ABSTRACT
Minimizing the use of antibiotics in the food production chain is essential for limiting the development and spread of antibiotic-resistant bacteria. One alternative intervention strategy is the use of probiotic bacteria, and bacteria of the marine Roseobacter clade are capable of antagonizing fish-pathogenic vibrios in fish larvae and live feed cultures for fish larvae. The antibacterial compound tropodithietic acid (TDA), an antiporter that disrupts the proton motive force, is key in the antibacterial activity of several roseobacters. Introducing probiotics on a larger scale requires understanding of any potential side effects of long-term exposure of the pathogen to the probionts or any compounds they produce. Here we exposed the fish pathogen Vibrio anguillarum to TDA for several hundred generations in an adaptive evolution experiment. No tolerance or resistance arose during the 90 days of exposure, and whole-genome sequencing of TDA-exposed lineages and clones revealed few mutational changes, compared to lineages grown without TDA. Amino acid-changing mutations were found in two to six different genes per clone; however, no mutations appeared unique to the TDA-exposed lineages or clones. None of the virulence genes of V. anguillarum was affected, and infectivity assays using fish cell lines indicated that the TDA-exposed lineages and clones were less invasive than the wild-type strain. Thus, long-term TDA exposure does not appear to result in TDA resistance and the physiology of V. anguillarum appears unaffected, supporting the application of TDA-producing roseobacters as probiotics in aquaculture.
IMPORTANCE It is important to limit the use of antibiotics in our food production, to reduce the risk of bacteria developing antibiotic resistance. We showed previously that marine bacteria of the Roseobacter clade can prevent or reduce bacterial diseases in fish larvae, acting as probiotics. Roseobacters produce the antimicrobial compound tropodithietic acid (TDA), and we were concerned regarding whether long-term exposure to this compound could induce resistance or affect the disease-causing ability of the fish pathogen. Therefore, we exposed the fish pathogen Vibrio anguillarum to increasing TDA concentrations over 3 months. We did not see the development of any resistance to TDA, and subsequent infection assays revealed that none of the TDA-exposed clones had increased virulence toward fish cells. Hence, this study supports the use of roseobacters as a non-risk-based disease control measure in aquaculture.
INTRODUCTION
Aquaculture has been one of the fastest growing protein-producing sectors for decades, providing high-quality protein to the growing world population (1); however, a key constraint in the aquaculture industry is microbial infectious diseases. More than 50% of such diseases are caused by bacteria, of which members of the Vibrio genus are some of the most prevalent (2, 3). The species Vibrio anguillarum alone is known to infect more than 50 different fish species (4). Antibiotics are used in the treatment of bacterial infections in aquaculture, and they have also been used as prophylactic measures, especially in smaller aquaculture facilities and in developing countries (5, 6). However, increases in antibiotic resistance and the associated health risks have led to a search for alternative means of treatment (6). Vaccination of juvenile and adult fish has reduced the use of antibiotics in the production of several finfish (7); however, it is not possible to vaccinate fish larvae, due to their immature immune systems, which makes them particularly vulnerable to infectious diseases (8).
The use of probiotic bacteria as sustainable controls for pathogenic bacteria in larviculture has been studied for several years (9–11), and two genera from the Roseobacter clade, i.e., Phaeobacter and Ruegeria, are especially promising as probiotics for marine larviculture (12–16). Phaeobacter and Ruegeria strains have repeatedly been isolated from aquaculture units and are harmless to fish larvae and their live feed (12–16). The probiotic (antibacterial) effects of Phaeobacter and Ruegeria are predominantly caused by their production of the dual-sulfur tropone-derived compound tropodithietic acid (TDA) (16–19). TDA-producing Phaeobacter and Ruegeria strains can prevent vibriosis in cod larvae (16) and reduce mortality rates for turbot larvae (15) and Artemia (20) challenged with V. anguillarum and V. harveyi, respectively.
TDA is a broad-spectrum antibacterial that is bactericidal and inhibits the growth of several human and fish pathogens (16, 21–23) but has very low toxicity in Caenorhabditis elegans and Artemia (24). Recently, it was demonstrated that TDA is cytotoxic against N2a cells and OLN-93 cells of the mammalian nervous system and several cancer cell lines, respectively (25, 26). Since TDA has pronounced antibacterial activity, the development of resistance to TDA could be a concern, especially following long-term use. Attempts to select for TDA resistance and tolerance in pathogenic bacteria have not been successful (22), suggesting that TDA resistance does not arise easily, likely due to a highly conserved target (or targets). Very recently, Wilson et al. demonstrated that cell membranes and the proton motive force (PMF) were targets of TDA, and TDA was suggested to function as an electroneutral proton antiporter, facilitating a one-to-one exchange of H+ for a single charged metal ion and thus disrupting the transmembrane proton gradient (ΔpH) (26). Also, genes conferring resistance to TDA were found in Phaeobacter inhibens, and Escherichia coli became resistant when harboring tdaR on a conjugative plasmid (26).
While those recent studies clearly indicated that resistance to TDA does not develop easily, e.g., by single point mutations, it is not known how long-term exposure to TDA can affect target cells. The long-term effects of other antibacterial compounds, such as antimicrobial peptides (AMPs), have been studied using adaptive laboratory evolution (ALE) experiments (27–31), and we chose such an approach to evaluate the long-term exposure of vibrios to TDA. Several studies have demonstrated that antibiotics can induce mutations in exposed strains (32, 33) and affect gene expression and phenotypes in exposed bacteria, e.g., inducing biofilm formation or the expression of virulence genes in pathogens (32). Trimethoprim, which at high concentrations is lethal for Burkholderia thailandensis E264, functioned as a global activator of its secondary metabolism, activating at least five biosynthetic gene clusters when added at subinhibitory concentrations (34). In several pathogenic bacteria, subinhibitory concentrations of antibiotics affect virulence genes and behavior (35–37); for example, subinhibitory concentrations of nafcillin were found to increase toxin production in Staphylococcus aureus, whereas clindamycin and linezolid decreased toxin production (38). Infection of fish by V. anguillarum depends on a series of virulence factors, including chemotaxis, motility, adhesion, and siderophore and exotoxin production (4); therefore, in the present study, we also determined whether long-term exposure of V. anguillarum to TDA affected the infectivity of the bacteria, as measured in fish cell lines.
Therefore, we exposed the fish pathogen V. anguillarum strain 90-11-286 to sublethal concentrations of TDA over the course of 220 to 280 generations, in an adaptive laboratory evolution experiment. Subsequently, we assessed phenotypes (resistance and infectivity) as well as genetic changes (single-nucleotide polymorphisms [SNPs]) and deletion-insertion polymorphisms [DIPs]).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All adaptive evolution experiments were performed with Vibrio anguillarum strain 90-11-286 (39, 40), serotype O1, isolated from diseased rainbow trout (Oncorhynchus mykiss) from a Danish fish farm. The strain was chosen due to its high level of pathogenicity for cod, halibut, and turbot larvae (A. Rønneseth, P. D'Alvise, Ø. Tønnesen, G. Haugland, T. Grotkjær, K. Engell-Sørensen, L. Nørremark, Ø. Bergh, L. Gram, and H. I. Wergeland, unpublished data). Strain 90-11-286 was grown in half-strength YTSS (½YTSS) (0.2% yeast extract, 0.125% tryptone, 2% Sigma sea salts) broth (41). Streaking and plating were performed on ½YTSS-agar plates at 25°C. Tropodithietic acid (BioViotica, Dransfeld, Germany) was dissolved in dimethyl sulfoxide (DMSO) (1.5 mg/ml) by vortex-mixing for 10 s and kept at −80°C. Daily working solutions were prepared from the TDA frozen stock solution by diluting the solution in sterile ½YTSS medium by vortex-mixing for 10 s, sonicating the mixture with ultrasound twice for 30 s, and vortex-mixing for 10 s. Stock cultures of the wild-type strain and isolates from the TDA-exposed lineage were stored at −80°C in a mixture of 4% (wt/vol) glycerol, 0.5% (wt/vol) glucose, 2% (wt/vol) skim milk powder, and 3% (wt/vol) tryptone soy powder (42), whereas lineage populations were stored at −80°C in 25% (wt/vol) glycerol.
Continuous-exposure experiment.
Continuous exposure to TDA was carried out in an adaptive evolution experimental setup, with the aim of increasing TDA concentrations to induce resistance or tolerance. Twenty microliters of a bacterial suspension of a single colony of V. anguillarum 90-11-286 was inoculated into 10 parallel 2-ml cultures. For 5 days, the lineages were diluted (1%) in fresh medium every 24 h. Seven of the lineages were subsequently diluted in ½YTSS medium supplemented with TDA, and the remaining three were unexposed control lineages (DMSO was added instead of TDA solution). After five dilutions at a constant TDA concentration (exposed lineages), the concentration was increased. The starting concentration was 1/16 of the MIC, i.e., 0.8 μg ml−1, and this was increased to 1.75× MIC during the course of the experiment. The concentration steps were 0.8 μg ml−1 (1/16× MIC), 3.1 μg ml−1 (1/4× MIC), 6.25 μg ml−1 (1/2× MIC), 12.5 μg ml−1 (1× MIC), 15.6 μg ml−1 (1.25× MIC), 18.75 μg ml−1 (1.5× MIC), and 21.9 μg ml−1 (1.75× MIC). Reinoculations were not performed for cultures exposed to TDA concentrations above 1× MIC if growth was not observed. In those cases, the reinoculations were postponed until turbidity was observed. Aliquots of the 10 lineages were deposited at −80°C prior to an increase in the TDA concentration. If a lineage failed to grow at an increased TDA concentration, even after repeated reinoculations from its last deposited stock, then it was terminated. The total numbers of passages were 33 to 42 for the 10 lineages, which were equivalent to approximately 220 to 280 generations, assuming six to seven generations per passage. At the end of the experiment, all lineages were plated to allow isolation of individual clones from each lineage. Five colony isolates were randomly selected from each lineage population at 1.5× MIC to 1.75× MIC and were preserved as frozen stocks.
Determination of MICs.
MICs were determined using the microdilution method (43). A 50 μg ml−1 TDA stock solution in DMSO was prepared and 1:2 serial dilutions were made, giving a final concentration range of 25 to 0.4 μg ml−1 in the wells. A control DMSO dilution series was prepared accordingly. MIC testing was performed with three biological replicates. The bacteria (lineages or clones) were revived from frozen cultures and grown overnight in ½YTSS medium at 20°C.
Stability of TDA tolerance.
Cultures were revived from frozen stocks in ½YTSS medium with 6.75 μg ml−1 TDA (one-half the wild-type MIC) at 20°C. The stability of the TDA tolerance was assessed by determining the MICs of bacterial populations as described above but with increased resolution. Four TDA-½YTSS working solutions were prepared, with 50 μg ml−1, 43.75 μg ml−1, 37.5 μg ml−1, and 31.25 μg ml−1 TDA, and 1:2 serial dilutions were made from the working solutions to give a final range of 25 to 4.7 μg ml−1 in the wells. The MIC assay results were read after 24 h and 72 h.
Whole-genome sequencing.
Twenty-eight isolates from TDA-exposed lineages (7 lineages and 21 clones) and 12 isolates from control lineages (3 lineages and 9 clones) were subjected to whole-genome sequencing. Genomic DNA was extracted from each using phenol-chloroform-isoamyl alcohol and then was precipitated with isopropanol. Samples were treated with RNase before quantification and quality analysis using a 1% agarose gel, a NanoDrop spectrophotometer (Saveen Werner, Sweden), and a Qubit 2.0 analyzer (Invitrogen, United Kingdom).
The genome of the ancestral wild-type V. anguillarum strain 90-11-286 was sequenced using a combined sequencing approach with PacBio and Illumina platforms, allowing the assembly of a fully closed high-quality genome. A SMRTbell template library was prepared according to the instructions from Pacific Biosciences (Menlo Park, CA, USA), following the procedure and checklist for 20-kb template preparation using the BluePippin size selection system. Briefly, for preparation of 15-kb libraries, 8 μg of genomic DNA was sheared using g-Tubes from Covaris (Woburn, MA, USA), according to the manufacturer's instructions. DNA was end repaired and ligated overnight to hairpin adapters by using components from DNA/polymerase binding kit P6 (Pacific Biosciences). Reactions were carried out according to the manufacturer's instructions. BluePippin size selection to 7,000 kb was performed according to the manufacturer's instructions (Sage Science, Beverly, MA, USA). Conditions for annealing of sequencing primers and binding of polymerase to the purified SMRTbell template were assessed with the calculator in RS Remote (Pacific Biosciences). SMRT sequencing was carried out with the PacBio RS II system (Pacific Biosciences), taking one 240-min movie for each SMRT cell. In total, two SMRT cells were run and a total of 56,839 reads, with a mean read length of 13,006 bp, were obtained. SMRT cell data were assembled using the RS_HGAP_Assembly.3 protocol included in SMRT Portal version 2.3.0, using default parameters. The assembly revealed one circular chromosome. The validity of the assembly was checked using the RS_Bridgemapper.1 protocol. Each of the replicons was circularized independently, and artificial redundancies at the ends of the contigs were removed; the chromosome was adjusted to dnaA as the first gene. Subsequently, each genome was error corrected by mapping of Illumina reads onto finished genomes using BWA software (44), with subsequent variant and consensus calling using CLC Genomics Workbench 7 (Qiagen, Aarhus, Denmark). A consensus concordance of QV60 could be confirmed for the genome. Finally, annotation was generated using Prokka 1.8 (45).
Five-hundred-base-pair insertion libraries were prepared for all 40 lineages and clones. These libraries were subjected to 100-bp paired-end sequencing, using the Illumina HiSeq2000 platform, at Beijing Genomic Institute. The 40 isolates were mapped onto the V. anguillarum 90-11-286 reference genome. Subsequently, single-nucleotide polymorphisms (SNPs) and deletion-insertion polymorphisms (DIPs) with frequencies above 70% in the reads were detected using CLC Genomic Workbench 8.0.2 (Qiagen).
Motility observations.
Several isolates had mutations in the fliM gene, which is important for flagellate motility in V. anguillarum. Therefore, motility was assessed by microscopy (×1,000 magnification, using an Olympus BX51 microscope) of broth cultures grown in ½YTSS medium for 24 h at 25°C.
Fish cell line infection assay.
The effects of the wild-type strain, eight TDA-exposed isolates, and four control isolates on the fish cell line CHSE-214 (catalogue no. 91041114; Sigma) (46) were assessed in three independent cell infection assays. CHSE-214 cells were grown for 7 days at 20°C in Leibovitz L-15 medium with glutamine (Sigma-Aldrich), with 10% heat-inactivated (56°C for 30 min) fetal bovine serum (FBS) (Sigma-Aldrich) and 1% antibiotic-antimycotic solution (Sigma-Aldrich), in a 75-cm2 flask until cells reached an 80% confluent cell layer. After removal of the growth medium, the cells were washed with 5 ml Hanks' balanced salt solution (HBSS) (catalogue no. H4641; Sigma). HBSS was removed, and the cells were trypsinized with 1 ml trypsin-EDTA (catalogue no. 59428C; Sigma) and incubated for 10 min. Cells were dislodged and suspended in 2.5 ml fresh medium. Two hundred microliters of CHSE-214 cell suspension (containing 1,000 to 10,000 cells ml−1) was transferred into each well of a six-well test plate with 3 ml fresh medium. After 7 to 9 days of incubation, when a complete confluent cell layer had formed, the cells were washed three times with fresh Hanks' balanced salt solution (HBSS) to remove the antibiotic-antimycotic solution. TDA-exposed isolates were grown for 24 h. Subsequently, the medium was removed, bacteria were washed with HBSS, and a bacterial suspension at an optical density at 600 nm (OD600) of 1 was prepared. Three milliliters of a 10−4 HBSS dilution of the bacterial suspension was transferred into each of the wells, producing a bacterial cell density of approximately 3 × 104 CFU ml−1. After 24 h of incubation at 20°C, the suspension was removed and the cells were trypsinized with 1 ml trypsin-EDTA. After 10 min, 250 μl FBS was added to inactivate the trypsin-EDTA, and the cells were dislodged. Cells were then washed three times in phosphate-buffered saline (PBS) (400 × g for 1 min at 5°C), and the pellet was resuspended in 500 μl PBS. Cell numbers were determined in a 100-μm Neubauer improved counting chamber (Assistent) and live/dead stained using a Cellstain double staining kit (Sigma-Aldrich), as described by the manufacturer. To assess CHSE-214 killing, microscopy of the live/dead-stained cells was performed at a magnification of ×100, using an Olympus BX51 fluorescence microscope. A counting chamber was used to define the area. All infection trials were performed in triplicate, assessing 70 to 400 cells per sample.
Statistics.
One-way analysis of variance (ANOVA) tests (n = 3, df = 12, α ≤ 0.05) were performed with the data obtained from each of the three infection assays, to estimate the significance of any observed differences between the treated strains (TDA-exposed or control strains) and the wild-type strain. Gaussian distributions were confirmed prior to the tests, and the analyses were performed using GraphPad Prism 6.
Accession number(s).
The complete genome of V. anguillarum strain 90-11-286 was deposited in NCBI GenBank under accession number CP011460 for chromosome I and accession number CP011461 for chromosome II. The whole-genome shotgun project was deposited in NCBI SRA under accession number SRP072381.
RESULTS
Attempts to select for TDA-resistant V. anguillarum.
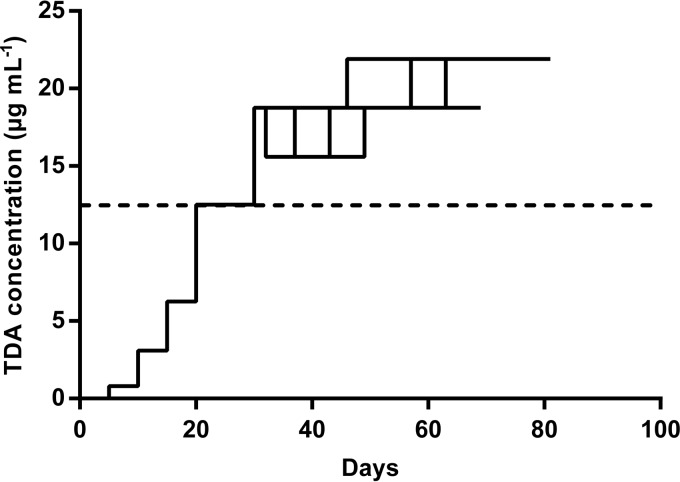
The adaptive evolution process was started at a TDA concentration of 1/16 times the wild-type MIC, which was measured to be 12.5 μg ml−1, and lineages were slowly taken to higher concentrations. After 33 to 42 reinoculations, corresponding to 220 to 280 generations, the maximum concentrations of TDA allowing growth were 1.5× MIC to 1.75× MIC (18.8 to 21.9 μg ml−1); it was not possible, despite repeated attempts, to grow the lineages at higher concentrations. Four of the seven TDA-exposed lineages (lineages 2, 3, 4, and 5) reached 1.75× MIC, and three (lineages 1, 6, and 7) reached 1.5× MIC (Fig. 1). The growth rates clearly decreased at TDA concentrations higher than the wild-type MIC, and periods of 2 to 3 days were required to reach visible turbidity, compared to 1 day at the MIC. Control lineages treated only with DMSO were reinoculated 30 times, corresponding to approximately 210 generations. The MIC values of clones from the control lineages were 12.5 μg ml−1 TDA, corresponding to the wild-type MIC.
FIG 1.
Attempt to select for TDA-tolerant or TDA-resistant V. anguillarum strains in an adaptive laboratory evolution experiment. Solid lines, individual lineages, of which 3 of 7 reached 1.5 times the wild-type MIC (18.75 μg/ml) and the rest reached 1.75 times the wild-type MIC (21.9 μg/ml). Dashed line, wild-type MIC (12.5 μg/ml TDA).
Stability of TDA tolerance.
To determine whether the slight TDA tolerance of the lineages was stable and whether clonal variation occurred, the MICs of the lineages and individual clones were determined. Twenty-four-hour MIC assays with TDA concentrations ranging from 2 times the wild-type MIC to 0.375 times the wild-type MIC revealed that all clones showed the same MIC, i.e., 12.5 μg ml−1, corresponding to the wild-type MIC. After an additional 48 h, a total of 26 clones in addition to the wild-type strain were able to grow at slightly higher concentrations than the wild-type MIC (Table 1). The MIC was based on two separate experiments, with three replicates per clone or lineage.
TABLE 1.
MICs of TDA after 24 h and 72 h for wild-type 90-11-286 strain and TDA-exposed (T) and DMSO-exposed (C) lineages and clones
| Clone | TDA MIC (μg/ml) |
|
|---|---|---|
| After 24 h | After 72 h | |
| 90-11-286 | 12.5 | 15.6 |
| C1 | 10.9–12.5 | 12.5 |
| C1-1 | 12.5 | 15.6 |
| C1-2 | 12.5 | 15.6 |
| C1-3 | 12.5 | 15.6 |
| C2 | 12.5 | 15.6 |
| C2-1 | 12.5 | 15.6 |
| C2-2 | 9.4–12.5 | 15.6 |
| C2-3 | 12.5 | 15.6 |
| C3 | 12.5 | 12.5 |
| C3-1 | 12.5 | 12.5 |
| C3-2 | 12.5 | 12.5–15.6 |
| C3-3 | 12.5 | 12.5 |
| T1 | 10.9–12.5 | 15.6 |
| T1-1 | 10.9–12.5 | 15.6 |
| T1-2 | 10.9–12.5 | 15.6 |
| T1-3 | 10.9–12.5 | 15.6 |
| T2 | 10.9–12.5 | 15.6 |
| T2-1 | 10.9–12.5 | 15.6 |
| T2-2 | 10.9–12.5 | 15.6 |
| T2-3 | 10.9–12.5 | 15.6 |
| T3 | 12.5 | 12.5 |
| T3-1 | 12.5 | 12.5–15.6 |
| T3-2 | 12.5 | 12.5 |
| T3-3 | 12.5 | 12.5 |
| T4 | 12.5 | 12.5–15.6 |
| T4-1 | 12.5 | 12.5 |
| T4-2 | 12.5 | 12.5 |
| T4-3 | 12.5 | 12.5 |
| T5 | 10.9–12.5 | 15.6 |
| T5-1 | 12.5 | 15.6 |
| T5-2 | 12.5 | 12.5 |
| T5-3 | 12.5 | 12.5 |
| T6 | 12.5 | 12.5 |
| T6-1 | 12.5 | 12.5–15.6 |
| T6-2 | 12.5 | 12.5 |
| T6-3 | 10.9–12.5 | 15.6 |
| T7 | 12.5 | 12.5 |
| T7-1 | 12.5 | 15.6 |
| T7-2 | 12.5 | 15.6 |
| T7-3 | 12.5 | 15.6 |
Whole-genome sequencing and SNP/DIP analyses.
The closed genome of wild-type V. anguillarum strain 90-11-286 consists of two chromosomes, of 3.0 Mbp and 1.3 Mbp, with a G+C content of 44.4%, and it has 3,817 coding sequences (CDSs), 4,003 genes, 31 rRNAs, and 106 tRNAs. This genome served as a reference for mapping and identification of single-nucleotide polymorphisms (SNPs) and deletion-insertion polymorphisms (DIPs) in the 40 genomes. All genomes had between 7.2 × 106 and 8.4 × 106 reads. The SNP and DIP analyses of the 40 genomes revealed that 11 different genes were affected by 15 different mutations with frequencies above 70% in the reads (Table 2). The 15 mutations included 11 SNPs and 4 DIPs. Each genome had between two and eight amino acid-changing mutations, with no distinct differences between TDA-exposed clones (0.007 to 0.033 mutations generation−1) and DMSO-exposed (control) clones (0.010 to 0.028 mutations generation−1) being observed. Clonal variations in mutations were seen within the populations of control lineages C2 and C3 and the TDA-exposed lineage T7 but not in the other lineages. Six of the 15 different mutations were found only in the genomes of TDA-exposed clones and lineages, whereas two were found only in the control genomes (Table 3). Three different mutations were found in the competence gene comM, which encodes a magnesium chelatase-related protein. All three comM mutations were found in genomes of 20 TDA-exposed clones and lineages and six control clones and lineages. A SNP in the kinE gene, encoding a two-component sensor histidine kinase, was found in two TDA-exposed clones and eight control clones and lineages. In lineage 1, a SNP was found in the dmlR gene, which codes for a helix-turn-helix (HTH)-type transcriptional regulator. In addition, a SNP was found in the transport permease protein gene yadH. This mutation was present in lineage 2 and two clones from lineage 7. A SNP mutation was also found in all isolates from lineage 5 in 90-11-286_01842, which encodes a 2-oxoglutarate/Fe(II)-dependent oxygenase. The isolates from lineage 6 had a SNP in the ompD gene, which encodes an outer membrane porin protein. Furthermore, a DIP resulting in a frameshift in the ftsI gene, which is involved in cell division, was found in two clones from control lineage 3. Finally, the fliM gene, which encodes a flagellar motor switch protein and thus is important for motility and potentially virulence, was affected. In the fliM gene, three different SNPs were found, with all clones and lineages harboring one of the three different SNPs.
TABLE 2.
Distribution of SNPs and DIPs in TDA-exposed lineages and clones of V. anguillaruma
| Gene name and type of mutation | Gene product | Control 1 |
Control 2 |
Control 3 |
Lineage 1b |
Lineage 2c |
Lineage 3c |
Lineage 4c |
Lineage 5c |
Lineage 6b |
Lineage 7b |
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | C1-1 | C1-2 | C1-3 | C2 | C2-1 | C2-2 | C2-3 | C3 | C3-1 | C3-2 | C3-3 | T1 | T1-1 | T1-2 | T1-3 | T2 | T2-1 | T2-2 | T2-3 | T3 | T3-1 | T3-2 | T3-3 | T4 | T4-1 | T4-2 | T4-3 | T5 | T5-1 | T5-2 | T5-3 | T6 | T6-1 | T6-2 | T6-3 | T7 | T7-1 | T7-2 | T7-3 | ||
| fliM, SNP1 | Flagellar motor switch protein | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||
| fliM, SNP2 | Flagellar motor switch protein | + | + | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||
| fliM, SNP3 | Flagellar motor switch protein | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||||||
| —,d DIP | Hypothetical protein | + | + | + | + | + | + | + | + | + | X | + | + | X | + | + | + | + | X | + | + | + | + | + | + | + | + | + | + | X | + | + | + | X | X | + | + | + | + | + | |
| comM, SNP1 | Mg chelatase activity | X | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| comM, DIP1 | Mg chelatase activity | X | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| comM, DIP2 | Mg chelatase activity | X | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| kinE, SNP | Two-component sensor histidine kinase | + | + | + | + | + | + | + | + | + | + | ||||||||||||||||||||||||||||||
| —, SNP1 | Hypothetical protein | + | + | + | + | + | + | + | + | + | + | + | X | + | + | + | + | + | + | ||||||||||||||||||||||
| dmlR, SNP | HTH-type transcriptional regulator | + | + | + | + | ||||||||||||||||||||||||||||||||||||
| yadH, SNP | Transport permease protein | + | + | + | + | + | + | ||||||||||||||||||||||||||||||||||
| 90-11-286_01842, SNP | 2-Oxoglutarate/Fe(II)-dependent oxygenase | + | + | + | + | ||||||||||||||||||||||||||||||||||||
| ompD, SNP | Outer membrane porin protein | + | + | + | + | ||||||||||||||||||||||||||||||||||||
| ftsI, DIP | Cell division protein | + | + | ||||||||||||||||||||||||||||||||||||||
| —, SNP2 | Hypothetical protein | + | + | ||||||||||||||||||||||||||||||||||||||
Nonsynonymous SNPs and DIPs with frequencies of occurrence above 70% in the sequencing reads are included. X, frequency between 70% and <80%; +, frequency between 80% and 100%.
Lineage that reached 1.5 times the wild-type MIC in the TDA exposure experiment.
Lineage that reached 1.75 times the wild-type MIC in the TDA exposure experiment.
—, hypothetical protein.
TABLE 3.
Mutational changes of SNPs and DIPs in TDA-exposed lineages and clones of V. anguillarum
| Gene name | Gene product | Mutation type | Amino acid position | Amino acid change | No. of affected clones |
|
|---|---|---|---|---|---|---|
| Control | Exposed | |||||
| fliM | Flagellar motor switch protein | SNP | 48 | Gly to Arg | 8 | 0 |
| fliM | Flagellar motor switch protein | SNP | 54 | Glu to Asp | 0 | 10 |
| fliM | Flagellar motor switch protein | SNP | 230 | Asp to Glu | 4 | 18 |
| —a | Hypothetical protein | DIP | Frameshift | 12 | 27 | |
| comM | Mg chelatase activity | SNP | 1 | Leu to Met | 6 | 20 |
| comM | Mg chelatase activity | DIP | 2 | Pro to frameshift | 6 | 20 |
| comM | Mg chelatase activity | DIP | 306 | Val to frameshift | 6 | 20 |
| kinE | Two-component sensor histidine kinase | SNP | 66 | Ile to Val | 8 | 2 |
| — | Hypothetical protein | SNP | 3 | 15 | ||
| dmlR | HTH-type transcriptional regulator | SNP | 228 | Lys to Arg | 0 | 4 |
| yadH | Transport permease protein | SNP | 157 | Gly to Val | 0 | 6 |
| 90-11-286_01842 | 2-Oxoglutarate/Fe(II)-dependent oxygenase | SNP | 32 | Ala to Glu | 0 | 4 |
| ompD | Outer membrane porin protein | SNP | 272 | Leu to Val | 0 | 4 |
| ftsI | Cell division protein | DIP | 4 | Asn to frameshift | 2 | 0 |
| — | Hypothetical protein | SNP | 0 | 2 | ||
—, hypothetical protein.
Effects of fliM gene mutations on motility.
The fliM gene is important for flagellate motility in V. anguillarum and potentially its virulence. Three different SNP mutations were found in the fliM gene and, since all lineages and clones harbored one of the mutations, the motility of the lineages and clones was assessed via phase-contrast microscopy. All clones and lineages were equally motile and similar to the wild-type strain (data not shown).
Fish cell infection assays.
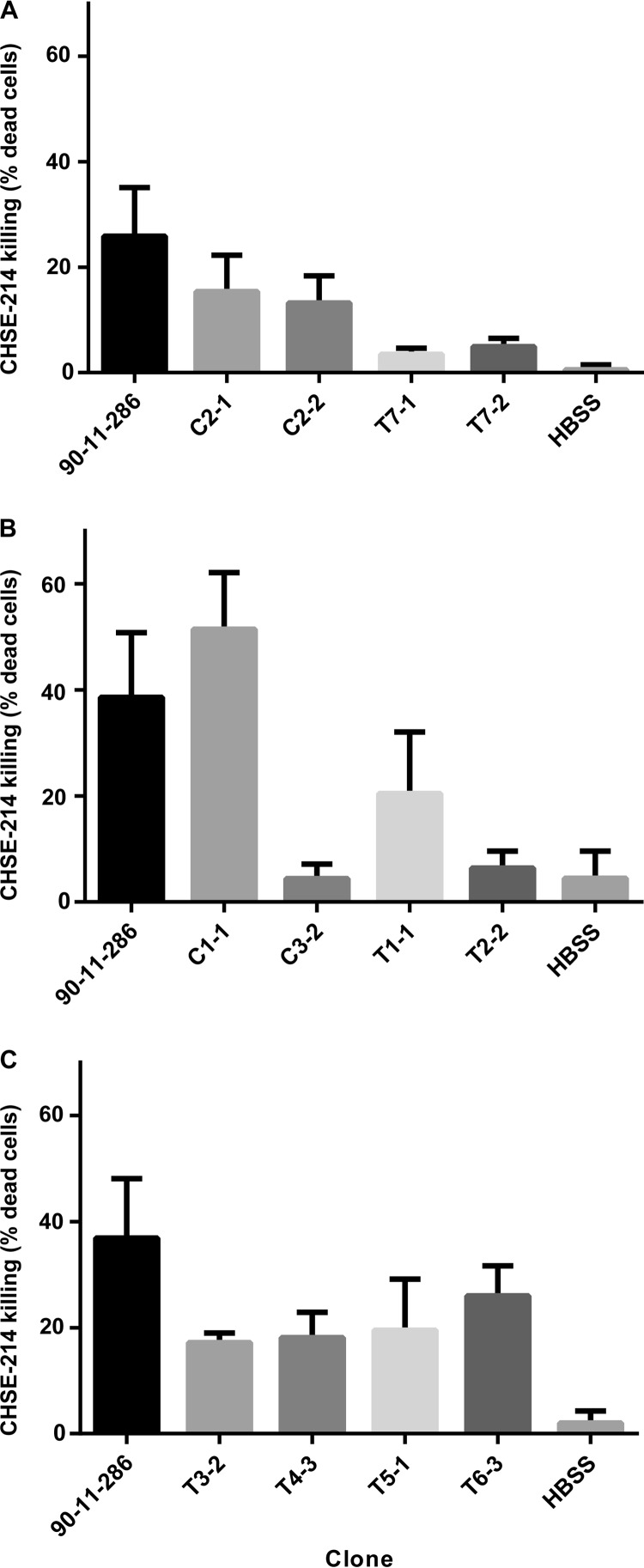
Since antimicrobial compounds can affect the virulence of pathogenic bacteria, the ability of 12 clones (8 TDA-exposed clones and 4 control clones) to kill fish cells was tested in CHSE-214 infection assays. The 12 clones were selected to cover all seven lineages and the different mutational patterns. None of the clones resulted in significantly higher mortality rates for the fish cells than did the wild-type strain; in fact, most clones appeared less infectious than the nonexposed wild-type strain (Fig. 2). The wild-type strain caused killing rates between 26% ± 8% and 39% ± 10%, based on three independent experiments (Fig. 2). Four of the clones (C2-1, C1-1, T1-1, and T6-3) resulted in the same mortality rates as the wild-type strain, killing 15% ± 6%, 51% ± 9%, 20% ± 9%, and 26% ± 5% of the fish cells, respectively (P values of 0.106, 0.294, 0.086, and 0.268, respectively). The remaining eight clones caused lower mortality rates. Two clones, C2-2 and T5-1, killed 13% ± 4% and 20% ± 8% of the fish cells, respectively; these findings were just below the significance level of 0.05, with both exhibiting P values of 0.044. The other six clones (C3-2, T2-2, T3-2, T4-3, T7-1, and T7-2) were significantly less virulent than the wild-type strain (P values of 0.002, 0.003, 0.022, 0.029, 0.001, and 0.002, respectively) and killed 4% ± 2%, 3% ± 6%, 17% ± 1%, 18% ± 4%, 3% ± 1%, and 5% ± 1% of the fish cells, respectively. Very few variations were seen between TDA-exposed and DMSO-exposed (control) clones, and TDA-exposed clones did not show higher mortality rates than DMSO-exposed clones.
FIG 2.
Killing of CHSE-214 fish cell lines by TDA-exposed V. anguillarum clones representing different mutational patterns in three independent experiments. All three experiments (A, B, and C) included the wild type and control. Each bar represents independent triplicates, and error bars are the standard deviation of the mean.
DISCUSSION
The successful application of TDA-producing roseobacters as probiotics in aquaculture requires detailed knowledge about the long-term effects of the roseobacters and TDA on the pathogens and on the general microbiota. Here we focused on TDA exposure and demonstrated that continuous exposure of V. anguillarum to TDA did not have any impact on the development of TDA resistance or tolerance. Furthermore, prolonged TDA exposure did not seem to increase the mutation rates of exposed strains or the infectivity of the bacteria toward CHSE-214 fish cell lines.
It has been known for some time that resistance to TDA does not arise easily in target pathogens, and single-point mutations do not seem to confer resistance (22). However, since the mechanism of action was not known, assessing the risk of resistance was difficult. Wilson et al. recently demonstrated that TDA acts as a neutral antiporter, upsetting the proton motive force; also, TDA resistance in P. inhibens could be genetically linked and proved to be controlled by the tdaR genes (26), which conferred TDA resistance in E. coli upon horizontal transfer. Tolerance and resistance to TDA have also been found in environmental marine bacteria; the majority (126 of 136 isolates) of non-TDA-producing bacteria isolated along with a TDA-producing Pseudovibrio sp. displayed tolerance to TDA (23). It is not known, however, whether this finding is related to the tdaR genes and/or whether these genes can be shared through horizontal gene transfer. In the present study, we were not able to adapt V. anguillarum to increasing TDA concentrations. The slight increase in tolerance, corresponding to 1.5 to 1.75 times the wild-type MIC, was likely caused by the extended incubation time (2 to 3 days) and disappeared when clones were tested in 24-h MIC assays.
As mentioned, the proton motive force is the target of TDA (26). This function is of course essential and highly conserved, which likely explains why resistance or tolerance does not develop, since mutational changes in the PMF apparatus would be lethal to the bacteria. It could be speculated that multiple cooccurring mutations could result in resistance, as has been shown for antimicrobial peptides (30, 31). It was assumed that resistance to AMPs could not develop; however, several adaptive laboratory evolution (ALE) experiments have demonstrated that long-term exposure does indeed result in the evolution of resistant strains (27, 30). Using this method, the MIC of the AMP pexiganan for Pseudomonas fluorescens increased to 128 times the wild-type MIC after approximately 700 generations (27), and the MICs of the synthetic AMP analogues α-peptide/β-peptoid peptidomimetics against E. coli increased to 16 to 32 times the wild-type MIC after approximately 500 generations (30). However, our ALE experiments did not result in TDA-resistant strains, and the experiment was terminated after two failed attempts to obtain growth in the presence of increased TDA concentrations. The experiment covered approximately 230 generations per lineage. Several antimicrobial compounds target the proton motive force, including food preservatives such as sorbic acid and benzoic acid (47–49). These weak acids, when nondissociated, diffuse across the cell membrane and the cell envelope and, to maintain homeostasis and a neutral intracellular pH, the bacteria use energy to pump out the hydrogen ions, thus depleting the PMF (50, 51). The antimicrobial effects of both compounds are dependent on environmental factors, especially pH, and the compounds are primarily effective under acidic conditions (52, 53). Similar to our observations with TDA, bacterial resistance to sorbic acid and benzoic acid does not seem to evolve easily (54, 55). To the best of our knowledge, mutational resistance to sorbic acid and benzoic acid in bacteria has not been reported, although several cases have been described for yeast (54, 55). However, adaptive resistance to benzoic acid was observed in E. coli strain O157:H7 after induction of the glutamate-dependent acid resistance (GDAR) response at pH 2.0 (56). The GDAR response decarboxylates glutamate, replacing the α-carboxyl group with a proton, and reduces glutamate to γ-aminobutyric acid (GABA), which is then exchanged for glutamate via the GadC antiporter (57). The proposed model for TDA resistance in P. inhibens relies on a GDAR-like system (26), and low pH combined with high glutamate availability could potentially induce a response in E. coli or other target bacteria, causing TDA tolerance. Furthermore, a similar response could possibly be achieved by a mutation causing continuous expression of the LuxR-like regulator GadE, the main regulator of GDAR (58, 59). Since access to glutamate would be a limiting factor, however, we consider it unlikely that TDA resistance would occur via simple mutations or deletions. Even so, horizontal gene transfer of TDA resistance genes could still be a risk.
The bacteriocin nisin is a cationic peptide that interferes with the PMF by forming pores in the cytoplasmic membrane, causing leakage of intracellular contents, loss of membrane potential, and disruption of ΔpH (60, 61). Four to 5 days of exposure to sub-MIC levels of nisin resulted in a 32-fold MIC increase for S. aureus SH1000 subcultures; for one mutant, the increase was attributed to two amino acid changes, in the purine operon repressor and a putative sensor histidine kinase (61). However, when the mutations' contributions to resistance were evaluated via allelic replacements, it was possible to reach only a 16-fold MIC increase, indicating that one or more additional unknown mutations contributed to the resistance (61). Thus, the development of resistance to a PMF-interfering compound is known; however, here modification of the membrane binding target is a potential resistance mechanism.
Mutation rates in bacteria are influenced by several factors, such as nutritional availability and environmental stress (62). Several antibiotics have been reported to increase mutation rates in exposed microorganisms (63–65), thus reducing the time it takes for resistance, or other genetic changes, to occur. Tetracycline increased Pseudomonas aeruginosa mutation rates 105 times after 4 days of incubation on selective plates (66), and ciprofloxacin and streptomycin increased the frequency of mutations causing rifampin resistance in Streptococcus pneumoniae (33). In the present study, no differences in mutation rates were seen between TDA-exposed clones and control strains; therefore, an increase in the mutation rate, resulting in potentially faster resistance development or changes in virulence genes, does not seem to be a concern in the application of TDA-producing probionts.
Antimicrobial compounds can affect the virulence of bacteria after prolonged exposure to the compound. When different fish pathogens were exposed to the antimicrobial peptide cecropin B, V. anguillarum strain 775 displayed increased adhesion to CHSE-214 fish cells and an increased mortality rate in challenged medaka (Oryzias latipes) (67). Induction of virulence genes by antimicrobial compounds has also been reported for human pathogens such as Listeria monocytogenes (37, 68) and Staphylococcus aureus (35, 38). In the present study, none of the TDA-exposed clones was more infective toward CHSE-214 fish cells than were nonexposed clones and the wild-type strain. In contrast, 8 of 12 clones, including 6 TDA-exposed clones and 2 control clones, were significantly less infective than the wild-type strain. Three different mutations were found in the virulence-related flagellar motor switch protein gene fliM. Since TDA affects the PMF and thereby motility (26), the mutations in fliM are likely a result of TDA exposure. However, no changes in motility could be observed in the mutants, and no clear patterns were seen for the mutations and the mortality rates in the fish cell trials. Since no differences in motility were observed and representatives from both the TDA-exposed and DMSO-exposed lineages resulted in fewer dead fish cells than did the wild type, we cannot conclude which of the factors (culturing or TDA exposure) had the strongest influence on the decreased infectivity. Somerville et al. (69) showed, for example, that the activity of aconitase, affecting the synthesis of several virulence factors and the expression of the global regulators RNA III and sarA, decreased 38% after 6 weeks of in vitro serial passage of a S. aureus strain isolated from a patient with toxic shock syndrome. Thus, it seems unlikely that TDA increases virulence postexposure.
Our study addressed the possible mutational changes caused by TDA; however, antimicrobials may also affect gene expression, and antibiotics at subinhibitory concentrations can activate “silent” gene clusters (34). Thus, studies addressing gene expression in TDA-exposed pathogenic bacteria will be relevant in assessments of risks and applications.
In conclusion, the present study demonstrates that prolonged TDA exposure does not result in TDA-resistant or TDA-tolerant strains. TDA exposure did not result in a changed mutational pattern and did not enhance virulence, as measured in fish cell lines. In fact, virulence appeared lower in TDA-exposed clones. Thus, based on the present study, the use of TDA-producing roseobacters would not likely pose risks of unwanted changes in the pathogen.
ACKNOWLEDGMENTS
This study was funded by grants from the Danish Council for Strategic Research Programme Commission on Health, Food, and Welfare (grant 12-132390; ProAqua), the European Union Seventh Framework Programme (FP7/2007–2013; grant agreement 311975), and the Villum Kahn-Rasmussen Foundation (grant VKR023285).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Food and Agriculture Organization, Fisheries and Aquaculture Department. 2014. The state of world fisheries and aquaculture: opportunities and challenges. Food and Agriculture Organization of the United Nations, Rome, Italy. [Google Scholar]
- 2.Kibenge FSB, Godoy MG, Fast M, Workenhe S, Kibenge MJT. 2012. Countermeasures against viral diseases of farmed fish. Antiviral Res 95:257–281. doi: 10.1016/j.antiviral.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Thompson FL, Iida T, Swings J. 2004. Biodiversity of vibrios. Microbiol Mol Biol Rev 68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frans I, Michiels CW, Bossier P, Willems KA, Lievens B, Rediers H. 2011. Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J Fish Dis 34:643–661. doi: 10.1111/j.1365-2761.2011.01279.x. [DOI] [PubMed] [Google Scholar]
- 5.Cabello FC. 2006. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol 8:1137–1144. doi: 10.1111/j.1462-2920.2006.01054.x. [DOI] [PubMed] [Google Scholar]
- 6.Defoirdt T, Sorgeloos P, Bossier P. 2011. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr Opin Microbiol 14:251–258. doi: 10.1016/j.mib.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Ringø E, Olsen RE, Jensen I, Romero J, Lauzon HL. 2014. Application of vaccines and dietary supplements in aquaculture: possibilities and challenges. Rev Fish Biol Fish 24:1005–1032. doi: 10.1007/s11160-014-9361-y. [DOI] [Google Scholar]
- 8.Sommerset I, Krossøy B, Biering E, Frost P. 2005. Vaccines for fish in aquaculture. Expert Rev Vaccines 4:89–101. doi: 10.1586/14760584.4.1.89. [DOI] [PubMed] [Google Scholar]
- 9.Skjermo J, Vadstein O. 1999. Techniques for microbial control in the intensive rearing of marine larvae. Aquaculture 177:333–343. doi: 10.1016/S0044-8486(99)00096-4. [DOI] [Google Scholar]
- 10.Gatesoupe FJ. 1999. The use of probiotics in aquaculture. Aquaculture 180:147–165. doi: 10.1016/S0044-8486(99)00187-8. [DOI] [Google Scholar]
- 11.Kesarcodi-Watson A, Kaspar H, Lategan MJ, Gibson L. 2008. Probiotics in aquaculture: the need, principles and mechanisms of action and screening processes. Aquaculture 274:1–14. doi: 10.1016/j.aquaculture.2007.11.019. [DOI] [Google Scholar]
- 12.Ruiz-Ponte C, Cilia V, Lambert C, Nicolas JL. 1998. Roseobacter gallaeciensis sp. nov., a new marine bacterium isolated from rearings and collectors of the scallop Pecten maximus. Int J Syst Bacteriol 48:537–542. doi: 10.1099/00207713-48-2-537. [DOI] [PubMed] [Google Scholar]
- 13.Hjelm M, Bergh O, Riaza A, Nielsen J, Melchiorsen J, Jensen S, Duncan H, Ahrens P, Birkbeck H, Gram L. 2004. Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst Appl Microbiol 27:360–371. doi: 10.1078/0723-2020-00256. [DOI] [PubMed] [Google Scholar]
- 14.Hjelm M, Riaza A, Formoso F, Gram L, Melchiorsen J. 2004. Seasonal incidence of autochthonous antagonistic Roseobacter spp. and Vibrionaceae strains in a turbot larva (Scophthalmus maximus) rearing system. Appl Environ Microbiol 70:7288–7294. doi: 10.1128/AEM.70.12.7288-7294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Planas M, Pérez-Lorenzo M, Hjelm M, Gram L, Uglenes Fiksdal I, Bergh Ø, Pintado J. 2006. Probiotic effect in vivo of Roseobacter strain 27-4 against Vibrio (Listonella) anguillarum infections in turbot (Scophthalmus maximus L.) larvae. Aquaculture 255:323–333. doi: 10.1016/j.aquaculture.2005.11.039. [DOI] [Google Scholar]
- 16.D'Alvise PW, Lillebø S, Prol-Garcia MJ, Wergeland HI, Nielsen KF, Bergh Ø, Gram L. 2012. Phaeobacter gallaeciensis reduces Vibrio anguillarum in cultures of microalgae and rotifers, and prevents vibriosis in cod larvae. PLoS One 7:e43996. doi: 10.1371/journal.pone.0043996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang L. 2003. Investigation of secondary metabolites of North Sea bacteria: fermentation, isolation, structure elucidation and bioactivity. Ph.D. thesis. Georg August University, Göttingen, Germany. [Google Scholar]
- 18.D'Alvise PW, Melchiorsen J, Porsby CH, Nielsen KF, Gram L. 2010. Inactivation of Vibrio anguillarum by attached and planktonic Roseobacter cells. Appl Environ Microbiol 76:2366–2370. doi: 10.1128/AEM.02717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prol García MJ, D'Alvise PW, Gram L. 2013. Disruption of cell-to-cell signaling does not abolish the antagonism of Phaeobacter gallaeciensis toward the fish pathogen Vibrio anguillarum in algal systems. Appl Environ Microbiol 79:5414–5417. doi: 10.1128/AEM.01436-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grotkjær T, Bentzon-Tilia M, D'Alvise P, Dourala N, Nielsen KF, Gram L. 2016. Isolation of TDA-producing Phaeobacter strains from sea bass larval rearing units and their probiotic effect against pathogenic Vibrio spp. in Artemia cultures. Syst Appl Microbiol 39:180–188. doi: 10.1016/j.syapm.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Brinkhoff T, Bach G, Heidorn T, Liang L, Schlingloff A, Simon M. 2004. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl Environ Microbiol 70:2560–2565. doi: 10.1128/AEM.70.4.2560-2565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porsby CH, Webber MA, Nielsen KF, Piddock LJV, Gram L. 2011. Resistance and tolerance to tropodithietic acid, an antimicrobial in aquaculture, is hard to select. Antimicrob Agents Chemother 55:1332–1337. doi: 10.1128/AAC.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington C, Reen FJ, Mooij MJ, Stewart FA, Chabot J-B, Guerra AF, Glöckner FO, Nielsen KF, Gram L, Dobson ADW, Adams C, O'Gara F. 2014. Characterisation of non-autoinducing tropodithietic acid (TDA) production from marine sponge Pseudovibrio species. Mar Drugs 12:5960–5978. doi: 10.3390/md12125960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neu AK, Månsson M, Gram L, Prol-García MJ. 2014. Toxicity of bioactive and probiotic marine bacteria and their secondary metabolites in Artemia sp. and Caenorhabditis elegans as eukaryotic model organisms. Appl Environ Microbiol 80:146–153. doi: 10.1128/AEM.02717-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wichmann H, Vocke F, Brinkhoff T, Simon M, Richter-Landsberg C. 2015. Cytotoxic effects of tropodithietic acid on mammalian clonal cell lines of neuronal and glial origin. Mar Drugs 13:7113–7123. doi: 10.3390/md13127058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson MZ, Wang R, Gitai Z, Seyedsayamdost MR. 2016. Mode of action and resistance studies unveil new roles for tropodithietic acid as an anticancer agent and the γ-glutamyl cycle as a proton sink. Proc Natl Acad Sci U S A 113:1630–1635. doi: 10.1073/pnas.1518034113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perron GG, Zasloff M, Bell G. 2006. Experimental evolution of resistance to an antimicrobial peptide. Proc R Soc B Biol Sci 273:251–256. doi: 10.1098/rspb.2005.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother 50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toprak E, Veres A, Michel J, Chait R, Hartl DL, Kishony R. 2012. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet 44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hein-Kristensen L, Franzyk H, Holch A, Gram L. 2013. Adaptive evolution of Escherichia coli to an α-peptide/β-peptoid peptidomimetic induces stable resistance. PLoS One 8:e73620. doi: 10.1371/journal.pone.0073620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jochumsen N. 2013. The causes and consequences of antibiotic resistance evolution in microbial pathogens. Ph.D. thesis. Technical University of Denmark, Kongens Lyngby, Denmark. [Google Scholar]
- 32.Romero D, Traxler MF, López D, Kolter R. 2011. Antibiotics as signal molecules. Chem Rev 111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henderson-Begg SK. 2006. Effect of subinhibitory concentrations of antibiotics on mutation frequency in Streptococcus pneumoniae. J Antimicrob Chemother 57:849–854. doi: 10.1093/jac/dkl064. [DOI] [PubMed] [Google Scholar]
- 34.Seyedsayamdost MR. 2014. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci U S A 111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subrt N, Mesak LR, Davies J. 2011. Modulation of virulence gene expression by cell wall active antibiotics in Staphylococcus aureus. J Antimicrob Chemother 66:979–984. doi: 10.1093/jac/dkr043. [DOI] [PubMed] [Google Scholar]
- 36.Yim G, De La Cruz F, Spiegelman GB, Davies J. 2006. Transcription modulation of Salmonella enterica serovar Typhimurium promoters by sub-MIC levels of rifampin. J Bacteriol 188:7988–7991. doi: 10.1128/JB.00791-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knudsen GM, Holch A, Gram L. 2012. Subinhibitory concentrations of antibiotics affect stress and virulence gene expression in Listeria monocytogenes and cause enhanced stress sensitivity but do not affect Caco-2 cell invasion. J Appl Microbiol 113:1273–1286. doi: 10.1111/j.1365-2672.2012.05435.x. [DOI] [PubMed] [Google Scholar]
- 38.Stevens DL, Ma Y, Salmi DB, McIndoo E, Wallace RJ, Bryant AE. 2007. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J Infect Dis 195:202–211. doi: 10.1086/510396. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen K, Larsen J. 1993. rRNA gene restriction patterns of Vibrio anguillarum serogroup O1. Dis Aquat Organ 16:121–126. doi: 10.3354/dao016121. [DOI] [Google Scholar]
- 40.Skov MN, Pedersen K, Larsen JL. 1995. Comparison of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar O1. Appl Environ Microbiol 61:1540–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sobecky PA, Mincer TJ, Chang MC, Helinski DR. 1997. Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl Environ Microbiol 63:888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibson LF, Khoury JT. 1986. Storage and survival of bacteria by ultra-freeze. Lett Appl Microbiol 3:127–129. doi: 10.1111/j.1472-765X.1986.tb01565.x. [DOI] [Google Scholar]
- 43.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard— 7th ed Publication M07-A7. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 44.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 46.Lannan CN, Winton JR, Fryer JL. 1984. Fish cell lines: establishment and characterization of nine cell lines from salmonids. In Vitro 20:671–676. doi: 10.1007/BF02618871. [DOI] [PubMed] [Google Scholar]
- 47.Eklund T. 1985. The effect of sorbic acid and esters of p-hydroxybenzoic acid on the protonmotive force in Escherichia coli membrane vesicles. J Gen Microbiol 131:73–76. [DOI] [PubMed] [Google Scholar]
- 48.Sofos JN, Pierson MD, Blocher JC, Busta FF. 1986. Mode of action of sorbic acid on bacterial cells and spores. Int J Food Microbiol 3:1–17. doi: 10.1016/0168-1605(86)90036-X. [DOI] [Google Scholar]
- 49.Krebs HA, Wiggins D, Stubbs M, Sols A, Bedoya F. 1983. Studies on the mechanism of the antifungal action of benzoate. Biochem J 214:657–663. doi: 10.1042/bj2140657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salmond CV, Kroll RG, Booth IR. 1984. The effect of food preservatives on pH homeostasis in Escherichia coli. J Gen Microbiol 130:2845–2850. [DOI] [PubMed] [Google Scholar]
- 51.Warth AD. 1985. Resistance of yeast species to benzoic and sorbic acids and to sulfur dioxide. J Food Prot 48:564–569. [DOI] [PubMed] [Google Scholar]
- 52.Eklund T. 1983. The antimicrobial effect of dissociated and undissociated sorbic acid at different pH levels. J Appl Bacteriol 54:383–389. doi: 10.1111/j.1365-2672.1983.tb02632.x. [DOI] [PubMed] [Google Scholar]
- 53.Eklund T. 1985. Inhibition of microbial growth at different pH levels by benzoic and propionic acids and esters of p-hydroxybenzoic acid. Int J Food Microbiol 2:159–167. doi: 10.1016/0168-1605(85)90035-2. [DOI] [Google Scholar]
- 54.Russell AD. 1991. Mechanisms of bacterial resistance to non-antibiotics: food additives and food and pharmaceutical preservatives. J Appl Bacteriol 71:191–201. doi: 10.1111/j.1365-2672.1991.tb04447.x. [DOI] [PubMed] [Google Scholar]
- 55.Beales N. 2004. Adaptation of microorganisms to cold temperatures, weak acid preservatives, low pH, and osmotic stress: a review. Compr Rev Food Sci Food Saf 3:1–20. doi: 10.1111/j.1541-4337.2004.tb00057.x. [DOI] [PubMed] [Google Scholar]
- 56.Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, Foster JW. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol 62:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foster JW. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol 2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 58.Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol 49:1309–1320. doi: 10.1046/j.1365-2958.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- 59.Hommais F, Krin E, Coppée JY, Lacroix C, Yeramian E, Danchin A, Bertin P. 2004. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 150:61–72. doi: 10.1099/mic.0.26659-0. [DOI] [PubMed] [Google Scholar]
- 60.Bruno MEC, Kaiser A, Montville TJ. 1992. Depletion of proton motive force by nisin in Listeria monocytogenes cells. Appl Environ Microbiol 58:2255–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blake KL, Randall CP, Neill AJO. 2011. In vitro studies indicate a high resistance potential for the lantibiotic nisin in Staphylococcus aureus and define a genetic basis for nisin resistance. Antimicrob Agents Chemother 55:2362–2368. doi: 10.1128/AAC.01077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martinez JL, Baquero F. 2000. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother 44:1771–1777. doi: 10.1128/AAC.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kohanski MA, DePristo MA, Collins JJ. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell 37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baharoglu Z, Mazel D. 2011. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob Agents Chemother 55:2438–2441. doi: 10.1128/AAC.01549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gutierrez A, Laureti L, Crussard S, Abida H, Rodríguez-Rojas A, Blázquez J, Baharoglu Z, Mazel D, Darfeuille F, Vogel J, Matic I. 2013. β-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat Commun 4:1610. doi: 10.1038/ncomms2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alonso A, Campanario E, Martínez JL. 1999. Emergence of multidrug-resistant mutants is increased under antibiotic selective pressure in Pseudomonas aeruginosa. Microbiology 145:2857–2862. doi: 10.1099/00221287-145-10-2857. [DOI] [PubMed] [Google Scholar]
- 67.Sallum UW, Chen TT. 2008. Inducible resistance of fish bacterial pathogens to the antimicrobial peptide cecropin B. Antimicrob Agents Chemother 52:3006–3012. doi: 10.1128/AAC.00023-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kastbjerg VG, Larsen MH, Gram L, Ingmer H. 2010. Influence of sublethal concentrations of common disinfectants on expression of virulence genes in Listeria monocytogenes. Appl Environ Microbiol 76:303–309. doi: 10.1128/AEM.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Somerville GA, Beres SB, Fitzgerald JR, DeLeo FR, Cole RL, Hoff JS, Musser JM. 2002. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J Bacteriol 184:1430–1437. doi: 10.1128/JB.184.5.1430-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]