ABSTRACT
Enterococcus faecalis, a common causative agent of hospital-acquired infections, is resistant to many known antibiotics. Its ability to acquire and transfer resistance genes and virulence determinants through conjugative plasmids poses a serious concern for public health. In some cases, induction of transfer of E. faecalis plasmids results from peptide pheromones produced by plasmid-free recipient cells, which are sensed by the plasmid-bearing donor cells. These plasmids generally encode an inhibitory peptide that competes with the pheromone and suppresses self-induction of donors. We recently demonstrated that the inhibitor peptide encoded on plasmid pCF10 is part of a unique quorum-sensing system in which it functions as a “self-sensing signal,” reducing the response to the pheromone in a density-dependent fashion. Based on the similarities between regulatory features controlling conjugation in pAD1 and pAM373 and those controlling conjugation in pCF10, we hypothesized that these plasmids are likely to exhibit similar quorum-sensing behaviors. Experimental findings indicate that for both pAD1 and pAM373, high donor densities indeed resulted in decreased induction of the conjugation operon and reduced conjugation frequencies. This effect was restored by the addition of exogenous inhibitor, confirming that the inhibitor serves as an indicator for donor density. Donor density also affects cross-species conjugative plasmid transfer. Based on our experimental results, we propose models for induction and shutdown of the conjugation operon in pAD1 and pAM373.
IMPORTANCE Enterococcus faecalis is a leading cause of hospital-acquired infections. Its ability to transfer antibiotic resistance and virulence determinants by sharing its genetic material with other bacteria through direct cell-cell contact via conjugation poses a serious threat. Two antagonistic signaling peptides control the transfer of plasmids pAD1 and pAM373: a peptide pheromone produced by plasmid-free recipients triggers the conjugative transfer in plasmid-containing donors, and an inhibitor peptide encoded on the plasmid and produced by donor cells serves to modulate the donor response in accordance with the relative abundance of donors and recipients. We demonstrate that high donor density reduces the conjugation frequency of both of these plasmids, which is a consequence of increased inhibitor concentration in high-donor-density cultures. While most antibiotic strategies end up selecting resistant strains and disrupting the community balance, manipulating bacterial signaling mechanisms can serve as an alternate strategy to prevent the spread of antibiotic resistance.
INTRODUCTION
Enterococcus faecalis, a bacterium normally present in the human intestinal tract, is a major cause of health care-associated infections. Treatment of these infections has become increasingly difficult with the emergence of E. faecalis strains that are resistant to multiple antibiotics, such as macrolides, tetracyclines, aminoglycosides, and glycopeptides, including vancomycin (1, 2). E. faecalis also possesses the ability to transfer these antibiotic resistances to other bacteria within and across species, facilitating the spread of resistance. Conjugative DNA transfer is particularly common among enterococci, and it frequently involves highly transmissible plasmids or conjugative transposons carrying antibiotic resistance (3). E. faecalis secretes a number of peptide sex pheromones that act as mating (conjugation) signals for donor bacteria harboring certain conjugative plasmids. Peptide signaling activates genes whose products mediate conjugative plasmid transfer. Enterococcal sex pheromones thus contribute directly to dissemination of antibiotic resistance (4, 5).
Plasmid pCF10 is a well-characterized conjugative plasmid that carries tetracycline resistance (6, 7). This plasmid encodes a DNA transfer machine whose expression is induced by the heptapeptide sex pheromone cCF10, which is secreted by plasmid-free (recipient) bacteria (8). In addition, pCF10 also encodes the peptide iCF10, which acts as a competitive inhibitor of cCF10 and functions in preventing self-induction by an endogenous pheromone produced by plasmid-containing cells (9). We have used pCF10 as a model system for analysis of control mechanisms and development of computational models that describe the regulation of conjugation functions (10–12). Recently, we demonstrated that iCF10 also serves as a classic quorum-sensing signal for donors, functioning to reduce conjugation at high donor densities (10).
Several families of conjugative plasmids that have mating responses to various peptide pheromones have been identified in E. faecalis clinical isolates (13, 14). Two of these conjugative plasmids are pAD1 and pAM373, which confer responses to pheromones cAD1 and cAM373, respectively (15, 16). Each plasmid also encodes a cognate small peptide (iAD1 or iAM373), which is secreted and acts as a competitive inhibitor of the corresponding pheromone (17, 18). Both pAD1 and pAM373 are clinically relevant due to the genetic features they encode. Plasmid pAD1 consists of elements that encode a hemolysin/bacteriocin and resistance to UV light (19). Derivatives of plasmid pAM373 often carry vancomycin resistance, and their mating response can be induced by peptides produced by Staphylococcus aureus, Streptococcus gordonii, and Enterococcus hirae (4, 20).
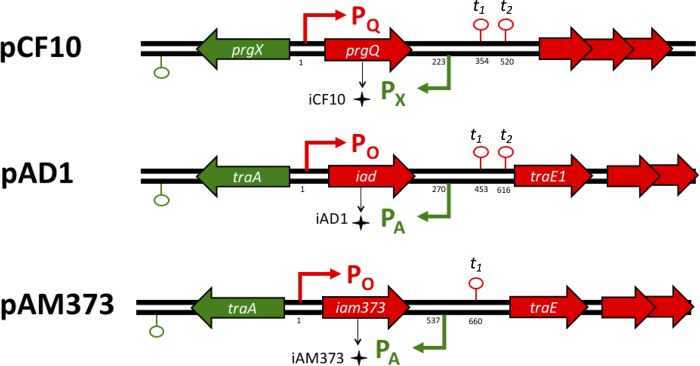
As shown in Fig. 1, there is substantial conservation of the critical regulatory regions in pCF10, pAD1, and pAM373. Plasmid pCF10 carries the prgX and prgQ operons on cDNA strands with an overlapping region at the 5′ end of each operon. This organization results in convergent transcription of ∼220 nucleotides of mRNA that can lead to reciprocal negative regulation by both antisense interactions (21–23) and transcription interference resulting from collisions between RNA polymerase elongation complexes (11). The overlapping region also encodes the inhibitor peptide iCF10 (22). Plasmids pAD1 and pAM373 also have convergent promoters in the regulatory region (24, 25), and the overlapping region between the two promoters encodes their respective peptide inhibitors, iAD1 and iAM373.
FIG 1.
Gene arrangement in pheromone-responsive plasmids. Comparison of the pheromone-responsive region of pCF10 to the sex pheromone plasmids pAD1 and pAM373 in their regulatory regions. The positions for the regulatory features, i.e., transcription start sites and transcriptional terminators, are indicated relative to the transcription start sites from PQ and PO (indicated as “1”).
Noting these similarities, we set out to examine whether a dual signaling system like that found in pCF10, with the pheromone indicating recipient density and the inhibitor indicating donor density, also functions in pAD1 and pAM373. Here we report that, as in the case of pCF10, high donor density has a suppressive effect on conjugation in both pAD1 and pAM373. The suppressive effect requires inhibitor production by the donor cells and is likely caused by the increased concentration of inhibitor in high-donor-density cultures. This confirms a broad role of inhibitor peptides as signals which regulate the social behavior of conjugative plasmid-harboring E. faecalis donor cells. To date, the only published studies of donor population density on expression of conjugation functions have been carried out with the Agrobacterium tumefaciens Ti plasmid system, where quorum sensing operates to enhance conjugation at high donor density (26, 27). Here, we report that the opposite situation holds for multiple enterococcal sex pheromone plasmids.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All liquid cultures were grown at 37°C in M9 medium containing 3 g/liter yeast extract, 10 g/liter Casamino Acids, 36 g/liter glucose, 0.12 g/liter MgSO4, and 0.011 g/liter CaCl2 or in Todd-Hewitt broth (THB; Difco). Antibiotics, when used for selection, were at the following concentrations: tetracycline (Tet), 10 μg/ml; chloramphenicol (Cm), 10 μg/ml; erythromycin (Erm), 20 μg/ml; streptomycin (Strep), 1,000 μg/ml; spectinomycin (Spec), 1,000 mg/ml; and rifampin (Rif), 25 μg/ml (for JH2-2) and 200 μg/ml (for OG1RF).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference(s) |
|---|---|---|
| Strains | ||
| E. faecalis | ||
| OG1RF | Rifr Far | 42 |
| OG1X | Strepr | 20, 43 |
| JH2-2 | Rifr Far | 20, 44 |
| OG1Sp | Specr | 45 |
| S. gordonii CH1 cm | Spontaneous Cmr derivative of strain Challis (CH1) | This study |
| Plasmids | ||
| pAM714 | pAD1 with Tn917 insert; Ermr | 20, 43 |
| pAM378 | pAM373 with Tn918 insert; Tetr | 23, 24 |
| pAMS470 | Shuttle vector which replicates in Enterococcus and Streptococcus; Ermr | 24 |
Rif, rifampin; Fa, fusidic acid; Strep, streptomycin; Spec, spectinomycin; Cm, chloramphenicol; Erm, erythromycin; Tet, tetracycline.
Conjugation assays.
Cultures of donor and recipient cells were grown overnight at 37°C in 4 ml of M9 medium, centrifuged, washed twice with 1 ml potassium phosphate-buffered saline (KPBS) containing 2 mM EDTA, resuspended in the original volume of fresh M9 medium, and adjusted to equal cell densities at an optical density of 600 nm (OD600). Plasmid-bearing donor cells (D) were diluted 1:10 or 1:1,000 in fresh M9 medium and added to equal volumes of a 1:10 dilution of plasmid-free recipient cells (R), resulting in D/R ratios of 1:1 and 0.01:1, respectively. Donor and recipient mating was carried out at 37°C, and 100 μl of each mixed culture was removed at 45 min and 90 min to monitor the emergence of transconjugants by using plate counts. Serial dilutions of these samples were plated on selective LB agar medium for donors, recipients, or transconjugants. The plates were incubated overnight before enumeration. Three or more biological replicates were done to show repeatability.
Filter matings were done to observe cross-species conjugation between E. faecalis and S. gordonii CH1 cm. Cultures of donor and recipient cells were grown overnight at 37°C in 4 ml of THB medium, washed twice, and resuspended in equal volumes of THB. E. faecalis JH2-2(pAM378, pASM470) donor cells were diluted 1:10 or 1:1,000 and added to equal volumes of 1:10 dilutions of recipient cells (S. gordonii CH1 cm), resulting in D/R ratios of 1:1 and 0.01:1, respectively. Fifty microliters of the mating mixture was pipetted onto the surface of a UV-treated 0.22-μm-pore-size membrane filter (Millipore, Bedford, MA, USA), placed on nonselective LB agar plates, and incubated at 37°C for 3 h. After mating, the cells were resuspended by vortexing the filter in 1 ml KPBS containing 2 mM EDTA, serially diluted, and plated on selective antibiotic plates for enumeration of donors, recipients, and transconjugants. Three biological replicates were done to show repeatability.
Quantification of induction of conjugation genes.
To examine the dynamic response of donor cells to pheromone (cAD1 and cAM373) at various cell densities, overnight cultures of donors in M9 medium were centrifuged and washed twice in 1 ml KPBS containing 2 mM EDTA. Cells were diluted 1:10 (high donor density) or 1:1,000 (low donor density) into 4.5 ml M9 medium. Cultures were incubated for 1 h at 37°C before the pheromone (50 ng/ml cAD1 or 500 ng/ml cAM373) was added. The cultures were incubated at 37°C, and samples were taken at various time intervals. Samples of cells were set on ice for 3 min, centrifuged, and washed once in 1 ml KPBS containing 2 mM EDTA. Samples were then treated with RNAprotect bacterial reagent (Qiagen, Inc., Valencia, CA) according to the manufacturer's instructions, flash frozen in ethanol and dry ice, and stored at −80°C until RNA extraction.
To examine the effect of the inhibitor on induction, overnight cultures of donors in M9 medium were centrifuged and washed twice in 1 ml KPBS containing 2 mM EDTA. Cells were diluted 1:1,000 (low donor density) into 4.5 ml M9 medium in duplicate sets and incubated for 1 h at 37°C. One set was treated with pheromone only (50 ng/ml cAD1 or 500 ng/ml cAM373). The other set was treated with the same concentration of pheromone (50 ng/ml cAD1 or 500 ng/ml cAM373) and an equal concentration of inhibitor (50 ng/ml iAD1 or 500 ng/ml iAM373). The cultures were incubated at 37°C, and samples were taken at various time points, treated with RNAprotect bacterial reagent, and flash frozen as described above.
Frozen cell pellets were thawed and treated with lysozyme (30 mg/ml) in Tris-EDTA (TE) buffer (10 mM Tris, 1 mM EDTA) with (500 U/ml) mutanolysin. RNA was extracted using an RNeasy kit (Qiagen, Inc.) per the manufacturer's instructions. The extracted RNA was subjected to DNase treatment with Turbo DNase (Ambion, Austin, TX) according to the manufacturer's instructions and normalized for RNA concentration across the samples. Subsequent reverse transcription into cDNA was carried out using a SuperScript III first-strand synthesis kit (Invitrogen Corp., Carlsbad, CA) with random hexamers. The cDNA was then used for quantitative reverse transcription-PCR (qRT-PCR) using SYBR green Supermix (Bio-Rad Laboratories Inc., Hercules, CA) and a CFX Connect real-time PCR detection system (Bio-Rad Laboratories Inc.) instrument. A total reaction volume of 15 μl, containing 2 μl of gene-specific primer mixture at a concentration of 10 μM, was used in each well. Each reaction was performed in triplicate, and threshold cycle (CT) values were obtained. A constitutive housekeeping gene, relA, was used as the reference gene to quantify expression, which was normalized relative to time zero. The sequences of primers are listed in Table 2. Three biological replicates were done to show repeatability.
TABLE 2.
Sequence of primers used for qPCR
| Primer target (name) | Sequence for forward primer | Sequence for reverse primer | Reference |
|---|---|---|---|
| relA | CAAGATTTACGGGTCATTATGG | GACTAATCCCTAAGCGATGTG | 51 |
| traE1 (pAD1) | CATGTTGGGTACTACGGCAA | TGACCAACTTCTCTTTCGCA | This study |
| traE (pAM373) | GCGGAAACTAGTTGGCTTCTT | TGCCTTATATGACTGTAAACCGAA | This study |
RESULTS
Plasmids pCF10, pAD1, and pAM373 show remarkable similarity in the gene arrangements of their regulatory regions (Fig. 1). However, they show variation in the nucleotide and amino acid sequences within these regions. The regulatory proteins (TraA, PrgX), which control induction, share only a 23 to 40% identity at the amino acid level. The three-dimensional structure of these regulatory TraA proteins was predicted using the online tool RaptorX (28). A remarkable similarity is seen in the predicted structure of the two TraA proteins and the PrgX crystal structure even though they have dissimilar amino acid sequences (see Fig. S2 in the supplemental material). PrgX was the first receptor protein for the signaling peptides whose structure was determined using X-ray crystallography (29). Based on the predicted structural homology of PrgX to the TraA proteins (see Fig. S2), it is likely that all three proteins are members of a growing family of peptide-modulated transcription factors of Gram-positive pathogens (RRNPP proteins) regulating diverse functions, including sporulation, biofilm formation, competent cell transformation, conjugation, and virulence (30–33).
Furthermore, the region between the inhibitor determinant and the downstream genes traE1 (pAD1), traE (pAM373), and prgR (pCF10) is highly conserved among the three pheromone-responding plasmids. This region is of great regulatory importance, as it is known to encode a small regulatory RNA that participates in the termination/antitermination decision controlling the transcription of conjugation genes downstream of the transcriptional terminator t1 shown in Fig. 1 (21, 24, 25). The secondary structures of the regulatory RNA transcribed from PX/PA promoters (Qa in pCF10 and mD in pAD1 and pAM373) were predicted using Sfold (34) (see Fig. S3 in the supplemental material), and they showed a remarkable similarity. These RNAs are known to act specifically on their respective targets (21). Along with the similar structures of the regulatory proteins (see Fig. S2), these findings suggest a high degree of conservation in the regulatory components of these pheromone-responsive plasmids. Hence we examined the effect of donor density and the role of the inhibitor peptide as an indicator for donor density for plasmids pAD1 and pAM373.
In spite of the overall similarity of the three sex pheromone plasmids, there are significant differences in them and in the biological activities of their corresponding inhibitor and pheromone peptides. First, pAM373 is only 37 kb in size, markedly smaller than pCF10 (65 kb) and pAD1 (58 kb) (35). Plasmid pAM373 also lacks a determinant resembling TraB of pAD1 or its equivalent PrgY in pCF10 (see Table S1 in the supplemental material) (20). PrgY prevents plasmid-bearing cells from responding to an endogenously produced pheromone by sequestering or inactivating the pheromone as it is released from the membrane (36). Furthermore, the adhesin encoded by pAM373 (Asa373) is much smaller than the adhesins of pCF10 and pAD1 (∼700 versus >1,200 amino acids) and does not show any sequence similarity to them (37). Plasmid pAM373 also lacks a surface exclusion protein (Sea1 in pAD1 and PrgA in pCF10) (38). These differences may allow the plasmids to have different induction and conjugation behaviors.
Donor density controls conjugation.
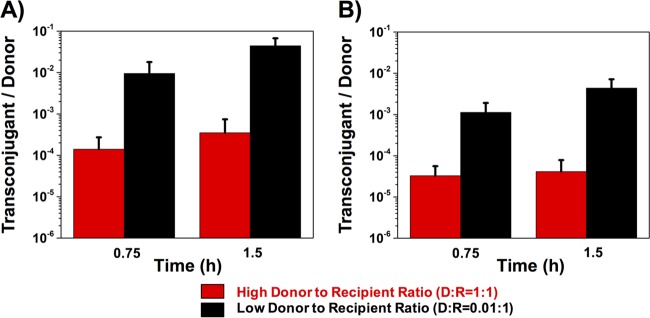
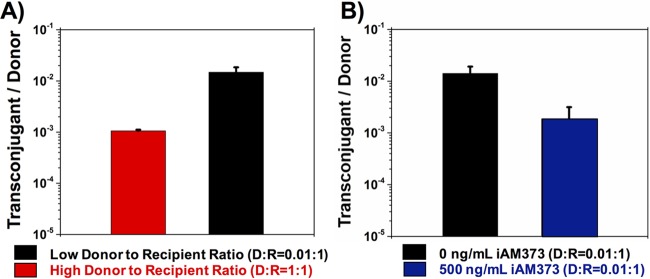
Overnight cultures of plasmid-free OG1RF recipient cells were mixed with different amounts of OGIX donor cells carrying plasmid pAM714 (a pAD1 derivative carrying a copy of Tn917 providing selectable erythromycin resistance [Table 1]) to generate donor-to-recipient (D/R) ratios of 1:1 and 0.01:1. Transconjugants, recipients, and donors were enumerated on selective agar medium after 0.75 and 1.5 h of coculture. The results shown in Fig. 2 indicate that decreasing the donor concentration to a donor-to-recipient ratio of 0.01 increased the conjugation frequency (number of transconjugants per donor) of pAM714 by about 2 orders of magnitude (Fig. 2A). A similar experiment was done using a JH2-2 strain harboring pAM378 (a pAM373 derivative carrying a selectable tetracycline resistance gene [Table 1]) as the donor and OG1Sp as the recipient strain. A similar trend was observed, i.e., the lower donor-to-recipient ratio resulted in a higher transfer frequency per donor (Fig. 2B). Thus, high donor concentrations have a suppressive effect on the conjugative transfer of both plasmids.
FIG 2.
Influence of donor density on conjugation frequency. Different concentrations of donors (D) and recipients (R) were mixed to achieve D/R ratios of 1:1 and 0.01:1 at time zero. Transconjugants, donors, and recipients were enumerated on selective agar at 0.75 h and 1.5 h of coculture, and the conjugation frequency is represented by the number of transconjugants per donor. (A) Liquid mating of pAD1; (B) liquid mating of pAM373. Data shown in panels A and B are averages of results of three biological replicates (error bars are SDs from mean values).
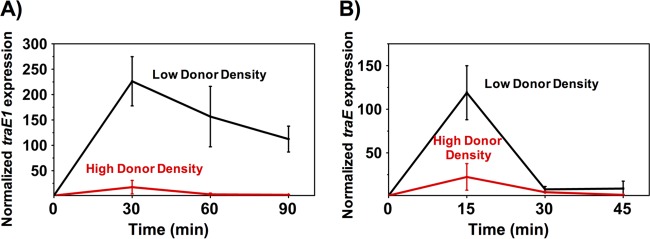
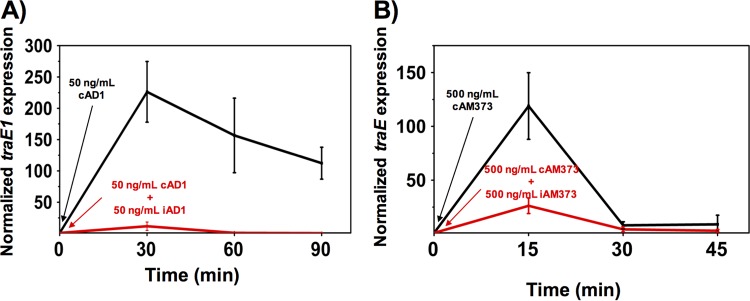
The dynamics of induction of the conjugation operon at high and low donor densities were analyzed to further examine how donor cell concentration affects conjugation. Overnight cultures of donor cells (the same two strains used in the mating experiments described above) diluted 1:10 (high donor density) or 1:1,000 (low donor density) in fresh medium were induced with their cognate pheromone peptide, and transcript levels of a conjugation gene (pAD1 traE1 or pAM373 traE [Fig. 1]) were measured using qRT-PCR. In preliminary experiments, we determined that higher levels of pheromone were required to induce pAM373 derivatives than pAD1 derivatives, so we used 500 ng/ml of cAM373 and 50 ng/ml cAD1 for induction. Both low-density donor cultures showed higher levels of induction and induction was sustained for longer periods of time than for the corresponding high-density cultures (Fig. 3). Both plasmid systems showed a rapid induction response, followed by a return of transcription to the preinduction level, as was previously shown for pCF10 (10). The pAD1 system showed higher and more sustained levels of induction than those shown by pAM373.
FIG 3.
Donor density affects expression levels of conjugation operons on plasmids pAD1 and pAM373 following induction. The low and high donor densities are derived from 1:1,000 and 1:10 dilutions of overnight cultures, respectively. A synthetic pheromone was added at time zero to induce the donor cells. qRT-PCR was used to measure the dynamic expression of conjugation determinants encoded by the plasmids. (A) Donors carrying pAD1 were induced with 50 ng/ml cAD1. (B) Donors carrying pAM373 were induced with 500 ng/ml cAM373. Data shown in panels A and B are averages of results of three biological replicates (error bars are SDs from mean values).
Role of inhibitor as a quorum sensor of donor density.
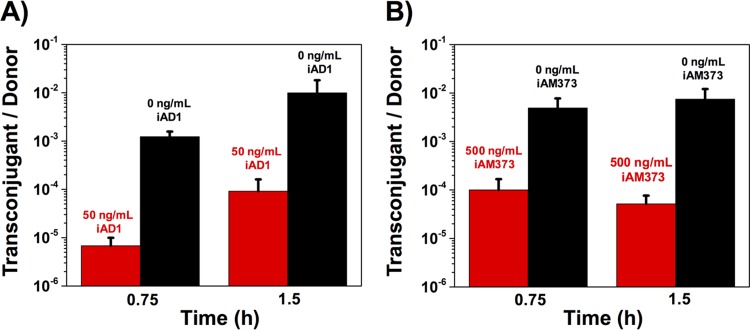
In order to examine if the effects of donor density on conjugation and expression of a gene in the conjugation machinery operon were a consequence of increased concentrations of the inhibitor peptide, mating experiments were carried out at a low donor-to-recipient ratio (0.01:1) with and without the addition of exogenously added inhibitor. In the presence of inhibitor, the conjugation frequency decreased by about 2 orders of magnitude for both plasmids (Fig. 4). The effect that the addition of exogenous inhibitor had on conjugation frequency was the same as that observed with high donor densities (Fig. 2), indicating the role of the inhibitor as a measure of donor cell density.
FIG 4.
Peptide inhibitor has a suppressive effect on conjugation frequency. Overnight cultures of donors and recipients were diluted 1:10 and mixed to achieve two sets with a D/R ratio of 0.01:1 (low donor-to-recipient ratio). One set of mating mixtures served as a control. The other set was treated with inhibitor peptide. The conjugation frequency is represented by the number of transconjugants per donor. (A) Liquid mating of pAD1 with and without 50 ng/ml of iAD1; (B) liquid mating of pAM373 with and without 500 ng/ml of iAM373. Data shown in panels A and B are averages of results of three biological replicates (error bars are SDs from mean values).
We also evaluated the effect of inhibitor concentration on gene expression by adding exogenous inhibitor peptides to low-density cultures. A 1:1,000 dilution of OG1X(pAM714) was induced using 50 ng/ml cAD1 peptide. An identical low-donor-density culture was exposed to 50 ng/ml of pheromone cAD1 and 50 ng/ml of the inhibitor iAD1. Transcript levels of traE1 in both cultures were measured over time using qRT-PCR (Fig. 5A). In the culture where both pheromone and inhibitor were added, the induction level was much lower than in the case where no inhibitor was added, restoring the trend observed in the high-donor-density culture shown in Fig. 3A. Similarly, the conjugative operon of plasmid pAM378 was induced with 500 ng/ml of cAM373 with and without iAM373 (500 ng/ml). Transcript levels of traE were much lower in the presence of the inhibitor (Fig. 5B), mirroring the trends observed in the high-donor-density culture (Fig. 3B).
FIG 5.
Peptide inhibitor suppresses expression of conjugation operons following induction. Overnight cultures of donor cells were diluted 1:1,000 to achieve two sets of low-donor-density cultures. A synthetic pheromone was added to induce the donor cells. One set of donor cells served as a control, and the other set was treated with inhibitor peptide. qRT-PCR was used to measure the dynamic expression of conjugation determinants encoded by the plasmids. (A) Donors carrying pAD1 were induced with 50 ng/ml cAD1 with and without 50 ng/ml of iAD1. (B) Donors carrying pAM373 were induced with 500 ng/ml cAM373 with and without 500 ng/ml of iAM373. Data shown in panels A and B are averages of results of three biological replicates (error bars are SDs from mean values).
The increase in transcript level of traE in pAM373 upon induction is lower than that seen in pAD1. The transcript level of traE also decreases rapidly, even without the addition of the inhibitor (Fig. 3B), and its peak induction time also varies somewhat. In comparison, the transcript level of traE1 in pAD1 does not decrease as rapidly after induction, making it possible to perform an experiment in which 10 ng/ml or 50 ng/ml of inhibitor was added 30 min after induction with 50 ng/ml of cAD1 and the traE1 transcript reached a high level (Fig. 6). With an increasing concentration of inhibitor, a higher rate of decrease of traE1 transcript was seen. These results strongly support the hypothesis that the inhibitor is responsible for the suppressive effect of donor density on conjugation, interfering with induction and increasing the rate at which the system is shut off after induction.
FIG 6.
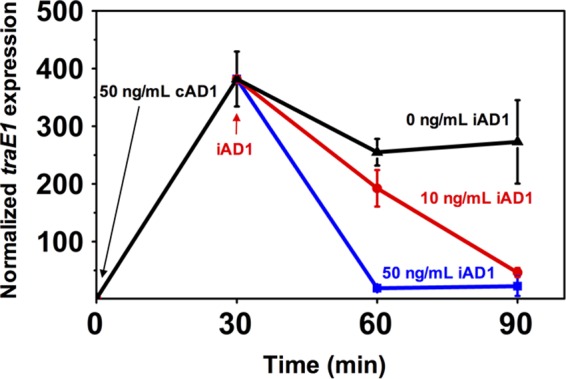
Peptide inhibitor shuts down induction. qRT-PCR was used to measure the dynamic expression of conjugation determinants encoded by traE1. Cells with plasmid pAD1 were induced by 50 ng/ml cAD1 at time zero. The inhibitor iAD1 was added at 30 min, with final concentrations of 0, 10 and 50 ng/ml. Data shown are the averages of results of three biological replicates (error bars are SDs from mean values).
Donor density affects cross-species plasmid transfer.
Numerous Staphylococcus aureus strains, as well as some Enterococcus faecium and Streptococcus gordonii strains, secrete peptides that solicit responses from Enterococcus faecalis strains harboring specific plasmids. Past studies have shown that some of the strains from the aforementioned bacterial species produce cAM373-like peptides, which can solicit a mating response from Enterococcus faecalis strains harboring plasmid pAM373 (39, 40) or vancomycin resistance plasmid pAM368 (4). In order to examine the effect of donor density on interspecies plasmid transfer, a chloramphenicol-resistant variant of Streptococcus gordonii Challis CH1 (Table 1) was used as the model recipient. E. faecalis JH2-2(pAM378, pASM470) was used as the donor strain (40). Because pAM378 can replicate only in E. faecalis, erythromycin (Erm)-resistant plasmid pASM470 was included in the donor strain to serve as a pAM378-dependent mobilizable vector due to its ability to replicate in both E. faecalis and S. gordonii. The donors and recipients were mixed at two different donor-to-recipient ratios (1:1 and 0.01:1) and placed on a filter membrane to mate for 3 h. Interspecies transfer was assayed by selecting for Erm-resistant S. gordonii, indicating acquisition of pAMS470. Results indicated that high donor density led to an overall decrease in conjugation frequency (number of transconjugants per donor) (Fig. 7). The mating mixture with a donor-to-recipient ratio of 1:1 showed a number of transconjugants per donor that was about an order of magnitude lower than that of the mating mixture with a low donor-to-recipient ratio of 0.01:1, reaffirming our hypothesis that high donor densities led to reduced conjugation frequencies, including in the context of cross-species conjugative plasmid transfer. To examine the role of the inhibitor, 500 ng/ml of iAM373 was exogenously added to the mating mixture with the low donor-to-recipient ratio (0.01:1), restoring the effect of increased donor density. An order-of-magnitude reduction in conjugation frequency was observed in the presence of exogenous inhibitor, demonstrating the role of the inhibitor as an indicator for donor density.
FIG 7.
Influence of donor density on interspecies conjugative transfer from E. faecalis to S. gordonii. (A) Different numbers of E. faecalis cells (donors) were mixed with S. gordonii cells (recipients) to achieve D/R ratios of 1:1 and 0.01:1 at time zero. Transconjugants, donors, and recipients were enumerated on selective agar after 3 h of coculture on a membrane filter, and the efficiency of conjugation is represented by the number of transconjugants per donor. (B) E. faecalis cells (donors) were mixed with S. gordonii cells (recipients) at a D/R ratio of 0.01:1 at time zero with and without 500 ng/ml iAM373. Transconjugants, donors, and recipients were enumerated on selective agar after 3 h of coculture on membrane filter, and the efficiency of conjugation is represented by the number of transconjugants per donor. Data shown in panels A and B are averages of results of three biological replicates (error bars are SDs from mean values).
DISCUSSION
The transfer of plasmids among bacteria is associated with bacterial adaptation and evolution and is particularly relevant to the increasing problem of antibiotic resistance. Genetic analysis of vancomycin-resistant isolates of Staphylococcus aureus indicated that the resistance was acquired from vancomycin-resistant E. faecalis via transfer of conjugative plasmids (41, 42). It has been shown in previous studies that donor cells harboring pAD1 and pAM373 can be induced by E. faecium, S. aureus, S. gordonii, and Escherichia coli cells (43, 44), and these plasmids may play a significant role in facilitating the transmission of virulence determinants and antibiotic resistance. Hence, a better understanding of the regulatory mechanisms controlling the conjugation of these plasmids would be highly beneficial. We demonstrated that donor cell densities affect the induction and subsequent shutting down of the conjugation operon in pAD1 and pAM373. Our data indicate that this behavior is a direct manifestation of inhibitor peptide accumulation.
In a mating mixture of donors and recipients, it might be expected that increasing the donor population density would increase the frequency of plasmid transfer. Indeed, it has been shown that Agrobacterium tumefaciens cells carrying conjugative Ti plasmids show a donor-density-dependent increase in transfer, which is mediated by an extracellular quorum-sensing acyl-homoserine lactone autoinducer synthesized by the plasmid-encoded TraI autoinducer synthetase (45, 46). In contrast, we have demonstrated that high donor density reduced the frequency of pheromone-controlled conjugative transfer of plasmids pAD1 and pAM373 in E. faecalis. The reduced frequency of conjugation is a consequence of the lower induction of the conjugation operon due to increased levels of the inhibitor peptide in high-donor-density cultures. This suppressive effect of high donor density on conjugation is similar to the quorum-sensing-controlled pCF10 conjugation (10). Our current results suggest that conjugation in at least 12 different families of pheromone-responsive plasmids (14) is likely to be controlled by two antagonistic signaling molecules, one of which (iCF10, iAD1, iAM373, etc.) serves as a classic quorum-sensing signal for donor population density. This pheromone system also utilizes a mate-sensing signaling molecule (cCF10, cAD1, cAM373, etc.) produced by recipient cells, which acts as a cue for donor cells to induce conjugation when recipients are present at high densities.
The pheromone peptides cCF10, cAD1, and cAM373 are very hydrophobic linear peptides consisting of seven or eight residues processed from chromosome-encoded lipoproteins (see Fig. S1 in the supplemental material). The cAM373-like peptides produced by S. aureus and S. gordonii have a few amino acid substitutions and are likely to show slightly different potencies than the analogous peptide produced by E. faecalis. The inhibitor peptides are the exact same sizes as their corresponding pheromones and are processed from 21- to 23-amino-acid peptide precursors resembling the precursor pheromone sequences (47). They lack any acidic or basic amino acid and are highly lipophilic (18). With the similarity of their gene organizations and overall regulatory structures, the mechanism of induction in the two plasmids is likely to be similar to that of pCF10. The general mechanism considered is depicted in Fig. S4 in the supplemental material. While the inducing concentration for pAD1 is very similar to that for pCF10, it is much higher for pAM373. Hence, we hypothesize that the binding behavior of peptides with TraA and the subsequent binding of the TraA complexes with the operator site of the conjugation operon are likely to be different between the two plasmids. We have recently measured the binding strength of cCF10 and iCF10 to PrgX and the peptide-PrgX complex to the operator and determined that both cCF10 and iCF10 have very strong binding constants to PrgX and that both peptide-PrgX complexes have similar binding affinities toward DNA (Y. Chen, A. Bandyopadhyay, B. K. Kozlowicz, H. A. H. Haemig, A. Tai, W.-S. Hu, and G. M. Dunny, unpublished data). This newly determined parameter value set is slightly different from that originally published (10). We applied the mathematical model that was developed for pCF10 to the other two plasmid systems. Considering the similarity of the overall dynamic behaviors and induction concentrations between pAD1 and pCF10, the values of the kinetic parameters of pCF10 are used for pAD1. The parameter values used in the simulation are shown in Table S2 in the supplemental material. Simulation results qualitatively described the induction behavior of pAD1 in response to 50 ng/ml cAD1, i.e., higher induction and slower shutdown (see Fig. S5A in the supplemental material).
By analyzing the two binding events of peptide to TraA and the peptide-TraA complex to the operator and comparing the model simulations and experimental observations for pAM373 (see Section S2 and Fig. S5B and C in the supplemental material), we postulate that the binding affinity of the cAM373-TraA complex to the operator site is likely to be 100 times lower than that for the pAD1 system, while the binding of cAM373 to TraA is at a level similar to that in the pAD1 system. The model simulation using the modified binding constant qualitatively described the fast reduction in conjugation gene expression postinduction as seen in the pAM373 system (see Fig. S5C). Interestingly, in simulating the effect of donor cell concentration, the model predicted a higher sensitivity to donor concentration in pAM373 than in the pAD1 system (see Fig. S6). With the same fold increase in donor cell density, pAM373 shuts down the induction faster than does pAD1. This is the same trend as shown in Fig. 3B.
We attribute the reduction in conjugation gene expression and the effect of donor density to the accumulation of the inhibitor peptides (iAD1 and iAM373) in culture. The presence of inhibitor peptide has been shown in all three systems (17, 18, 49). Additionally, iAD1 has been shown to accumulate in culture over time (50), although at the time it was not known to mediate the donor density effect. A codon deletion in the inhibitor coding sequence producing an inactive form of iCF10 in the pCF10 system exhibited a derepressed phenotype with a high expression of conjugation transcript and abolished the effect of donor density, and the addition of the inhibitor progressively restored the normal induction behavior and the effect of donor density (10). Given the similarity of the three systems, a similar role of the inhibitor is likely to be at play in pAD1 and pAM373.
This study is an illustration of the versatility of the regulatory architecture of the three conjugative plasmid systems. The organization of the regulatory elements, formed by the quartet of inducer peptide, antagonistic peptide, the repressor protein, and the operator, are similar in all three systems at structural levels. However, they are rather different at DNA and protein sequence levels. Interestingly, through differences in their binding constants among the four players in the quartet, the system can also have different kinetic behaviors.
Conclusions.
Conjugative plasmid transfer in E. faecalis has been studied for more than 3 decades, and the primary role of the plasmid-encoded inhibitor peptide has been considered to be the blocking of self-induction by an endogenous pheromone in donor cells. Here, we demonstrated a critical role for the inhibitor peptides (iAD1 and iAM373) in reducing the ability of donors harboring plasmids pAD1 and pAM373 to initiate a mating response at high donor density. The concentration of the inhibitor peptide serves as an indicator for donor density and functions as a “self-sensing” signaling molecule used by the donor cells to calibrate its mating response, by titrating the levels of pheromone and inhibitor. This kind of dual sensing mechanism may possibly help maintain two subpopulations of bacterial cells: a plasmid-free subpopulation with reduced metabolic burden and a subpopulation of plasmid-bearing cells with increased metabolic burden but an added fitness benefit. By manipulating their signaling mechanisms, thus perturbing the balance of subpopulations, one may be able to devise a strategy of controlling the population to combat the spread of antibiotic resistance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Keith Weaver, University of South Dakota, and Susan Flannagan, University of Michigan, for supplying isolates of E. faecalis harboring the plasmids of interest. We also acknowledge the help of Justinus Hartoyo, Siyu Zhu, and Qinnan Zhang for their help and assistance with the experiments.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00363-16.
REFERENCES
- 1.Murray BE. 1997. Vancomycin-resistant enterococci. Am J Med 102:284–293. doi: 10.1016/S0002-9343(99)80270-8. [DOI] [PubMed] [Google Scholar]
- 2.Klevens RM, Edwards JR, Richards CL, Horan TC, Gaynes RP, Pollock DA, Cardo DM. 2007. Estimating health care-associated infections and deaths in US hospitals, 2002. Public Health Rep 122:160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clewell DB, Francia MV, Flannagan SE, An FY. 2002. Enterococcal plasmid transfer: sex pheromones, transfer origins, relaxases, and the Staphylococcus aureus issue. Plasmid 48:193–201. doi: 10.1016/S0147-619X(02)00113-0. [DOI] [PubMed] [Google Scholar]
- 4.Showsh SA, De Boever EH, Clewell DB. 2001. Vancomycin resistance plasmid in Enterococcus faecalis that encodes sensitivity to a sex pheromone also produced by Staphylococcus aureus. Antimicrob Agents Chemother 45:2177–2178. doi: 10.1128/AAC.45.7.2177-2178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewell DB. 1990. Movable genetic elements and antibiotic-resistance in enterococci. Eur J Clin Microbiol Infect Dis 9:90–102. doi: 10.1007/BF01963632. [DOI] [PubMed] [Google Scholar]
- 6.Carniol K, Gilmore MS. 2004. Signal transduction, quorum-sensing, and extracellular protease activity in Enterococcus faecalis biofilm formation. J Bacteriol 186:8161–8163. doi: 10.1128/JB.186.24.8161-8163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres OR, Korman RZ, Zahler SA, Dunny GM. 1991. The conjugative transposon Tn925—enhancement of conjugal transfer by tetracycline in Enterococcus faecalis and mobilization of chromosomal genes in Bacillus subtilis and E. faecalis. Mol Gen Genet 225:395–400. [DOI] [PubMed] [Google Scholar]
- 8.Dunny GM, Leonard BA. 1997. Cell-cell communication in gram-positive bacteria. Annu Rev Microbiol 51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama J, Ruhfel RE, Dunny GM, Isogai A, Suzuki A. 1994. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J Bacteriol 176:7405–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee A, Cook LCC, Shu CC, Chen YQ, Manias DA, Ramkrishna D, Dunny GM, Hu WS. 2013. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc Natl Acad Sci U S A 110:7086–7090. doi: 10.1073/pnas.1212256110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee A, Johnson CM, Shu CC, Kaznessis YN, Ramkrishna D, Dunny GM, Hu WS. 2011. Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc Natl Acad Sci U S A 108:9721–9726. doi: 10.1073/pnas.1101569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook L, Chatterjee A, Barnes A, Yarwood J, Hu WS, Dunny G. 2011. Biofilm growth alters regulation of conjugation by a bacterial pheromone. Mol Microbiol 81:1499–1510. doi: 10.1111/j.1365-2958.2011.07786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francia MV, Clewell DB. 2002. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol Microbiol 45:375–395. doi: 10.1046/j.1365-2958.2002.03007.x. [DOI] [PubMed] [Google Scholar]
- 14.Clewell D, Dunny G. 2002. Conjugation and genetic exchange in enterococci, p 265–300. In Gilmore M, Clewell D, Courvalin P, Dunny G, Murray B, Rice L (ed), The enterococci. ASM Press, Washington, DC. doi: 10.1128/9781555817923.ch7. [DOI] [Google Scholar]
- 15.Mori M, Sakagami Y, Narita M, Isogai A, Fujino M, Kitada C, Craig RA, Clewell DB, Suzuki A. 1984. Isolation and structure of the bacterial sex pheromone, Cad1, that induces plasmid transfer in Streptococcus faecalis. FEBS Lett 178:97–100. doi: 10.1016/0014-5793(84)81248-X. [DOI] [PubMed] [Google Scholar]
- 16.Flannagan SE, Clewell DB. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol Microbiol 44:803–817. doi: 10.1046/j.1365-2958.2002.02922.x. [DOI] [PubMed] [Google Scholar]
- 17.Clewell DB, Pontius LT, An FY, Ike Y, Suzuki A, Nakayama J. 1990. Nucleotide sequence of the sex pheromone inhibitor (Iad1) determinant of Enterococcus faecalis conjugative plasmid Pad1. Plasmid 24:156–161. doi: 10.1016/0147-619X(90)90019-9. [DOI] [PubMed] [Google Scholar]
- 18.Nakayama J, Ono Y, Suzuki A. 1995. Isolation and structure of the sex pheromone inhibitor, iAM373, of Enterococcus faecalis. Biosci Biotechnol Biochem 59:1358–1359. doi: 10.1271/bbb.59.1358. [DOI] [PubMed] [Google Scholar]
- 19.Clewell DB. 2007. Properties of Enterococcus faecalis plasmid pAD1, a member of a widely disseminated family of pheromone-responding, conjugative, virulence elements encoding cytolysin. Plasmid 58:205–227. doi: 10.1016/j.plasmid.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 20.De Boever EH, Clewell DB, Fraser CM. 2000. Enterococcus faecalis conjugative plasmid pAM373: complete nucleotide sequence and genetic analyses of sex pheromone response. Mol Microbiol 37:1327–1341. doi: 10.1046/j.1365-2958.2000.02072.x. [DOI] [PubMed] [Google Scholar]
- 21.Shokeen S, Johnson CM, Greenfield TJ, Manias DA, Dunny GM, Weaver KE. 2010. Structural analysis of the Anti-Q-Qs interaction: RNA-mediated regulation of E. faecalis plasmid pCF10 conjugation. Plasmid 64:26–35. doi: 10.1016/j.plasmid.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson CM, Manias DA, Haemig HA, Shokeen S, Weaver KE, Henkin TM, Dunny GM. 2010. Direct evidence for control of the pheromone-inducible prgQ operon of Enterococcus faecalis plasmid pCF10 by a countertranscript-driven attenuation mechanism. J Bacteriol 192:1634–1642. doi: 10.1128/JB.01525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson CM, Haemig HH, Chatterjee A, Wei-Shou H, Weaver KE, Dunny GM. 2011. RNA-mediated reciprocal regulation between two bacterial operons is RNase III dependent. mBio 2:e00189-11. doi: 10.1128/mBio.00189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozawa Y, De Boever EH, Clewell DB. 2005. Enterococcus faecalis sex pheromone plasmid pAM373: analyses of TraA and evidence for its interaction with RpoB. Plasmid 54:57–69. doi: 10.1016/j.plasmid.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 25.do Carmo de Freire Bastos M, Tomita H, Tanimoto K, Clewell DB. 1998. Regulation of the Enterococcus faecalis pAD1-related sex pheromone response: analyses of traD expression and its role in controlling conjugation functions. Mol Microbiol 30:381–392. doi: 10.1046/j.1365-2958.1998.01074.x. [DOI] [PubMed] [Google Scholar]
- 26.Piper KR, Vonbodman SB, Farrand SK. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 27.Fuqua C, Winans SC. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J Bacteriol 178:435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. 2012. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi K, Brown CK, Gu ZY, Kozlowicz BK, Dunny GM, Ohlendorf DH, Earhart CA. 2005. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc Natl Acad Sci U S A 102:18596–18601. doi: 10.1073/pnas.0506163102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocha-Estrada J, Aceves-Diez AE, Guarneros G, de la Torre M. 2010. The RNPP family of quorum-sensing proteins in Gram-positive bacteria. Appl Microbiol Biotechnol 87:913–923. doi: 10.1007/s00253-010-2651-y. [DOI] [PubMed] [Google Scholar]
- 31.Parashar V, Aggarwal C, Federle MJ, Neiditch MB. 2015. Rgg protein structure-function and inhibition by cyclic peptide compounds. Proc Natl Acad Sci U S A 112:5177–5182. doi: 10.1073/pnas.1500357112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleuchot B, Gitton C, Guillot A, Vidic J, Nicolas P, Besset C, Fontaine L, Hols P, Leblond-Bourget N, Monnet V, Gardan R. 2011. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol Microbiol 80:1102–1119. doi: 10.1111/j.1365-2958.2011.07633.x. [DOI] [PubMed] [Google Scholar]
- 33.Cook LC, Federle MJ. 2014. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev 38:473–492. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding Y, Lawrence CE. 2003. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res 31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirth R. 1994. The sex pheromone system of Enterococcus faecalis. More than just a plasmid-collection mechanism? Eur J Biochemistry 222:235–246. [DOI] [PubMed] [Google Scholar]
- 36.Chandler JR, Flynn AR, Bryan EM, Dunny GM. 2005. Specific control of endogenous cCF10 pheromone by a conserved domain of the pCF10-encoded regulatory protein PrgY in Enterococcus faecalis. J Bacteriol 187:4830–4843. doi: 10.1128/JB.187.14.4830-4843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muscholl-Silberhorn A. 1999. Cloning and functional analysis of Asa373, a novel adhesin unrelated to the other sex pheromone plasmid-encoded aggregation substances of Enterococcus faecalis. Mol Microbiol 34:620–630. doi: 10.1046/j.1365-2958.1999.01631.x. [DOI] [PubMed] [Google Scholar]
- 38.De Boever EH, Clewell DB. 2001. The Enterococcus faecalis pheromone-responsive plasmid pAM373 does not encode an entry exclusion function. Plasmid 45:57–60. doi: 10.1006/plas.2000.1494. [DOI] [PubMed] [Google Scholar]
- 39.Clewell DB, An FY, White BA, Gawron-Burke C. 1985. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J Bacteriol 162:1212–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vickerman MM, Flannagan SE, Jesionowski AM, Brossard KA, Clewell DB, Sedgley CM. 2010. A genetic determinant in Streptococcus gordonii Challis encodes a peptide with activity similar to that of enterococcal sex pheromone cAM373, which facilitates intergeneric DNA transfer. J Bacteriol 192:2535–2545. doi: 10.1128/JB.01689-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flannagan SE, Chow JW, Donabedian SM, Brown WJ, Perri MB, Zervos MJ, Ozawa Y, Clewell DB. 2003. Plasmid content of a vancomycin-resistant Enterococcus faecalis isolate from a patient also colonized by Staphylococcus aureus with a VanA phenotype. Antimicrob Agents Chemother 47:3954–3959. doi: 10.1128/AAC.47.12.3954-3959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 43.Firth N, Fink PD, Johnson L, Skurray RA. 1994. A lipoprotein signal peptide encoded by the staphylococcal conjugative plasmid pSK41 exhibits an activity resembling that of Enterococcus faecalis pheromone Cad1. J Bacteriol 176:5871–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berg T, Firth N, Skurray RA. 1997. Enterococcal pheromone-like activity derived from a lipoprotein signal peptide encoded by a Staphylococcus aureus plasmid. Adv Exp Biol Med 418:1041–1044 . doi: 10.1007/978-1-4899-1825-3_245. [DOI] [PubMed] [Google Scholar]
- 45.White CE, Winans SC. 2007. Cell-cell communication in the plant pathogen Agrobacterium tumefaciens. Philos Trans R Soc Lond B Biol Sci 362:1135–1148. doi: 10.1098/rstb.2007.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lang J, Faure D. 2014. Functions and regulation of quorum-sensing in Agrobacterium tumefaciens. Front Plant Sci 5:14. doi: 10.3389/fpls.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clewell DB, An FY, Flannagan SE, Antiporta M, Dunny GM. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol Microbiol 35:246–247. doi: 10.1046/j.1365-2958.2000.01687.x. [DOI] [PubMed] [Google Scholar]
- 48.Reference deleted.
- 49.Buttaro BA, Antiporta MH, Dunny GM. 2000. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J Bacteriol 182:4926–4933. doi: 10.1128/JB.182.17.4926-4933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clewell DB, An FY, Mori M, Ike Y, Suzuki A. 1987. Streptococcus faecalis sex pheromone (Cad1) response—evidence that the peptide inhibitor excreted by Pad1-containing cells may be plasmid determined. Plasmid 17:65–68. doi: 10.1016/0147-619X(87)90011-4. [DOI] [PubMed] [Google Scholar]
- 51.Frank KL, Colomer-Winter C, Grindle SM, Lemos JA, Schlievert PM, Dunny GM. 2014. Transcriptome analysis of Enterococcus faecalis during mammalian infection shows cells undergo adaptation and exist in a stringent response state. PLoS One 9(12):e115839. doi: 10.1371/journal.pone.0115839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.