M, Steensma HY, Verrips CT, Vindelov J, Pronk JT. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol 26:706–714. doi: 10.1016/S0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
- 56.Formosa C, Schiavone M, Martin-Yken H, Francois JM, Duval RE, Dague E. 2013. Nanoscale effects of caspofungin against two yeast species, Saccharomyces cerevisiae and Candida albicans. Antimicrob Agents Chemother 57:3498–3506. doi: 10.1128/AAC.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osumi M. 1998. The ultrastructure of yeast: cell wall structure and formation. Micron 29:207–233. doi: 10.1016/S0968-4328(97)00072-3. [DOI] [PubMed] [Google Scholar]
- 58.Zlotnik H, Fernandez MP, Bowers B, Cabib E. 1984. Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J Bacteriol 159:1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Backhaus K, Heilmann CJ, Sorgo AG, Purschke G, de Koster CG, Klis FM, Heinisch JJ. 2010. A systematic study of the cell wall composition of Kluyveromyces lactis. Yeast 27:647–660. doi: 10.1002/yea.1781. [DOI] [PubMed] [Google Scholar]
- 60.Klis FM, Mol P, Hellingwerf K, Brul S. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol Rev 26:239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x. [DOI] [PubMed] [Google Scholar]
- 61.Dupres V, Dufrene YF, Heinisch JJ. 2010. Measuring cell wall thickness in living yeast cells using single molecular rulers. ACS Nano 4:5498–5504. doi: 10.1021/nn101598v. [DOI] [PubMed] [Google Scholar]
- 62.Dinh TN, Nagahisa K, Hirasawa T, Furusawa C, Shimizu H. 2008. Adaptation of Saccharomyces cerevisiae cells to high ethanol concentration and changes in fatty acid composition of membrane and cell size. PLoS One 3:e2623. doi: 10.1371/journal.pone.0002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lloyd D, Morrell S, Carlsen HN, Degn H, James PE, Rowlands CC. 1993. Effects of growth with ethanol on fermentation and membrane fluidity of Saccharomyces cerevisiae. Yeast 9:825–833. doi: 10.1002/yea.320090803. [DOI] [PubMed] [Google Scholar]
- 64.Rowe ES. 1992. Effects of ethanol on membrane lipids, p 239–268. In Watson RR. (ed), Alcohol and neurobiology receptors, membranes and channels. CRC Press, Boca Raton, FL. [Google Scholar]
- 65.Vanegas JM, Contreras MF, Faller R, Longo ML. 2012. Role of unsaturated lipid and ergosterol in ethanol tolerance of model yeast biomembranes. Biophys J 102:507–516. doi: 10.1016/j.bpj.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamaguchi M, Namiki Y, Okada H, Mori Y, Furukawa H, Wang J, Ohkusu M, Kawamoto S. 2011. Structome of Saccharomyces cerevisiae determined by freeze-substitution and serial ultrathin-sectioning electron microscopy. J Electron Microsc (Tokyo) 60:321–335. doi: 10.1093/jmicro/dfr052. [DOI] [PubMed] [Google Scholar]
- 67.Chen S, Wang J, Muthusamy BP, Liu K, Zare S, Andersen RJ, Graham TR. 2006. Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic 7:1503–1517. doi: 10.1111/j.1600-0854.2006.00485.x. [DOI] [PubMed] [Google Scholar]
- 68.Hao B, Cheng S, Clancy CJ, Nguyen MH. 2013. Caspofungin kills Candida albicans by causing both cellular apoptosis and necrosis. Antimicrob Agents Chemother 57:326–332. doi: 10.1128/AAC.01366-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chi Z, Arneborg N. 1999. Relationship between lipid composition, frequency of ethanol-induced respiratory deficient mutants, and ethanol tolerance in Saccharomyces cerevisiae. J Appl Microbiol 86:1047–1052. doi: 10.1046/j.1365-2672.1999.00793.x. [DOI] [PubMed] [Google Scholar]
- 70.Henderson CM, Lozada-Contreras M, Jiranek V, Longo ML, Block DE. 2013. Ethanol production and maximum cell growth are highly correlated with membrane lipid composition during fermentation as determined by lipidomic analysis of 22 Saccharomyces cerevisiae strains. Appl Environ Microbiol 79:91–104. doi: 10.1128/AEM.02670-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Francois JM, Formosa C, Schiavone M, Pillet F, Martin-Yken H, Dague E. 2013. Use of atomic force microscopy (AFM) to explore cell wall properties and response to stress in the yeast Saccharomyces cerevisiae. Curr Genet 59:187–196. doi: 10.1007/s00294-013-0411-0. [DOI] [PubMed] [Google Scholar]
- 72.Levin DE. 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189:1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jung US, Sobering AK, Romeo MJ, Levin DE. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol Microbiol 46:781–789. doi: 10.1046/j.1365-2958.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- 74.Martin-Yken H, Dagkessamanskaia A, Basmaji F, Lagorce A, Francois J. 2003. The interaction of Slt2 MAP kinase with Knr4 is necessary for signalling through the cell wall integrity pathway in Saccharomyces cerevisiae. Mol Microbiol 49:23–35. doi: 10.1046/j.1365-2958.2003.03541.x. [DOI] [PubMed] [Google Scholar]
- 75.Parrou JL, Teste MA, François J. 1997. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology 143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- 76.Yoshimoto H, Saltsman K, Gasch AP, Li HX, Ogawa N, Botstein D, Brown PO, Cyert MS. 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem 277:31079–31088. doi: 10.1074/jbc.M202718200. [DOI] [PubMed] [Google Scholar]
- 77.Garcia R, Bermejo C, Grau C, Perez R, Rodriguez-Pena JM, Francois J, Nombela C, Arroyo J. 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J Biol Chem 279:15183–15195. doi: 10.1074/jbc.M312954200. [DOI] [PubMed] [Google Scholar]
- 78.Lewis JA, Elkon IM, McGee MA, Higbee AJ, Gasch AP. 2010. Exploiting natural variation in Saccharomyces cerevisiae to identify genes for increased ethanol resistance. Genetics 186:1197–1205. doi: 10.1534/genetics.110.121871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eng WK, Faucette L, McLaughlin MM, Cafferkey R, Koltin Y, Morris RA, Young PR, Johnson RK, Livi GP. 1994. The yeast FKS1 gene encodes a novel membrane protein, mutations in which confer FK506 and cyclosporin A hypersensitivity and calcineurin-dependent growth. Gene 151:61–71. doi: 10.1016/0378-1119(94)90633-5. [DOI] [PubMed] [Google Scholar]
- 80.Mouyna I, Fontaine T, Vai M, Monod M, Fonzi WA, Diaquin M, Popolo L, Hartland RP, Latge JP. 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J Biol Chem 275:14882–14889. doi: 10.1074/jbc.275.20.14882. [DOI] [PubMed] [Google Scholar]
- 81.Jungmann J, Rayner JC, Munro S. 1999. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J Biol Chem 274:6579–6585. doi: 10.1074/jbc.274.10.6579. [DOI] [PubMed] [Google Scholar]
- 82.Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 83.Estruch F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev 24:469–486. doi: 10.1111/j.1574-6976.2000.tb00551.x. [DOI] [PubMed] [Google Scholar]
- 84.Bolard J. 1986. How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim Biophys Acta 864:257–304. doi: 10.1016/0304-4157(86)90002-X. [DOI] [PubMed] [Google Scholar]
- 85.Brajtburg J, Elberg S, Bolard J, Kobayashi GS, Levy RA, Ostlund RE Jr, Schlessinger D, Medoff G. 1984. Interaction of plasma proteins and lipoproteins with amphotericin B. J Infect Dis 149:986–997. doi: 10.1093/infdis/149.6.986. [DOI] [PubMed] [Google Scholar]
- 86.Li H, Ma ML, Luo S, Zhang RM, Han P, Hu W. 2012. Metabolic responses to ethanol in Saccharomyces cerevisiae using a gas chromatography tandem mass spectrometry-based metabolomics approach. Int J Biochem Cell Biol 44:1087–1096. doi: 10.1016/j.biocel.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 87.Dufrene YF, Pelling AE. 2013. Force nanoscopy of cell mechanics and cell adhesion. Nanoscale 5:4094–4104. doi: 10.1039/c3nr00340j. [DOI] [PubMed] [Google Scholar]
- 88.Kock C, Dufrene YF, Heinisch JJ. 2015. Up against the wall: is yeast cell wall integrity ensured by mechanosensing in plasma membrane microdomains? Appl Environ Microbiol 81:806–811. doi: 10.1128/AEM.03273-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mrsa V, Klebl F, Tanner W. 1993. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-beta-1,3-glucanase. J Bacteriol 175:2102–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kubota S, Takeo I, Kume K, Kanai M, Shitamukai A, Mizunuma M, Miyakawa T, Shimoi H, Iefuji H, Hirata D. 2004. Effect of ethanol on cell growth of budding yeast: genes that are important for cell growth in the presence of ethanol. Biosci Biotechnol Biochem 68:968–972. doi: 10.1271/bbb.68.968. [DOI] [PubMed] [Google Scholar]
- 91.Sahin A, Daignan-Fornier B, Sagot I. 2008. Polarized growth in the absence of F-actin in Saccharomyces cerevisiae exiting quiescence. PLoS One 3:e2556. doi: 10.1371/journal.pone.0002556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Caro LH, Tettelin H, Vossen JH, Ram AF, van den EH, Klis FM. 1997. In silico identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477–1489. doi:. [DOI] [PubMed] [Google Scholar]
- 93.Larriba G, Basco RD, Andaluz E, Luna-Arias JP. 1993. Yeast exoglucanases. Where redundancy implies necessity. Arch Med Res 24:293–299. [PubMed] [Google Scholar]
- 94.Roemer T, Bussey H. 1995. Yeast Kre1p is a cell surface O-glycoprotein. Mol Gen Genet 249:209–216. doi: 10.1007/BF00290368. [DOI] [PubMed] [Google Scholar]
- 95.Karreman RJ, Dague E, Gaboriaud F, Quiles F, Duval JF, Lindsey GG. 2007. The stress response protein Hsp12p increases the flexibility of the yeast Saccharomyces cerevisiae cell wall. Biochim Biophys Acta 1774:131–137. doi: 10.1016/j.bbapap.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 96.Rodriguez-Pena JM, Perez-Diaz RM, Alvarez S, Bermejo C, Garcia R, Santiago C, Nombela C, Arroyo J. 2005. The ‘yeast cell wall chip’—a tool to analyse the regulation of cell wall biogenesis in Saccharomyces cerevisiae. Microbiology 151:2241–2249. doi: 10.1099/mic.0.27989-0. [DOI] [PubMed] [Google Scholar]
- 97.Straede A, Corran A, Bundy J, Heinisch JJ. 2007. The effect of tea tree oil and antifungal agents on a reporter for yeast cell integrity signalling. Yeast 24:321–334. doi: 10.1002/yea.1478. [DOI] [PubMed] [Google Scholar]
- 98.Nishida N, Jing D, Kuroda K, Ueda M. 2014. Activation of signaling pathways related to cell wall integrity and multidrug resistance by organic solvent in Saccharomyces cerevisiae. Curr Genet 60:149–162. doi: 10.1007/s00294-013-0419-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.