ABSTRACT
Although Escherichia coli is generally considered to be predominantly a commensal of the gastrointestinal tract, a number of recent studies suggest that it is also capable of long-term survival and growth in environments outside the host. As the extraintestinal physical and chemical conditions are often different from those within the host, it is possible that distinct genetic adaptations may be required to enable this transition. Several studies have shown a trade-off between growth and stress resistance in nutrient-poor environments, with lesions in the rpoS locus, which encodes the stress sigma factor RpoS (σS). In this study, we investigated a unique collection of long-term soil-persistent E. coli isolates to determine whether the RpoS-controlled general stress response is altered during adaptation to a nutrient-poor extraintestinal environment. The sequence of the rpoS locus was found to be highly conserved in these isolates, and no nonsense or frameshift mutations were detected. Known RpoS-dependent phenotypes, including glycogen synthesis and γ-aminobutyrate production, were found to be conserved in all strains. All strains expressed the full-length RpoS protein, which was fully functional using the RpoS-dependent promoter reporter fusion PgadX::gfp. RpoS was shown to be essential for long-term soil survival of E. coli, since mutants lacking rpoS lost viability rapidly in soil survival assays. Thus, despite some phenotypic heterogeneity, the soil-persistent strains all retained a fully functional RpoS-regulated general stress response, which we interpret to indicate that the stresses encountered in soil provide a strong selective pressure for maintaining stress resistance, despite limited nutrient availability.
IMPORTANCE Escherichia coli has been, and continues to be, used as an important indicator species reflecting potential fecal contamination events in the environment. However, recent studies have questioned the validity of this, since E. coli has been found to be capable of long-term colonization of soils. This study investigated whether long-term soil-persistent E. coli strains have evolved altered stress resistance characteristics. In particular, the study investigated whether the main regulator of genes involved in stress protection, the sigma factor RpoS, has been altered in the soil-persistent strains. The results show that RpoS stress protection is fully conserved in soil-persistent strains of E. coli. They also show that loss of the rpoS gene dramatically reduces the ability of this organism to survive in a soil environment. Overall, the results indicate that soil represents a stressful environment for E. coli, and their survival in it requires that they deploy a full stress protection response.
INTRODUCTION
Escherichia coli is a Gram-negative, facultative anaerobe, belonging to the Enterobacteriaceae family, which inhabits the intestinal tracts of humans, warm-blooded animals, and reptiles (1, 2). It can be transferred through water and sediments via feces and is almost universally used as an indicator of fecal contamination in drinking and recreational water. The use of E. coli as a fecal indicator is based, at least in part, on the assumption that it exists transiently outside the host gastrointestinal tract (3) and does not survive for a long time in the external environment. Though several authors have isolated E. coli from various natural environments, such as freshwater (4, 5), beach water (6, 7), beach sand (7), tropical and subtropical soils (8–12), coastal temperate forest soils (13), riverine temperate soil (14), and sediments (15), it is difficult to determine unequivocally whether these isolates originated from recent contamination or whether they represent long-term residents in those environments. In 2010, Brennan et al. (16) reported the recovery of E. coli populations from intact soil monoliths maintained in lysimeter units (previously described by Ryan and Fanning [17]), which have been protected from fecal contamination since 1998. These long-term soil-persistent E. coli isolates are the subject of the present study.
Soil-persistent E. coli strains from the lysimeters are genotypically diverse and possess unique growth and metabolic characteristics, suggesting adaptation to conditions present in soils (18, 19). These strains are assumed to have developed mechanisms that could help them survive in soil. For example, it was shown that a soil-persistent E. coli strain was nutritionally versatile and metabolized more substrates at 15°C than E. coli K-12 (19). Furthermore, E. coli strains have been shown to survive and grow in both amended (8, 20) and unamended (14) soil. The capacity of these E. coli strains to survive for long periods of time and grow in the external environment raises questions about the validity of its continued use as indicator of water quality (16).
The ability to survive environmental stresses has been shown to be controlled by the general stress response regulator, RpoS, in E. coli and other related bacteria. RpoS (σs) is an alternative sigma factor that is involved in E. coli resistance to stresses typically encountered in the external environment, such as cold stress, osmotic stress, oxidative stress, and desiccation (21–23). When environmental stresses are encountered, cellular RpoS levels increase dramatically; the resulting RNA polymerase-RpoS holoenzyme complex produces the appropriate transcriptional response (24). A number of studies have shown that, in low-nutrient conditions, particularly when growth rates are very slow, mutations can accumulate within the rpoS open reading frame (ORF), resulting in the partial or complete loss of RpoS function and reduced stress tolerance (25, 26). Mutations in rpoS have been reported in strains obtained from laboratory growth conditions as well as among natural isolates of E. coli (7, 26–34), Salmonella (35–37), Cronobacter (38), and Citrobacter (39). These mutations are thought to provide a selective advantage to microorganisms undergoing nutrient starvation due to a trade-off between stress resistance and growth (40). Indeed, mutations that confer a growth and survival advantage in stationary-phase cultures of E. coli (so-called growth advantage in stationary phase, or GASP, mutants) are frequently found to map to the rpoS locus (41). Thus, the long-term soil-persistent E. coli isolates present a unique opportunity to study the evolution of the rpoS locus in a natural nutrient-limited environment.
Soil can be considered a highly competitive environment where nutrient sources are significantly less abundant than in the host gastrointestinal tract (42). We hypothesized that low-nutrient microniches within the soil environment may select for rpoS mutations during long-term soil adaptation, perhaps providing a growth rate advantage. These rpoS lesions could also confer a competitive advantage through increased nutritional competence, a phenotype associated with the loss of rpoS (26, 40). Knowledge of how E. coli responds to chemical and physical stresses has been derived almost exclusively from studying laboratory strains; thus, the available collection of soil-persistent isolates of E. coli (16; F. Abram, unpublished data) from a closed system represents a rare opportunity to understand the role that stress responses play in the survival of E. coli in an environment outside the host, where environmental conditions, nutrient availability, and competing microorganisms are different from those present in the gastrointestinal tract.
In the present study, we have characterized the RpoS-dependent stress response in phylogenetically distinct soil-persistent E. coli strains and compared this to responses in commensal and laboratory E. coli strains. The rpoS locus was sequenced, and Western blotting and green fluorescent protein (GFP) reporter fusion (using the RpoS-dependent gadX promoter) assays were used to determine the presence and activity of the RpoS protein in each of the isolates. Soil survival assays were also performed to investigate the role of an intact rpoS locus in soil survival. We show that a functional RpoS is retained in long-term soil-persistent isolates of E. coli and demonstrate that RpoS is essential for long-term survival in the soil.
MATERIALS AND METHODS
Strains used and growth conditions.
Five long-term (>9 years) soil-persistent Escherichia coli strains, belonging to five distinct phylogenetic groups, as described by Clermont et al. (43), were studied (here named COB583 to COB587) (Table 1). These strains were leached from lysimeter units (Luvic Stagnosol; C-to-N ratio, 10.1) (44), to which no fecal material had been applied since at least 1998 (16). The lysimeter unit was enclosed within a mesh netting to prevent contamination from birds and small mammals. The soil isolates were considered long-term soil-persistent isolates, since the experimental soil columns from which they were derived had not been exposed to fecal material for at least 9 years prior to leachate collection. E. coli strains were periodically isolated from these soil columns over a period of 4 years, indicating that the strains were resident in the soil. Two commensal strains (SE11 and SE15) and a well-studied laboratory strain (BW25113) with its corresponding rpoS deletion mutant (strain BW25113ΔrpoS) were used for comparative purposes. The strains are described in Table 1. An rpoS deletion mutant strain of soil-persistent E. coli COB583 was constructed by the one-step inactivation method using a λ Red recombinase-assisted approach, which replaces target gene sequence with a kanamycin resistance (Kmr) cassette described by Datsenko and Wanner (45), with minor modifications. Primers COB740R and COB743F were used to amplify the kanamycin cassette flanked by homologous sequences covering 915 bp upstream and 582 bp downstream of rpoS to generate PCR linear fragments, using the kanamycin cassette from a Keio collection mutant as the template. The λ Red recombinase plasmid used was pKOBEGA (46), and 500 ng of linear DNA was electroporated into the λ Red recombinase-expressing COB583 strain at 3.0 kV. The kanamycin resistance cassette was removed from the ΔrpoS mutant strains by FLP-FRT recombination, and removal of the cassette was confirmed by plating on Luria-Bertani (LB) agar with 50 μg · ml−1 kanamycin (LBKan). E. coli K-12 strain carrying a gadX-GFP promoter fusion on pUA66 (PgadX) was obtained from David Clarke (University College Cork, Ireland). gadX is a transcriptional activator of the glutamate-dependent acid resistance system in E. coli and is RpoS dependent (47). PgadX was obtained from overnight (∼16 h) culture in LBKan at 37°C, and the resulting plasmids were transformed into all nine strains used in the present study.
TABLE 1.
Description of strains used in study
| Strain | Phylogenetic group | Habitat | Source or reference |
|---|---|---|---|
| COB583 (isolate 3; Lys9) | C | Soil | Brennan et al., 2010 (16) |
| COB584 (Lys24) | B1 | Soil | F. Abram (unpublished) |
| COB585 (Lys25) | E | Soil | F. Abram (unpublished) |
| COB586 (Lys28) | B1 | Soil | F. Abram (unpublished) |
| COB587 (Lys36) | B1 | Soil | F. Abram (unpublished) |
| E. coli SE11 | B1 | Commensal | JCM16574 (Riken BRC, Japan) |
| E. coli SE15 | B2 | Commensal | JCM16575 (Riken BRC, Japan) |
| E. coli BW25113 | A | Lab strain | Baba et al., 2006 (80) |
| E. coli BW25113 ΔrpoS | Baba et al., 2006 (80) | ||
| COB583 ΔrpoS | C | Soil | This study |
Motility at 15°C and 37°C.
Since motility and biofilm formation are traits that might be subject to selection during niche adaptation (48, 49), we measured these traits at 15°C and 37°C for each strain. In order to evaluate cell motility, each strain was grown in LB overnight (∼16 h) with agitation at 37°C. Overnight cultures were then spot-inoculated onto LB containing 0.25% (wt/vol) agar plates, and radial motility was measured after 16 h of incubation at 37°C and 40 h at 15°C. The experiment was conducted with three independent biological replicates.
Biofilm assay.
This was done according to the method described by O'Toole (50), with slight modifications. Each strain was grown overnight in LB with agitation at 37°C, and then 1 ml of the overnight culture was centrifuged at 8,000 × g for 6 min at room temperature (23°C) to recover the cells. The cell pellets were washed in 1 ml sterile phosphate-buffered saline (PBS) (pH 7.3) (Oxoid, United Kingdom) and resuspended in 1 ml sterile PBS. The culture was diluted 1:1,000 into the respective media—LB broth and succinate minimal medium (SMM)—after which 200 μl was added into a round-bottomed well in 96-well microtiter plates. Each strain was inoculated in eight replicate wells, and the assay was repeated at least twice for each condition. Strains were assigned at random to the wells of a 96-well microtiter plate. Plates were incubated statically at 37°C for 84 h and 15°C for 138 h. After incubation, the optical density at 600 nm (OD600) was determined, and the medium was removed from each well carefully with a pipette so as not to disrupt the biofilm. Each well was washed three times with 200 μl PBS by inverting the plates in order to remove all nonadhering bacterial cells, after which the plates were dried at room temperature (23°C) for 45 min. Biofilm was stained by adding 150 μl of 1% (wt/vol) crystal violet solution to each well, and the plates were incubated for 30 min at 37°C. After staining, excess crystal violet was removed from the wells, and the wells were rinsed four times with 200 μl PBS. Destaining was done by adding 160 μl of 95% (vol/vol) ethanol to each well for 30 min, after which the absorbance was determined using a Sunrise microplate absorbance reader (Tecan, Austria).
Survival under starvation at 15°C.
Strains were inoculated in 10 ml LB and incubated overnight at 37°C with agitation before washing at 9,000 × g for 10 min at room temperature (23°C) in 10 ml PBS twice. The washed cells were resuspended in 15 ml PBS and used as inoculum. One milliliter of the inoculum was added to 15 ml PBS in sterile 50-ml tubes in triplicates and incubated at 15°C with shaking. This temperature was chosen to approximate the mean Irish soil temperature. Aliquot (200-μl) samples were taken at days 0, 7, 14, 21, 28, and 35 for enumerating culturable cells. Samples were serially diluted in PBS, and 10 μl was spot-plated in triplicates onto LB agar and incubated overnight at 37°C.
Soil survival assay.
This was done according to the method described by Ma et al. (51), with some modifications. Two types of silty loam soils were used for this analysis: soil A (sand, 47%; silt, 52%; clay, <1%) (pH 7.2) (with total organic carbon, 16.53% of dry solids; total nitrogen, 1.56% of dry solids; C-to-N ratio, 10.6; and organic matter, 31.3%) was collected from Ballyvaughan (53°07′15.6″N 9°09′24.8″W) in the west of Ireland, and soil B (sand, 43%; silt, 53.9%; clay, 3.1%) (pH 5.19) (with total organic carbon, 4.7%; total nitrogen, 0.35%; C-to-N ratio, 13.43; and organic matter, 9.1%) was collected from Kilfergus (53°07′15.6″N 9°09′24.8″W) in the midwest of Ireland. The soils were sieved with a 2-mm sieve and kept in a sealed bag with airspace at a constant temperature (15°C) until used. One colony each of the BW25113 and BW25113ΔrpoS strains was inoculated into 10 ml LB and incubated overnight at 37°C with agitation. The overnight cultures were harvested by centrifugation at 9,000 × g for 10 min at room temperature (23°C), washed twice with sterile PBS, and resuspended in PBS to give an OD600 equivalent to 2 × 108 CFU/ml, which served as the inoculum. Then, 50 μl of the inoculum was added into 1 g of soil A in a series of 15-ml sterile tubes inverted 10 times by hand, slightly capped to allow air exchange, and incubated at 15°C. As a control, 50 μl of sterile PBS was added to 1 g of soil A. The experiment was set up in triplicate. Inoculated soils were destructively sampled (i.e., PBS was added and could not be reused at another time point) on days 0, 1, 2, 7, 14, 21, 28, and 35 to determine survival of the wild-type and rpoS mutant strains. For cell recovery, 2 ml of PBS was added to each tube and mixed by inverting three times followed by vortexing for 2 × 20 s. The resulting soil slurry was allowed to settle for 2 min, and 20 μl was collected from the supernatant and serially diluted. Two minutes was adopted as the standard time for this protocol, because there was no significant difference in the recoverable cell numbers when soil slurry was allowed to settle for a longer period (10 min). A total of 10 μl of all dilutions was plated in triplicate on MacConkey agar and incubated overnight at 37°C. The soil used had no detectable background levels of coliforms or E. coli. Preliminary experiments showed an average of 91% to 102% of the added E. coli was recovered at 10 min after inoculation into soil A, while recovery was 91% to 97% in soil B. A subsequent soil assay was arranged with all nine test strains and sampled on days 0, 7, 14, 28, 42, 55, 84, 122, 154, and 230. The wild-type COB583 strain and its corresponding constructed rpoS deletion mutant strain (COB583ΔrpoS) were subjected to the soil survival assay with the silty loam soils A and B to investigate the effect of RpoS on soil survival in a soil-persistent strain.
PCR amplification and sequencing.
Whole-colony PCR targeting rpoS and its flanking genes was carried out on the soil-persistent strains. Single colonies were picked and resuspended in 500 μl of sterile nuclease-free water (Ambion, USA). A 1-μl aliquot was then transferred to the PCR reagent mix. The primers used for amplification are listed in Table 2. PCR was performed with high-fidelity Velocity DNA polymerase (Bioline, Inc., USA). PCR mixture (50 μl) consisted of 10 μl of 5× hi-fi buffer (containing 10 mM Mg2+), 5 μl of 10 μM deoxynucleoside triphosphate (dNTP) mix, 1.5 μl of dimethyl sulfoxide (Bioline, Inc., USA), 29.5 μl of nuclease-free H2O, 2 U (1 μl) of Velocity DNA polymerase (Bioline, Inc., USA), 1 μl of template DNA, and 1 μl of each primer (25 μM). The PCR was performed with a Primus DNA cycler (MWG-Biotech, Inc.) with the following steps: initial denaturation step at 98°C for 2 min, 30 cycles of 30 s at 98°C, 30 s at 54°C, and 3 min at 72°C, and a terminal extension step at 72°C for 7 min. The amplified PCR products were examined by agarose electrophoresis on 1% gels, purified using the GenElute PCR cleanup kit (Sigma-Aldrich, USA), and visualized on a 1% agarose gel for quantification prior to sequencing. Samples were sequenced by Source Bioscience (Dublin, Ireland), while assembly and analysis were carried out using DNABaser version 4 (Heracle BioSoft, USA). Sequencing was performed on both strands of the PCR product.
TABLE 2.
List of primers used in this study
Phylogenetic analysis.
Nucleotide sequences of E. coli SE11, SE15, and BW25113 were retrieved from the National Center for Biotechnology Information (NCBI) database with GenBank accession numbers AP009240.1, AP009378.1 and CP009273.1, respectively. Nucleotide sequences of rpoS were translated, and the resulting amino acid sequences were analyzed. Sequences were aligned using Clustal Omega (52), and a phylogenetic tree was generated using the maximum likelihood method based on the Kimura 2-parameter model (53) with bootstrap analysis (1,000 iterations) using MEGA6 (54).
Acid survival at 37°C.
Cultures of bacteria were grown to the stationary phase at 37°C in LB medium with agitation. The pH of these cultures was lowered to a pH of 2.5 with 3 M HCl. Samples were taken at 20, 40, and 60 min and serially diluted in PBS. Aliquots of 10 μl of the serial dilutions of the samples were plated in triplicate onto LB agar and incubated overnight at 37°C. Colonies were counted to enumerate the culturable cells.
GABase assay.
Intracellular γ-aminobutyrate (GABA) and extracellular GABA (GABAi and GABAe, respectively) were measured as previously described (55). Briefly, strains were grown to the stationary phase at 37°C with agitation in LB medium. Prior to the GABA measurements, the pH of the cultures was lowered to 4.0 with 3 M HCl. Extractions were made after 1 h of acid treatment. Non-HCl-treated cultures were used as negative controls. GABase from Pseudomonas fluorescens (Sigma-Aldrich, Steinheim, Germany) was used in the enzymatic assay, and the OD340 was measured using the Sunrise microplate absorbance reader (Tecan, Austria).
Glycogen accumulation test.
Levels of glycogen accumulated in the strains were determined by iodine staining, as described previously (26) with some modifications. Strains were streaked onto LB agar and incubated overnight at 37°C and then left at 4°C for 48 h, after which they were flooded with 0.05 M iodine solution (Sigma-Aldrich, USA). The glycogen level is indicated by the intensity of brown coloration and is an indirect measure of the level of RpoS. Images were captured on an HP Scanjet 5400c at 600 dpi.
Western blot for RpoS.
As a direct measure of RpoS levels, we detected RpoS protein in the test strains by Western blotting. Stationary-phase cells were inoculated into 25 ml LB starting at an OD600 of 0.05 and incubated overnight (∼16 h) at 37°C and 36 h at 15°C with continuous shaking. Then, 1 ml of culture was taken and centrifuged at 12,000 × g for 10 min. Each cell pellet was resuspended in 100 μl of BugBuster cell lysis reagent (Novagen, USA) supplemented with 1% (vol/vol) DNase I (Thermo Scientific, USA), 1% (vol/vol) Halt protease inhibitor cocktail (Thermo Scientific, USA), and 1% (wt/vol) lysozyme (Sigma-Aldrich, USA). Each cell suspension was then incubated at room temperature (23°C) for 20 min with agitation. Cell lysates were centrifuged at 16,000 × g for 10 min at 4°C. Protein concentrations were determined using the Bio-Rad DC protein assay (Bio-Rad). An equal amount of each protein (25 μg) was resolved on 10% SDS-PAGE at 100 V for 1.5 h at 4°C. After electrophoresis, proteins were blotted onto a polyvinylidene difluoride (PVDF) membrane using a semidry system (Jencons, United Kingdom) at 3 V for 1 h. A blocking step with 5% skim milk in Tris-buffered saline with 0.05% Tween 20 (TBST) was performed, and the membrane was incubated with a 1,000-fold-diluted mouse monoclonal anti-RpoS antibody (Santa Cruz). Blots were washed in TBST three times for 10 min each and incubated in 3,000-fold diluted peroxidase conjugated anti-mouse IgG (Pierce). The ECL Prime Western blotting detection reagent (GE Healthcare) was used to detect the RpoS bands, as recommended by the manufacturer, with exposure to X-ray films.
RpoS-dependent GFP expression.
RpoS activity was determined by the ability of the strains to transcribe the gadX promoters in the reporter fusion (PgadX) indicated by green fluorescence. Overnight culture of all strains carrying PgadX was prepared in LBKan broth and incubated at 37°C and 15°C. Stationary-phase cells were inoculated into 25 ml LBKan at a starting OD600 of 0.05 and incubated with continuous shaking at 37°C and 15°C. To determine the induction of fluorescence, samples were taken at the stationary phase (17 h at 37°C and 36 h at 15°C), fixed with an ethanol- methanol (1:1) solution, and resuspended in PBS, and 2 μl was placed on a slide and imaged with Leica DMI3000 B microscope. Green fluorescent protein (GFP) was detected by Western blotting according to the method described above, but with rabbit polyclonal GFP antibody (Santa Cruz; SC-8334) diluted 5,000-fold in TBST and 20,000-fold-diluted peroxidase-conjugated anti-rabbit IgG (Santa Cruz).
Statistical analysis.
Analysis of variance (ANOVA) was performed to investigate the differences in motility, biofilm formation, GABA levels, acid survival, and soil survival at the specified time points using SPSS 21.0 for Windows (SPSS, Inc., Chicago, IL). All assumptions of the test were met. Statistical comparisons among the means were made using the Duncan multiple range test at a 5% probability level.
Accession number(s).
The accession numbers KU948321 to KU948325 were assigned by GenBank for rpoS nucleotide sequences COB583 to COB587, respectively.
RESULTS
Soil-persistent E. coli displayed heterogeneous phenotypes.
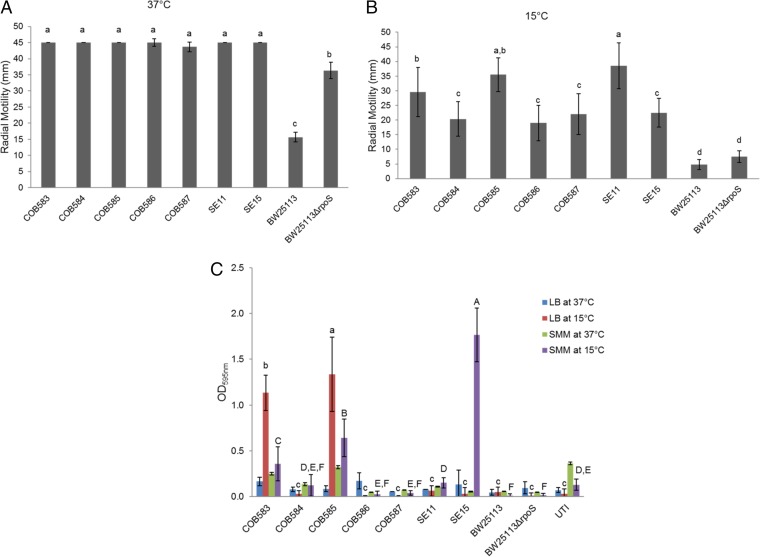
All strains were motile to a similar extent at 37°C (Fig. 1A), with the exception of BW25113, which showed poor motility at this temperature. In contrast, motility was found to be more variable at 15°C, with three of the wild isolates (COB583, COB585, and SE11) displaying significantly greater motility than the other strains (Fig. 1B). Very low levels of biofilm were detected for all strains at 37°C in both LB broth and a minimal medium with succinate as the sole carbon source. Two of the five soil isolates (COB583 and COB585) produced significant biofilm at 15°C in both media tested (Fig. 1C), and SE15 produced high levels of biofilm (OD595 of 1.77) in the minimal medium, while the others produced very little biofilm. Together, these data demonstrate the phenotypic diversity of E. coli soil residents, consistent with the fact that these isolates belong to different phylogenetic groups.
FIG 1.
Motility of E. coli strains on 0.25% (wt/vol) LB agar at 37°C for 16 h (A) and at 15°C for 40 h (B), and biofilm formation in LB broth and SMM (C). All soil-persistent and commensal strains were significantly (P < 0.0001) more motile than the laboratory strain BW25113 at 15°C and 37°C. The wild-type BW25113 strain was significantly (P < 0.0003) less motile than the BW25113ΔrpoS strain at 37°C but not at 15°C. Biofilm was produced in rich and minimal media, with more biofilm produced at 15°C than at 37°C. UTI, urinary tract infection-associated E. coli isolate. Error bars represent standard deviations from three independent replicates. Data with similar lowercase or uppercase letters are not significantly different (P > 0.05).
RpoS is required for survival in soil at 15°C.
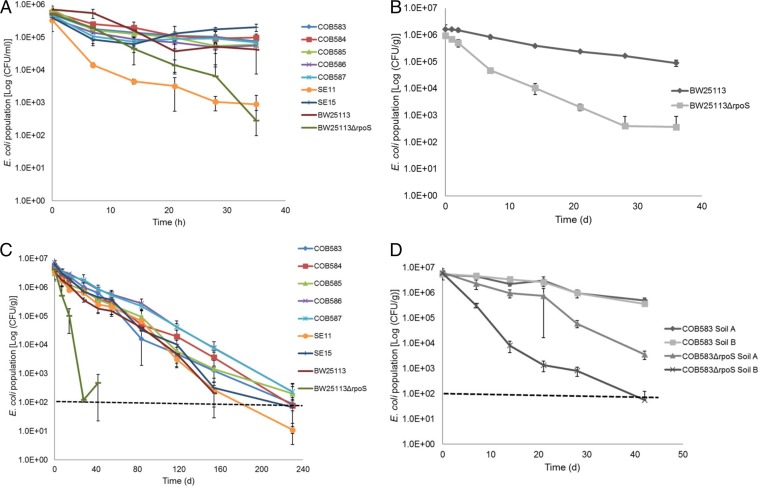
All the soil-persistent and commensal isolates of E. coli were found to survive well in a nutrient-poor environment at 15°C over 35 h, with the exception of SE11, which showed approximately 2-log reduction in culturable cell numbers (Fig. 2A). To investigate whether RpoS was required for survival in soil, the survivals of BW25113 and its rpoS mutant strain (BW25113ΔrpoS) were compared following the inoculation of a live soil sample with 106 CFU · g−1. The parent strain lost less than a 1-log cycle of viability, while the BW25113ΔrpoS strain was significantly (P = 0.0004) impaired in its soil survival, dropping to less than 103 CFU · g−1 after 35 days (Fig. 2B). Soil-persistent and commensal strains had similar survival patterns which were not significantly different (P > 0.05) after 230 days in live soil (Fig. 2C). The laboratory strain BW25113 also survived well under these conditions in soil, but the BW25113ΔrpoS strain was again severely impaired in its survival, with no culturable survivors detected after 42 days. In order to directly test the role that RpoS plays in the soil-persistent strains, an rpoS deletion was constructed in COB583, and the survival of this mutant (strain COB583ΔrpoS) was compared to the parental strain in silty loam soils A and B. In both cases, loss of rpoS was associated with a faster loss of viability in the soil, and a stronger effect was observed in soil B (Fig. 2D). The effect of the rpoS deletion on soil survival was similar in both the BW25113 and COB583 backgrounds for silty loam soil A (Fig. 2B compared to Fig. 2D).
FIG 2.
Survival of E. coli under various environmental conditions at 15°C. (A) All of the soil-persistent, commensal strains and E. coli BW25113 survived significantly (P < 0.0001) better in PBS than the BW25113ΔrpoS strain did, except SE11, after 35 h. (B) The E. coli BW25113 parental strain survived significantly better than the BW25113ΔrpoS strain in soil. Time is shown in days (d) on the x axis. (C) Long-term soil survival experiment showed that RpoS is required for survival in silty loam soil A, as there were no detectable BW25113ΔrpoS strains after 42 days in soil, whereas all other test strains survived long term. (D) The E. coli COB583 parental strain survived significantly better (P = 0.002) than the COB583ΔrpoS strain in silty loam soils A and B. Error bars represent standard deviations from three independent replicates. Dashed lines represent the detection limit of the soil survival assay.
The rpoS locus is conserved in soil-persistent E. coli.
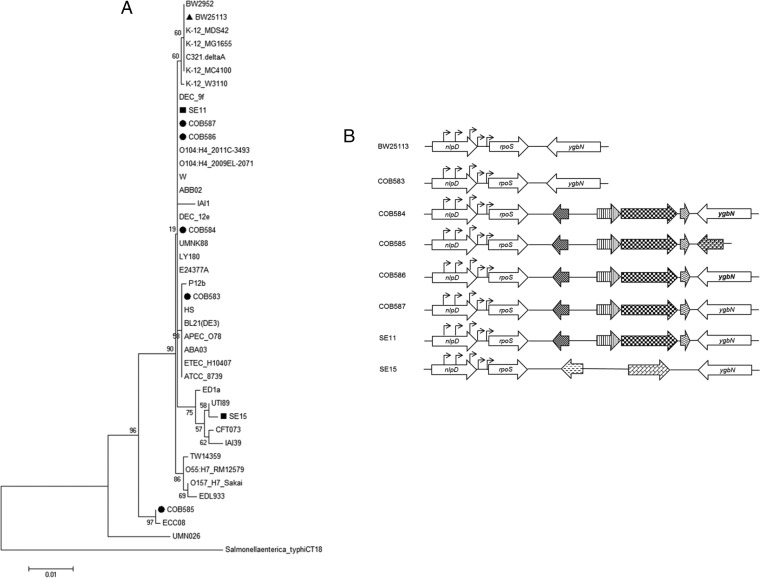
Although there were some sequence differences between the isolates, all of the soil-persistent strains were found to have an intact rpoS ORF (Table 3). Nucleotide substitution C→G at position 97 was present in all of the soil-persistent and commensal strains, and this resulted in a corresponding amino acid change from glutamine (Q) to glutamic acid (E) at codon 33. Although there were other nucleotide changes in the test strains, the change at codon 33 was the only nucleotide change that resulted in an amino acid change; the others were silent mutations. The sequence of rpoS in COB585 diverged significantly from the other strains, which was reflected in the phylogenetic tree generated when these sequences were compared to other sequenced E. coli strains (Fig. 3A). The region downstream from the rpoS ORF was conserved in three soil-persistent strains (COB584, COB586, and COB587) and in one of the commensal strains (SE11), but the locus had a different gene order in each of the other strains (Fig. 3B). The regions carrying the known rpoS promoter sequences, located in the upstream nlpD ORF and in the intergenic region, were compared and were found to be conserved in all soil-persistent strains (Fig. 3B). These results confirm that the rpoS gene and regulatory elements are conserved in the soil-persistent strains but that differences exist in the genetic structure of this chromosomal locus, highlighting the genetic diversity that exists within this collection of soil isolates.
TABLE 3.
Mutations within the rpoS gene of soil-persistent and commensal E. coli strains compared to MG1655
| Strain | Nucleotide change |
Corresponding amino acid change (codon position) | |
|---|---|---|---|
| Position | Change | ||
| COB583 | 97 | C→G | Gln (Q) to Glu (E) (33) |
| 942 | C→T | —a | |
| COB584 | 97 | C→G | Gln (Q) to Glu (E) (33) |
| COB585 | 42 | T→C | — |
| 81 | A→G | — | |
| 84 | G→A | — | |
| 93 | A→T | — | |
| 97 | C→G | Gln (Q) to Glu (E) (33) | |
| 123 | C→T | — | |
| 132 | A→G | — | |
| 144 | G→A | — | |
| 147 | A→G | — | |
| 819 | G→A | — | |
| 927 | C→T | — | |
| 972 | C→A | — | |
| 990 | C→A | — | |
| COB586 | 97 | C→G | Gln (Q) to Glu (E) (33) |
| COB587 | 97 | C→G | Gln (Q) to Glu (E) (33) |
| SE11 | 97 | C→G | Gln (Q) to Glu (E) (33) |
| SE15 | 97 | C→G | Gln (Q) to Glu (E) (33) |
| 163 | T→C | — | |
| 357 | T→C | — | |
| 462 | T→C | — | |
| 573 | T→C | — | |
| 699 | G→A | — | |
| 732 | C→T | — | |
| 990 | G→A | — | |
| BW25113 | No change | No change | — |
—, silent.
FIG 3.
(A) Phylogenetic tree of rpoS performed using the maximum likelihood method based on the Kimura 2-parameter model with bootstrap analysis (1,000 iterations) using MEGA6 showed that rpoS in soil-persistent strains is similar to previously known E. coli strains. (B) Gene outline of rpoS and its flanking genes in the soil-persistent and commensal strains shows four distinct patterns. PCR and sequencing of genes flanking rpoS revealed insertion of open reading frames (ORFs) between rpoS and inner membrane permease (ygbN). Pattern I (COB583), which is similar to the reference strain (BW25113), shows rpoS directly flanked by murein hydrolase activator (nlpD) and ygbN. Pattern II (COB584, COB586, COB587, and SE11) shows insertion of 4 ORFs between rpoS and ygbN. Pattern III (COB585) shows insertion of 5 ORFs after rpoS, with ygbN absent, while pattern IV (SE15) had insertion of 2 ORFs between rpoS and ygbN. The region between nlpD (which carried rpoS promoters) and rpoS was conserved in all the strains. Block arrows with similar patterns indicate the same ORF, while arrowheads represent rpoS promoter sites.
Phenotypes under the control of RpoS are retained in soil-persistent E. coli.
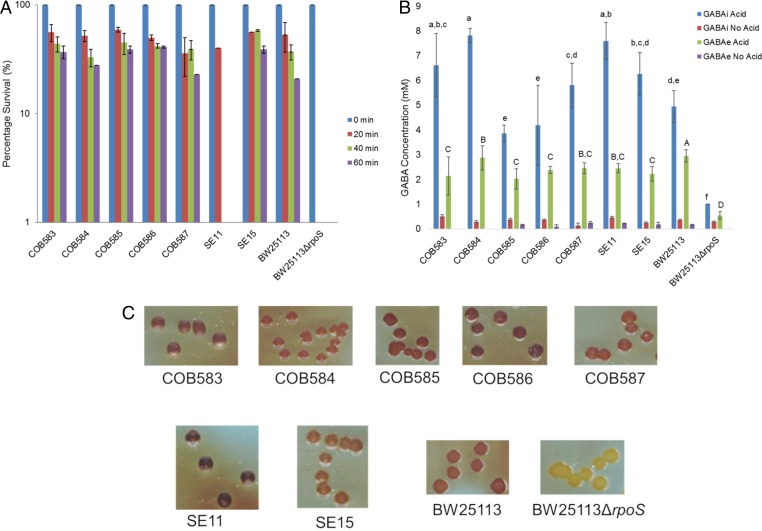
Acid tolerance and GABA levels following acidification were measured for each of the soil-persistent and commensal isolates. Both acid tolerance and GABA production were confirmed to be under RpoS control, since the BW25113ΔrpoS mutant strain was exquisitely acid sensitive (Fig. 4A) and produced only very low levels of GABA in response to acidification compared to the parental control strain BW25113 (Fig. 4B). With the exception of SE11, all of the other strains survived a pH of 2.5 for 1 h (Fig. 4A). Furthermore, they all produced significant quantities of GABA both intracellularly and extracellularly in response to acidification of the culture media (pH 4.0), although there were some strain differences in the amounts produced (Fig. 4B). The accumulation of intracellular glycogen in E. coli is another trait known to be under RpoS control (56). An iodine-based staining assay was used to determine if the soil-persistent and reference strains could accumulate glycogen. The BW25113ΔrpoS mutant strain was found to stain negative (yellow-white) for glycogen, while all other strains gave a glycogen-positive stain (red-brown) (Fig. 4C). Together, these results indicated that traits under the control of RpoS were retained during long-term soil adaptation, suggesting the presence of a general stress response in these isolates.
FIG 4.
RpoS-dependent phenotypes show soil-persistent strains have functional RpoS. All E. coli strains survived significantly better than the BW25113ΔrpoS strain at pH 2.5 (A), produced significantly higher (P < 0.0001) amounts of gamma-aminobutyric acid (GABA) in response to acid stress (pH 4) compared to the BW25113ΔrpoS strain (B), and turned brown to dark brown upon iodine staining, showing the accumulation of glycogen compared to the BW25113ΔrpoS strain (C). Error bars represent standard deviations from three independent replicates. Data with similar lowercase or uppercase letters are not significantly different (P > 0.05).
RpoS is present and functional in long-term soil-persistent E. coli.
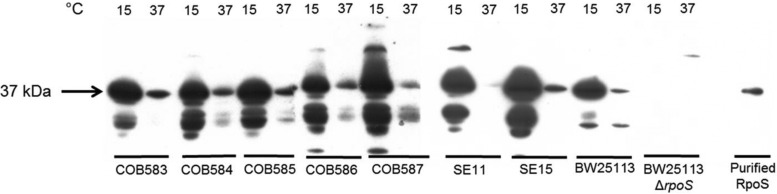
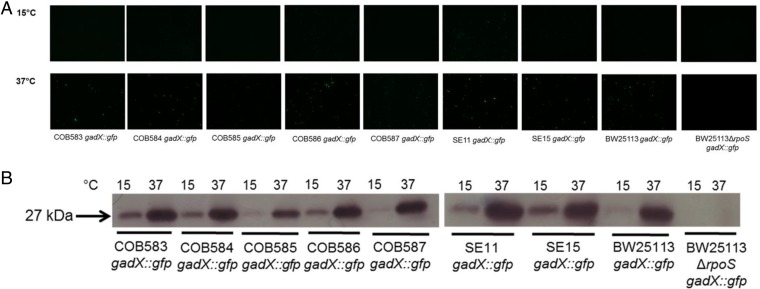
While the above data suggested the presence of an active RpoS-mediated stress response in the soil-persistent strains, it did not confirm that RpoS was expressed and active. The expression of RpoS was investigated in all strains at both 15°C (mimicking summer-time soil temperature in the east of Ireland, where isolates were collected) and 37°C using Western blot analyses with anti-RpoS monoclonal antibodies. A 37-kDa band corresponding to full-length RpoS was detected in all strains, with the exception of the BW25113ΔrpoS mutant strain, which confirmed the specificity of the antibodies. Strikingly, there was a large increase in RpoS levels at 15°C compared to 37°C in all strains (Fig. 5). RpoS degradation products of similar sizes were also detected in most strains at 15°C (but not in the BW25113ΔrpoS mutant strain), except for COB587 and BW25113. To assess the activity of RpoS, a plasmid-based GFP reporter (gadX::gfp) was transformed into all strains and used to record the transcription from the gadX promoter region, which is known to be highly RpoS dependent and has been used by others as reporter of RpoS activity (57–59). A microscopic analysis of stationary-phase cultures grown at 37°C revealed that all strains showed significant GFP expression, with the exception of the BW25113ΔrpoS mutant strain, which showed no detectable fluorescence. The fluorescence levels detected in the strains were not identical; COB585 reproducibly (n = 2; with at least 3 fields each time) had lower levels of fluorescence under these conditions. At 15°C, however, the fluorescence levels detected for all strains were greatly reduced compared to levels detected at 37°C, with COB585 again showing the least fluorescence (Fig. 6A). One possibility was that decreased fluorescence might be caused by failure of the GFP protein to fold or mature properly (60) at 15°C. To test whether the levels of GFP were indeed reduced at 15°C, the levels were assessed by Western analyses using anti-GFP antibodies. The results showed that there was a strong correlation between the GFP levels and the levels of fluorescence detected by microscopy, suggesting that transcription of the gadX::gfp reporter was reduced at 15°C (Fig. 6B). RpoS activity, based on the reporter activity, was indeed decreased at 15°C despite the increased levels of the RpoS protein detected at this temperature. Taken together, these data confirm that RpoS is both present and active in the soil-persistent and commensal isolates of E. coli.
FIG 5.
A higher level of RpoS is expressed in the stationary phase at 15°C than at 37°C in E. coli. Protein preparations were obtained from E. coli strains grown in LB broth with agitation to the stationary phase at 15°C and 37°C. The same amount of protein (25 μg) from each strain at both temperatures was run on SDS-PAGE, and RpoS was detected by immunoblotting. Purified RpoS protein was used as the control. More RpoS was produced at 15°C than at 37°C in all of the strains, whereas RpoS was not detected in the BW25113ΔrpoS strain at either 15°C or 37°C. The image shown is representative of three independent replicates.
FIG 6.
RpoS-dependent GFP expression of gadX::gfp promoter fusions in the stationary phase at 15°C and 37°C by fluorescence microscopy (A) and Western blotting using anti-GFP antibody (B). E. coli strains containing the reporter fusion were cultured in LB broth supplemented with kanamycin (50 μg/ml) with agitation. Samples were taken at stationary phase (17 h at 37°C and 36 h at 15°C), fixed with ethanol-methanol (1:1) solution, and resuspended in PBS, and 2 μl was placed on a slide and imaged with a Leica DMI3000 B microscope. RpoS activity indicated by fluorescence was higher at 37°C than at 15°C (A), and this correlated with GFP detection by immunoblotting (B). COB585 carrying the reporter fusion had the lowest RpoS activity among strains with intact RpoS. Fluorescence was not detected in the BW25113ΔrpoS strain either by microscopy or immunoblotting. Fluorescent images presented are representatives of 2 independent experiments with >3 field captures in each experiment. Western blot image is representative of 3 independent experiments.
DISCUSSION
Many studies in recent years have isolated and described E. coli from nonhost environments (7, 12, 16), but not much is known on the mechanisms of adaptation of E. coli to the soil. In this study, we characterized the phenotypic properties of some long-term soil-persistent E. coli, established that the general stress response regulated by RpoS is conserved in them, and showed that RpoS is required for long-term survival in soil. This collection of long-term soil-persistent isolates represents an opportunity to understand the role that stress responses play in allowing E. coli to thrive in an environment outside the host, where environmental conditions, nutrient availability, and competing microorganisms are different from those present in the gastrointestinal tract.
All soil-persistent strains are motile at 37°C and 15°C, suggesting that motility is likely to be important for life in the soil environment. Though the individual strains showed different levels of motility (Fig. 1B), there was no significant difference (P > 0.05) in motility when soil-persistent and commensal strains were compared at 37°C and 15°C. In an environment where nutrients are spatially and temporally heterogeneous, such as the soil, active motility could also be important in terms of a strategy for growth and survival. Brennan et al. (61) reported some evidence of active movement of E. coli within soil profiles. Furthermore, motility is important for initiating biofilm formation, because flagella help the initial attachment and movement along surfaces (62, 63). We determined biofilm formation, which is known to be RpoS dependent (64), and showed that the strains produced various amounts of biofilm. COB583 and COB585 produced the most biofilm in rich medium (LB), while the commensal strain SE15 produced the highest biofilm in minimal medium (i.e., SMM) (Fig. 1C). We initially hypothesized that high biofilm formation may be an important phenotype for survival in the soil; however, our results showed that only two soil-persistent isolates produced high levels of biofilm under the conditions investigated, suggesting that the ability to form biofilm may not be an essential phenotype for long-term soil survival. The result of the heterogeneity in biofilm production observed in our study is consistent with the study of Skyberg et al. (65), which showed that biofilm production is strain specific and not significantly influenced by phylogenetic group. Similar to previous studies that have shown that biofilm formation is higher at lower temperatures (48, 66) and that biofilm-related genes are upregulated at 23°C compared to 37°C (67), we demonstrate that E. coli strains can produce high levels of biofilm at 15°C.
The survival data suggested that the general stress response was conserved in the soil-persistent strains, but we sought to investigate this further by sequencing the rpoS locus in each of the five soil isolates and comparing this to the rpoS locus from other sequenced E. coli genomes. E. coli strains have been shown to accumulate mutations in the rpoS gene when grown long term in the laboratory, in batch culture, in stab cultures, in chemostats run with poor carbon sources, and even in natural commensal, pathogenic, and environmental isolates from fermented sausage, human feces and urine, beaches, wastewater effluent, and animals (7, 26–34). When we compared the rpoS locus in the test strains with E. coli K-12 MG1655, various nucleotide substitution mutations were observed, but none of these resulted in a premature stop codon (Table 3). This shows that rpoS is conserved in the soil-persistent and commensal strains. E. coli BW25113 had the same rpoS sequence as E. coli K-12 MG1655, as expected, since it is a derivative of the K-12 strain. We analyzed rpoS of sequenced E. coli strains in the NCBI (http://www.ncbi.nlm.nih.gov/) database and observed that this glutamine (Q) at codon 33 was conserved only in K-12-derived laboratory-adapted strains (data not shown). Atlung et al. (28) had previously reported a similar observation when comparing K-12 strains with six non-K-12 strains, and they proposed that GAG (glutamic acid at codon 33) was present in the E. coli common ancestor and that it evolved to TAG (STOP) in the process of laboratory evolution and then mutated into CAG (glutamine at codon 33). Polymorphism at codon 33 has been reported among E. coli strains in the literature (33, 34, 38), but most of these studies used the original K-12 sequence (33Q) as their wild-type reference sequence, when the polymorphism occurred only in the K-12 lineage. It has been shown that there is no difference in RpoS activity with either glutamic acid (E) or glutamine (Q) at codon 33 (28), suggesting that this residue is not critical for the functioning of RpoS.
Our data show that the soil environment does not preferentially select for rpoS mutations, and this is consistent with recent studies reporting that loss-of-function RpoS mutations are rare in a large collection of natural isolates of commensal, pathogenic, and environmental E. coli (32, 68). Bleibtreu et al. (68) reported no variation in the amino acid sequence of RpoS in the E. coli strains which had minimal laboratory handling before being sequenced and showed that storage and successive transfers resulted in the rpoS mutations. Spira et al. (69) also reported that transfer of E. coli on LB stabs between laboratories led to mutations in rpoS. Retention of an intact stress response may be important for E. coli in soil, since it is a dynamic environment where E. coli may encounter multiple stresses. E. coli must overcome stresses such as UV radiation (if close to soil surface), low nutrients, low temperature, desiccation, competition, predation, and more to thrive in the soil. Furthermore, using phylogenetic analysis, we showed that RpoS is highly conserved in E. coli. The clustering of rpoS in the E. coli strains reflects phylogenetic diversity in the long-term soil-persistent E. coli strains we used for this study (Fig. 3A). It has been recently shown that the phylogenetic grouping based on rpoS is highly consistent with phylogenetic clustering based on multilocus sequence typing (MLST), thus suggesting that rpoS is a good indicator of evolutionary history of E. coli strains (68). On this basis, strain COB585 is the most divergent of the soil-persistent strains used in study, and it is most closely related to ECC08, which is a strain collected from a beach water sample at Bayfront Park Beach, Hamilton, Canada (7).
Having established that the rpoS gene is conserved in the long-term soil-persistent strains, it was important to measure the RpoS level and its activity, since its role in the general stress response is complex, being regulated at multiple levels (reviewed in reference 70). We showed that all the strains with an intact rpoS gene produced detectable RpoS protein (Fig. 5). Besides the 37-kDa RpoS protein observed in the RpoS-positive strains, there were other bands detected in all of the strains which were not found in the BW25113ΔrpoS strain. These are likely to be degradation products specific to RpoS, as one of the major regulatory mechanisms of RpoS level is proteolysis by ClpXP (71). All of the soil-persistent strains, the commensal strains, and BW25113 displayed known RpoS-dependent phenotypes that were absent in the BW25113ΔrpoS mutant strain, albeit with some small differences, suggesting that RpoS is active in these strains (Fig. 4). For example, RpoS plays an important role in acid tolerance in E. coli through its influence on the glutamate decarboxylase (GAD) system; specifically RpoS controls the transcription of two operons encoding components of the GAD system (gadA and gadBC) and controls a positive regulator of this system, GadE (reviewed in reference 72). GABA was produced by all strains with intact RpoS in order to overcome acid stress. E. coli SE11, a commensal strain, had an attenuated response to extreme acidity at 37°C (Fig. 4A), as it survived a pH of 2.5 for only 20 min. Interestingly, the RpoS level in SE11 at 37°C was lowest among the strains (Fig. 5), and it is possible that this contributed to the attenuated acid stress response and reduced survival under starvation at 37°C (data not shown). However, this was not the case at 15°C, where SE11 produced high levels of RpoS and survived extreme acidity for 1 h (data not shown), similar to other soil-persistent and commensal strains, but still survived rather poorly in PBS compared to the other RpoS-positive strains (Fig. 2A). The reason for reduced stress response in E. coli SE11 under these conditions is not clear at present.
RpoS activity as determined by the gadX reporter fusions shows that RpoS was active in all the long-term soil-persistent strains at 15°C and 37°C. However, expression of GFP reporters at the stationary phase was higher at 37°C than at 15°C (Fig. 5B). This low GFP expression correlated with lower fluorescence at 15°C (Fig. 6A). This is contrary to the fact that RpoS levels, as determined by Western blotting, were higher at 15°C (Fig. 5A). For example, E. coli SE11, which had low levels of RpoS at 37°C, was active in transcribing the RpoS-dependent promoter and leading to high GFP expression (Fig. 6A). These data show that RpoS protein levels did not always correlate with reporter activity. E. coli COB585, the most divergent of the strains, had the lowest RpoS activity among the test strains at 37°C. Similar attenuated RpoS activity was also observed in E. coli ECC08, which is closely related to COB585 (Fig. 3A) (7). No RpoS activity was observed in exponential phase at 15°C and 37°C in all of the strains (data not shown). This growth-phase-dependent RpoS activity was also reported by Sledjeski et al. (73) and shows that increasing RpoS levels at low temperature does not necessarily lead to the physiological response observed when cells enter into the stationary phase or when they encounter other environmental stresses. Though gadX is strongly RpoS dependent in the stationary phase, it was shown that the RpoS dependence may be reduced or abolished under acid stress (24). It will be interesting to further investigate if RpoS activity reduces when exposed to cold stress and other similar stresses encountered in the environment.
We speculate that low activity of RpoS in the stationary phase at 15°C may be due to the upregulation of factors that inhibit promoter recognition by RpoS (74), increased negative feedback inhibition/autoregulation of RpoS (74), or an increase in induction of anti-sigma factors (75). The response regulator RssB, which is important for regulating intracellular RpoS levels at the exponential phase and under low stress, can also serve as an anti-sigma factor and inhibit the RpoS-dependent gene expression in the presence of high RpoS levels (75). Though Becker et al. (75) could not identify in vivo growth conditions that resulted in such high RssB induction, we speculate that growth in LB at 15°C may be an example of such conditions, based on the high RpoS degradation observed. There is also evidence that, besides its role in RpoS proteolysis, RssB directly interacts with the turnover element in RpoS, thus blocking promoter recognition by RpoS and transcription initiation of RpoS-dependent promoters, and thus inhibiting transcriptional activity of RpoS (74). Further work will be necessary to establish whether RssB contributes to the reduced RpoS activity that we observed at 15°C.
The heterogeneity in RpoS activity may reflect the genetic diversity of E. coli lineages present in the soil, perhaps suggesting that they have evolved to occupy different microniches in the soil. We have shown that long-term soil adaptation does not select for the rpoS mutation. The soil survival data clearly support the conclusion that it is important to retain a functional RpoS-mediated stress response in order to survive long-term in the soil (Fig. 2C and D). These data demonstrate that, regardless of gut, soil, or laboratory origin, E. coli can persist in soil for long periods, and the data also suggest that the general stress response is intrinsic to this trait. Since RpoS controls multiple stress responses and since the soil environment is very dynamic, an intact stress response was retained as E. coli encountered UV radiation from sunlight, lower temperature, hyperosmotic and hypo-osmotic stress, nutrient availability, competition, and predation (76–78). Thus, it seems clear that the selective pressure to maintain stress resistance within the soil environment outweighs the potential growth advantage that might arise from loss of RpoS function.
Overall, the phenotypic characteristics and stress response of long-term soil-persistent E. coli were found to be very similar to those of commensal E. coli, suggesting that an intact stress response may be required by E. coli to ensure environmental persistence prior to availability of a new host. Our results also suggest that the long-term residence of E. coli strains in a soil niche does not select for consistently high levels of motility or biofilm formation. We speculate that the huge genetic diversity found in E. coli as a species could be driven by the fluctuations within environmental niches rather than by the comparatively homogenous environment of the gastrointestinal tract. The variety of microniches available within the soil could provide a variety of different selective pressures that each result in phenotypically different populations. The idea that intraspecies and interspecies diversities are driven largely by evolutionary trade-offs that arise between different properties of the cell (e.g., biofilm formation, metabolism, motility, stress resistance) when they are subject to the constraints of different environments has recently been reviewed comprehensively (79).
Since soil-persistent strains have been reported to have unique growth and metabolic characteristics compared to the common laboratory reference strain (E. coli K-12 and its derivatives) (19), there is the need to utilize wild isolates of E. coli when studying mechanisms involving growth and metabolic capacities in E. coli under different conditions. In summary, our findings show that loss-of-function mutations are absent in the rpoS gene of long-term soil-persistent E. coli strains and that RpoS is highly conserved in these strains. Using RpoS-dependent phenotypes and reporter activity measurements, we confirm that a functional RpoS response is retained among long-term soil-persistent strains and that RpoS is important for long-term survival of E. coli in soil.
ACKNOWLEDGMENTS
We thank colleagues in the Bacterial Stress Response group for useful comments and discussions.
We are grateful to David Clarke (University College Cork) for supplying the reporter fusion used in this study.
Funding Statement
This work was supported by an NUI Galway College of Science Ph.D. Fellowship and Thomas Crawford Hayes research awards to Y.S.
REFERENCES
- 1.Berg R. 1996. The indigenous gastrointestinal microflora. Trends Microbiol 4:430–435. doi: 10.1016/0966-842X(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 2.Gordon DM, Cowling A. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575–3586. doi: 10.1099/mic.0.26486-0. [DOI] [PubMed] [Google Scholar]
- 3.Ishii S, Sadowsky MJ. 2008. Escherichia coli in the environment: implications for water quality and human health. Microbes Environ 23:101–108. doi: 10.1264/jsme2.23.101. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Torres AJ, Hazen TC, Toranzos GA. 1987. Distribution and in situ survival and activity of Klebsiella pneumoniae and Escherichia coli in a tropical rain forest watershed. Curr Microbiol 15:213–218. doi: 10.1007/BF01577533. [DOI] [Google Scholar]
- 5.Jiménez L, Muñiz I, Toranzos GA, Hazen TC. 1989. Survival and activity of Salmonella typhimurium and Escherichia coli in tropical freshwater. J Appl Bacteriol 67:61–69. doi: 10.1111/j.1365-2672.1989.tb04955.x. [DOI] [PubMed] [Google Scholar]
- 6.McLellan SL, Salmore AK. 2003. Evidence for localized bacterial loading as the cause of chronic beach closings in a freshwater marina. Water Res 37:2700–2708. doi: 10.1016/S0043-1354(03)00068-X. [DOI] [PubMed] [Google Scholar]
- 7.Chiang SM, Dong T, Edge TA, Schellhorn HE. 2011. Phenotypic diversity caused by differential RpoS activity among environmental Escherichia coli isolates. Appl Environ Microbiol 77:7915–7923. doi: 10.1128/AEM.05274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byappanahalli M, Fujioka R. 1998. Evidence that tropical soil environment can support the growth of Escherichia coli. Water Sci Technol 38:171–174. doi: 10.1016/S0273-1223(98)00820-8. [DOI] [Google Scholar]
- 9.Desmarais TR, Solo-Gabriele HM, Palmer CJ. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl Environ Microbiol 68:1165–1172. doi: 10.1128/AEM.68.3.1165-1172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujioka RS. 2001. Monitoring coastal marine waters for spore-forming bacteria of fecal and soil origin to determine point from nonpoint source pollution. Water Sci Technol 44:181–188. [PubMed] [Google Scholar]
- 11.Solo-Gabriele HM, Wolfert MA, Desmarais TR, Palmer CJ. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl Environ Microbiol 66:230–237. doi: 10.1128/AEM.66.1.230-237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byappanahalli MN, Yan T, Hamilton MJ, Ishii S, Fujioka RS, Whitman RL, Sadowsky MJ. 2012. The population structure of Escherichia coli isolated from subtropical and temperate soils. Sci Total Environ 417–418:273–279. doi: 10.1016/j.scitotenv.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 13.Byappanahalli MN, Whitman RL, Shively DA, Sadowsky MJ, Ishii S. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ Microbiol 8:504–513. doi: 10.1111/j.1462-2920.2005.00916.x. [DOI] [PubMed] [Google Scholar]
- 14.Ishii S, Ksoll WB, Hicks RE, Sadowsky MJ. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl Environ Microbiol 72:612–621. doi: 10.1128/AEM.72.1.612-621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson KL, Whitlock JE, Harwood VJ. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl Environ Microbiol 71:3041–3048. doi: 10.1128/AEM.71.6.3041-3048.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan FP, O'Flaherty V, Kramers G, Grant J, Richards KG. 2010. Long-term persistence and leaching of Escherichia coli in temperate maritime soils. Appl Environ Microbiol 76:1449–1455. doi: 10.1128/AEM.02335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan M, Fanning A. 1996. Effects of fertiliser N and slurry on nitrate leaching: lysimeter studies on 5 soils. Irish Geogr 29:126–136. doi: 10.1080/00750779609478655. [DOI] [Google Scholar]
- 18.Brennan FP, Abram F, Chinalia FA, Richards KG, O'Flaherty V. 2010. Characterization of environmentally persistent Escherichia coli isolates leached from an Irish soil. Appl Environ Microbiol 76:2175–2180. doi: 10.1128/AEM.01944-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennan FP, Grant J, Botting CH, O'Flaherty V, Richards KG, Abram F. 2013. Insights into the low-temperature adaptation and nutritional flexibility of a soil-persistent Escherichia coli. FEMS Microbiol Ecol 84:75–85. doi: 10.1111/1574-6941.12038. [DOI] [PubMed] [Google Scholar]
- 20.Byappanahalli M, Fujioka R. 2004. Indigenous soil bacteria and low moisture may limit but allow fecal bacteria to multiply and become a minor population in tropical soils. Water Sci Technol 50:27–32. [PubMed] [Google Scholar]
- 21.Coldewey SM, Hartmann M, Schmidt DS, Engelking U, Ukena SN, Gunzer F. 2007. Impact of the rpoS genotype for acid resistance patterns of pathogenic and probiotic Escherichia coli. BMC Microbiol 7:21. doi: 10.1186/1471-2180-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stasic AJ, Lee Wong AC, Kaspar CW. 2012. Osmotic and desiccation tolerance in Escherichia coli O157:H7 requires rpoS (σ38). Curr Microbiol 65:660–665. doi: 10.1007/s00284-012-0210-8. [DOI] [PubMed] [Google Scholar]
- 23.Hryckowian AJ, Welch RA. 2013. RpoS contributes to phagocyte oxidase-mediated stress resistance during urinary tract infection by Escherichia coli CFT073. mBio 4:e00023-13. doi: 10.1128/mBio.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King T, Ishihama A, Kori A, Ferenci T. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J Bacteriol 186:5614–5620. doi: 10.1128/JB.186.17.5614-5620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notley-McRobb L, King T, Ferenci T. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J Bacteriol 184:806–811. doi: 10.1128/JB.184.3.806-811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alvarez-Ordóñez A, Alvseike O, Omer MK, Heir E, Axelsson L, Holck A, Prieto M. 2013. Heterogeneity in resistance to food-related stresses and biofilm formation ability among verocytotoxigenic Escherichia coli strains. Int J Food Microbiol 161:220–230. doi: 10.1016/j.ijfoodmicro.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Atlung T, Nielsen HV, Hansen FG. 2002. Characterisation of the allelic variation in the rpoS gene in thirteen K-12 and six other nonpathogenic Escherichia coli strains. Mol Genet Genomics 266:873–881. doi: 10.1007/s00438-001-0610-0. [DOI] [PubMed] [Google Scholar]
- 29.King T, Seeto S, Ferenci T. 2006. Genotype-by-environment interactions influencing the emergence of rpoS mutations in Escherichia coli populations. Genetics 172:2071–2079. doi: 10.1534/genetics.105.053892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferenci T, Galbiati HF, Betteridge T, Phan K, Spira B. 2011. The constancy of global regulation across a species: the concentrations of ppGpp and RpoS are strain specific in Escherichia coli. BMC Microbiol 11:62. doi: 10.1186/1471-2180-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bleibtreu A, Gros P-A, Laouénan C, Clermont O, Le Nagard H, Picard B, Tenaillon O, Denamur E. 2013. Fitness, stress resistance, and extraintestinal virulence in Escherichia coli. Infect Immun 81:2733–2742. doi: 10.1128/IAI.01329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder E, Gordon DM, Stoebel DM. 2012. Escherichia coli lacking RpoS are rare in natural populations of nonpathogens. G3 (Bethesda) 2:1341–1344. doi: 10.1534/g3.112.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong T, Chiang SM, Joyce C, Yu R, Schellhorn HE. 2009. Polymorphism and selection of rpoS in pathogenic Escherichia coli. BMC Microbiol 9:118. doi: 10.1186/1471-2180-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spira B, Hu X, Ferenci T. 2008. Strain variation in ppGpp concentration and RpoS levels in laboratory strains of Escherichia coli K-12. Microbiology 154:2887–2895. doi: 10.1099/mic.0.2008/018457-0. [DOI] [PubMed] [Google Scholar]
- 35.Jørgensen F, Leach S, Wilde SJ, Davies A, Stewart GSAB, Humphrey T. 2000. Invasiveness in chickens, stress resistance and RpoS status of wild-type Salmonella enterica subsp. enterica serovar Typhimurium definitive type 104 and serovar Enteritidis phage type 4 strains. Microbiology 146:3227–3235. doi: 10.1099/00221287-146-12-3227. [DOI] [PubMed] [Google Scholar]
- 36.Shah DH, Casavant C, Hawley Q, Addwebi T, Call DR, Guard J. 2012. Salmonella enteritidis strains from poultry exhibit differential responses to acid stress, oxidative stress, and survival in the egg albumen. Foodborne Pathog Dis 9:258–264. doi: 10.1089/fpd.2011.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robbe-Saule V, Algorta G, Rouilhac I, Norel F. 2003. Characterization of the RpoS status of clinical isolates of Salmonella enterica. Appl Environ Microbiol 69:4352–4358. doi: 10.1128/AEM.69.8.4352-4358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez-Ordóñez A, Begley M, Hill C. 2012. Polymorphisms in rpoS and stress tolerance heterogeneity in natural isolates of Cronobacter sakazakii. Appl Environ Microbiol 78:3975–3984. doi: 10.1128/AEM.07835-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong T, Coombes BK, Schellhorn HE. 2009. Role of RpoS in the virulence of Citrobacter rodentium. Infect Immun 77:501–507. doi: 10.1128/IAI.00850-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferenci T. 2005. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol Microbiol 57:1–8. doi: 10.1111/j.1365-2958.2005.04649.x. [DOI] [PubMed] [Google Scholar]
- 41.Zambrano MM, Siegele DA, Almirón M, Tormo A, Kolter R. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 42.Savageau MA. 1983. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am Nat 122:732–744. doi: 10.1086/284168. [DOI] [Google Scholar]
- 43.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylotyping method revisited: improvement of specificity and detection of new phylogroups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 44.Kramers G, Holden NM, Brennan F, Green S, Richards KG. 2012. Water content and soil type effects on accelerated leaching after slurry application. Vadose Zone J 11:1. doi: 10.2136/vzj2011.0059. [DOI] [Google Scholar]
- 45.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaveroche MK, Ghigo JM, d'Enfert C. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res 28:E97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tramonti A, Visca P, De Canio M, Falconi M, De Biase D. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J Bacteriol 184:2603–2613. doi: 10.1128/JB.184.10.2603-2613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ingle DJ, Clermont O, Skurnik D, Denamur E, Walk ST, Gordon DM. 2011. Biofilm formation by and thermal niche and virulence characteristics of Escherichia spp. Appl Environ Microbiol 77:2695–2700. doi: 10.1128/AEM.02401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao K, Liu M, Burgess RR. 2007. Adaptation in bacterial flagellar and motility systems: from regulon members to foraging-like behavior in E. coli. Nucleic Acids Res 35:4441–4452. doi: 10.1093/nar/gkm456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Toole G. 2011. Microtiter dish biofilm formation assay. J Vis Exp 47:pii=2437. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma J, Ibekwe AM, Yi X, Wang H, Yamazaki A, Crowley DE, Yang C-H. 2011. Persistence of Escherichia coli O157:H7 and its mutants in soils. PLoS One 6:e23191. doi: 10.1371/journal.pone.0023191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 54.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Byrne CP, Feehily C, Ham R, Karatzas KAG. 2011. A modified rapid enzymatic microtiter plate assay for the quantification of intracellular γ-aminobutyric acid and succinate semialdehyde in bacterial cells. J Microbiol Methods 84:137–139. doi: 10.1016/j.mimet.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 56.Hengge-Aronis R, Fischer D. 1992. Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol Microbiol 6:1877–1886. doi: 10.1111/j.1365-2958.1992.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 57.Chattopadhyay MK, Keembiyehetty CN, Chen W, Tabor H. 2015. Polyamines stimulate the level of the σ38 subunit (RpoS) of Escherichia coli RNA polymerase resulting in the induction of the glutamate decarboxylase dependent acid response system via the gadE regulon. J Biol Chem 290:17809–17821. doi: 10.1074/jbc.M115.655688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Biase D, Tramonti A, Bossa F, Visca P. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol 32:1198–1211. doi: 10.1046/j.1365-2958.1999.01430.x. [DOI] [PubMed] [Google Scholar]
- 59.Ma Z, Richard H, Foster JW. 2003. pH-dependent modulation of cyclic AMP levels and GadW-dependent repression of RpoS affect synthesis of the GadX regulator and Escherichia coli acid resistance. J Bacteriol 185:6852–6859. doi: 10.1128/JB.185.23.6852-6859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu S, Wei X, Dong X, Xu L, Liu J, Jiang B. 2015. Structural plasticity of green fluorescent protein to amino acid deletions and fluorescence rescue by folding-enhancing mutations. BMC Biochem 16:17. doi: 10.1186/s12858-015-0046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brennan FP, Kramers G, Grant J, O'Flaherty V, Holden NM, Richards K. 2012. Evaluating E. coli transport risk in soil using dye and bromide tracers. Soil Sci Soc Am J 76:663–673. doi: 10.2136/sssaj2011.0250. [DOI] [Google Scholar]
- 62.González Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J Bacteriol 188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 64.Schembri MA, Kjaergaard K, Klemm P. 2003. Global gene expression in Escherichia coli biofilms. Mol Microbiol 48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- 65.Skyberg JA, Siek KE, Doetkott C, Nolan LK. 2007. Biofilm formation by avian Escherichia coli in relation to media, source, and phylogeny. J Appl Microbiol 102:548–554. doi: 10.1111/j.1365-2672.2006.03076.x. [DOI] [PubMed] [Google Scholar]
- 66.Else TA, Pantle CR, Amy PS. 2003. Boundaries for biofilm formation: humidity and temperature. Appl Environ Microbiol 69:5006–5010. doi: 10.1128/AEM.69.8.5006-5010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White-Ziegler CA, Um S, Pérez NM, Berns AL, Malhowski AJ, Young S. 2008. Low temperature (23 degrees C) increases expression of biofilm-, cold-shock-, and RpoS-dependent genes in Escherichia coli K-12. Microbiology 154:148–166. doi: 10.1099/mic.0.2007/012021-0. [DOI] [PubMed] [Google Scholar]
- 68.Bleibtreu A, Clermont O, Darlu P, Glodt J, Branger C, Picard B, Denamur E. 2014. The rpoS gene is predominantly inactivated during laboratory storage and undergoes source-sink evolution in Escherichia coli species. J Bacteriol 196:4276–4284. doi: 10.1128/JB.01972-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spira B, de Almeida Toledo R, Maharjan RP, Ferenci T. 2011. The uncertain consequences of transferring bacterial strains between laboratories: rpoS instability as an example. BMC Microbiol 11:248. doi: 10.1186/1471-2180-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lange R, Hengge-Aronis R. 1994. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev 8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 72.Lund P, Tramonti A, De Biase D. 2014. Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol Rev 38:1091–1125. doi: 10.1111/1574-6976.12076. [DOI] [PubMed] [Google Scholar]
- 73.Sledjeski DD, Gupta A, Gottesman S. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J 15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 74.Becker G, Klauck E, Hengge-Aronis R. 1999. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc Natl Acad Sci U S A 96:6439–6444. doi: 10.1073/pnas.96.11.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Becker G, Klauck E, Hengge-Aronis R. 2000. The response regulator RssB, a recognition factor for σs proteolysis in Escherichia coli, can act like an anti-σs factor. Mol Microbiol 35:657–666. doi: 10.1046/j.1365-2958.2000.01736.x. [DOI] [PubMed] [Google Scholar]
- 76.Arrange AA, Phelps TJ, Benoit RE, Palumbo AV, White DC. 1993. Bacterial sensitivity to UV light as a model for ionizing radiation resistance. J Microbiol Methods 18:127–136. doi: 10.1016/0167-7012(93)90029-H. [DOI] [Google Scholar]
- 77.Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. 2003. Physiological and community responses of established grassland bacterial populations to water stress. Appl Environ Microbiol 69:6961–6968. doi: 10.1128/AEM.69.12.6961-6968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ibekwe AM, Poss JA, Grattan SR, Grieve CM, Suarez D. 2010. Bacterial diversity in cucumber (Cucumis sativus) rhizosphere in response to salinity, soil pH, and boron. Soil Biol Biochem 42:567–575. doi: 10.1016/j.soilbio.2009.11.033. [DOI] [Google Scholar]
- 79.Ferenci T. 2016. Trade-off mechanisms shaping the diversity of bacteria. Trends Microbiol 24:209–223. doi: 10.1016/j.tim.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 80.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2016. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]