Abstract
Several pathogens including Brucella spp. are shed in semen of infected bulls and can be transmitted to cows through contaminated semen during artificial insemination. The present study reports omp2a and bcsp31 gene based loop-mediated isothermal amplification (LAMP) assays for detection of Brucella genomic DNA in semen from infected bulls. The positive results could be interpreted visually by change in colour of reaction mixture containing hydroxyl naphthol blue (HNB) dye from violet to sky blue. LAMP assays based on omp2a and bcsp31 could detect as little as 10 and 100 fg of B. abortus S19 genomic DNA, respectively. Sensitivity of omp2a and bcsp31 LAMP assays for direct detection of organisms in bovine semen was 2.28 × 101 CFU and 2.28 × 102 CFU of B. abortus S19 in spiked bovine semen, respectively. The omp2a LAMP assay was found equally sensitive to TaqMan probe based real-time PCR and 100 times more sensitive than conventional PCR in identifying Brucella in spiked semen. The diagnostic applicability of the omp2a LAMP assay was evaluated with seventy-nine bovine semen samples and results were re-evaluated through TaqMan probe based real-time PCR and conventional PCR. Taken together, the omp2a LAMP assay is easy to perform, rapid and sensitive in diagnosis of Brucella spp. in bovine semen.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-015-0563-3) contains supplementary material, which is available to authorized users.
Keywords: Loop-mediated isothermal amplification assay, Bovine brucellosis, Bovine semen, omp2a, bcsp31
Introduction
Brucellosis is an infectious and zoonotic disease of animals and humans caused by bacteria belonging to the genus Brucella. In cattle, brucellosis is usually caused by Brucella abortus (B. abortus), less frequently by B. melitensis and occasionally by B. suis [1]. The disease is characterised by abortion, retention of placenta, orchitis and epididymitis [1]. The Brucella pathogens are excreted through semen of infected bulls and the disease may be transmitted to healthy cows through Brucella contaminated semen used in artificial insemination (AI) [2]. Thus, transmission of Brucella pathogen, through frozen semen may be a threat to national cattle herd, where AI is used as a major breeding tool for genetic improvement. This is also a matter of concern for countries involved in international trading of frozen semen for overseas distribution of superior livestock germplasm. Diagnostic methods like microbiological isolation and identification of organisms and nucleic acid amplification based techniques have been used for detection of Brucella pathogens in bull semen [1]. Although microbiological isolation is the gold standard for Brucella detection in semen, this method is time consuming and there is risk to laboratory personnel. A sensitive nucleic acid based method, polymerase chain reaction (PCR) has been used in detection of Brucella genomic DNA in bovine semen [2–4]. Minimal requirements like expensive PCR machines and skilled personnel (for reaction set up and result analysis) limit PCR technique being used as a routine diagnostic test for Brucellosis under field condition and less equipped laboratories at breeding bull stations.
Loop-mediated isothermal amplification (LAMP) is a novel nucleic acid sequence amplification based assay, amplifies target DNA under isothermal condition in the range of 60–65 °C with high specificity, sensitivity and rapidity [5]. LAMP amplified products can be detected either visually with naked eyes by observing post amplification turbidity of reaction mixture or by using nucleic acid intercalating dyes [6]. Because of high target specificity, sensitivity, simplicity and rapidity, LAMP assay has been widely used in laboratories as a nucleic acid sequence based test to detect microorganisms causing infectious diseases in humans, animals and plants, identification of genetically modified products and tumours and embryo sexing [7].
LAMP assays have been reported for detection of Brucella organisms in milk, blood and organs of aborted foetuses [8–13]. In this study, we report LAMP assays targeting omp2a and bcsp31 genes for detection of Brucella in bull semen. The omp2a (Outer membrane protein 2a) gene encodes porin, a pore forming protein and responsible for efficient diffusion of sugar into cells [14]. The bcsp31 (Brucella cell surface protein 31) gene encodes a 31 kDa immunogenic protein. The bcsp31 is conserved among many Brucella spp [15]. Diagnosis sensitivity of standardised LAMP assays (omp2a and bcsp31) were compared with that of conventional PCR and TaqMan probe based real-time PCR. This is the first report of visual detection of Brucella spp. in the bovine semen by omp2a LAMP.
Materials and Methods
Bacterial Strains and Semen Samples
Four Brucella species and thirteen non-Brucellaspecies of bacteria are used in this study (Table S1). Trypticase soy agar (Difco, USA) was used to grow B. abortus S19.
A total of seventy-nine semen samples were collected from breeding bulls housed in semen collection centres and organised farms. All bulls were found sero-negative to anti-Brucella antibodies in rose bengal plate test (RBPT).
Primer Design
The coding sequences for Brucella spp. specific genes, namely, omp2a (GenBank accession no. M26034) and bcsp31 (GenBank accession no. M20404) were aligned with available sequences in GenBank. The nucleotide sequence region of genes found conserved among Brucella genus members were selected as targets for designing LAMP primers. Online software program, “Primer Explorer V4” (http://primerexplorer.jp/e) was used for designing primers. For each target, six primers, including outer primers (F3 and B3), loop primers (LF and LB) and inner primers (FIP and BIP) were designed (Fig S1a and S1b, Table S2). All primers were purchased from Integrated DNA Technologies (IDT, USA).
Bacterial Genomic DNA Isolation
Reaction conditions of LAMP assays were optimised by using genomic DNA (as template) isolated from inactivated B. abortus S19 using CTAB with slight modifications [16]. The cell pellet from 1.5 mL culture was re-suspended in 567 µL of TE buffer (pH 8.0). The cells were incubated for 1 h at 37 °C with 30 µL of sodium dodecyl sulphate (10 %) and 3.0 µL of Proteinase K (20 mg/mL). The cell lysate was further incubated at 65 °C for 10 min with 100 µL of 5 M NaCl and 80 µL of CTAB/NaCl solution. Following incubation, extracts were purified using phenol: chloroform: isoamyl alcohol (25:24:1). Then after, genomic DNA was precipitated from aqueous phase by adding 1/10 volume of 7.5 % ammonium acetate and double volume of absolute ethanol, washed twice with 70 % ethanol, air-dried and dissolved in 100 µL of TE buffer.
To get pure form of genomic DNA, QIAamp DNA Mini Kit (Qiagen, Hilden) was used to isolate total genomic DNA from bovine semen samples and B. abortus S19 spiked bovine semen as per manufacturer’s protocol. A volume of 10 µL bovine semen was used for genomic DNA isolation and DNA was eluted in 20 µL elution buffer.
Optimisation of LAMP Assays
Both omp2a and bscp31 LAMP assays were optimized by varying concentration of reaction components and reaction conditions. The assays were evaluated at different amplification temperatures (60–68 °C), reaction time (15–60 min), concentration of MgSO4 (0–6.4 mM) and dNTPs (0 to 2 mM). The LAMP assay reactions were performed in a PCR machine (S-24 Satellite thermal cycler, Quanta Biotech). The optimized reaction conditions for omp2a constituted of the following components: 0.2 µM each of outer primers (F3 and B3), 1.6 µM each of inner primers (FIP and BIP), 0.8 µM each of loop primers (LF and LB), 1.2 M betain (Sigma), 3.2 mM MgSO4 (Amresco), 1.2 mM dNTPs mix (Fermentas), 1X ThermoPol reaction buffer (20 mM Tris–HCl, 10 mM KCl, 2 mM MgSO4, 10 mM (NH4)2SO4 and 0.1 % Triton X-100) (NEB), 120 µM hydroxynaphthol blue (HNB) dye (Sigma) and target DNA (50–100 ng) in a 25 µL reaction volume. The reaction mixture was heated at 95 °C for 5 min for genomic DNA (template) denaturation and immediately cooled on ice for 2 min. The reaction tubes were briefly centrifuged and 8 units large fragment of Bst DNA polymerase (NEB) was added to the reaction mixture and incubated at 63 °C for 60 min for amplification reaction to happen. Finally the reaction was terminated by heating at 80 °C for 5 min. The optimised condition for bcsp31 LAMP assay was similar to omp2a assay except for 2.4 mM MgSO4 and 0.8 mM dNTPs.
Detection of Amplified LAMP Products
The amplified products of LAMP reactions were analysed visually by naked eye as well as by agarose gel electrophoresis. With HNB dye, the colour of the reaction mixture changes from violet to sky blue in case of positive samples and remains unchanged (i.e. violet) for negative samples. The results were further confirmed by resolving 3 µL of LAMP product on 2 % agarose gel.
Sensitivity and Specificity of LAMP Assays
The sensitivity of omp2a and bcsp31 LAMP assays was performed with and without HNB dye in the reaction mixture. The detection limits of omp2a and bcsp31 LAMP assays were determined by limiting dilution method. The purified B. abortus S19 genomic DNA was serially diluted (tenfold) with nuclease free water and each dilution was put to LAMP reaction to determine detection limit. Further to determine the sensitivity of LAMP assays, semen from a healthy bull (Brucella negative) was spiked with known number of B. abortus S19 cells. Diagnosis sensitivity of the LAMP assays were compared with reported conventional PCR and TaqMan probe based real time PCR methods.
The specificity of omp2a and bcsp31 LAMP assays was analysed with total genomic DNA isolated from Brucella and non-Brucella spp. (Table S1). The sensitivity and specificity experiments were repeated thrice.
Conventional PCR and TaqMan Probe Based Real-Time PCR Assays
The conventional PCR was performed by using BMEI0535f and BMEI0536r primers from reported Bruce-ladder multiplex PCR assay [1]. PCR reactions were carried out in a thermal cycler (S-24 Satellite, Quanta Biotech), in 50 µL of reaction volume consisting of primers (each of 10 pico moles), 1X PCR master mix (ThermoFischer Scientific) and genomic DNA template (at various concentrations). The reaction mixture was subjected to initial denaturation at 94 °C for 3 min followed by 30 cycles of denaturation, primer annealing and extension at 94 °C for 1 min, 55 °C for 1 min and 72 °C for 1 min respectively and final extension at 72 °C for 5 min. The PCR products were analysed on 1.5 % agarose gel.
The TaqMan probe based real-time PCR was carried out as reported earlier [17]. In brief, the 25 µL reaction volume consisted of 0.2 µM of probe, 0.4 µM each of primers and 0.5 µL of isolated DNA in 1X probe Mastermix (Sigma, EU). The reaction mixture was initially denatured at 94 °C for 2 min. followed by amplification reactions included 40 cycles of denaturation at 94 °C for 15 s, annealing and extension at 60 °C for 1 min. The amplification was carried out in a real-time PCR machine (Mx3005P, Agilent).
Screening of Bovine Semen Samples
Seventy-nine bovine semen samples along with positive and negative controls were subjected to omp2a LAMP assay, conventional PCR and TaqMan probe based real-time PCR. Genomic DNA isolated from spiked semen of dilutions 2.28 × 101 CFU and 2.28 × 103 CFU were used as positive controls for omp2a LAMP assay/TaqMan probe based real time PCR and conventional PCR respectively. Genomic DNA isolated from semen of Brucella negative bulls were used as template for negative controls. Each test was performed three times for all semen samples in order to avoid false positive and false negative results.
Results and Discussion
The present study was aimed at developing a diagnostic test for rapid detection of Brucella spp. in bovine semen. Two LAMP assays targeting conserved regions from omp2a and bcsp31 genes of Brucella spp were analysed and optimized. Amplification was observed from 60–68 °C. However, 63 °C was chosen as optimum temperature for LAMP assays (data not shown). The amplified products could be visualized as early as 30 min of incubation at 63 °C, however, higher amplification was observed after 60 min (data not shown). LAMP assay targeting omp2a gene produced best result with 1.2 mM dNTPs and 3.2 mM MgSO4 while bcsp31 LAMP was optimised with 0.8 mM of dNTPs and 2.4 mM of MgSO4 (data not shown). Positive LAMP reactions were visually identified by change in colour of reaction mixture from violet to sky blue. In LAMP negative reactions, the colour of reaction mixture remained violet (Fig S2a). Amplified products in positive reactions exhibited typical DNA laddering pattern on agarose gel (Fig S2b). A positive LAMP reaction can also be detected by visualising turbidity generated due to accumulation of reaction by-product magnesium pyrophosphate [18]. But detection of turbidity requires some skill and intense observation to distinguish positive from negative LAMP results. In this study, inclusion of HNB dye in reaction mixture before amplification reaction has facilitated visual distinction between positive and negative LAMP reactions. There is no requirement of post-amplification electrophoretic analysis of samples. Gradual depletion of magnesium ion during amplification reaction is the reason behind change of pre-amplification violet colour of reaction mixture to post-amplification sky blue colour in a positive LAMP reaction [6]. DNA intercalating dyes like SYBR green I and picogreen can be used for detecting amplified product [19, 20]. But it requires a UV light source for visualisation. Moreover, these dyes are added to reaction mixture on completion of amplification reaction thus increasing the possibility of laboratory contamination [21]. HNB dye based omp2a and bcsp31 LAMP assays are more of closed tube approach. There is no need of opening the tubes after reaction to add dye and thereby, minimizes cross contamination.
The specificity of the LAMP assays was analysed by using genomic DNA isolated from Brucella and non-Brucella species as targets for amplification reactions. LAMP reaction mixtures were turned sky blue for all Brucella strains and remained violet for non-Brucella species. The results were also confirmed through agarose gel electrophoresis. The non-Brucella bacterial pathogens like Leptospira spp., Coxiella burnetii, Mycobacterium avium subsp paratuberculosis and Mycoplasma mycoides ssp. Mycoides can be transmitted through semen [22]. These organisms didn’t show cross reactivity in both omp2a and bcsp31 LAMP assays (Fig S3a and S3b), confirming their specificity to Brucella species.
The sensitivity of omp2a and bcsp31 LAMP assays was remained same with and without HNB dye in the reaction mixture (data not shown). This indicated that HNB dye didn’t interfere in amplification process. Sensitivity of the LAMP assays was determined by using Brucella genomic DNA as template. The omp2a and bcsp31 LAMP assays could detect as little as 10 fg and 100 fg of Brucella genomic DNA respectively (data not shown). In conventional PCR, the detection limit was found 10 pg of Brucella genomic DNA (data not shown). Thus, sensitivity of both LAMP assays was found 100 times higher to that of conventional PCR when pure genomic DNA was used as template.
Sensitivity of the LAMP assays for direct detection of Brucella organisms in semen was determined by spiking bovine semen with B. abortus S19. The omp2a and bcsp31 LAMP assays could detect 2.28 × 101 CFU (Fig. 1a) and 2.28 × 102 CFU (Fig. 1b) of B. abortus S19 respectively in spiked semen. The results evaluated omp2a LAMP assay as ten times more sensitive than bcsp31 LAMP assay in detecting Brucella in spiked semen. The sensitivity of omp2a LAMP assay suggests its usefulness in detecting as little as 23 Brucella organisms in semen. The sensitivity of omp2a and bcsp31 LAMP assays was also compared with conventional PCR and TaqMan probe based real-time PCR using total genomic DNA isolated from each dilutions of B. abortus S19 spiked semen. The TaqMan probe based real-time PCR could detect 2.28 × 101 CFU of B. abortus S19 (Fig. 1c) while the sensitivity of the conventional PCR was to detect 2.28 × 103 CFU of B. abortus S19 in spiked semen (Fig. 1d). These results indicated that the LAMP assays were 10–100 times more sensitive than conventional PCR in detecting Brucella in semen. Presence of amplification inhibitory components in bovine semen [2, 3] or presence of large amount of host genomic DNA [25] or lesser number of Brucella organisms in higher dilutions of spiked semen (below detection limit) could be reasons for lower sensitivity of conventional PCR. However, sensitivity of the omp2a LAMP assay was equivalent to TaqMan probe based real time PCR. The IS711 TaqMan probe based real time PCR has been reported as a highly sensitive method in detecting less than 10 fg of genomic DNA (three genome copies) of Brucella organisms [17, 23, 24].
Fig. 1.
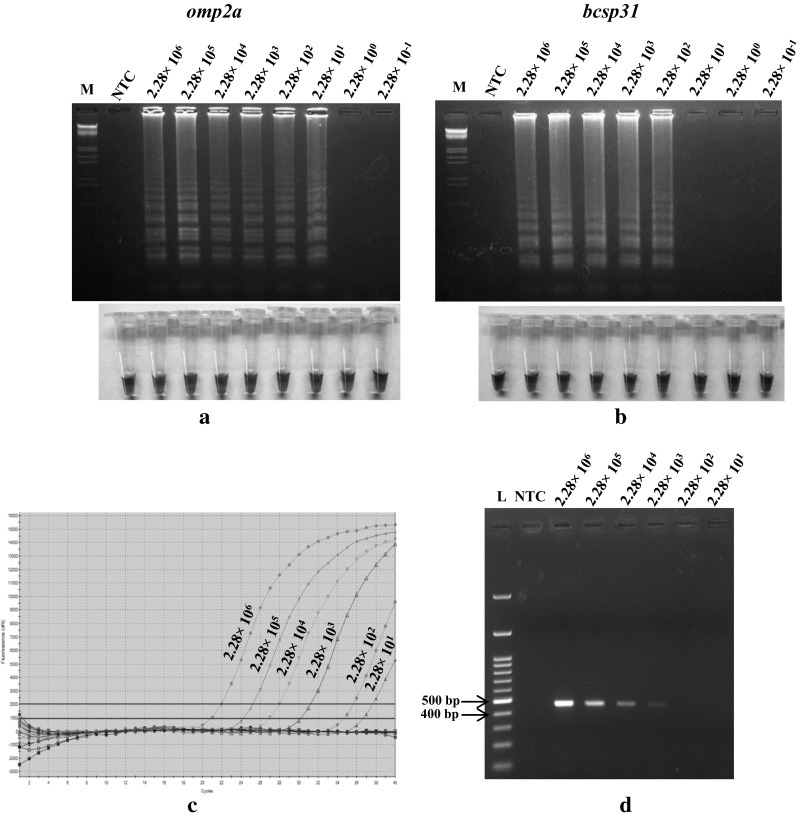
Analytical sensitivity of omp2a and bcsp31 LAMP assays (a, b) in comparison with TaqMan probe based real-time PCR (c) and conventional PCR (d) for detection of Brucella abortus S19 organism. The bovine semen spiked with B. abortus S19 (from 2.28 × 106 to 2.28 × 10−1 CFU) was used for total genomic DNA isolation and used for amplification. Lane L GeneRuler 100 bp plus ladder. Lane M λ DNA double digested with EcoRI and HindIII Marker. Lane NTC No template control
Seventy-nine semen samples collected from breeding bulls were tested by omp2a LAMP assay. All samples were found negative with LAMP assay as evident from violet colour of the reaction mixture after amplification reaction and gel electrophoresis analysis of LAMP products. These samples were also found negative with conventional PCR and TaqMan probe based real-time PCR. Brucella organisms could not be isolated on trypticase soy agar (TSA) from all the seventy-nine semen samples. This might be due to regular screening and segregation of infected bulls at germ plasm centres where bulls are used for semen collection. The negative result further proves the high specificity of omp2a LAMP assay to Brucella spp.
Conclusions
In summary, HNB dye based omp2a LAMP assay could be used as sensitive, specific and user-friendly test to detect Brucella spp. in bovine semen at organized farms and semen collection centres with limited laboratory facilities. In future more number of field semen samples need to be subjected to omp2a LAMP assay in order to validate the test for detection of Brucella spp. in bovine semen samples.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was funded by a project of NFBSFARA scheme, ICAR, Government of India. We sincerely thank Indian Veterinary Research Institute (IVRI) for providing necessary facilities to carry out this work. We acknowledge Division of Bacteriology, Division of Biological Products and Division of Veterinary Public Health of our institute for providing Brucella and non-Brucella spp. for this study.
Compliance with Ethical Standards
Conflict of interest
Authors declare that they have no conflict of interest.
Contributor Information
Bikash R. Prusty, Phone: +91 581 2301584, Email: dr.bprusty@gmail.com
Praveen K. Gupta, Email: praveen.indian@gmail.com
References
- 1.OIE (2009) Bovine brucellosis. OIE Terrestrial Manual 2009, Chapter 2.4.3
- 2.Amin AS, Hamdy MER, Ibrahim AK. Detection of Brucella melitensis in semen using the polymerase chain reaction. Vet Microbiol. 2001;83:37–44. doi: 10.1016/S0378-1135(01)00401-1. [DOI] [PubMed] [Google Scholar]
- 3.Vinodh R, Raj GD, Govindarajan R, Thiagarajan V. Detection of Leptospira and Brucella genomes in bovine semen using polymerase chain reaction. Trop Anim Health. 2008;40:323–329. doi: 10.1007/s11250-007-9110-5. [DOI] [PubMed] [Google Scholar]
- 4.Junqueira Junior DG, Rosinha GMS, Carvalho CEG, Oliveira CE, Sanches CC, Lima-Ribeiro AMC. Detection of Brucella spp. DNA in the semen of seronegative bulls by polymerase chain reaction. Transbound Emerg Dis. 2013;6:376–377. doi: 10.1111/j.1865-1682.2012.01347.x. [DOI] [PubMed] [Google Scholar]
- 5.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goto M, Honda E, Ogura A, Nomoto A, Hanak KI. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 7.Fu S, Qu G, Guo S, Ma L, Zhang N, Zhang S, Gao S. Applications of loop-mediated isothermal DNA amplification. Appl Microbiol Biotechnol. 2011;163:845–850. doi: 10.1007/s12010-010-9088-8. [DOI] [PubMed] [Google Scholar]
- 8.Karthik K, Rathore R, Thomas P, Arun TR, Viswas KN, Dhama K, Agarwal RK. New closed tube loop mediated isothermal amplification assay for prevention of product cross-contamination. MethodsX. 2014;1:137–143. doi: 10.1016/j.mex.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karthik K, Rathore R, Thomas P, Arun TR, Viswas KN, Manjunathachar HV, Agarwal RK, Dhama K. Loop mediated isothermal amplification (LAMP) test for specific and rapid detection of Brucella abortus in cattle. Vet Q. 2014;34:174–179. doi: 10.1080/01652176.2014.966172. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Sancho M, Garcia-Seco T, Arrogante L, Garcia N, Martinez I, Diez-Guerrier A, Perales A, Goyache J, Dominguez L, Alvarez J. Development and evaluation of an IS711-based loop mediated isothermal amplification method (LAMP) for detection of Brucella spp. on clinical samples. Res Vet Sci. 2013;95:489–494. doi: 10.1016/j.rvsc.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Song L, Li J, Hou S, Li X, Chen S. Establishment of loop-mediated isothermal amplification (LAMP) for rapid detection of Brucella spp. and application to milk and blood samples. J Microbiol Methods. 2012;90:292–297. doi: 10.1016/j.mimet.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 12.Lin GZ, Zheng FY, Zhou JZ, Gong XW, Wang GH, Cao XA, Qiu C. Loop-mediated isothermal amplification assay targeting the omp25 gene for rapid detection of Brucella spp. Mol Cell Probes. 2011;25:126–129. doi: 10.1016/j.mcp.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Ohtsuki R, Kawamoto K, Kato Y, Shah MM, Ezaki T, Makino SI. Rapid detection of Brucella spp. by the loop-mediated isothermal amplification method. J Appl Microbiol. 2008;104:1815–1823. doi: 10.1111/j.1365-2672.2008.03732.x. [DOI] [PubMed] [Google Scholar]
- 14.Marquis H, Ficht TA. The omp2 gene locus of Brucella abortus encodes two homologous outer membrane proteins with properties characteristic of bacterial porins. Infect Immun. 1992;61:3785–3790. doi: 10.1128/iai.61.9.3785-3790.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halling SM, Detilleux PG, Tatum FM, Judge BA, Mayfield JE. Deletion of the BCSP31 gene of Brucella abortus by replacement. Infect Immun. 1991;59:3863–3868. doi: 10.1128/iai.59.11.3863-3868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson K (2001) Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol 00: I: 2.4:2.4.1–2.4.5. doi: 10.1002/0471142727.mb0204s56 [DOI] [PubMed]
- 17.Hinic V, Brodar I, Thomann A, Cvetnic Z, Makaya PV, Frey J, Abril C. Novel identification and differentiation of Brucella melitensis, B. abortus, B. suis, B. ovis, B. canis, and B.neotomae suitable for both conventional and real-time PCR systems. J Microbiol Methods. 2008;75:375–378. doi: 10.1016/j.mimet.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 19.Dukes JP, King DP, Alexandersen S. Novel reverse transcription loop-mediated isothermal amplification for rapid detection of foot-and-mouth disease virus. Arch Virol. 2006;151:1093–1106. doi: 10.1007/s00705-005-0708-5. [DOI] [PubMed] [Google Scholar]
- 20.Parida M, Horioke K, Ishida H, Dash PK, Saxena P, Jana AM, Islam MA, Inoue S, Hosaka N, Morita K. Rapid detection and differentiation of dengue virus serotypes by a real time reverse transcription loop-mediated isothermal amplification assay. J Clin Microbiol. 2005;43:2895–2903. doi: 10.1128/JCM.43.6.2895-2903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almasi MA, Ojaghkandi MA, Hemmatabadi A, Hamidi F, Aghaei S. Development of colorimetric loop-mediated isothermal amplification assay for rapid detection of the tomato yellow leaf curl virus. J Plant Pathol Microb. 2013;4:153. [Google Scholar]
- 22.Daniel Givens M, Marley MSD. Pathogens that cause infertility of bulls or transmission via semen. Theriogenology. 2008;70:504–507. doi: 10.1016/j.theriogenology.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 23.Bounaadja L, Albert D, Chenais B, Henault S, Zygmunt MS, Poliak S, Garin-Bastuji B. Real-time PCR for identification of Brucella spp.: a comparative study of IS711, bcsp31 and per target genes. Vet Microbiol. 2009;137:156–164. doi: 10.1016/j.vetmic.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 24.Newby DT, Hadfield TL, Roberto FF. Real-time PCR detection of Brucella abortus: a comparative study of SYBR green I, 59-exonuclease, and hybridization probe assays. Appl Environ Microbiol. 2003;69:4753–4759. doi: 10.1128/AEM.69.8.4753-4759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cogswel FB, Banta CE, Hughes TG, Gu Y, Philipp MT. Host DNA can interfere with detection of Borrelia burgdorferi in skin biopsy specimens by PCR. J Clin Microbiol. 1996;34:980–982. doi: 10.1128/jcm.34.4.980-982.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


