Abstract
The disruption of the energy or nutrient balance triggers endoplasmic reticulum (ER) stress, a process that mobilizes various strategies, collectively called the unfolded protein response (UPR), which reestablish homeostasis of the ER and cell. Activation of the UPR stress sensor IRE1α (inositol-requiring enzyme 1α) stimulates its endoribonuclease activity, leading to the generation of the mRNA encoding the transcription factor XBP1 (X-box binding protein 1), which regulates the transcription of genes encoding factors involved in controlling the quality and folding of proteins. We found that the activity of IRE1α was regulated by the ER oxidoreductase PDIA6 (protein disulfide isomerase A6) and the microRNA miR-322 in response to disruption of ER Ca2+ homeostasis. PDIA6 interacted with IRE1α and enhanced IRE1α activity as monitored by phosphorylation of IRE1α and XBP1 mRNA splicing, but PDIA6 did not substantially affect the activity of other pathways that mediate responses to ER stress. ER Ca2+ depletion and activation of store operated Ca2+ entry reduced the abundance of the microRNA miR-322, which increased PDIA6 mRNA stability and consequently IRE1α activity during the ER stress response. In vivo experiments with mice and worms showed that the induction of ER stress correlated with decreased miR-322 abundance, increased PDIA6 mRNA abundance, or both. Together these findings demonstrated that ER Ca2+, PDIA6, IRE1α, and miR-322 function in a dynamic feedback loop modulating the UPR under conditions of disrupted ER Ca2+ homeostasis.
Introduction
The endoplasmic reticulum (ER) is involved in the production of newly synthesized secretory and membrane proteins, where several mechanisms control proper folding and posttranslational modifications of these proteins. Many different intrinsic and extrinsic factors may disrupt ER homeostasis leading to the activation of ER stress coping responses and multiple corrective strategies (1). A strategy to restore homeostasis is the activation of the unfolded protein response (UPR) (2, 3). The UPR is a dynamic signal transduction pathway that reduces unfolded protein load by attenuating protein synthesis, increasing protein chaperone production and augmenting ER-associated degradation (ERAD) and autophagy (3–6). The UPR signals through activating transcription factor 6 (ATF6); inositol-requiring element 1α (IRE1α), a bi-functional protein kinase and endoribonuclease; and dsRNA-activated protein kinase-like ER kinase (PERK) which phosphorylates and inactivates eukaryotic translation initiation factor 2 on the alpha subunit (eIF2α). These sensors are maintained in an inactive state through interaction with the ER chaperone immunoglobulin binding protein (BiP) (4–6). As misfolded proteins in the ER accumulate, BiP binds to them to prevent aggregation and in the process is released from the sensors, permitting their activation. Each sensor activates downstream factors that transcriptionally regulate genes that enable adaptation to stress or trigger the induction of apoptosis. For example, activated IRE1α undergoes autophosphorylation and oligomerization, leading to the conformational activation of the endoribonuclease domain, which splices the mRNA encoding the transcription factor XBP1. This processing event removes a 26 base intron in the coding region that changes the reading frame, producing the transcription factor XBP1s (7). XBP1s binds to ER stress elements (ERSE) and UPR elements (UPRE) to transcriptionally activate genes encoding proteins involved in protein folding, transport and ERAD (8, 9). Depending on the intensity and the duration of the stress stimuli, UPR signaling events may trigger cell adaptation or the induction of apoptosis through complementary mechanisms including BCL-2 family members, miRNAs and other factors (1, 10, 11).
Depletion of ER Ca2+ stores results in the activation of store-operated Ca2+ entry (SOCE), an important Ca2+ signaling pathway (12). Prolonged ER Ca2+ depletion, in addition to the induction of SOCE, is also a potent inducer of ER stress, resulting in disrupted ER homeostasis, accumulation of misfolded proteins and activation of the three branches of the UPR (1, 6). Fine-tuning the UPR response is fundamental to determine whether cells survive or undergo apoptosis under ER stress and increasing evidence indicates that the activity of the UPR sensors may be modulated through the direct binding of specific regulators (6). In this study, we focused on identifying ER stress coping responses induced by disruption of the ER homeostasis by depletion of ER Ca2+ stores. We employed a small interfering RNA (siRNA) library screen combined with deep sequencing microRNA (miRNA) analysis to identify factors that mediate UPR modulation. We discovered that silencing of the gene encoding PDIA6, an ER resident oxidoreductase, affected ER Ca2+ depletion-dependent activation of the IRE1α signaling branch. Deep sequencing analysis identified miR-322 as one of the miRNAs that was significantly decreased in abundance after ER Ca2+ store depletion-induced ER stress. We also showed that Ca2+ store depletion and SOCE activation-dependent activation of IRE1α by PDIA6 was affected by Ca2+ and miR-322. The PDIA6 gene was a target of miR-322, and miR-322 abundance was sensitive to changes in ER and cytosolic Ca2+ concentrations. This work identified PDIA6 as a component of the UPR and demonstrated interplay between ER and cytosolic Ca2+, PDIA6, IRE1α and miR-322 as a part of a coping mechanism activated by disrupted ER Ca2+ homeostasis and activation of SOCE as an adaptive response to cope with ER stress.
Results
An siRNA screen identifies a role for PDIA6 in Ca2+ store depletion-induced UPR
To identify the molecular factors involved in the ER luminal Ca2+ depletion-dependent modulation of the UPR, we performed a genome-wide siRNA screen for genes required for IRE1α activation or inactivation. We used NIH-3T3 cells transfected with the pRL-IXFL XBP1 mRNA splicing reporter plasmid (Suppl. Fig. S1) (13).
To identify genes required for Ca2+ store depletion-induced ER stress, reporter cells transfected with the siRNA library were treated with thapsigargin, a sarcoplasmic/endoplasmic reticulum Ca2+ ATPase (SERCA) inhibitor, to induce ER Ca2+ depletion and activation of SOCE (14). The library included internal controls such as a scrambled siRNA, an siRNA targeting IRE1α as a negative control, and an siRNA targeting BiP as a positive control for ER stress (Suppl. Fig. S2). Analysis of approximately 6600 genes identified five gene candidates whose knockdown produced the highest increase and four genes whose knockdown produced the greatest decrease in IRE1α reporter activity in response to ER stress due to ER Ca2+ store depletion (Table 1). One of the genes in the latter group was PDIA6, which encoded an ER luminal oxidoreductase. This protein was selected for further analysis based on its subcellular localization and statistical analysis.
Table 1.
Gene candidates identified by a genome-wide siRNA screen.
| Ref Seq | Gene ID | Gene Symbol | Ratio Firefly/Renilla |
IRE1α reporter activity | |
|---|---|---|---|---|---|
| Normalized value |
High or low | ||||
| NMJ45360 | 319554 | Idi1 | 0.173948 | 8.71E-06 | low |
| NM_027959 | 71853 | PDIA6 | 0.173993 | 3.68E-07 | low |
| NM_145561 | 231382 | BC020151 | 0.174441 | 3.75E-06 | low |
| NM_007898 | 13595 | Ebp | 0.195196 | 5.17E-06 | low |
| NM_008323 | 15929 | Idh3g | 0.376557 | 0.999998 | high |
| NM_172780 | 236794 | Slc9a6 | 0.389342 | 0.999976 | high |
| NM_019420 | 54218 | B3galt4 | 0.395039 | 1.0 | high |
| NM_130864 | 113868 | Acaa1 | 0.444224 | 0.99983 | high |
| NM_007515 | 11989 | Slc7a3 | 0.541056 | 0.999997 | high |
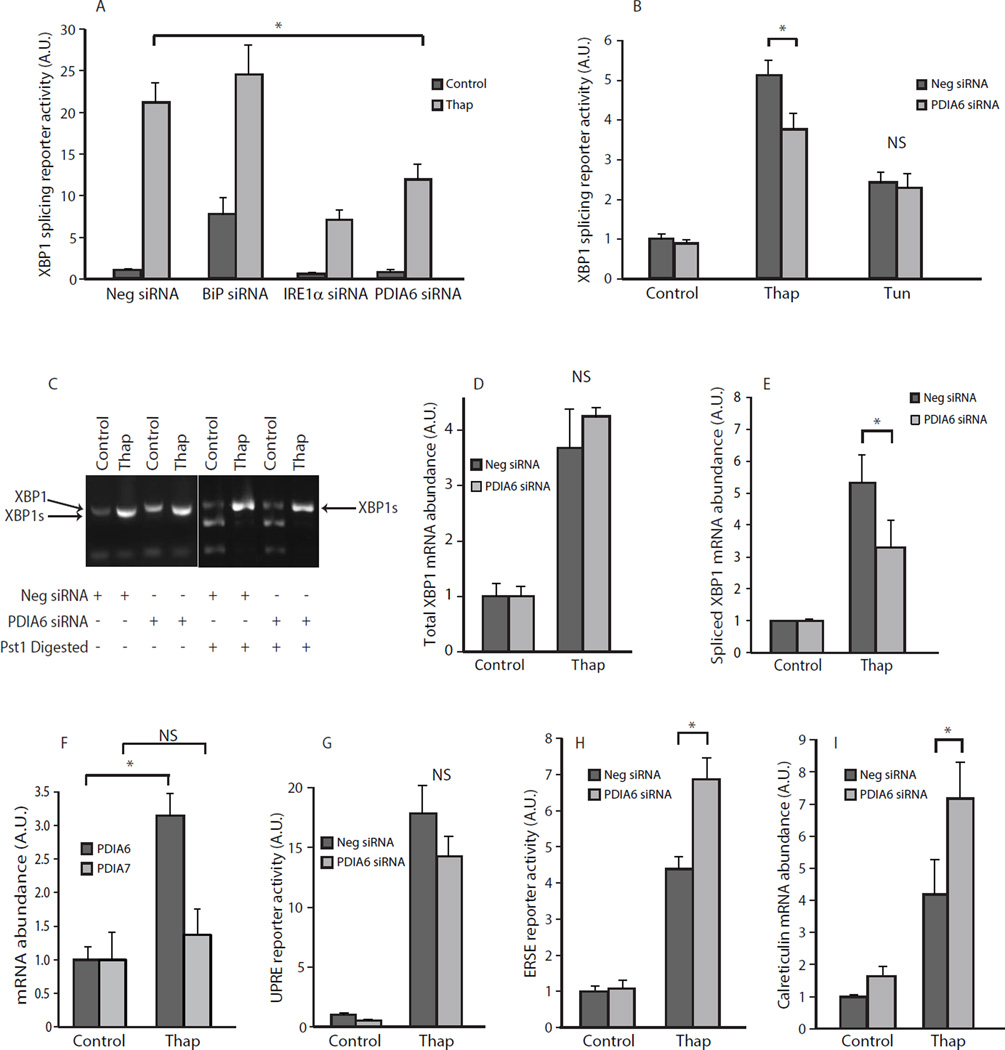
We validated a role for PDIA6 in ER stress responses by transfecting the reporter cell line with siRNA directed against PDIA6 (Suppl. Fig. S3A). Cell growth was not affected by siRNA transfection (Suppl. Fig. S2D). Quantitative (Q)-PCR and Western blot analyses confirmed that the siRNA was effective in silencing PDIA6, at the mRNA and protein level (up to 95%) under both control and thapsigargin-treated conditions (Suppl. Fig. S3A,B). PDIA6 abundance can be increased by pharmacological induction of ER stress (15) or during cardiac ischemia (16); therefore we also monitored PDIA6 mRNA abundance under ER stress conditions. Thapsigargin stimulation led to a 4-fold increase in PDIA6 mRNA abundance, which was prevented by siRNA-dependent silencing (Suppl. Fig. S3B). In our system, thapsigargin treatment triggered a 20-fold increase in the activity of the XBP1 reporter (Fig. 1A). Under these conditions, the PERK pathway was also activated as measured by phosphorylation of eIF2α, confirming that thapsigargin activated other UPR pathways (Suppl. Fig. S4). As expected, silencing of the ER chaperone BiP (Suppl. Fig. S3A) resulted in the robust induction of reporter activity under unstimulated conditions (Fig. 1A), and silencing of IRE1α (Suppl. Fig. S3A) caused a 4-fold reduction in reporter activity with thapsigargin treatment (Fig. 1A). Silencing of PDIA6 significantly reduced IRE1α reporter activity in response to thapsigargin (Fig. 1C), to a similar extent as silencing of IRE1α (Fig. 1A). This reduction in IRE1α reporter activity was recapitulated by transfection of a PDIA6 siRNA pool as well as with four independent PDIA6 specific siRNAs (Suppl. Fig. S3C,D). Next, we used tunicamycin, an inhibitor of N-linked protein glycosylation (17) that induces protein misfolding and activates XBP1 splicing. Tunicamycin did not affect the activity of the IRE1α reporter activity at the concentration and time point tested (Fig. 1B), suggesting that PDA6 may specifically regulate IRE1α under conditions of ER Ca2+ depletion.
Figure 1. Silencing of PDIA6 modulates IRE1α activity.
(A), (B). NIH-3T3 cells were transfected with XBP1 splicing reporter and siRNA directed against PDIA6, BiP, IRE1α or negative scrambled control (Neg) and treated with thapsigargin (Thap) (A,B) or tunicamycin (Tun) (B). In (A)* P-value = 0.002598. In (B) * P-value = 0.002598. NS, not significant. (C) RT-PCR analysis of XBP1 splicing in control cells and cells treated with siRNA against PDIA6. Thap, thapsigargin. Right panel, PCR products were digested with Pst1. XBP1s, spliced XBP1. Neg, negative scrambled control. (D), (E) Q-PCR analysis of total XBP1 (D) and spliced XBP1 (E) in control and PDIA6 silenced cells treated with thapsigargin (Thap). P-value = 0.0124. NS, not significant. Neg, negative scrambled control. (G). UPRE splicing reporter activity in NIH-3T3 fibroblasts transfected with PDIA6 siRNA. (H). ERSE splicing reporter activity in NIH-3T3 fibroblasts transfected with PDIA6 siRNA. *P-value = 0.004284. (I). NIH-3T3 fibroblasts were transfected with PDIA6 siRNA followed by treatment with thapsigargin (Thap). *P-value = 0.0098. All data in the figure are representative of more than 3 biological replicates.
Next we asked if PDIA6 affected splicing of endogenous XBP1 mRNA. Endogenous XBP1 was efficiently spliced in cells in response to thapsigargin (Fig. 1C). Because the XBP1 amplicon fragment in the spliced intron contains a unique PstI restriction site, we expected that Pst1 would digest the unspliced XBP1 but not the spliced variant of XBP1, which would enable quantitative analysis of the splicing event (Fig. 1C). Total XBP1 mRNA abundance was not affected in PDIA6 silenced and thapsigargin treated cells (Fig. 1D). Using Q-PCR we confirmed that knocking down PDIA6 reduced the splicing of endogenous XBP1 mRNA in response to thapsigargin treatment (Fig. 1E). Therefore, we concluded that silencing of PDIA6 attenuates IRE1α signaling as measured by XBP1 mRNA splicing in response to ER Ca2+ depletion.
Because the PDIA6 gene contains several ERSEs in its proximal promoter region, we tested whether the PDIA6 gene was sensitive to thapsigargin-induced ER stress. Thapsigargin treatment induced an increase in the mRNA abundance of PDIA6 but not that of PDIA7, another ER associated oxidoreductase (Fig. 1F). PDIA6 mRNA was increased in cells treated with thapsigargin, tunicamycin, brefeldin A and the ER luminal Ca2+ chelator TPEN, but not when ER stress was induced by oxidative stress through the addition of dithiothreitol, treatment with cyclosporine A, or staurosporine (Suppl. Fig. S5). These results suggest that PDIA6 mRNA abundance is selectively regulated by specific ER stress stimuli.
PDIA6 differentially affects the IRE1α and ATF6 pathways
Next, we tested whether PDIA6 silencing influenced other branches of the UPR using the UPRE and ERSE reporters. The UPRE reporter contains an UPRE element that responds to the transcriptional activities of XBP1 and ATF6α (18). The ERSE reporter contains multiple ERSEs that report ATF6 transcriptional activity (19). ATF6 exhibits low affinity for the UPRE, but high affinity for the ERSE, whereas XBP1 has high affinity for the UPRE and low affinity for the ERSE (19). The induction of the UPRE reporter in response to thapsigargin was not affected by PDIA6 silencing under these conditions (Fig. 1G). In contrast, the response of the ERSE to thapsigargin was significantly increased upon PDIA6 silencing (Fig. 1H). Furthermore, these conditions also increased the expression of the gene encoding calreticulin (Fig. 1I), a Ca2+ sensitive ERSE responsive gene (20). These data suggest that in the absence of PDIA6, Ca2+ store depletion resulted in activation of the ATF6 pathway. Next, we tested for the effect of PDIA6 on the PERK pathway by analyzing the phosphorylation of Ser51 in eIF2α. Western blot analysis showed that thapsigargin treatment induced phosphorylation of Ser51 in eIF2α that was not affected by silencing PDIA6 with the time point and concentration of thapsigargin used (Suppl. Fig. S4). Combined with the XBP1 reporter data, we concluded that PDIA6 silencing did not affect the PERK pathway but suppressed the IRE1α activity and increased ATF6 activity in response to ER Ca2+ store depletion-induced ER stress.
PDIA6 forms complexes with BiP and IRE1α
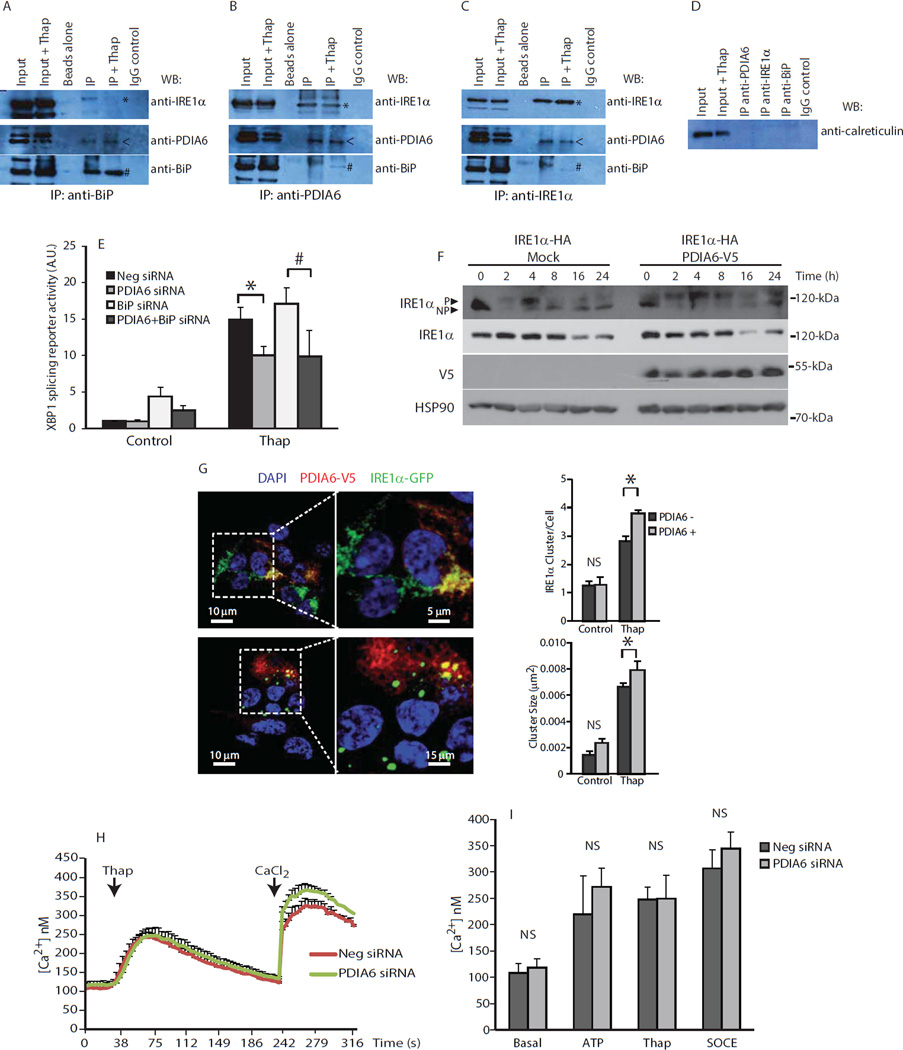
We hypothesized that PDIA6 could regulate UPR signaling through physical interactions with ER stress sensors and/or ER luminal modulators. As previously reported (21, 22), BiP coimmunoprecipitated with PDIA6 (Fig. 2A). PDIA6 also coimmunoprecipitated with IRE1α (Fig. 2B,C) and PDIA6-IRE1α complex formation did not appear to be altered when immunoprecipitation was carried out from cells treated with thapsigargin (Fig. 2B,C). We also showed that calreticulin, another ER luminal resident protein, was not present in BiP, PDIA6 or IRE1α immunocomplexes (Fig. 2D).
Figure 2. PDIA6 interacts with BiP and IRE1α and controls IRE1α activity.
(A to C). Top blots: To detect associations of IRE1 with proteins involved in ER stress responses, immunoprecipitations (IP) using anti-BiP, anti-PDIA6, or anti-IRE-1α were performed in COS-1 cells expressing IRE1-NLD and immunoblotted with anti-IRE1α antibodies. Middle and bottom blots: immunoprecipitations (IP) using anti-BiP, anti-PDIA6, or anti-IRE-1α were performed in NIH-3T3 fibroblasts and immunoblotted with anti-PDIA6 or anti-BiP. *, depicts the location of IRE1-NLD protein band; <, depicts the location of PDIA6 protein band; #, depicts the location of BiP protein band. (D) Immunoprecipitations (IP) using anti-BiP, anti-PDIA6, or anti-IRE-1α were performed in NIH-3T3 fibroblasts and immunoblotted with anti-calreticulin. Data for (A) to (D) is representative of more than 3 biological replicates. (E). Activity of the XBP1 splicing reporter in NIH-3T3 fibroblasts transfected with siRNA for PDIA6, BiP *Neg siRNA/PDIA6 siRNA. *P-value = 0.00176; #BiP siRNA/PDIA6+BiP siRNA. Data are representative of more than 3 biological replicates. P-value = 0.04988. (F). Phosphorylation of IRE1α-HA was analyzed by a Phostag™ assay in HEK293 cells expressing IRE1α-HA or IRE1α-HA and PDIA6-V5. P, phosphorylated protein band; NP, non-phosphorylated protein band. Total abundance of IRE1α-HA and PDAI-V5 proteins was analyzed by Western blot. Data is representative of 3 biological replicates. (G). HEK293 cells expressing IRE1-GFP were transiently transfected with expression vector encoding PDIA6-V5 or control vector (control) and treated with thapsigargin (Thap). Left panel; IRE1α-GFP positive clusters were analyzed by immunofluorescence in cells expressing PDIA6-V5. Right panel: IRE1α clusters per cell and cluster size. For cluster/cell P-value = 0.0417; for cluster size P-value = 0.0274. A representative experiment of three independent experiments is presented. Sixty cells were analyzed for each independent experiment. (H), (I). Ca2+ measurements in cells with silenced PDIA6. SOCE was initiated by the addition of CaCl2. SOCE, store-operated Ca2+ entry. Data is representative of more than 3 biological replicates.
Three additional techniques provided evidence for a potential interaction between PDIA6 and IRE1α. His-tagged IRE1α ER luminal domain (IRE1-NLD) pulled down PDIA6 in the absence or presence of ER stress (Suppl. Fig. S6A). Using surface plasmon resonance (BIACore) and thermophoresis we showed that PDIA6, but not calreticulin, tightly bound to the immobilized IRE1α with a relatively high Kd value of approximately 22 nM (Suppl. Fig. S6B,E). Thus, PDIA6 could directly associate with the ER luminal domain of IRE1α with high affinity and form a stable complex.
Because PDIA6 is an ER luminal oxidoreductase, we considered whether Cys109, Cys148 and Cys332, in the ER luminal portion of IRE1α could be involved in binding to PDIA6. Cys109 and Cys148 are highly conserved in the IRE1α proteins (23). We generated C109A, C148A and C332A mutations to create the triple IRE1-NLD mutant (C109,148,332A–IRE1-NLD). PDIA6 binding to the C109,148,332A–IRE1-NLD mutant was greatly reduced as assessed by surface plasmon resonance analysis (Suppl. Fig. S6C), suggesting that cysteine residues were involved in the binding of PDIA6 to IRE1α. In addition, surface plasmon resonance indicated that PDIA6 did not interact with wild-type IRE1-NLD domain treated with NEM, which alkylates cysteine thiols, thereby irreversibly blocking the cysteines (Suppl. Fig. S6D). Taken together, these results suggested that cysteine residues in the IRE1α ER luminal domain were required for binding to PDIA6. Furthermore, binding of PDIA6 to IRE1-NLD was abolished in the presence of EGTA (Suppl. Fig. S6D), indicating that under BIACore conditions there was a requirement for Ca2+ for the PDIA6-IRE1-NLD interaction. Finally, MicroScale Thermophoresis (MST) showed that PDIA6 bound IRE1-NLD at a similar Kd (20 nM) to that determined by surface plasmon resonance (Suppl. Fig. S6B,E). Taken together these results indicated that PDIA6 formed complexes with both BiP and IRE1α and these interactions may be at least partially responsible for PDIA6-dependent effects on IRE1α activity.
Considering that PDIA6 and BiP form complexes, we asked whether PDIA6 directly affected the activity of BiP towards the activation of IRE1α-mediated ER stress responses. The increase in IRE1α reporter activity was reduced by silencing of PDIA6 and enhanced by silencing of BiP (Fig. 2E). When both genes were silenced, PDIA6 silencing blunted the increase in the ER stress that was caused by BiP silencing (Fig. 2E). Thus, in the absence of BiP, IRE1α activity was modulated by the silencing of PDIA6, but BiP was not necessary for PDIA6-dependent regulation of IRE1α.
PDIA6 regulates the inactivation of IRE1α
Because the effects of PDIA6 were observed in cells exposed to prolonged treatments with thapsigargin, we monitored the early stage kinetics of phosphorylation of IRE1α. Time course experiments indicated that the early increase in the phosphorylation of IRE1α in thapsigargin-treated wild-type cells was transient (Fig. 2F). In cells overexpressing PDIA6, phosphorylation of IRE1α was sustained upon thapsigargin treatment, even up to 24 hours of treatment (Fig. 2F). Autophosphorylated and dimerized IRE1α form large clusters that enhance its activity (24). In a cell line expressing doxycycline-inducible IRE1α-GFP (24), the number and size of IRE1α clusters in cells was increased by expression of PDIA6-V5 and treatment with thapsigargin (Fig. 2G). We concluded that increased PDIA6 abundance promoted the sustained activation of IRE1α signaling.
To process the mRNA encoding for XBP1, the RNAse activity of IRE1α mediates the rapid degradation of a subset of mRNAs that encode ER membrane associated or secreted proteins, a process referred to as regulated IRE1α-dependent decay (RIDD) (25–27). We measured the mRNA abundance of col6 and scara, which are IRE1α RIDD substrates (Suppl. Fig. S7). As expected, wild-type cells exposed to the thapsigargin-induced ER stress exhibited a time-dependent decay of col6 and scara mRNA (Suppl. Fig. S7). In the early phase of IRE1α activation, PDIA6 overexpression in HEK293T cells resulted in increased decay of scara mRNA, but not of col6 mRNA (Suppl. Fig. S7). Taken together, these results suggested that increased PDIA6 abundance may affect different IRE1α signaling outputs by modulating the kinetics of IRE1α activation.
PDIA6 does not disrupt ER Ca2+ homeostasis
PDIA6 resides in the lumen of the ER, and contains acidic amino acid residues near the C-terminus that may be involved in Ca2+ binding that could enable PDIA6 to play a role in buffering ER luminal Ca2+ (28). We asked whether PDIA6 affected Ca2+ buffering of the ER and consequently ER Ca2+ homeostasis. In cells treated with thapsigargin or ATP to induce Ca2+ release from the ER, PDIA6 silencing did not alter the amount of Ca2+ released from the ER or the amount of SOCE (Fig. 2H,I). These results suggest that PDIA6 modulated IRE1α activity through a direct interaction between the two proteins, rather than by directly altering ER Ca2+ homeostasis.
PDIA6 is a target for miR-322
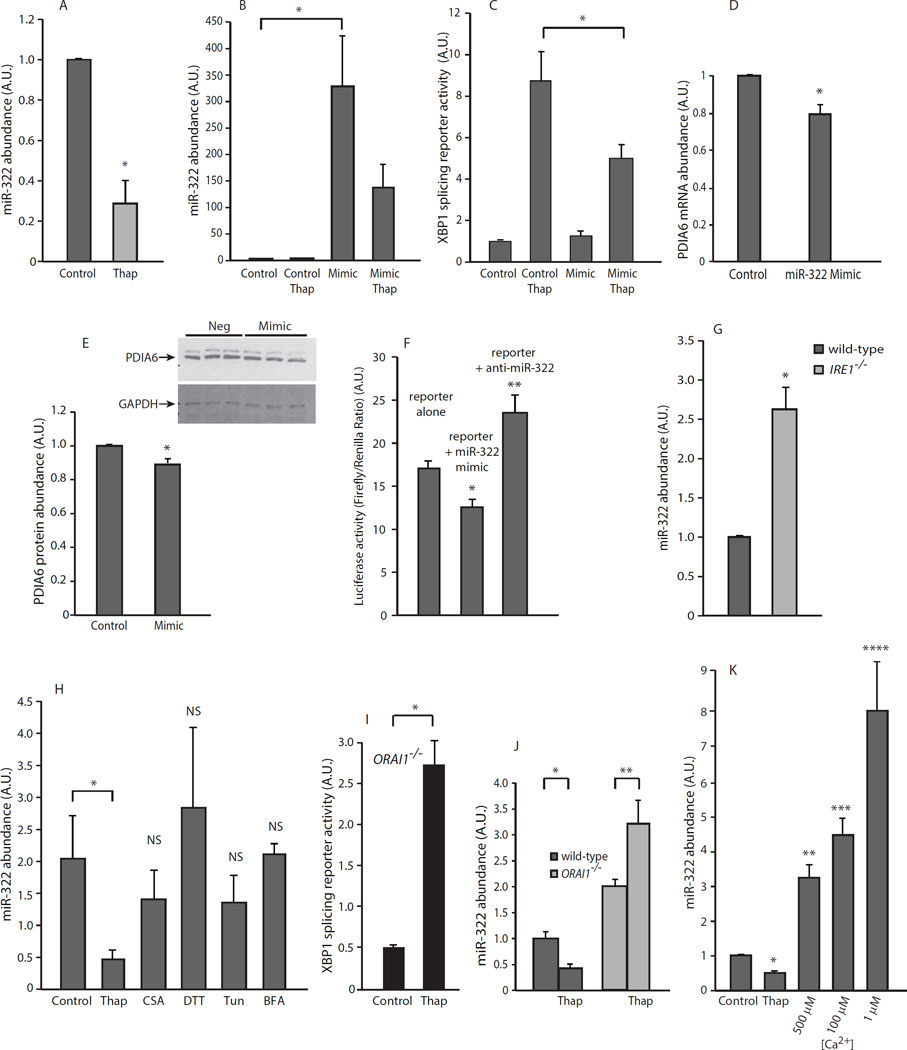
Thapsigargin-dependent activation of the UPR by Ca2+-store depletion involves changes in miRNA abundance (29); therefore, we carried out deep sequencing analysis of thapsigargin-treated NIH-3T3 cells and identified 13 miRNAs showing differential expression. Eight of these miRNAs showed increased expression (miR-217, miR-216, miR-217*, miR-216b, miR-92a-1, miR-708, miR-1937a, and miR-1O1b) whereas five exhibited decreased expression (miR-671-5p, miR-503, miR-669f-3p, miR-322, and miR-143) in response to thapsigargin treatment (Suppl. Fig. S8). We used multiple target prediction programs, including TargetScan (30, 31), DIANAmicroT (32) and RepTar (33), to generate a list of candidate transcripts with putative miRNA binding sites. Using Ingenuity Pathways Analysis (IPA), we determined that each miRNA had the potential to target different cellular pathways. Direct comparison of the two screens (Suppl. Fig. S9) revealed an overlap between miRNA targets and the top candidates in the siRNA library screen. Bioinformatics analysis indicated that miR-322 (miR-424 in the human miRNA database) might target the PDIA6 gene.
Q-PCR analysis of thapsigargin-treated NIH-3T3 cells showed that miR-322 abundance was reduced over 60% by depletion of Ca2+ stores (Fig. 3A), thus confirming the deep sequencing results. To determine whether PDIA6 was a target for miR-322, we used chemically synthesized miRNA “mimics” (34) to ectopically increase miR-322 abundance (Fig. 3B). The miR-322 mimic decreased the activity of the IRE1α reporter (Fig. 3C) in a manner that was dependent on ER Ca2+ depletion. It also decreased the mRNA and protein abundance of PDIA6 (Fig. 3D, E). Next we measured miR-322-mediated translational repression of PDIA6 mRNA using a luciferase reporter containing the predicted miR-322 targeted PDIA6 3’ UTR sequence. Luciferase reporter activity was decreased by transfection of the miR-322 mimic and increased by transfection of anti-miR-322 (Fig. 3F). These results suggested that miR-322 targeted the 3’ UTR of PDIA6 mRNA.
Figure 3. miR-322 abundance is regulated by ER stress.
(A). Q-PCR analysis of miR-322 abundance. *P-value = 0.0073. (B). Q-PCR analysis of miR-322 abundance in NIH-3T3 fibroblasts transfected with miR-322 mimic (Mimic) and treated with thapsigargin (Thap). *P-value = 2.31E-06. Data is representative of more than 3 biological replicates. (C). NIH-3T3 fibroblasts were transfected with miR-322 mimic (Mimic) and XBP1 splicing reporter to monitor IRE1α activity. *P-value = 0.0383. (D). NIH-3T3 fibroblasts transfected with miR-322 mimic (Mimic) followed by treatment with thapsigargin (Thap). PDIA6 mRNA abundance was assessed by Q-PCR. *P-value = 0.0002. (E). NIH-3T3 fibroblasts were transfected with miR-322 mimic (Mimic) followed by treatment with thapsigargin (Thap). *P-value = 0.0366. Inset, Western blot representing three independent experiments was first probed with anti-PDIA6 antibodies (upper panel), striped and then re-probed with anti-GAPDH antibodies (lower panel). Neg, negative scrambled control siRNA; Mimic, miR-322-mimic siRNA. (F). PDIA6 3’ UTR reporter activity in the presence of miR-322 mimic or inhibitor. *P-value=0.0001 (mimic), **P-value=0.029 (inhibitor). (G). miR-322 abundance in wild-type and IRE1−/− mouse embryonic fibroblasts. *P-value=0.0045. Data is representative of more than 3 biological replicates. (H). NIH-3T3 fibroblasts were treated with thapsigargin (Thap), cyclosporine A (CSA), dithiothreitol (DTT), tunicamycin (Tun) or brefeldin A (BFA). *P-value = 0.0073. (I). XBP1 splicing in ORAI1-deficient cells. ORAI1−/− fibroblasts; *P-value = 0.0061. (J). miR-322 in ORAI1−/− fibroblasts. *P-value = 0.0061, *P-value = 0.0123. (K). miR-322 abundance in cells treated with thapsigargin (Thap) or with different extracellular Ca2+ concentrations. *P-value = 0.0036; **P-value = 0.0006; ***P-value = 0.001; ****P-value = 0.0022. All data in the figure is representative of more than 3 biological replicates.
IRE1α can act as an endoribonuclease for specific miRNAs (35) and many miRNAs are regulated by the UPR (36). Therefore, we tested whether the abundance of miR-322 depended on IRE1α. miR-322 abundance was increased 2.5-fold in IRE1α-deficient cells compared with control cells (Fig. 3G), suggesting that IRE1α affected miR-322 abundance. It remains to be determined if miR-322 abundance is directly controlled by the RNAse activity of IRE1α or through downstream signaling responses. Taken together, these findings indicated that miR-322 abundance was sensitive to ER Ca2+ depletion-induced ER stress and modulated by IRE1α, thereby directly affecting the downstream expression of PDIA6 mRNA at the transcriptional and translational level.
miR-322 abundance is regulated by ER Ca2+ depletion and activation of SOCE
Although miR-322 abundance was robustly reduced in cells treated with thapsigargin, it was not significantly affected in cells treated with CSA, DTT, tunicamycin, or brefeldin A (Fig. 3H). We concluded that miR-322 abundance was not affected by ER stress induced by redox or folding environment changes but that it was sensitive to ER stress induced by thapsigargin.
To examine whether SOCE activity and consequently changes in the cytosolic Ca2+ concentration in thapsigargin-treated cells also played a role in the regulation of miR-322 abundance, we used cells deficient in ORAI1, a plasma membrane Ca2+ channel that is responsible for SOCE (12), Depletion of ER Ca2+ stores with thapsigargin results in activation of ORAI1, Ca2+ entry and a rapid increase in the cytosolic Ca2+ concentration (12), In the absence of ORAI1, Ca2+ depletion of ER stores with thapsigargin does not promote SOCE and cytosolic Ca2+ remains at the resting concentration (12, 14). In ORAI1−/− cells, thapsigargin induced ER stress as monitored by XBP1 mRNA splicing was increased (Fig. 3I). In addition, these cells had increased miR-322 abundance as compared to the wild-type cells and this was further increased with thapsigargin (Fig. 3J). These results indicated that thapsigargin-induced ER Ca2+ depletion at resting cytosolic Ca2+ concentrations in the absence of SOCE may have been ineffective in suppressing miR-322 abundance. An increase in miR-322 abundance in ORAI1-deficient cells pointed at a potential additional role of cytosolic Ca2+ in processing this microRNA. Next, we varied extracellular Ca2+ concentration in the absence of thapsigargin to reduce both ER stores and cytosolic Ca2+ concentrations. Lowering the extracellular Ca2+ concentration from 500 µM to 1 µM, a condition that reduces the cytosolic Ca2+ concentration but may also modify ER Ca2+ content, resulted in a concomitant increase in miR-322 abundance (Fig. 3K). Taken together, these data suggested that under conditions of ER stress and UPR activation due to disrupted ER Ca2+ homeostasis and activation of SOCE, miR-322 abundance was reduced. In contrast, reduction of cytosolic Ca2+ in the absence of Ca2+ store depletion and SOCE led to an increase in miR-322 abundance, although there is the possibility that ER Ca2+ was altered by these experimental conditions.
PDIA6 and miR-322 are regulated by ER stress in vivo
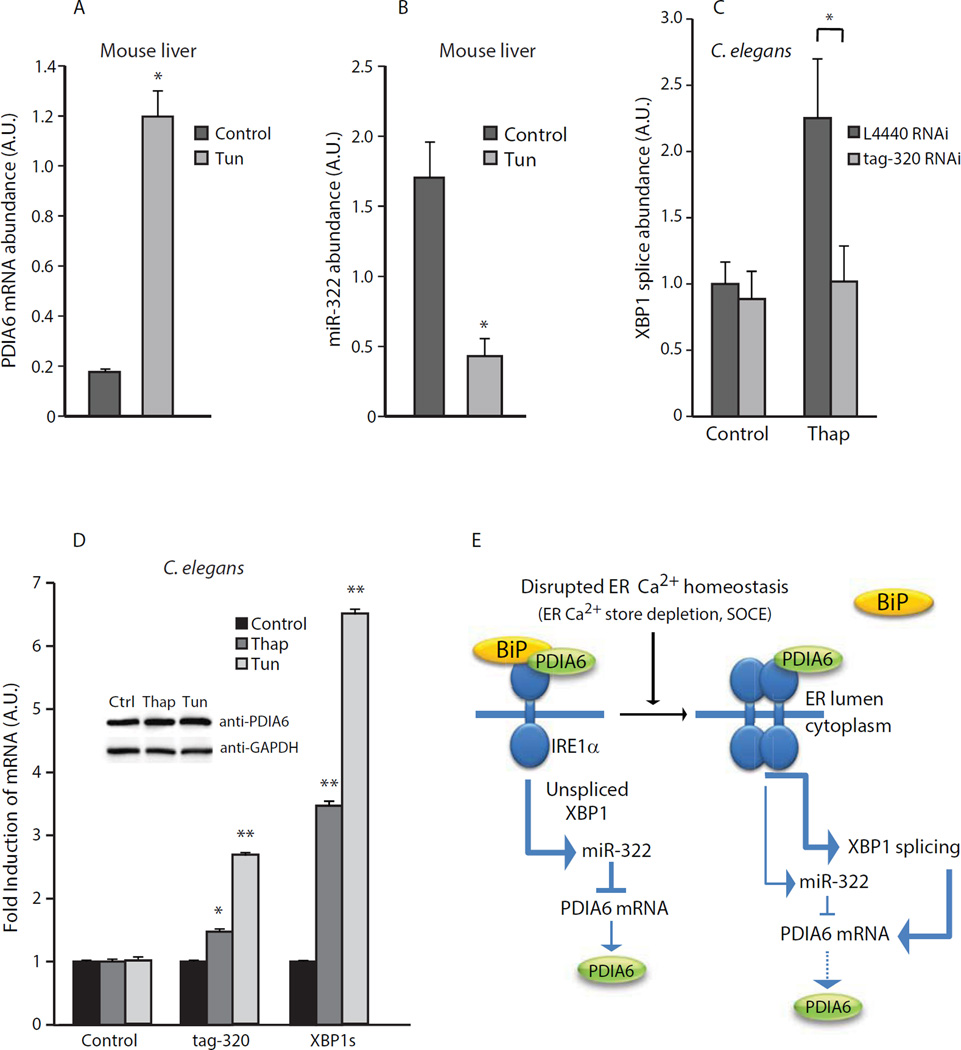
To establish the effect of PDIA6 on XBP1 splicing in vivo, we induced ER stress in mice and C. elegans and analyzed PDIA6 protein and mRNA abundance, miR-322 abundance and PDIA6-dependent XBP1 splicing. In agreement with the in vitro analysis (Fig. 1C), PDIA6 abundance in the livers of mice injected with tunicamycin to induce ER stress was significantly increased (Fig. 4A). Full activation of XBP1 mRNA splicing and increases in the abundance of CHOP and BiP occur in this in vivo animal model (37). We also observed a concomitant reduction in miR-322 abundance in the livers of mice with induced ER stress (Fig. 4B) although miR-322 abundance was only somewhat reduced in tunicamycin-treated NIH-3T3 cells (Fig. 3H).
Figure 4. In vivo analysis of PDIA6, ER stress and miR-322 in mouse liver and C. elegans.
(A). PDIA6 mRNA abundance in liver from mice with tunicamycin-induced ER stress. *P-value = 0.0001. (B). miR-322 abundance in liver from mice with tunicamycin-induced ER stress. Tun, tunicamycin. *P-value = 0.01. In (A) and (B) data is representative of 3 mice per condition. (C). Spliced XBP1 abundance in C. elegans treated with indicated RNAi in the absence (Control) or presence of thapsigargin (Thap). L4440 is the control RNAi. tag-320 is a C. elegans PDIA6 homolog. Thap, thapsigargin. *P < 0.05. (D). tag-320 (PDIA6) and spliced XBP1 (XBP1s) abundance in C. elegans treated with thapsigargin (Thap) or tunicamycin (Tun). *P < 0.01, *P<0.005. In (C) and (D) data is representative of more than 3 animals per condition. Insert, Western blot of C. elegans protein extracts probed with anti-PDIA6 antibodies. (E). A schematic representation of interplay between ER Ca2+ depletion induced-ER stress, Ca2+, PDIA6 and miR-322. Disruption of ER Ca2+ homeostasis induces the unfolded protein response (UPR) and dissociation of BiP from IRE1α, the IRE1α endoribonuclease splices the XBP1 mRNA and the spliced XBP1 mRNA encodes a transcription factor that activates ER stress responsive genes including the PDIA6 gene. In parallel, miR-322 abundance is reduced and PDIA6 interacts with IRE1α to support and sustain endoribonuclease activity to deal with disrupted ER homeostasis-induced ER stress. Dashed lines indicate a modest effect.
To test whether PDIA6 affected XBP1 mRNA splicing, we measured XBP1s transcript abundance in C. elegans treated with RNAi against tag-320, the C. elegans homolog of PDIA6. Knock-down of tag-320 reduced the thapsigargin-induced increase in XBP1s mRNA abundance (Fig 4C). Furthermore, tag-320 abundance was increased at the mRNA level when worms were under ER stress induced by thapsigargin or tunicamycin (Fig 4D). These results suggested that tag-320 was functionally required for XBP1 mRNA splicing, and that tag-320 expression was also inducible in nematodes challenged with ER stress.
Discussion
A critical function of the UPR is the recovery of normal functions of the cell by halting protein translation and activating signaling pathways that lead to increased production of molecular chaperones. The disruption of energy and/or nutrient balance is a fundamental cause of ER stress, and induces various corrective strategies. The ER stress coping responses help to reestablish ER and cellular proteostasis and ensure cell survival. In this study, we focused on the identification of ER stress coping responses induced by alterations of ER Ca2+ homeostasis, and consequently by changes in cellular Ca2+ homeostasis induced by the ER Ca2+ stores depletion. We performed siRNA arrays and miRNA profiling arrays to search for ER coping components. Our findings identified two unexpected levels of UPR modulation and point at a potential regulatory network involving a specific microRNA and SOCE. We discovered that the ER oxidoreductase PDIA6 interacted with IRE1α. We demonstrated that PDIA6 was required for maintenance of IRE1α activity as monitored by IRE1α phosphorylation, cluster formation and downstream XBP1 mRNA splicing. ER Ca2+ depletion and activation of SOCE promoted changes in the abundance of different miRNAs, including miR-322, which targeted PDIA6 and regulated downstream IRE1α activity. We showed that in mouse and C. elegans animal models PDIA6 and miR-322 abundance was modulated by ER stress and that PDIA6 influenced IRE1α signaling in C. elegans. It remains to be determined why ER stress induced by tunicamycin decreased miR-322 abundance in mice but not in cells. Silencing PDIA6 also induced global ER stress suggesting that the protein has an important function in maintaining protein folding in the ER. Together, these findings suggested that interplay between ER Ca2+, PDIA6, IRE1α and miR-322 was part of a regulatory network activated by disrupted ER Ca2+ homeostasis that fine-tuned the UPR and probably the survival of cells undergoing ER stress. However, it will be necessary to perform additional, more precise manipulations to firmly establish the roles of Ca2+ in the cytosolic and ER compartments in the modulation of this regulatory network.
Figure 4E shows a schematic representation of the relationship between disrupted ER Ca2+ homeostasis, ER stress, IRE1α, PDIA6 and miR-322. BiP is a centrally located modulator and sensor of the ER stress response, and regulates all branches of the UPR. Under basal conditions, BiP binding to IRE1α desensitizes IRE1α to low amounts of stress, and promotes its deactivation when favorable folding conditions are maintained or restored to the ER. BiP forms complexes with PDIA6 [this work, (21, 22)], and this study further showed that PDIA6 also bound to IRE1α. We propose a model in which under basal conditions, IRE1α is not activated when complexed with BiP and low amounts of PDIA6, thus resulting in little splicing of XBP1 mRNA. The relatively high miR-322 abundance under these conditions results in low PDIA6 mRNA abundance. Disruption of ER Ca2+ homeostasis and activation of SOCE results in dissociation of BiP from IRE1α and rapid dimerization of IRE1α, leading to activation of its kinase and endoribonuclease activities. Under stress conditions, PDIA6 binding to IRE1α may sustain its long term activity by stabilizing the dimeric or oligomeric state of the protein. This stabilization, combined with increased abundance of XBP1, increases the abundance of the PDIA6 protein, and may allow the UPR to function robustly over time. Under the conditions of disrupted ER Ca2+ homeostasis, the interplay between Ca2+, PDIA6 and miR-322 creates a reciprocal regulatory loop to promote sustained IRE1α activity to support corrective strategies to restore ER homeostasis.
The ER stress coping response is composed of distinct pathways controlled by common regulatory components (IRE1, PERK, ATF6, ATF4, and CHOP). BiP appears to function as a ligand for IRE1α, PERK and ATF6. Here we showed that other components of the ER luminal environment may regulate ER stress responses. The ability of PDIA6 to specifically affect IRE1α function supports the concept that IRE1α, PERK and AFT6 branches of ER stress are selectively modulated by additional mechanisms. This is in line with the identification of specific regulators of IRE1α that form a protein complex referred to as the UPRosome, of which PDIA6 may be a new component (38). It is likely that UPR pathways may function together as a single entity (“regulon”) that would be controlled by the same regulatory system and would respond to ER stress in a coordinated fashion (1). Silencing PDIA6 during ER stress induced by Ca2+ store depletion had opposite effects on the IRE1α and ATF6 branches of ER stress responses, without affecting PERK signaling. Activation of ATF6 may compensate for the inhibition of IRE1α and would support a concept of ER stress induced pathways (IRE1α, PERK and ATF6), functioning in a coordinated fashion to respond to ER stress.
Disrupted ER Ca2+ homoeostasis decreased miR-322 abundance, thereby allowing increased PDIA6 abundance under conditions that stimulate ER stress. Approximately 60% of the mRNAs of the cell are predicted to be regulated by miRNA function (38). At present, more than 1000 miRNAs have been identified in the human genome, with each miRNA targeting numerous mRNAs. In addition, one target mRNA may be regulated by multiple miRNAs (39). This work and other evidence points to a role for miRNAs during ER stress (40), and indicates that miRNAs regulate the cellular coping response under various stress conditions including ER stress. For example, miRNA profiles are changed during oxidative stress (41, 42), nutrient deficiency (43, 44), DNA damage (45–47), and oncogenic stress (48). Therefore, in response to stress, the cell may alter the gene expression program through regulation of miRNAs, which does not involve de novo synthesis of protein and therefore is a quicker response. Disruptions of specific miRNAs may not present a noticeable phenotype unless the system is stressed (40). For example, mice lacking miR-208 do not have an overt phenotype unless stressed with cardiac overload (49). As well, Drosophila lacking miR-7 show a breakdown in eye development when subjected to alternating temperatures (50), and inactivation of miR-8 in Danio rerio prevents responses to osmotic stress (51).
One way in which ER stress regulates miRNA abundance could be through direct regulation of miRNA expression by members of the ER stress pathway. ER stress-dependent activation of ATF6 decreases the abundance of miR-455 as a coping mechanism, contributing to the protective effects of ATF6 in the heart (52). Other coping mechanisms involve the transcription factors CHOP and XBP1 which regulate miR-708 and miR-346, respectively (29, 53). The Dicer splicing machinery itself is activated by calpain and Ca2+ (54). Dicer is involved in processing small hairpin precursors (pre-miRs) into mature miRNAs that become associated with Argonaute to form RNA-induced silencing complexes (RISC). Exposure of a neuronal cell line to increased extracellular Ca2+ results in the appearance of the active form of Dicer, as well as the full length form of Argonaute, both cleaved by activated calpain, possibly leading to direct cleavage of pri-miRNAs (55). The endoribonuclease activity of IRE1α directly cleaves the pri-RNA cluster complex of miR-17 to antagonize classical Dicer processing of miR-17, thereby promoting apoptosis (35). IRE1α may regulate miR-322 in a similar manner as miR-17 because miR-322 has nucleotide sequences similar to the IRE1α endonuclease target sites in the IRE1α endoribonuclease substrates miR-17 and XBP1 mRNA. In support of this possibility, we showed that the IRE1α-deficient cells have significantly increased miR-322 abundance. This study suggests that the decreased abundance of miR-322 during disrupted ER Ca2+ homeostasis could rely on changes in the cytosolic Ca2+ concentration specifically provided by SOCE. ER Ca2+ depletion and activation of SOCE were at least partially necessary for the decreased abundance of miR-322 under ER stress conditions. We showed that miR-322 abundance was increased in ORAI1-deficient cells which do not have SOCE and by reduced cytosolic Ca2+ in the absence of SOCE. Taken together, these findings suggest that the disruption in ER Ca2+ homeostasis due to depletion of ER Ca2+, activation of SOCE and SOCE-independent changes in the cytosolic Ca2+ may differentially control the transcriptional regulation of this miRNA.
Based on the increasing relevance of the UPR for the development of several human diseases, including cancer, neurodegeneration and diabetes (1), as well as advances in therapeutic strategies to target the UPR in diseases (56), our study provides potential points of manipulation of the UPR through the IRE1α, PDIA6 and miR-322 axis.
Materials and Methods
Plasmids and site-specific mutagenesis
The pRL-IXFL XBP1 splicing reporter contained an internal Renilla control and the nucleotide sequence encoding XBP1 followed by firefly luciferase separated by an internal ribosomal entry site (IRES) initiation region (13). This reporter will only generate Firefly luciferase activity if the XBP1 cDNA is spliced in frame with the cassette encoding firefly luciferase. The Cignal Luciferase Reporter Assays were from Qiagen (Cat #CCS-9031L, CCS-2032L). Expression vector IRE1-NLD was constructed as described in (23) and used for site specific mutagenesis. The following DNA primers were used for site specific mutagenesis of IRE1-NLD: for C109A mutation, forward primer 5’-GAATTGGTGCAGGCATCCCCAGCCCGAAGTTCAGATGGAATCC-3’, reverse primer 5’-GGATTCCATCTGAACTTCGGGCTGGGGATGCCTGCACCAATTC-3’; for C148A mutation, forward primer 5’-GGCCTTTGCAGATAGTCTCGCCCCATCAACCTCTCTTCTG-3’ and reverse primer 5’-CAGAAGAGAGGTTGATGGGGCGAGACTATCTGCAAAGGCC-3’; for C332A mutation, forward primer 5’-GGGACAAGGGGGAGGCTGTGATCACGCCC-3’ and reverse primer 5’-GGGCGTGATCACAGCCTCCCCCTTGTCCC-3’.
Cell culture, transfection and siRNA library screen
NIH-3T3 mouse fibroblasts, ORAI1-deficient mouse fibroblasts, IRE1α-deficient mouse embryonic fibroblasts and COS-1 cells were maintained under standard tissue culture conditions, including 5% CO2 with high humidity. Tissue culture media included 10% fetal bovine serum (FBS) in Dulbecco’s Modified Eagle Medium (DMEM) (both from Sigma). The siRNA library (Ambion) was pooled and aliquoted into 96 well full skirt PCR plates (Axygen) (57). Reverse transfection was performed which involves preparation of a mixing plate containing the siRNA, plasmid DNA, Dharmafect Duo (ThermoFisher) and Optimem (Life Technologies), followed by addition of cells and plating into white 96 well plates (Corning) for 48 hours. Briefly, mixing plates were prepared containing 22 µl of 200 nM siRNA per well in a deep well format followed by addition of a mixture of 42 µl Optimem (Invitrogen), 22 µl of 20 µg/ml XBP1 splicing reporter, 1.76 µl Dharmafect Duo, and 352 µl of 1.2×105 cells/ml per well. The plates were mixed with four mixing cycles using the PerkinElmer robot and then 100 µl was aliquoted into the wells of four white 96 well plates (four technical replicates). The plates were then incubated. Drug treatment consisted of removal of 50 µl of the media, followed by addition of 50 µl of 1 µM thapsigargin in DMEM with 10% FBS, bringing the final concentration of thapsigargin to 0.5 µM. Cells were incubated with thapsigargin for 24 hours followed by removal of media. Cell lysis was performed in plate with 20 µl Passive Lysis Buffer™ (Promega) followed by the luciferase assay, and analyzed with a PerkinElmer EnVision 2104 Multilabel Plate Reader (PerkinElmer). Briefly, 100 µl of luciferase buffer (20 mM Tricine, pH 7.2, 1.07 mM MgCO3, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM DTT, 270 µM coenzyme A, 470 µM luciferin, 530 µM ATP) was added to each well. Luciferase activity was monitored within 30 seconds from the addition of the luciferase buffer to minimize loss of signal.
The secondary screen involved computer directed and robot generated picking of statistically significant siRNA for 400 genes, including siRNA that either increased or decreased ER stress. siRNAs for each gene were randomly placed in 96 well full skirt PCR plates (Axygen). These plates also included the water, negative and positive control samples. A similar transfection was performed as above, using mixing plates to combine the ingredients for the transfection, except volumes were calculated for technical triplicates, followed by 48 hours incubation to allow silencing to occur. Drug application was performed in a similar manner as above, with harvesting after 24 hours using 20 µl Passive Lysis Buffer™ according to the manufacturer’s protocol. Dual Luciferase Assay analysis (Promega) was performed by adding 50 µl of LARII buffer that gives the Firefly luciferase signal, followed by addition of 50 µl of the Stop and Glo buffer to generate the Renilla signal according to the manufacturer’s protocol (58). Cell viability assays were carried out three times in biological triplicate in a 96 well format using the pooled siRNA for each gene selected as described above, including a negative siRNA control and a non-silencing control. Results are presented as normalized to the internal Renilla expression and untreated negative scrambled control siRNA set at 1. Cell growth was analyzed using the MTS Cell Proliferation Assay (Promega) according to the manufacturer’s protocol. Briefly, the tetrazolium compound MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H–tetrazolium] is biologically reduced by dehydrogenase enzymes found in metabolically active cells into a formazan product that is soluble. The amount of formazan measured at an absorbance of 490 nm is directly proportional to the number of living cells.
To validate our screening results, Dharmacon siGENOME Smartpools were used to silence the selected genes. Manual large format siRNA transfections were performed more than three times in technical triplicate using 48 or 24 well plates with siRNA for each selected gene. Conditions were maintained according to the primary library screen and scaled up 2-fold or 4-fold to account for 48 or 24 well plates respectively. Results were obtained using the Dual Luciferase Assay analysis according to manufacturer’s protocol. The following are the nucleotide sequences of the siRNAs employed in this study: PDIA6 (Thermo Scientific, siGENOME Smartpool and set of 4 upgrade): 5’-UCGAUUUGUUCUCUGAUAA-3’ (siRNA #1), 5’-GCCUGUGGCUUGUAGAAUU-3’ (siRNA #2), 5’- GAUAAUCAACGAAGACAUA-3’ (siRNA #3), 5’-GGUGAUAGUUCAAGUAAGA-3’ (siRNA #4); BiP (Silencer, Ambion) 5’-GGACAUCAAGUUCUUGCCATT-3’, 5’- GGUGGUUGAAAAGAAAACUTT-3’, 5’-GGUUACCCAUGCAGUUGUUTT-3’; IRE1α (Silencer, Ambion), 5’-CCUUGUUGUUUGUCUCGACTT-3’, 5’-GGCCUGACGAAACUUCCCUTT-3’, 5’-GCAAGCUGAACUACUUGAGTT-3’, and negative control siRNA (Ambion Cat #: AM4635).
Statistical analysis of primary and secondary siRNA library screen
Before statistical analyses of the primary and secondary library screens, values on the plate were shifted by the average water readout. Then for each siRNA, the average was computed from four technical replicates; the value of the replicate that was the furthest away from the average was removed as an outlier. The average was then computed using only the values of the three remaining replicates and was used as a representative value. Next, for each treatment, all representative siRNA values were converted into P-values, using the SN package (59) for R platform to fit the skewed normal distribution. Finally, for each treatment, the 400 most extreme P-values (containing a mix of the lowest and the highest values) were selected and used in the secondary screen with the dual luciferase reporter assay.
The results from the secondary screen were normalized as previously described (60). As each plate contained a different set of siRNAs, the 16 wells which had the same content along all plates were used as a normalization baseline. The Firefly/Renilla (F/R) fold changes for the above mentioned 16 wells across all plates were computed, and these values were used to normalize all plates using the first plate as reference. After normalization, the average from two replicates was used as the representative F/R readout for an siRNA. Similar to the first round, the CDF (cumulative distribution function) values were generated using skewed normal distributions which were fitted to the data.
Protein purification, pull-down and BIACore analyses
PDIA6 was isolated from mouse liver microsomal fraction. In brief, livers were homogenized using a buffer containing 37.5 mM Tris Maleate, pH 6.4, 150 mM NaCl, 5 mM MgCl2, and 250 mM sucrose. The resulting mixture was centrifuged at 10,000 xg for 30 min at 4°C. The supernatant was further centrifuged at 100,000 xg for 90 min at 4°C. The pellet containing microsomes was washed with 10 mM Tris, pH 7.4, and re-suspended in 10 mM Tris, pH 7.4 and 20% glycerol and stored at −80°C. The microsomes were permeabilized with a buffer containing 1% Triton X-100, 25 mM KCl, 5 mM MgCl2, and 50 mM Tris pH 7.5, incubated on ice for 30 min and centrifuged at 100,000 xg for 90 min at 4°C. The supernatant was subjected to 50% ammonium sulfate precipitation followed by centrifugation at 10,000 xg for 15 min. Ammonium sulfate was added to the supernatant to 93% saturation, followed by 30 min incubation and centrifugation at 10,000 xg for 15 min. The 93% ammonium sulfate cut pellet containing PDIA6, was dialyzed against a buffer containing 20 mM Hepes, pH 7.5, 25 mM KCl, and 5 mM MgCl2. The dialyzed protein was then loaded onto a 5 ml Concanavalin A affinity column (to remove glycosylated proteins). The flow through containing PDIA6 was heat treated at 54°C for 15 min followed by centrifugation at 30,000 ×g for 40 min to remove non-heat stable proteins (PDIA6 is heat stable). The supernatant was then applied in the presence of Ca2+ to a Phenyl-Sepharose column (many Ca2+ binding proteins in the ER bind to Phenyl-Sepharose in the presence of Ca2+). The fractions containing PDIA6 was collected and proteins were separated by Sephadex G-50 chromatography. PDIA6 co-eluted with BiP. BiP was separated from PDIA6 by ATP-Agarose chromatography in a buffer containing 20 mM Hepes, pH 7.5, 135 mM KCl, 20 mM MgCl2 and 1 mM DTT. PDIA6 was eluted in the void volume whereas BiP, an ATP binding protein, was retained on the column. BiP was eluted from the ATP-Agarose with 10 mM ATP. Samples purified using this protocol resulted in greater than 95% pure BiP and approximately 90% pure PDIA6.
IRE1α ER luminal domain (IRE1-NLD) or IRE1-NLD cysteine triple mutant were expressed in COS-1 cells and purified by Ni-NTA-Agarose (23). COS-1 cells were transfected with IRE1-NLD expression vector, harvested and lysed in a buffer containing 25 mM Tris-Cl, pH 8.0, 150 mM NaCl, and 1% Nonidet P-40. Cell lysates were centrifuged at 20,000 ×g for 15 min, and cell extracts were used for protein purification. Ni-NTA-Agarose chromatography was carried out using a buffer containing 50 mM Tris-Cl, pH 8.0, 500 mM NaCl, and 5 mM imidazole. The IRE1-NLD protein was eluted with 100 mM imidazole and concentrated.
For pull-down experiments, COS-1 cells were transfected with pED-IREl-NLD-His6 expression vector. Cells were further treated with 0.5 µM thapsigargin for an additional 24 hours. Formaldehyde was added directly to the culture to 1% (final concentration) and incubated for 20 minutes. Cross-linking was quenched by addition of 0.5 M glycine. Cells were harvested by washing with cold Tris-buffered saline followed by scraping into cold RIPA buffer (50 mM Tris, pH 7.2, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40, protease inhibitors). The lysate was incubated on ice for 30 min followed by centrifugation and overnight incubation at 4°C with 100 µl of 10% slurry of Ni-NTA-Agarose beads (Qiagen). The beads were centrifuged briefly to pellet, and washed three times with 1.5 ml RIPA buffer, three times with 1.5 ml of the RIPA buffer with 20 mM imidazole, and twice with 1.5 ml of a buffer containing 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA. The beads were pelleted, boiled in SDS-PAGE sample buffer, separated on SDS-PAGE (10% acrylamide), and followed by Western blot analysis using anti-IRE1α, anti-PDIA6 and anti-tubulin antibodies.
For BIACore analysis, the carboxymethylated dextran (CMD) surface of a CM5 chip was activated using N-hydroxysuccinimide (NHS)/1-ethyl-3(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC). IRE1-NLD or IRE1-NLD cysteine triple mutant (C109,148,332A–IRE1-NLD ) were captured at a flow rate of 5 µl/min to a total of −2000 response units. Uncoupled amine reactive sites on the CMD surface were then blocked by an injection of ethanolamine. The CM5 chip was normalized and prepared for kinetic analysis. PDIA6 in a buffer containing 10 mM Hepes, pH 7.4, 150 mM NaCl, and 0.005% P20 at concentrations from 0 to 5000 nM, was passed over the sensor surface at a flow rate of 30 µl/min. The BIACore analysis was also performed in a buffer containing 2 mM CaCl2 or 2 mM EGTA. Kinetic analysis was performed in biological triplicate. All experiments were conducted on either a BIACore 3000 instrument or BIACore T200 instrument (GE). For Surface Plasmon Resonance analysis with cysteine blocking, 200 µM N-ethylmaleimide (NEM) was injected for 60 seconds at a flow rate of 30 µl/min over the captured IRE1-NLD. Addition of the N-ethylmaleimide alone did not result in a binding response. PDIA6 was then injected at a flow rate of 30 µl/min for 30 seconds over the captured IRE1-NLD. The cysteine blocking experiment was performed three times.
MicroScale Thermophoresis (MST)
MST assays were carried out with a Monolith NT.115 instrument (Nano Temper, Munich, Germany) (61). To evaluate PDIA6 binding to IRE1-NLD, an increasing concentration of purified IRE1-NLD (0–1 µM) was used against FITC-labeled PDIA6. Experiments were carried out in a buffer containing 10 mM Hepes, pH 7.4, 100 mM NaCl. Data evaluation was performed with the Monolith software.
XBP1 splicing and RIDD analysis
The quantitative analysis of spliced and total XBP1 transcripts in mammalian cells protocol was utilized to identify mouse XBP1 specific splicing with a pair of real-time PCR primers designed for quantification of mouse XBP1 mRNA splicing (62). The forward primer sequence was 5’-GAGTCCGCAGCAGGTG-3’ (mouse), and the reverse primer sequence was 5’-GTGTCAGAGTCCATGGGA-3’ (mouse). Pairs of realtime PCR primers were also designed for quantification of mouse total XBP1 mRNA. The forward primer was 5’-AAGAACACGCTTGGGAATGG-3’ (mouse). The reverse primer was 5’-ACTCCCCTTGGCCTCCAC-3’ (mouse). These pairs of real-time PCR primers amplified both the unspliced and spliced forms of XBP1 mRNA transcripts. SYBR green PCR master mix (Bio-Rad 170–8880S) was used to set up the quantitative real-time PCR: the reaction (20 µl) contained 500 nM forward and reverse primers, 10–100 ng cDNA templates made from murine total RNA, and l× SYBR Green Supermix (50 mM KCl, 20 mM Tris-HCl, pH 8.4, 0.2 mM of each dNTP, 25 U/ml iTaq DNA polymerase, 3 mM MgCl2, SYBR Green 1, 10 nM fluorescein and stabilizers). The thermal cycling parameters were: step 1, 95°C for 10 min; step 2, 95°C for 20 s, 58°C 15 s, 72°C for 15 s. Step 2 was repeated for 40 cycles. Specificity of the amplification product from each primer pair was confirmed by melting curve analysis of the PCR product. Quantification was performed by expressing the threshold for each gene as a cycle number (Ct) and normalizing it to a housekeeping gene such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH), Actin, or β-tubulin using the equation 1/2^(Ct(gene)-Ct(gapdh)) and subsequently to the untreated negative siRNA control.
To measure the mRNA abundance of unspliced and spliced XBP1, RT-PCR was performed using total RNA isolated by TRIzol (Invitrogen) and the RNeasy kit (Qiagen) followed by PCR using specific primers. The PCR product was purified, and then cleaved with Pst1 to generate two smaller fragments. Only the unspliced XBP1 can be cleaved by Pst1, while the spliced XBP1 has the Pst1 site removed by the splicing event. Samples were separated on a 2% agarose gel, with densitometry performed on the digested and undigested fragments, to determine the amount of splicing occurring in endogenous XBP1. Mouse sequence specific primers for XBP1 splicing were: forward: 5’-CCTTGTGGTTGAGAACCAGG-3’; and reverse: 5’-CTAGAGGCTTGGTGTATAC-3’.
For measuring RIDD activity, total RNA was prepared from cells using TRIzol (Invitrogen), and cDNA was synthesized with SuperScript III (Invitrogen) using random primers p(dN)6 (Roche). Quantitative real-time PCR reactions were done employing Brilliant® II Sybr® Green qPCR Master Mix (Agilent Technologies). The relative amounts of mRNAs were calculated from the values of comparative threshold cycle by using rpl19 as control. The following primers were used for human cells: scara, 5’-GAGCCGTGTGTAGTTCTGCC-3 ‘and 5’-TCACCCAGGAGTGCTACGAT-3’; rpl19, 5’-TCAGGTACAGGCTGTGATACA-3’ and 5’-GGGCATAGGTAAGCGGAAGG-3’; and col6, 5’-GATAGCGCAGTCGGTGTAGG-3 ‘and 5 ‘-ACAGTGACGAGGTGGAGATCA-3’.
Western blot analysis, immunoprecipitation and Ca2+ measurements
Large format siRNA transfection was performed a minimum of three times in duplicate using 24 well plates with the siRNA for the selected genes. Cells were harvested at day three after siRNA transfection using RIPA lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate) containing protease inhibitors (63). Protein assays were performed using the BioRad protein assay, and 20 µg of protein was loaded in each lane on an SDS-PAGE gel and after electrophoresis, transferred to nitrocellulose. Antibodies used were rabbit anti-PDIA6 (Abcam, ab11432), rabbit anti-BiP (Abcam, ab21685), rabbit anti-IRE1α (Abcam, ab37073), goat anti-calreticulin, rabbit anti-GAPDH (Abcam, ab9483) and rabbit anti-tubulin antibodies (Abcam, ab6046). Antibodies were used at a dilution of 1:500 except: 1:300 for anti-GAPDH, anti-calreticulin and 1:2000 for anti-tubulin. Western blot images were scanned, and densitometry was performed using ImageJ and plotted in Excel.
Immunoprecipitations were performed with lysates of NIH-3T3 fibroblasts, except when it was necessary to detect IRE1α in the immunoprecipitates. In this case, it was necessary to perform immunoprecipitations with lysates of COS-1 cells expressing IRE1α-NLD. Both NIH-3T3 fibroblasts and transfected COS-1 cells were treated with 0.5 µM thapsigargin for 24 hours. Proteins were cross-linked using 4% formaldehyde for 12 min, followed by 15 minutes of quenching with 100 mM glycine in phosphate buffered saline (PBS) and washed extensively with PBS. Lysates were harvested with RIPA buffer. Protein assays were performed, and the lysate was diluted to 1 mg/ml. Lysates were pre-cleared with 60 µl of a 10% slurry of protein A:G Sepharose beads (Roche). Two µl of antibody (control IgG, rabbit anti-PDIA6, rabbit anti-IRE1α, or rabbit anti-BiP) was added, and lysates were incubated overnight at 4°C with rotation. 100 µl of a 10% slurry of protein A:G Sepharose was added, and incubated for 4 hours with rotation at 4°C. Beads were pelleted and washed with RIPA buffer 5 times. Pellets were re-suspended in 60 µl of sample buffer and loaded on a SDS-PAGE gel followed by protein transfer. Western Blot analysis was carried out using specific antibodies as indicated above.
Ca2+ measurements were performed as previously described (14, 64). Fura-2-acetomethylester fluorescence was monitored in a scanning spectrofluorometer (Photon Technology International). ER Ca2+ release was induced by 200 nM thapsigargin or 100 µM ATP, with SOCE induced by addition of 4 mM CaCl2. To chelate extracellular Ca2+, EGTA was added to DMEM at concentrations from 1 to 4 mM. The EGTA calculator, MaxChelator (maxchelator.stanford.edu), was used to determine free Ca2+ concentration in growth media. The initial (prechelator) [Ca2+]total in DMEM (Invitrogen) was assumed to be 1.8 mM. FBS contains approximately 3.7 mM Ca2+. Addition of 50 ml of FBS added 0.37 mM Ca2+ to 500 ml DMEM, giving a final concentration of 2.17 mM Ca2+ in complete media. EGTA was added at various concentrations to reach three final Ca2+ concentrations: 1.67 mM EGTA for 500 µM Ca2+, 2.08 mM EGTA for 100 µM Ca2+, and 3.25 mM EGTA for 1 µM Ca2+. Cells were grown for 24 hours in regular media, followed by washing with PBS and addition of varying amounts of Ca2+ concentrations for 24 hours.
Reverse transcriptase-PCR (RT-PCR) and Q-PCR for mRNA and miRNA
To monitor mRNA abundance, cells were harvested at day three after large format siRNA transfection using TRIzol (Invitrogen). RNA was isolated using the RNeasy kit (Qiagen) and total RNA (200 ng) was subsequently used in RT-PCR to generate cDNA for each sample. To monitor mRNA abundance, the cDNA was diluted 10-fold, with 2 µl of cDNA used in subsequent PCR reactions with primers targeting controls or selected genes. Q-PCR was performed using a LightCycler rapid thermal cycler system (Corbett Research) according to the manufacturer’s instructions. Reactions were performed in a 20 µl volume with 0.5 µM primers. Other reagents including nucleotides, Taq DNA polymerase and buffer, were used as provided in the SYBR Green Master Mix (BioRad). The amplification protocol included: 10 min 95°C denaturation; forty cycles with 95°C denaturation for 15 s, 58°C annealing for 15 s and 72°C extension for 15 s. Detection of the fluorescent product occurred at the end of the 72°C extension periods. Specificity of the amplification product from each primer pair was confirmed by a melting curve analysis of the PCR product. Quantification was performed as described above. The following nucleotide primers were used for RT-PCR and Q-PCR analyses: for PDIA6, forward: 5’-TCTGGTGAGCTGCACCTTCTTTCT-3’, reverse: 5’-AGCCGTTGCTGCTTTCTTCCATTC-3’; for GAPDH, forward: 5’-TTCACCACCATGGAGAAGGC-3’, reverse: GGCATGGACTGTGGTCATGA-3’; for BiP, forward: 5’-AAGCTCAAAGAGCGCATTGACACC-3’, reverse: AGTCTTCAATGTCCGCATCCTGGT-3’; for PDIA7, forward: 5’-AAATGCAGGGTGCTGTTACC-3’, reverse: 5’-AAGCCCTTCAATGGTGTGTTC-3’; for IRE1α, forward: 5’- TATGCCTCTCCCTCAATGGTGCAT-3’, reverse: 5’-TCAAACTTGAGGTCTGTGCTGGGA-3’; for calreticulin, forward: 5’-AAGACTGGGATGAACGAGCCAAGA-3’, reverse: 5’- AATTTGACGTGGTTTCCACTCGCC-3’.
IRE1α cluster quantification and phosphorylation
T-REx293 IRE1-3F6HGFP cells (24) were transiently transfected with pcDNA3.1-PDIA6-V5 or pcDNA3.1 as control. 24 hours post-transfection cells were split and re-seeded on 25 mm diameter coverslips in DMEM medium with 5% FBS and treated with 5 µg/ml of doxycycline for 24 hours to induce IRE1α-GFP expression followed by addition of 1 µM thapsigargin. Cells were fixed in 4% paraformaldehyde for 6 hours and followed by incubation with a blocking solution (0.25% bovine serum albumin, 10% horse serum in PBS) for 10 min. Cells were permeabilized with 0.5% Nonidet P-40 in 0.25% bovine serum albumin, 10% horse serum in PBS for 10 min at room temperature. Samples were incubated sequentially with primary antibodies (mouse anti-V5, 1:500 dilution, Invitrogen) and secondary antibody (goat anti-mouse Alexa Fluor 594, Life Technologies) for 60 min at room temperature. Nuclei were stained using DAPI. Images were acquired using an Olympus Fluoview FV1000 confocal laser-scanning microscope. Image stacks were captured using a 63x/1.4 objective with constant parameters for all conditions of each type of experiment guaranteeing that the image was not saturated and that image background was slightly above zero. Twenty different fields were analyzed with a total of 150–200 cells per group in three independent experiments. The numbers of IRE1α clusters and cluster size were quantified using ImageJ for PDIA6-V5 positive and negative cells.
A Phostag™ assay was performed to monitor IRE1α phosphorylation (37). HEK293T cells were transiently transfected with PMSCV-IRE1α-HA and pcDNA3.1 or PMSCV-IRE1-HA and pcDNA3.1-PDIA6-V5 for 48 hours. Cells were treated with 50 nM of thapsigargin for indicated time points and total cell extracts were analyzed by Western blot. Phostag™ assay was performed using 50 mg of total protein loaded in 8% SDS-PAGE minigel containing 25 mM of Phostag™ in the presence of 25 mM MnCl2. The following antibodies and dilutions were used: anti-HA 1:1000 (Covance), anti-V5 1:10000 (Sigma) and anti-HSP90 1:5000 (Santa Cruz).
In vivo ER stress analysis
Wild-type mice were given a single 50 ng/g body weight intraperitoneal injection of a 0.05 mg/ml suspension of tunicamycin in 150 mM dextrose (65). After 16 hours, mice were euthanized and liver extracts were prepared for immunoblot or Q–PCR analyses. All animal experiments were performed according to procedures approved by the Animal Use Committee of the Faculty of Medicine of the University of Chile.
The wild-type N2 Caenorhabditis elegans strain (Bristol) was obtained from the Caenorhabditis Genetics Center at the University of Minnesota. Worm breeding and handling were conducted following standard methods as described previously (66). For RNA interference experiments, synchronized first larval stage (L1) worms were placed on Nematode Growth Medium agar medium which contained 100 µg/ml ampicillin and 1 mM isopropyl-thio-β-D-galactoside (IPTG), and was seeded with an overnight culture of bacteria containing each RNAi clone (www.geneservice.co.uk). The worms were used for analysis three days after RNAi application. The worms were treated with 3 µg/ml thapsigargin for 5 hrs. Total mRNA was extracted from worms using the RNeasyPlus Mini Kit (Qiagen), cDNA was synthesized using Prime Script RT reagent kit (TaKaRa), and the quantitative RT-PCR analysis was performed using the KAPA SYBR FAST qPCR kit (KAPA Biosystems) and a Thermal Cycler Dice Real Time System (TaKaRa) according to the manufacturers’ protocols.
For analysis of PDIA6 expression in C. elegans, the worms were cultured in the RNAi-treated plates containing 3 µg/ml thapsigargin or 5 µg/ml tunicamycin for 5 hours. RNA was isolated for Q-PCR analysis and worm extracts were used for Western blot analysis with anti-PDIA6 antibodies (Abcam, ab11432) and anti-GAPDH (AbCam8C2). The following pairs of real-time PCR primers were used for quantification of C. elegans genes: for XBP-1s, forward primer 5’-TGCCTTTGAATCAGCAGTGG-3’ and reverse primer 5’-ACCGTCTGCTCCTTCCTCAATG-3’; for PDI-1, forward primer 5’-GAAAGCCAACGAAATACACCG-3’ and reverse primer 5’-ACAACTCTGGTCTTTCCCTTC-3’; for PDI-2, forward primer 5’-AGAGATCAAGTCGCACAACC-3’ and reverse primer 5’-CGGTGTTGATGTAGACGAAGAG-3’; for PDI-3, forward primer 5’- GTTGACAATCTCCAGCAATTCG-3’ and reverse primer 5’-AACGTCTCTTGTTCATCTGG-3’; for tag-320, forward primer 5’-CCCAACCATCAAGTATTTCGC-3’ and reverse primer 5’-GCATGTTTTCTTGAGCACGAG-3’.
Deep sequencing analysis and selection of miRNA
Deep sequencing was performed by Plant BioSys (University of Lethbridge). NIH-3T3 fibroblasts were treated with 0.5 µM thapsigargin for 24 hours and harvested with TRIzol (Invitrogen). The RNA was isolated using the RNeasy kit (Qiagen) and total RNA was used for deep sequencing. Statistical analysis was performed on the raw data. Short reads generated using Illumina GAIIx were cleaned up using FastQC program (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc), continuous poly-A/T/C/G were removed, and adapters were trimmed at the 3'-end using the Btrim program (67). Sequences between 18 and 29 nucleotides in length were retained. The leading N base was trimmed at the 5' end. The remaining short RNAs were filtered to find both known and putative (novel) miRNAs. Reads that did not match the NCBI’s mouse genome (using build 37.2 from http://www.ncbi.nlm.nih.gov/projects/genome/guide/mouse/) were discarded. Sequence alignment was performed using the Bowtie program (68) assuming perfect match. Reads that matched repetitive DNAs from Repbase (69) (uploaded from http://www.girinst.org/server/RepBase/) and non-coding RNAs, including tRNAs, rRNAs, snRNAs, and snoRNAs, from Rfam (70) (using build 10.0 from http://rfam.janelia.org/) were removed. Known miRNAs were tagged using release 17 of miRBase (71) and set aside. The remaining short RNAs were processed to find putative miRNAs. First, they were aligned to: (i) mRNAs using RefSeq database (72); the matching reads were tagged as mRNA-matching; and (ii) expressed sequence tags (ESTs) using dbEST (73); the matching sequences were tagged as EST-matching. Two putative miRNA precursor sequences of the mRNA-matching, EST-matching and the remaining short reads (one with 10 nucleotides upstream and 70 nucleotides downstream, assuming that miRNA is at the 5’ arm of the RNA hairpin and the with 70 nucleotides upstream and 10 nucleotides downstream, assuming that miRNA is at the 3’ arm) were processed by the MIREAP program (http://sourceforge.net/projects/mireap/) to select those that have hairpin structure. The hairpin-like reads were folded using RNAfold (74) to select those with a minimum free energy below −25Kcal/mol. Finally, the remaining short RNAs were clustered to group similar reads. Both miRNAs and miRNAs* were considered. Each cluster represents one putative miRNA, and its sequence was set to be the most frequent or abundant sequence in a given cluster. The abundance for each putative miRNA was calculated as a sum of abundance of all (similar) reads in this cluster.
Annotation of differentially expressed miRNAs
All known miRNAs and miRNAs*, which were annotated using miRBase, and putative miRNA and miRNAs* were combined together, and those with abundance (counts) below 5 were removed. Bioconductor package edgeR (75) was applied to determine whether a given miRNA was differentially expressed between the wild-type and knockout groups. The miRNAs were sorted by the adjusted P-values, which were computed utilizing trimmed mean of M-values (TMM) normalization and tagwise dispersion. All miRNAs with the adjusted P-values >0.5 were annotated with their (putative) target genes and considered for experimental validation. The experimentally validated targets were collected using miRecords database (76). Since the number of experimental annotations was relatively low, we used three target predictors (77): TargetScan (30, 31), DIANAmicroT (32), and RepTar (33). Targets that were associated with multiple annotations were considered to be more reliable. Statistically significant miRNAs were submitted to IPA (IngenuitySystems, www.ingenuity.com), generating a network of bioactive systems affected by these miRNAs.
miR-322 expression, treatment with mimic and PDIA6 3’ UTR reporter analysis
The miR-322 sequence specific mimic and inhibitor (Qiagen) were used at a final concentration of 20 nM. To measure the amount of miRNA present in samples, the samples were isolated with TRIzol (Invitrogen) and subjected to RNA purification using the RNeasy kit (Qiagen). To monitor the miRNA, a specific miRNA RT-PCR to generate cDNA was performed using the miRCURY LNA Universal RT miRNA PCR kit (Exiqon). This RT-PCR generates cDNA that corresponds to miRNA, and contains a particular motif that can be recognized by specific primers for miRNA analysis (Exiqon). The cDNA was diluted 20-fold, and miRNA specific primers were used in the subsequent Q-PCR. The Q-PCR was prepared as follows; 4 µl of cDNA, 2 µl of PCR primer mix (prepared by diluting forward and reverse primers 1:1), 4 µl water and 10 µl of SYBR Green Master Mix (BioRad). The amplification protocol included: 10 min 95°C denaturation; forty cycles with 95°C denaturation for 15 s, 58°C annealing for 15 s and 72°C extension for 15 s. Detection of the fluorescent product occurred at the end of the 72°C extension periods. Specificity of the amplification product from each primer pair was confirmed by a melting curve analysis of the PCR product. Quantification was performed as described above, except that 5S ribosomal RNA was used as the housekeeping control (Exiqon Cat# 203906). The mmu-miR-322 PCR Primer Set (Cat #:205182 Exiqon) and mmu-miR-322 mimic (Cat #:MSY0000548, Qiagen) and inhibitor (Cat #: MIN0000548, Qiagen) were based on 5’-CAGCAGCAAUUCAUGUUUUGGA-3’. Results were normalized to the 5S control with untreated control set at 1.
For PDIA6 3’ UTR reporter analysis, 3T3 fibroblasts were reverse transfected with PDIA6 3’ UTR Reporter (OriGene) vector (pMirTarget) with either miR-322 mimic or anti-miR-322 (Qiagen) to monitor miR-322 activity towards PDIA6 3’UTR. Luciferase activity was monitored using the Dual Luciferase Activity Assay (Promega).
Statistical analysis
As suggested in (78), if the measurements were normal, as evaluated with the Anderson-Darling test at 0.05 significance (79), then we utilized the t-test; otherwise we used the non-parametric Wilcoxon rank sum test (80). A difference was determined to be statistically significant if the P-value < 0.05.
Supplementary Material
Acknowledgments
We thank members of the Michalak lab for valuable comments and suggestions. We thank Rob Maranchuk at the RNAi screening core in the Department of Medical Microbiology and Immunology, University of Alberta for his expert technical help. We thank Bart Hazes (University of Alberta) for help with the analysis of the RNAi screen data. We thank Tullio Pozzan (University of Padova) for invaluable help, suggestions and critical analysis of the data. We thank Guennadi Kozlov and Kalle Gehring (McGill University) for a generous gift of purified PDIA6.
Funding: Supported by the Canadian Institutes of Health Research grants MOP-15291, MOP-15415, MOP-53050 to M.M. Supported by US National Institutes of Health grants DK042394, DK088227, HL052173 and HL057346 to RJ.K. Supported by US National Institute Health grant, AI083432 to Y.G. and by American Heart Association (12SDG12040188) to S.S. Supported by FONDEF D11I1007, Ring Initiative ACT1109, Millennium Institute No. P09-015-F, the Alzheimer’s Association, FONDECYT No. 1140549, ECOS CONICYT C13S02 the Muscular Dystrophy Association, and ALS Therapy Alliance, CONICYT-EEUU collaboration grant: USA2013-0003 to C.H. and FONDECYT No. 3130365 Post-doctoral grant to D.R-R E.D., H.U., D.S. were supported by CONICYT PhD fellowships. 2013 GIST Systems Biology Infrastructure Establishment Grant to D.H.K. Korean Ministry of Science, ICT & Future Planning (the Women Scientist program, 2013R1A1A3A04006010) to S-K.L. Korean Ministry of Education (Basic Science Research Program 2013R1A1A2005836) to JA. E.D. is supported by the Alberta Innovates-Health Solutions Postdoctoral Fellowship.
Footnotes
Author contributions: J.G. designed experiments, analyzed data, performed biochemical and cell biological, siRNA screening and validation experiments, and wrote the paper; E.D., K.B., E.D., H.U., D.S., and D.R-R. designed experiments, analyzed data, and performed experiments; Z.P., X.F., M.M., and L.K. designed experiments and performed bioinformatics and computational analyses; K.B. analyzed ORAI1-deficient cells; S.S. and Y.G. developed and characterized ORIA1-deficient cells; JA. and D.H.K. provided reagents for C. elegans experiments; Y.L. performed C. elegans experiments; S-K.L. designed experiments and wrote the manuscript; RJ.K. designed experiments and developed and characterized IRE1 research tools; L.K., C.H., M.M. designed experiments, analyzed data, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data availability: Deep sequencing data have been deposited with the NCBI GEO Sequence Database, accession number GSE57138.
Supplementary Materials
Figure S1. The XBP1 splicing reporter system
Figure S2. A genome-wide siRNA screen.
Figure S3. Silencing of the PDIA6 gene.
Figure S4. Phosphorylation of eIF2α in PDIA6-silenced cells
Figure S5. PDIA6 mRNA abundance under ER stress conditions.
Figure S6. PDIA6 interacts with the luminal domain of IRE1.
Figure S7. PDIA6 silencing and IRE1α RIDD activity.
Figure S8. Selected miRNAs identified by deep sequencing analysis.
Figure S9. Schematic representation of multiple target prediction analysis.
References and Notes
- 1.Groenendyk J, Agellon LB, Michalak M. Coping with Endoplasmic Reticulum Stress in the Cardiovascular System. Annu. Rev. Physiol. 2013;75:49. doi: 10.1146/annurev-physiol-030212-183707. [DOI] [PubMed] [Google Scholar]
- 2.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J. Cell Biol. 2012;197:857. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimata Y, Kohno K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr. Opin. Cell Biol. 2011;23:135. doi: 10.1016/j.ceb.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Kraskiewicz H, Fitzgerald U. InterfERing with endoplasmic reticulum stress. Trends Pharmacol. Sci. 2012;33:53. doi: 10.1016/j.tips.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nature Rev. Mol. Cell Biol. 2012;13:89. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 7.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J. Biochem. 2004;136:343. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- 9.Acosta-Alvear D, et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell. 2007;27:53. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nature Cell Biol. 2011;13:184. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urra H, Dufey E, Lisbona F, Rojas-Rivera D, Hetz C. When ER stress reaches a dead end. Biochim. Biophys. Acta. 2013;1833:3507. doi: 10.1016/j.bbamcr.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nature Rev. Mol. Cell Biol. 2012;13:549. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Back SH, Lee K, Vink E, Kaufman RJ. Cytoplasmic IRE1 alpha-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress. J. Biol. Chem. 2006;281:18691. doi: 10.1074/jbc.M602030200. [DOI] [PubMed] [Google Scholar]
- 14.Prins D, Groenendyk J, Touret N, Michalak M. Modulation of STIM1 and capacitative Ca2+ entry by the endoplasmic reticulum luminal oxidoreductase ERp57. EMBO Rep. 2011;12:1182. doi: 10.1038/embor.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kantor L, et al. A point mutation in translation initiation factor 2B leads to a continuous hyper stress state in oligodendroglial-derived cells. PLoS ONE. 2008;3:e3783. doi: 10.1371/journal.pone.0003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vekich JA, Belmont PJ, Thuerauf DJ, Glembotski CC. Protein disulfide isomerase-associated 6 is an ATF6-inducible ER stress response protein that protects cardiac myocytes from ischemia/reperfusion-mediated cell death. J. Mol. Cell. Cardiol. 2012;53:259. doi: 10.1016/j.yjmcc.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heifetz A, Lennarz WJ. Biosynthesis of N-glycosidically linked glycoproteins during gastrulation of sea urchin embryos. J. Biol. Chem. 1979;254:6119. [PubMed] [Google Scholar]
- 18.Yoshida H, et al. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 2003;4:265. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J. Biochem. 2004;136:343. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- 20.Waser M, Mesaeli N, Spencer C, Michalak M. Regulation of calreticulin gene expression by calcium. J. Cell Biol. 1997;138:547. doi: 10.1083/jcb.138.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell. 2002;13:4456. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jessop CE, Watkins RH, Simmons JJ, Tasab M, Bulleid NJ. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J. Cell Sci. 2009;122:4287. doi: 10.1242/jcs.059154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu CY, Wong HN, Schauerte JA, Kaufman RJ. The protein kinase/endoribonuclease IRE1 alpha that signals the unfolded protein response has a luminal N-terminal ligand-independent dimerization domain. J. Biol. Chem. 2002;277:18346. doi: 10.1074/jbc.M112454200. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Korennykh AV, Behrman SL, Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16113. doi: 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 26.Han D, et al. IRElalpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollien J, et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009;186:323. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coe H, Michalak M. Calcium binding chaperones of the endoplasmic reticulum. Gen. Physiol Biophys. 2009;28:F96. [PubMed] [Google Scholar]
- 29.Behrman S, Acosta-Alvear D, Walter P. A CHOP-regulated microRNA controls rhodopsin expression. J Cell Biol. 2011;192:919–927. doi: 10.1083/jcb.201010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 31.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Vergoulis T, Koziris N, Sellis T, Tsanakas P, Hatzigeorgiou AG. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elefant N, Berger A, Shein H, Hofree M, Margalit H, Altuvia Y. RepTar: a database of predicted cellular targets of host and viral miRNAs. Nucleic Acids Res. 2011;39:Dl88–D194. doi: 10.1093/nar/gkq1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouyang Z, Snyder MP, Chang HY. SeqFold: Genome-scale reconstruction of RNA secondary structure integrating high-throughput sequencing data. Genome Res. 2012;23:377. doi: 10.1101/gr.138545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, Truitt M, McManus MT, Ruggero D, Goga A, Papa FR, Oakes SA. IRE1 alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chitnis N, Pytel D, Diehl JA. UPR-inducible miRNAs contribute to stressful situations. Trends Biochem. Sci. 2013;38:447. doi: 10.1016/j.tibs.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez DA, Zamorano S, Lisbona F, Rojas-Rivera D, Urra H, Cubillos-Ruiz JR, Armisen R, Henriquez DR, Cheng EH, Letek M, Vaisar T, Irrazabal T, Gonzalez-Billault C, Letai A, Pimentel-Muinos FX, Kroemer G, Hetz C. BH3-only proteins are part of a regulatory network that control the sustained signalling of the unfolded protein response sensor IRE1 alpha. EMBO J. 2012;31:2322. doi: 10.1038/emboj.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol. Cell. 2010;40:205. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M. A microRNA signature of hypoxia. Mol. Cell Biol. 2007;27:1859. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 43.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 44.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 45.Sokolov MV, Panyutin IV, Neumann RD. Unraveling the global microRNAome responses to ionizing radiation in human embryonic stem cells. PLoS ONE. 2012;7:e31028. doi: 10.1371/journal.pone.0031028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan G, Shi Y, Wu ZH. MicroRNA-22 promotes cell survival upon UV radiation by repressing PTEN. Biochem. Biophys. Res. Commun. 2012;417:546. doi: 10.1016/j.bbrc.2011.11.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, Lu X. Posttranscriptional regulation of miRNAs in the DNA damage response. RNA Biol. 2011;8:960. doi: 10.4161/rna.8.6.17337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 49.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 50.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flynt AS, Thatcher EJ, Burkewitz K, Li N, Liu Y, Patton JG. miR-8 microRNAs regulate the response to osmotic stress in zebrafish embryos. J. Cell Biol. 2009;185:115. doi: 10.1083/jcb.200807026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belmont PJ, Chen WJ, Thuerauf DJ, Glembotski CC. Regulation of microRNA expression in the heart by the ATF6 branch of the ER stress response. J. Mol. Cell. Cardiol. 2012;52:1176. doi: 10.1016/j.yjmcc.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartoszewski R, Brewer JW, Rab A, Crossman DK, Bartoszewska S, Kapoor N, Fuller C, Collawn JF, Bebok Z. The unfolded protein response (UPR)-activated transcription factor X-box-binding protein 1 (XBP1) induces microRNA-346 expression that targets the human antigen peptide transporter 1 (TAP1) mRNA and governs immune regulatory genes. J. Biol. Chem. 2011;286:41862. doi: 10.1074/jbc.M111.304956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 2005;94:896. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- 55.Smalheiser NR, Lugli G. microRNA regulation of synaptic plasticity. Neuromolecular Med. 2009;11:133. doi: 10.1007/s12017-009-8065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hetz C, Chevet E, Harding HP. Targeting the unfolded protein response in disease. Nature Rev. Drug Discov. 2013;12:703. doi: 10.1038/nrd3976. [DOI] [PubMed] [Google Scholar]
- 57.Groenendyk J, Michalak M. A genome-wide siRNA screen identifies novel phospho-enzymes affecting Wnt/beta-catenin signaling in mouse embryonic stem cells. Stem Cell Rev. 2011;7:910. doi: 10.1007/s12015-011-9265-3. [DOI] [PubMed] [Google Scholar]
- 58.Coe H, Jung J, Groenendyk J, Prins D, Michalak M. ERp57 modulates STAT3 signaling from the lumen of the endoplasmic reticulum. J. Biol. Chem. 2010;285:6725. doi: 10.1074/jbc.M109.054015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Azzalini A. R package 'sn': The skew-normal and skew-t distributions (version 0.4-16) 2010 [Google Scholar]
- 60.Schagat T, Paguio A, Kopish K. Normalizing genetic reporter assays: approaches and considerations for increasing consistency and statistical significance. Cell Notes. 1996;17:9. [Google Scholar]
- 61.Seidel SA, Dijkman PM, Lea WA, van den Bogaart G, Jerabek-Willemsen M, Lazic A, Joseph JS, Srinivasan P, Baaske P, Simeonov A, Katritch I, Melo FA, Ladbury JE, Schreiber G, Watts A, Braun D, Duhr S. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59:301. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Back SH, Schroder M, Lee K, Zhang K, Kaufman RJ. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods. 2005;35:395. doi: 10.1016/j.ymeth.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 63.Milner RE, Baksh S, Shemanko C, Carpenter MR, Smillie L, Vance JE, Opas M, Michalak M. Calreticulin, and not calsequestrin, is the major calcium binding protein of smooth muscle sarcoplasmic reticulum and liver endoplasmic reticulum. J. Biol. Chem. 1991;266:7155. [PubMed] [Google Scholar]
- 64.Mesaeli N, Nakamura K, Zvaritch E, Dickie P, Dziak E, Krause K-H, Opas M, MacLennan DH, Michalak M. Calreticulin is essential for cardiac development. J. Cell Biol. 1999;144:857. doi: 10.1083/jcb.144.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1 alpha. Science. 2006;312:572–576. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- 66.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kong Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics. 2011;98:152. doi: 10.1016/j.ygeno.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 70.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boguski MS, Lowe TM, Tolstoshev CM. dbEST—database for "expressed sequence tags". Nature Genet. 1993;4:332. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 74.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan Gana NH, Victoriano AF, Okamoto T. Evaluation of online miRNA resources for biomedical applications. Genes Cells. 2012;17:11. doi: 10.1111/j.1365-2443.2011.01564.x. [DOI] [PubMed] [Google Scholar]
- 78.Ramsey FL, Schafer DW. The statistical sleuth: a course in methods of data analysis. 3rd. Australi; Boston: Brooks/Cole, Cengage Learning; 2013. p. 760. [Google Scholar]
- 79.Anderson T, Darling D. Asymptotic Theory of Certain Goodness of Fit Criteria Based on Stochastic Processes. Ann. Math. Stat. 1952;23:193. [Google Scholar]
- 80.Wilcoxon F. Individual Comparisons by Ranking Methods. Biometrics Bulletin. 1945;1:80. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.