Abstract
Plasmid-dependent overexpression of enzyme(s) aims to divert carbon flux toward a desired compound. One drawback of this strategy is compromise of growth due to massive consumption of host resources. Here we show that replenishment of sigma factor rpoE improves the growth of Klebsiella pneumoniae. The gene rpoE was expressed alone or coexpressed with Ald4 (an aldehyde dehydrogenase from Saccharomyces cerevisiae) in K. pneumoniae. We found that the Ald4 activity was higher in the strain coexpressing Ald4 and rpoE (32.3 U/mg) than that expressing Ald4 alone (29.9 U/mg). Additionally, under shake-flask conditions, the strain coexpressing Ald4 and rpoE produced 0.5 g 3-hydroxypropionic acid (3-HP) and 9.8 g 1,3-propanediol (1,3-PD) per liter in 24 h, which were 1.6- and 0.85-fold enhancement, respectively, compared to those expressing Ald4 alone. Notably, under non-optimized bioreactor conditions, the strain coexpressing Ald4 and rpoE produced 13.5 g 3-HP and 37.8 g 1,3-PD per liter with glycerol conversion ratio of 0.45 mol/mol. These results indicate that replenishment of rpoE enhanced promoter activity and stimulated glycerol consumption.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-016-0576-6) contains supplementary material, which is available to authorized users.
Keywords: Klebsiella pneumoniae, rpoE, 3-Hydroxypropionic acid, Aldehyde dehydrogenase, Glycerol
Introduction
To date, glycerol-based biosynthesis of chemicals has attracted much attention due to ample glycerol derived from biodiesel production. Klebsiella pneumoniae can naturally convert glycerol into a series of economically important chemicals, including 1,3-propanediol (1,3-PD) [1], 2,3-butanediol (2,3-BD) [2], and 3-hydroxypropionic acid (3-HP) [3]. In K. pneumoniae, glycerol metabolism is mediated by the dha regulon which governs the parallel oxidation and reduction pathways [4]. In the oxidation pathway, glycerol is converted into dihydroxyacetone phosphate which subsequently enters the glycolic pathway. In the reduction pathway, glycerol is converted into 3-hydroxypropionaldehyde (3-HPA) by glycerol dehydratase (dhaB) [5]. 3-HPA is catalyzed into 1,3-PD by 1,3-propanediol oxidoreductase (dhaT) [4]. 3-HPA can also be naturally catalyzed into 3-HP by aldehyde dehydrogenases (AldHs) [6]. Since native AldHs show lower activities than dhaT, 3-HP is less than 1,3-PD in wild type K. pneumoniae. Although AldH overexpression benefits 3-HP production, the lack of appropriate promoter restricts the metabolic engineering of K. pneumoniae [7].
Previously, the endogenous promoter (named pk) of dhaB1 gene (GenBank: U30903) in K. pneumoniae was employed to drive the expression of AldHs for enhancing 3-HP production. However, pk showed low activity [3, 8]. The reasons behind might be multifaceted. One reason is the inherent low activity of pk because it is a constitutive promoter tailored for governing glycerol metabolism in K. pneumoniae. The second reason may be cofactor imbalance caused by AldH overexpression [3]. Apart from above reasons, RNA polymerases especially sigma factors (σ factors) may be related to pk activity, because sigma factors recognize and bind promoters to initiate transcription. Among sigma factors reported to date, rpoS has been mostly studied for its versatile roles in transcription, signal transduction and gene regulation [9–11]. It was reported that an engineered sigma factor library could reprogram metabolic flux and thus led to diverse phenotypes, and some of mutant strains could be used for industrial production purposes [12]. Additionally, sigma factors were shown to be associated with the cell survival under stress conditions [13–15]. Based on above information, we speculated that sigma factors may affect promoter activity.
Following this assumption, in this study, three native sigma factors including rpoE (GenBank, KPN_02898), rpoS (GenBank, KPN_03103) and dksA (GenBank, KPN_00145) were expressed alone or coexpressed with Ald4 in K. pneumoniae to investigate the influences of sigma factors on pk activity. Fluorescence-activated cell sorting (FACS) was performed to monitor the expression level of green fluorescent protein (GFP) which was employed to indicate promoter activity. Comprehensive analysis of the cell growth, glycerol consumption, enzymatic activity, and metabolites production was to systematically assess the affects of sigma factors on the activity of pk promoter.
Materials and Methods
Strains, Vectors, Medium and Reagents
The vector pET-28a (Novagen) was used in this study with minor modification. The original T7 promoter was replaced by the native pk promoter of dhaB1 gene, the first subunit of dhaB gene cluster in K. pneumoniae DSM 2026. The resultant vector was designated as pET-pk [8]. Based on this vector, a series of recombinant vectors were constructed, including pET-pk-GFP-Pkana-rpoE, pET-pk-GFP-Pkana-rpoS, pET-pk-Pkana-dksA, and pET-pk-ald4-Pkana-rpoE, where Pkana refers to the promoter of kanamycin resistance gene, GFP indicates green fluorescent protein, while rpoE, rpoS and dksA are distinct sigma factor genes from K. pneumoniae. The strains of K. pneumoniae DSM 2026, Escherichia coli DH5α, and Saccharomyces cerevisiae (baker’s yeast) were from DSMZ GmbH, Germany. E. coli DH5α was grown in Luria–Bertani (LB) medium. S. cerevisiae was grown in Yeast Extract Peptone Dextrose medium (g/L): yeast extract, 10; peptone, 20; glucose, 20. The medium for producing 3-HP by recombinant K. pneumoniae was the same as the previously reported [3]. The recombinant strain was microaerobically grown in 50 mL Erlenmeyer flasks containing 25 mL medium and 50 μg kanamycin/mL at 37 °C and 150 rpm continuous shaking.
Construction of the Recombinants
The vectors for tandem co-expression were constructed based on vector pET-pk. The gfp gene was chemically synthesized and the ald4 gene was amplified by PCR from the genomic DNA of S. cerevisiae. The native pk promoter of gene dhaB1 was used to drive gene expression. The promoter of kanamycin resistance gene (Pkana) was amplified by PCR from vector pET-28a to drive the expression of sigma factor genes. Three sigma factor genes including rpoE, rpoS and dksA were amplified from the genomic DNA of K. pneumoniae. The following are PCR procedures: initial denaturation at 94 °C for 4 min; followed by 30 cycles of 94 °C for 1 min, 55 °C for 45 s, 72 °C for 90 s, and 72 °C for 10 min. The PCR products were cloned into vector pET-pk, resulting in recombinant vectors pET-pk-GFP, pET-pk-GFP-Pkana-rpoE, pET-pk-GFP-Pkana-rpoS, and pET-pk-GFP-Pkana-dksA. To construct a recombinant vector that would decipher the influence of sigma factor on Ald4 activity, the gfp gene was replaced by the ald4 gene on above recombinant vector in which the GFP was highly expressed. After these recombinant vectors were transformed into K. pneumoniae, the positive recombinants were screened by LB kanamycin plate and further confirmed by sequencing. All the primers and restriction enzymes used in this study were listed in supplementary Table 1.
Promoter Activity Assay by FACS
In this study, GFP was applied to indicate promoter activity. Five strains including wild type K. pneumoniae, K. pneumoniae(pET-pk-GFP), K. pneumoniae(pET-pk-GFP-Pkana-rpoE), K. pneumoniae(pET-pk-GFP-Pkana-rpoS), and K. pneumoniae(pET-pk-GFP-Pkana-dksA) were grown in medium with 50 μg/mL kanamycin at 37 °C and 150g continuous shaking. The recombinant strains were harvested by centrifugation at 9000g and washed twice by phosphate buffered saline (PBS, pH 7.0). Cells were resuspended in PBS. Fluorescence-activated cell sorter (FACS) analysis was performed with FACS Aria II (BD).
Enzyme Activity Assay
The strains were grown in fermentation medium containing kanamycin of 50 μg/mL. After 9 h cultivation, cells were centrifuged at 10,000g for 10 min and washed by 5 mL PBS. 50 µL phenylmethylsulfonyl fluoride (10 mg/mL) was used to inhibit protease activity. The cells were sonicated and the resultant solution was centrifuged at 17,000g for 15 min. Protein concentration was determined using Bradford assay (BioRad). 200 µL cell-free solution was added into 5 mL Bradford solution. After 5 min reaction, absorbance was measured by spectrophotometer at a wavelength of 595 nm. Another 200 µL cell-free solutions were added into 2 mL centrifugation tubes containing 0.2 mM NADH (or NADPH) and 2 mM propionaldehyde, and incubated at 37 °C for 5 min. The amount of consumed NADH (or NADPH) was determined by measuring the decrease of absorbance at 340 nm wavelength in a spectrophotometer. The unit of ALDH activity was defined as the amount of enzyme used in consuming 1 μmol NADH per minute.
Flask and Bioreactor Cultivation
In shake-flask cultivation, the recombinants were grown in LB medium containing the following components per liter: yeast extract 5 g, peptone 10 g, NaCl 10 g, and kanamycin 50 mg. 1 % of overnight culture was inoculated to medium containing the same concentration of antibiotics. The flasks were plugged with an oxygen-permeable cotton stopper and incubated in an orbital incubator shaker at an agitation speed of 180g, 37 °C. Samples were taken out every 3 h to measure biomass, residual glycerol and metabolites (see “Analytical methods”).
Fed-batch cultivation of the recombinant strain K. pneumoniae(pET-pk-ald4-pkana-rpoE) was performed in a 5 L bioreactor (Baoxing, China) containing 3 L medium. The experiments were performed in glycerol fed-batch mode. The fermentation conditions were the same as those previously reported [16]. The strain was pre-cultivated in a 100 mL fermentation medium overnight at 37 °C and then added into the bioreactor. The agitation speed was 400g and the air was supplied at 1.5 vvm. During the entire fermentation process, pH was maintained at 7.0 by automatic addition of 5 M NaOH. Residual glycerol was maintained at 25 g/L. Dissolved oxygen (DO) was monitored automatically. Samples were taken out every 3 h to measure cell concentration, residual glycerol, and metabolites.
SDS-PAGE Analysis
The ald gene in recombinant strain K. pneumoniae(pET-pk-ald4-Pkana-rpoE) could be continuously expressed during entire fermentation because pk is a constitutive promoter. The expression level of Ald4 was analyzed by 12 % (w/v) polyacrylamide gel electrophoresis (PAGE) with cell-free extract under denaturing conditions. Mini-Protein III Electrophoresis System (Bio-Rad, USA) was used to perform the operation. Coomassie Brilliant Blue R-250 (0.2 %, w/v) was used to stain protein on the gel and the protein concentration was measured by Bradford method with bovine serum albumin (BSA) as standard protein.
Analytical Methods
Cell concentrations were measured by using a microplate reader at 600 nm with 200 µL fermentation broth added in the cuvette. 3-HP, lactic acid and acetic acid were monitored by a high performance liquid chromatography (HPLC) system (Shimazu, Kyoto, Japan) at 210 nm equipped with a C18 column and a SPD-20A UV detector. The column temperature was 30 °C, and the mobile phase was 0.05 % phosphoric acid at a flow rate of 0.8 mL/min. 1,3-PD and 2,3-butanediol (2,3-BD) were quantitatively analyzed by HPLC (Shimazu, Japan) equipped with a column of Aminex HPX-87H Ion Exclusion particles (300 mm × 7.8 mm, Bio-Rad, Hercules, CA, USA) using a differential refractive index detector. The column was maintained at 65 °C and the mobile phase was 5 mM sulfuricacid (in Milli-Q water) at a flow rate of 0.6 mL/min. Residual glycerol concentration was measured every 3 h by a titration method with NaIO4 (for control of glycerol). All samples were filtered through 0.22-µm membrane filter.
Results
Characterization of the Recombinants
To investigate the influence of sigma factor on gfp expression which was harnessed to indicate promoter activity, we engineered several recombinant strains, including K. pneumoniae(pET-pk), K. pneumoniae(pET-pk-gfp), K. pneumoniae(pET-pk-gfp-Pkana-rpoE), K. pneumoniae(pET-pk-gfp-Pkana-rpoS), and K. pneumoniae(pET-pk-gfp-Pkana-dksA) (Fig. 1). The strain K. pneumoniae(pET-pk) served as the control because it harbored the empty vector pET-pk. In other recombinant strains, the gfp gene was expressed alone or in combination with sigma factor gene rpoE, rpoS, or dksA, respectively. Among all recombinant strains, K. pneumoniae(pET-pk-gfp-Pkana-rpoE) showed highest level of gfp expression (Table 1), indicating that rpoE was the most favorable sigma factor for gfp expression. To determine the effects of rpoE on Ald4 activity and carbon flux allocation, the gfp gene on vector pET-pk-gfp-Pkana-rpoE was replaced by ald4, resulting in recombinant vector pET-pk-ald4-Pkana-rpoE. Sequencing result verified that all genes were correctly arranged on vector. The corresponding recombinant strain K. pneumoniae(pET-pk-ald4-Pkana-rpoE) was grown in medium for analysis of both the Ald4 activity and the production of metabolites including 3-HP, 1,3-PD, lactic acid and acetic acid.
Fig. 1.
Schematic diagram of the recombinant plasmids. Kan, kanamycin resistance gene; Ori, the replication origin of plasmids; Ald4, the aldehyde dehydrogenase from S. cerevisiae; PK, the native promoter of dhaB gene in K. pneumoniae; Pkana, the promoter of kanamycin resistance gene from plasmid pET-28a; rpoE, rpoS; dksA, sigma factor genes from K. pneumoniae
Table 1.
Flow cytometric analysis of GFP expression in different strains
| WT | Kp(pET-pk-GFP) | Kp(pET-pk-GFP-Pkana-rpoE) | Kp(pET-pk-GFP-Pkana-rpoS) | Kp(pET-pk-GFP-Pkana-dksA) |
|---|---|---|---|---|
| 0 | 14 | 37 | 5 | −13 |
WT wild type K. pneumoniae, Kp(pET-pk-GFP) recombinant K. pneumoniae harboring vector pET-pk-GFP, Kp(pET-pk-GFP-Pkana-rpoE) recombinant K. pneumoniae harboring vector pET-pk-GFP-Pkana-rpoE, Kp(pET-pk-GFP-Pkana-rpoS) recombinant K. pneumoniae harboring vector pET-pk-GFP-Pkana-rpoS, Kp(pET-pk-GFP-Pkana-dksA) recombinant K. pneumoniae harboring vector pET-pk-GFP-Pkana-dksA
Effects of Sigma Factors on GFP Expression
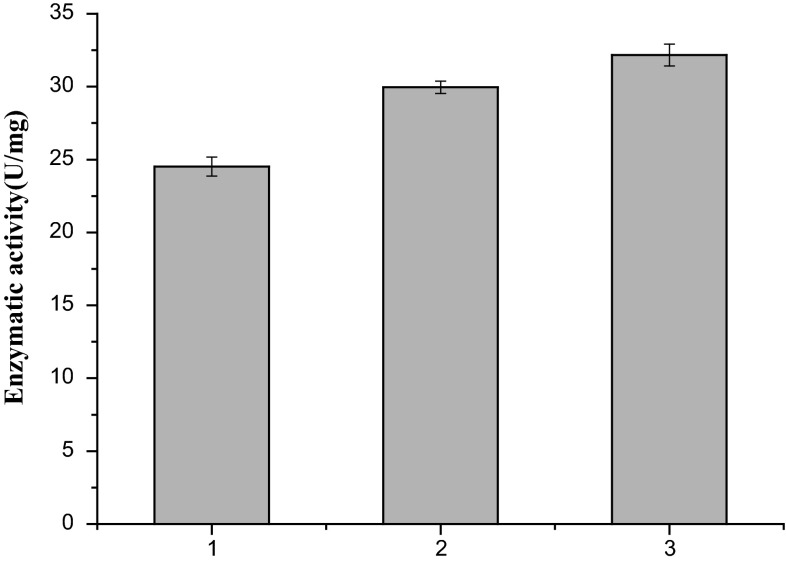
Considering that sigma factors bind promoters and initiate transcription, we reasoned that the amount of sigma factors might affect promoter activity. To verify this hypothesis, in this study, gfp gene was expressed alone or coexpressed with rpoE, rpoS, or dksA, the three distinct sigma factor genes from K. pneumoniae. As shown in Table 1, the GFP values of recombinants were higher than that of wild type K. pneumoniae. Notably, the GFP value of K. pneumoniae(pET-pk-gfp-Pkana-rpoE) was 41, which was nearly threefold of the strain K. pneumoniae(pET-pk-gfp), implying that the supplement of rpoE significantly benefited GFP expression. To explore the influences of rpoE expression on Ald4 activity, the enzyme activity was analyzed. As shown in Fig. 2, the Ald4 activity of the strain K. pneumoniae(pET-pk-ald4-pkana-rpoE) was higher than that of the strain K. pneumoniae(pET-pk-ald4), indicating that the overexpression of rpoE elevated Ald4 activity. Moreover, Ald4 exhibited different activities when coexpressed with distinct sigma factors. This may be attributable to the differential affinity of sigma factors with promoters. Overall above results indicate that rpoE expression increased the Ald4 activity.
Fig. 2.
The Ald4 activities of the recombinant strains. (1) Recombinant strain K. pneumoniae(pET-pk). pET-pk: an empty vector carrying pk promoter, (2) recombinant strain K. pneumoniae(pET-pk-ald4). (3) Recombinant strain K. pneumoniae(pET-pk-ald4-Pkana-rpoE), where ald4 was coexpressed with rpoE
Protein Expression Analysis and Enzymatic Activity Assay
Three recombinant K. pneumoniae strains including K. pneumoniae(pET-pk), K. pneumoniae(pET-pk-ald4), and K. pneumoniae(pET-pk-ald4-Pkana-rpoE) were analyzed for the protein expression and Ald4 activity. The former two strains served as the control strains. SDS-PAGE analysis showed that the ald4 gene was expressed in both K. pneumoniae(pET-pk-ald4) and K. pneumoniae(pET-pk-ald4-Pkana-rpoE) (supplementary Figure 2). To elucidate the effects of sigma factors on Ald4 activity, the above three strains were investigated for their Ald4 activity. The wild type K. pneumoniae showed 24.7 U/mg of activity toward propionaldehyde. This is because wild type K. pneumoniae genome harbors nearly 20 copies of aldehyde dehydrogenases. In contrast, K. pneumoniae(pET-pk-ald4) exhibited 29.9 U/mg of Ald4 activity, and K. pneumoniae(pET-pk-ald4-Pkana-rpoE) showed 32.3 U/mg of Ald4 activity, clearly indicating that the enhanced Ald4 activity was ascribed to the coexpression of rpoE and Ald4 (Fig. 2). These results were consistent with the GFP expression values (Table 1).
Cell Growth and Glycerol Consumption
Shake-flask cultivation was performed to investigate glycerol consumption and biomass accumulation. A total of three strains, including wild type K. pneumoniae, K. pneumoniae(pET-pk) and K. pneumoniae(pET-pk-ald4), were used as the control strains, while K. pneumoniae(pET-pk-ald4-pkana-rpoE) was analyzed for the coexpression of Ald4 and rpoE. The strain K. pneumoniae(pET-pk) grew slower than wild type K. pneumoniae (Fig. 3). Similarly, K. pneumoniae(pET-pk-ald4) grew slower than K. pneumoniae(pET-pk). On the contrary, the strain K. pneumoniae(pET-pk-ald4-pkana-rpoE) grew faster compared to the control strain K. pneumoniae(pET-pk-ald4), indicating that the expression of rpoE in K. pneumoniae(pET-pk-ald4-pkana-rpoE) stimulated cell growth (Fig. 3a). Consistent with the enhanced cell growth, glycerol consumption was accordingly increased presumably due to the supplement of sigma factor (Fig. 3b).
Fig. 3.
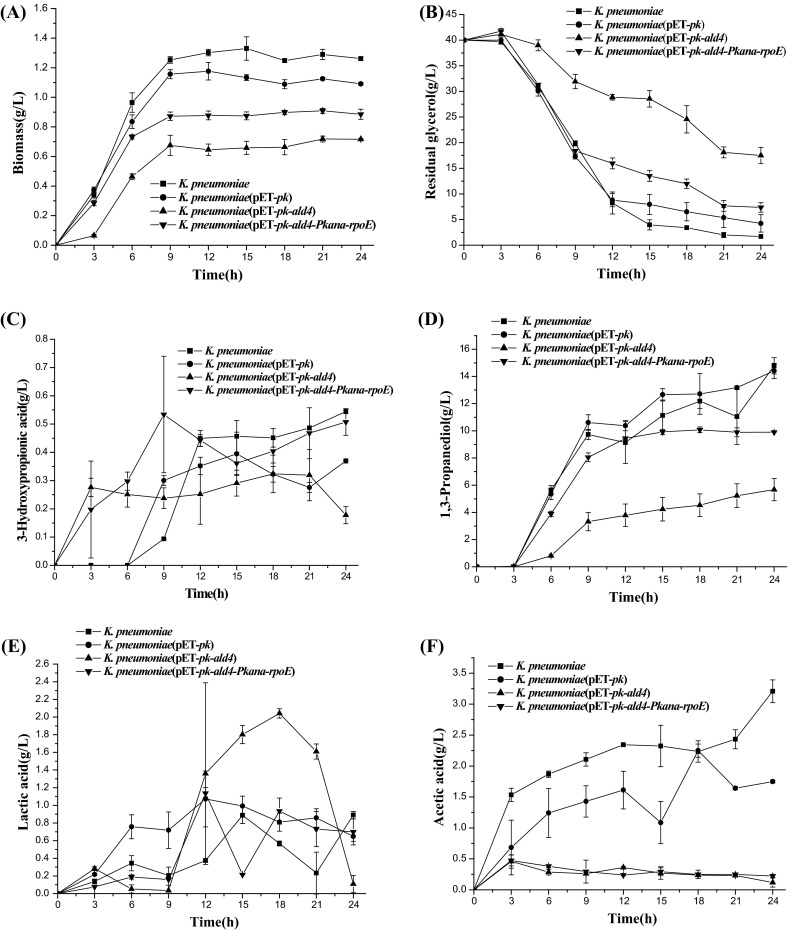
Shake-flask cultivation of the strains. a Cell concentration; b residual glycerol; c 3-hydroxypropionic acid; d 1,3-propanediol; e lactic acid; f acetic acid. K. pneumoniae, wild-type K. pneumoniae; Kp(pET-pk-ald4), recombinant K. pneumoniae harboring vector pET-pk-ald4; Kp(pET-pk-ald4-Pkana-rpoE), recombinant K. pneumoniae coexpressing ald4 and rpoE genes
Flask Cultivation of the Recombinant Strains
Under shake-flask culture conditions, the recombinant strain K. pneumoniae(pET-pk-ald4) produced 0.3 g 3-HP per liter in 21 h. However, this 3-HP concentration dramatically decreased to 0.15 g/L in 24 h. The strain K. pneumoniae(pET-pk-ald4-pkana-rpoE) produced more 3-HP and 1,3-PD since 6 h fermentation compared with K. pneumoniae(pET-pk-ald4). As shown in Fig. 3c, d, the strain K. pneumoniae(pET-pk-ald4-pkana-rpoE) produced 9 g/L 1,3-PD in 24 h, which was nearly twofold of that of control K. pneumoniae(pET-pk-ald4). The overall glycerol conversion ratio of the strain K. pneumoniae(pET-pk-ald4-pkana-rpoE) was much higher than that of K. pneumoniae(pET-pk-ald4). In addition, compared with K. pneumoniae(pET-pk-ald4), K. pneumoniae(pET-pk-ald4-pkana-rpoE) produced less lactic acid during the late phase of fermentation particularly in 18 h (Fig. 3e). Furthermore, all recombinant strains produced less acetic acid than that of wild type K. pneumoniae. In particular, K. pneumoniae(pET-pk-ald4-pkana-rpoE) synthesized similar amount of acetic acid to K. pneumoniae(pET-pk-ald4) (Fig. 3f). Collectively, the expression of rpoE not only benefited the production of 3-HP and 1,3-PD, but also attenuated the formation of lactic acid and acetic acid.
Bioreactor Cultivation of the Recombinant Strains
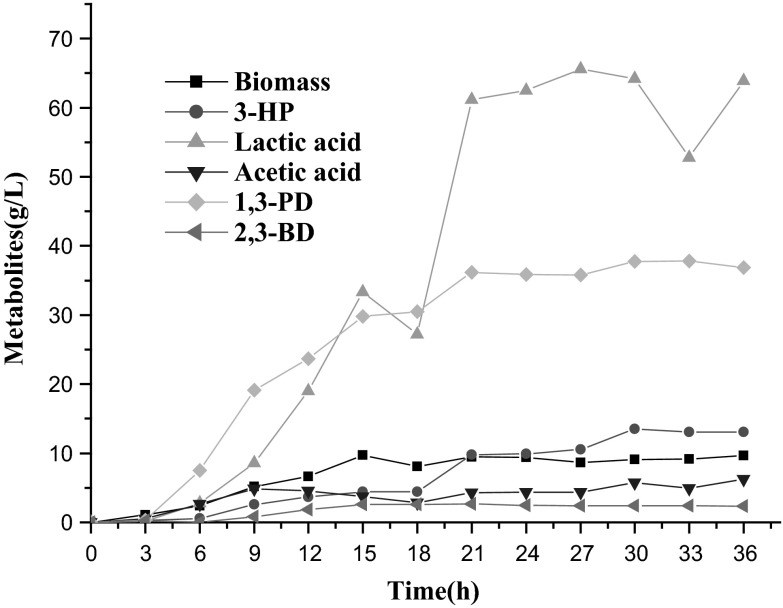
To further investigate the production of 3-HP and 1,3-PD by the strain K. pneumoniae(pET-pk-ald4-pkana-rpoE), fed-batch fermentation was carried out in a 5 L bioreactor containing 3 L medium. The glycerol was fed-batch added and pH value was maintained at 7.0. This recombinant strain yielded 3-HP at 13.5 g/L and 1,3-PD at 37.8 g/L in 30 h. The overall conversion ratio from glycerol to 3-HP and 1,3-PD was 0.45 mol/mol. Moreover, this strain produced 60 g/L of lactic acid, 6.2 g/L of acetic acid and 2.4 g/L of 2,3-BD (Fig. 4). Overall, 3-HP and 1,3-PD in the strain K. pneumoniae(pET-pk-ald4-pkana-rpoE) were highly produced.
Fig. 4.
Bioreactor cultivation of the recombinant K. pneumoniae(pET-pk-ald4-pkana-rpoE). 3-HP, 3-hydroxypropionic acid; 1,3-PD, 1,3-propanediol; 2,3-BD, 2,3-butanediol. Biomass, cell dry weight
Discussion
Although plasmid-dependent overexpression of key enzymes can push carbon flux into desired pathways, it consumes host resources such as RNA polymerase and cofactors, and thus halts cell growth especially when a strong promoter like T7 is employed [17]. Previously, pk promoter was harnessed to drive the expression of ALDH, however, 3-HP failed to be overproduced [3, 8]. One major reason is that pk is a constitutive promoter and its intrinsic activity is lower than T7 promoter. In this study, rpoE was demonstrated to be positively associated with pk activity and glycerol consumption. This conclusion is evidenced by the enhanced ALDH activity, glycerol consumption and 3-HP production. Compared with K. pneumoniae(pET-pk-ald4), K. pneumoniae(pET-pk-ald4-Pkana-rpoE) exhibited higher ALDH activity (Fig. 2), consumed more glycerol (Fig. 3b) and therefore produced more 3-HP (Fig. 3c). As for the less glycerol consumption and slower cell growth compared to wild type strain, these results could be explained by plasmid burden and the complexity of the dha regulon which mediates glycerol metabolism. As depicted in Fig. 1, in comparison to wild type K. pneumoniae, K. pneumoniae(pET-pk-ald4-Pkana-rpoE) harbors the heavy vector pET-pk-ald4-Pkana-rpoE, whereby ald4 and rpoE were coexpressed. In addition, the expression of ald4 consumed cofactor NAD+, which in turn affected a series of biochemical reactions that were closely related to glycerol dissimilation. In fact, glycerol metabolism in K. pneumoniae is mediated by the dha regulon which involves multiple factors, including cofactor availability, energy provision, and carbon flux allocation. Presumably, the lack of NAD+ caused by ald4 expression may hinder glycerol consumption and cell growth.
Although the supplement of rpoE was shown to be favorable for promoter activity, the contributions of other subunits to the transcription efficiency remain unknown. Hence, further study is needed. Recently, Voigt group in MIT developed a “resource allocator”, whereby RNA polymerase was fragmented into “core fragment” and “sigma fragment”. Adjusting the concentration of “core fragment” could set the maximum transcription efficiency [18]. This study suggests the potential applications of RNA polymerase in metabolic engineering of industrial strains. In fact, other studies also highlight the crucial role of RNA polymerase in allocating metabolic flux. In addition to RNA polymerase, other elements also participate in transcription and translation processes, and the underlying mechanisms remain ambiguous. Among questions to be addressed, the interplay between promoters and sigma factors is most attractive. Based on the differential binding of sigma factors to promoters, a sigma factor gene libraries were constructed and transformed into cells for reprogramming global transcription and thus leading to diverse phenotypes [12, 19]. Among numerous mutant clones, the strain manifesting desired phenotype could be obtained via high throughput screening. Since pk promoter worked well in K. pneumoniae, sigma factors could be rationally designed to specifically bind desired promoters. Given the extensive studies on K. pneumoniae, it is foreseeable that an efficient K. pneumoniae expression system could be developed. Therefore, this work may be insightful for metabolic engineering of other species.
Conclusion
Considering sigma factors bind promoters and initiate global transcription, we speculated that the sigma factors might be critical for pk activity. To verify this hypothesis, three sigma factors including rpoE, rpoS and dksA were expressed alone or in combination with Ald4 in K. pneumoniae. GFP expression analysis showed that the Ald4 activity in the strain coexpressing Ald4 and rpoE was higher than that expressing Ald4 alone. Moreover, under shake-flask conditions, the concentration of 3-HP and 1,3-PD in the strain that coexpressing Ald4 and rpoE increased by 1.6- and 0.85-fold, respectively, compared with that expressing Ald4 alone. Furthermore, under non-optimized bioreactor conditions, the strain coexpressing Ald4 and rpoE produced 13.5 g 3-HP and 37.8 g 1,3-PD per liter, and the glycerol conversion ratio reached 0.45 mol/mol. All these results indicate that the supplement of rpoE elevated Ald4 activity and stimulated glycerol consumption.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by grants from National Basic Research Program of China (973 Program) (2012CB725200), National High Technology Research and Development Program (863 Program) (No. 2015AA021003), National Natural Science Foundation of China (Nos. 21276014, 21476011), and the Fundamental Research Funds for the Central Universities (YS1407).
References
- 1.Kumar V, Ashok S, Park S. Recent advances in biological production of 3-hydroxypropionic acid. Biotechnol Adv. 2013;31:945–961. doi: 10.1016/j.biotechadv.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Ji XJ, Huang H, Ouyang PK. Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol Adv. 2011;29:351–364. doi: 10.1016/j.biotechadv.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Su MY, Ge XZ, Tian PF. Enhanced aldehyde dehydrogenase activity by regenerating NAD+ in Klebsiella pneumoniae and implications for the glycerol dissimilation pathways. Biotechnol Lett. 2013;35:1609–1615. doi: 10.1007/s10529-013-1243-1. [DOI] [PubMed] [Google Scholar]
- 4.Forage RG, Lin EC. DHA system mediating aerobic and anaerobic dissimilation of glycerol in Klebsiella pneumoniae NCIB 418. J Bacteriol. 1982;151:591–599. doi: 10.1128/jb.151.2.591-599.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forage RG, Foster MA. Glycerol fermentation in Klebsiella pneumoniae: functions of the coenzyme B12-dependent glycerol and diol dehydratases. J Bacteriol. 1982;149:413–419. doi: 10.1128/jb.149.2.413-419.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashok S, Raj SM, Rathnasingh C, Park S. Development of recombinant Klebsiella pneumoniae∆dhaT strain for the co-production of 3-hydroxypropionic acid and 1, 3-propanediol from glycerol. Appl Microbiol Biotechnol. 2011;90:1253–1265. doi: 10.1007/s00253-011-3148-z. [DOI] [PubMed] [Google Scholar]
- 7.Ma Z, Rao Z, Zhuge B, Fang H, Liao X, Zhuge J. Construction of a novel expression system in Klebsiella pneumoniae and its application for 1,3-propanediol production. Appl Biochem Biotechnol. 2010;162:399–407. doi: 10.1007/s12010-009-8743-4. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Sa N, Wang FH, Tian PF. Engineered constitutive pathway in Klebsiella pneumoniae for 3-hydroxypropionic acid production and implications for decoupling glycerol dissimilation pathways. Curr Microbiol. 2013;66:293–299. doi: 10.1007/s00284-012-0271-8. [DOI] [PubMed] [Google Scholar]
- 9.Loewen PC, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–717. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 10.Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heydorn A, Ersboll B, Kato J, Hentzer M, Parsek MR, Tolker-Nielsen T, Givskov M, Molin S. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl Environ Microbiol. 2002;68:2008–2017. doi: 10.1128/AEM.68.4.2008-2017.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alper H, Stephanopoulos G. Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab Eng. 2007;9:258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrikh H, Ferrazzoli AE, Bougdour A, Olivier-Mason A, Lovett ST. A DNA damage response in Escherichia coli involving the alternative sigma factor, RpoS. Proc Natl Acad Sci USA. 2009;106:611–616. doi: 10.1073/pnas.0803665106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riordan JT, Tietjen JA, Walsh CW, Gustafson JE, Whittam TS. Inactivation of alternative sigma factor 54 (RpoN) leads to increased acid resistance, and alters locus of enterocyte effacement (LEE) expression in Escherichia coli O157: H7. Microbiology. 2010;156:719–730. doi: 10.1099/mic.0.032631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Li Z, Shimizu K, Ye Q. Co-production of 3-hydroxypropionic acid and 1,3-propanediol by Klebseilla pneumoniae expressing aldH under microaerobic conditions. Bioresour Technol. 2013;128:505–512. doi: 10.1016/j.biortech.2012.10.143. [DOI] [PubMed] [Google Scholar]
- 17.Seoane J, Sin G, Lardon L, Gernaey KV, Smets BF. A new extant respirometric assay to estimate intrinsic growth parameters applied to study plasmid metabolic burden. Biotechnol Bioeng. 2010;105:141–149. doi: 10.1002/bit.22518. [DOI] [PubMed] [Google Scholar]
- 18.Segall-Shapiro TH, Meyer AJ, Ellington AD, Sontag ED, Voigt CA. A ‘resource allocator’ for transcription based on a highly fragmented T7 RNA polymerase. Mol Syst Biol. 2014;10:742. doi: 10.15252/msb.20145299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.