Abstract
Background:
Epigenetic deregulation is considered as a new hallmark of cancer. The long non-coding RNA MALAT1 has been implicated in several cancers; however, its role in breast cancer is still little known.
Methods:
We used RT–PCR, in situ hybridisation, and RPPA methods to quantify (i) the full-length (FL) and an alternatively spliced variant (Δsv) of MALAT1, and (ii) a panel of transcripts and proteins involved in MALAT1 pathways, in a large series of breast tumours from patients with known clinical/pathological status and long-term outcome.
Results:
MALAT1 was overexpressed in 14% (63/446) of the breast tumours. MALAT1-overexpressed tumour epithelial cells showed marked diffuse nuclear signals and numerous huge nuclear speckles. Screening of the dbEST database led to the identification of Δsv-MALAT1, a major alternatively spliced MALAT1 transcript, with a very different expression pattern compared with FL-MALAT1. This alternative Δsv-MALAT1 transcript was mainly underexpressed (18.8%) in our breast tumour series. Multivariate analysis showed that alternative Δsv-MALAT1 transcript is an independent prognostic factor. Δsv-MALAT1 expression was associated with alterations of the pre-mRNAs alternative splicing machinery, and of the Drosha-DGCR8 complex required for non-coding RNA biogenesis. Alternative Δsv-MALAT1 transcript expression was associated to YAP protein status and with an activation of the PI3K-AKT pathway.
Conclusions:
Our results reveal a complex expression pattern of various MALAT1 transcript variants in breast tumours, and suggest that this pattern of expressions should be taken into account to evaluate MALAT1 as predictive biomarker and therapeutic target.
Keywords: MALAT1, alternatively spliced transcript, breast cancer, prognostic value, signalling pathways
Several studies have recently shown that expression of long non-coding RNAs (lncRNAs) are dysregulated in various cancers and that these lncRNAs have important roles in tumourigenesis and tumour progression (Spizzo et al, 2012). One example of such oncogenic lncRNA is HOTAIR, which is highly expressed in breast cancer and is a predictor for metastasis formation and associated with a poor prognosis (Gupta et al, 2010). Among these lncRNAs, MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), also referred as NEAT2 (nuclear-enriched abundant transcript 2) was discovered 10 years ago by using a subtractive hybridisation approach. MALAT1 was originally identified as a transcript showing significant expression in non-small cell lung tumours at high risk for metastasis (Ji et al, 2003). MALAT1 gene has a length of 8708 bp (NR_002819.2) and is localised in chromosome 11q13.1. Unlike most of lncRNAs, MALAT1 is extremely abundant, ubiquitously expressed and highly conserved among mammals, with potentially major functional roles in mammalian cells. MALAT1 is a nuclear-retained lncRNA, suggesting both structural and functional properties, for example, nuclear architecture and organisation, splicing, or gene-expression regulation (Gutschner et al, 2013a). MALAT1 has been implicated in alternative splicing regulation, showing interactions with several splicing factors, such as SRSF1 (Tripathi et al, 2010). MALAT1 has also been linked to transcriptional control of genes involved in cell cycle, cell motility and EMT (Gutschner et al, 2013a). MALAT1 could act as a transcription activator by mediating assembly of Polycomb repressive complexes (Yang et al, 2011).
MALAT1 upregulation has been reported in several tumour types and is also a negative prognostic factor in lung, pancreas, colorectal and bladder cancers (Zhang et al, 2015). Molecular mechanisms involved in MALAT1 dysregulation are still unclear. Activation of MALAT1 by gene amplification seems unlikely because MALAT1 is located in a chromosomal region (11q13.1) not recurrently amplified in human cancers (Curtis et al, 2012). Mutations in the MALAT1 gene were recently discovery in human cancers (The Cancer Genome Atlas studies; Kandoth et al, 2013). MALAT1 seems the most frequently lncRNA mutated in human cancers. Rare cases of chromosomal translocations involving MALAT1 have also been reported in mesenchymal harmatomas and renal cell carcinomas (Davis et al, 2003; Mathews et al, 2013). MALAT1 epigenetic dysregulation mediated by CpG island methylation was not reported. A post-transcriptional MALAT1 regulation mechanism mediated by one microRNA (Hsa-miR-125b) has been only reported in bladder cancer (Han et al, 2013).
Few studies concerning MALAT1 in breast cancer are available. Guffanti et al (2009) identified MALAT1 as an abundantly expressed lncRNA in breast tumours. Rare mutations were recently described in luminal breast cancer (Ellis et al, 2012). Clinical prognostic value of MALAT1 dysregulation in breast cancer is little known at this time (Xu et al, 2015).
To obtain further insight concerning involvement of MALAT1 in molecular pathogenesis of breast cancer, we used quantitative real-time reverse-transcriptase–polymerase chain reaction (qRT–PCR) assay, to quantify the full-length (FL) and an alternatively spliced variant (Δsv) of MALAT1 mRNA expression in a series of 446 patients with unilateral invasive breast tumours and known long-term outcome. We sought links between MALAT1 mRNA expression pattern and classical clinical and pathological parameters, including patient outcome. We also sought relationships between MALAT1 and genes and proteins expression known to be involved in different steps of MALAT1 pathway dysregulation observed in others types of human cancers.
Materials and methods
Patients and samples
Samples of 446 unilateral invasive primary breast tumours excised from women managed at Institut Curie–Hôpital René Huguenin (Saint-Cloud, France) from 1978 to 2008 have been analysed. All patients cared in our institution before 2007 were informed that their tumour samples might be used for scientific purposes and had the opportunity to decline. Since 2007, patients treated in our institution have given their approval by signed inform consent. This study was approved by the local ethics committee (Breast Group of René Huguenin Hospital). Samples were immediately stored in liquid nitrogen until RNA extraction. A tumour sample was considered suitable for our study if the proportion of tumour cells exceeded 70%.
All patients (mean age 61.8 years, range 31–91 years) met the following criteria: primary unilateral nonmetastatic breast carcinoma for which complete clinical, histological and biological data were available; no radiotherapy or chemotherapy before surgery; and full follow-up at Institut Curie–Hospital René Huguenin.
Treatment (information available for 438 patients) consisted of modified radical mastectomy in 278 cases (63.9%) and breast-conserving surgery plus locoregional radiotherapy in 160 cases (36.1%). The patients had a physical examination and routine chest radiotherapy every 3 months for 2 years, then annually. Mammograms were done annually. Adjuvant therapy was administered to 360 patients, consisting of chemotherapy alone in 87 cases, hormone therapy alone in 172 cases and both treatments in 101 cases. The histological type and the number of positive axillary nodes were established at the time of surgery. The malignancy of infiltrating carcinomas was scored according to Scarff Bloom Richardson's histoprognostic system.
Hormone receptor (HR; i.e., oestrogen receptor-alpha (ERα), progesterone receptor (PR) and human epidermal growth factor receptor 2 (ERBB2) statuses were determined at the protein level by using biochemical methods (dextran-coated charcoal method, enzyme immunoassay or immunohistochemistry) and confirmed by qRT–PCR assays (Bieche et al, 1999, 2001).
The population was divided into four groups according to HRs (ERα and PR) and ERBB2 status, as follows: two luminal subtypes (HR+/ERBB2+ (n=45)) and (HR+/ERBB2− (n=195)); an ERBB2+ subtype (HR−/ERBB2+ (n=46)) and a triple-negative subtype (HR−/ERBB2− (n=64)). Standard prognostic factors of this tumour set are presented in (Supplementary Table 1). During a median follow-up of 9.1 years (range 4.3 months to 33.2 years), 176 patients developed metastasis.
Ten specimens of adjacent normal breast tissue from breast cancer patients and normal breast tissue from women undergoing cosmetic breast surgery were used as sources of normal RNA.
RNA extraction
Total RNA was extracted from breast tissue samples by using acid–phenol guanidium. RNA quality was determined by electrophoresis through agarose gels, staining with ethidium bromide and visualisation of the 18S- and 28S-RNA bands under ultraviolet light.
qRT–PCR
Quantitative values were obtained from the cycle number (Ct value) at which the increase in the fluorescence signal associated with exponential growth of PCR products started to be detected by the laser detector of the ABI Prism 7900 sequence detection system (Perkin-Elmer Applied Biosystems, Foster City, CA, USA), using PE biosystems analysis software according to the manufacturer's manuals.
The precise amount of total RNA added to each reaction mix (based on optical density) and its quality (i.e., lack of extensive degradation) are both difficult to assess. Therefore, transcripts of the TBP gene (Genbank accession NM_003194) encoding the TATA box-binding protein (a component of the DNA-binding protein complex TFIID) were also quantified as an endogenous RNA control. Each sample was normalised on the basis of its TBP content. TBP was selected as an endogenous control due to the absence of known TBP retropseudogenes (retropseudogenes lead to co-amplification of contaminating genomic DNA and thus interfere with qRT–PCR, despite the use of primers in separate exons; Bieche et al, 1999).
Results, expressed as N-fold differences in target-gene expression relative to the TBP gene and termed ‘Ntarget', were determined as Ntarget=2ΔCtsample, where the ΔCt value of the sample was determined by subtracting the average Ct value of target gene from the average Ct value of TBP gene.
The target-gene values of the breast tumour samples were subsequently normalised such that the median of the target-gene values for the 10 normal breast tissues was 1.
The primers for TBP, MALAT1 and others target genes were chosen with the assistance of the Oligo 6.0 program (National Biosciences, Plymouth, MN, USA; Supplementary Table 2). dbEST and nr databases were scanned to confirm the total gene specificity of the nucleotide sequences chosen for the primers and the absence of single-nucleotide polymorphisms. To avoid amplification of contaminating genomic DNA, one of the two primers was placed at the junction between two exons or on two different exons. Agarose gel electrophoresis was used to verify the specificity of PCR amplicons. The conditions of cDNA synthesis and PCR were as previously described (Bieche et al, 1999).
In situ hybridisation
We used Stellaris FISH Probes, Human MALAT1 with Quasar 570 Dye (Biosearch Technologies, Petaluma, CA, USA). First, paraffin-embedded tissue sliced at 4–5 μm thickness were obtained from normal and tumour tissues by using a microtome (Thermo scientific Sandom HE 340 E, Walldorf, Germany). Formalin-fixed paraffin-embedded breast tissue sections were deparaffinized by using 100% xylene, 100% ethanol, 95% ethanol, 70% ethanol and RNase-free PBS. Then, slides were incubated for 20 min at 37 °C, and washed twice with PBS. We created a working probe solution at 125 nM (probe diluted in hybridisation buffer). We immersed tissue sections in a wash buffer for 2–5 min, while assembling the humidified chamber. Then, we dispensed 100 μl of working probe solution onto tissue sections, placed them in the humidified chamber, and covered them with parafilm. We incubated tissue sections in the dark at 37 °C for at least 4 h. After decanting wash buffer, we added DAPI nuclear stain and immersed tissue sections in SSC. Finally, we added a small drop of antifade onto tissue sections and covered with a cover glass and proceeded to imaging.
RPPA
Samples were disrupted in Laemmli buffer (50 mM Tris pH=6.8, 2% SDS, 5% glycerol, 2 mM DTT, 2.5 mM EDTA, 2.5 mM EGTA, 1 × HALT phosphatase inhibitor (Perbio, Villebon-sur-Yvette, France; 78420), protease inhibitor cocktail complete MINI EDTA-free (Roche, Basel, Switzerland; 1836170, 1 tablet per 10 ml), 4 mM Na3VO4 and 20 mM NaF) qsp 5 ml H2O, using a Tissue Lyser (Qiagen, Venlo, Netherlands) and two 5 mm stainless beads per sample. Extracts were then boiled for 10 min at 100 °C, passed through a fine needle to reduce viscosity and centrifuged for 15 min at 13 000 r.p.m. The supernatant was collected and stored at −80 °C. Protein concentration was determined (Pierce BCA reducing agent compatible kit, Pierce, Waltham, MA, USA; ref 23252). Samples were deposited onto nitrocellulose covered slides (Fast slides, Maine Manufacturing, Sanford, ME, USA) using a dedicated arrayer (2470 arrayer, Aushon Biosystems, Billerica, MA, USA). Five serial dilutions, ranging from 1000 to 62.5 μg ml−1, and two technical replicates per dilution were printed for each sample. Arrays were labelled with specific antibodies or without primary antibody (negative control), using an Autostainer Plus (Dako, Glostrup, Denmark). Briefly, slides were incubated with avidin, biotin and peroxides blocking reagents (Dako) before saturation with TBS containing 0.1% Tween-20 and 5% BSA (TBST-BSA). Slides were then probed overnight at 4 °C with primary antibodies diluted in TBST-BSA. After washes with TBST, arrays were probed with horseradish peroxidase-coupled secondary antibodies (Jackson ImmunoResearch Laboratories, New market, UK) diluted in TBST-BSA for 1 h at room temperature (RT). To amplify the signal, slides were incubated with Bio-Rad Amplification Reagent (Bio-Rad, Hercules, CA, USA) for 15 min at RT. The arrays were washed with TBST, probed with Alexa647-Streptavidin (Molecular Probes, Eugene, OR, USA) diluted in TBST-BSA for 1 h at RT and washed again in TBST. For staining of total protein, arrays were incubated 15 min in 7% acetic acid and 10% methanol, rinsed twice in water, incubated 10 min in Sypro Ruby (Invitrogen, Carlsbad, CA, USA) and rinsed again. The processed slides were dried by centrifugation and scanned using a GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA, USA). Spot intensity was determined with MicroVigene software (VigeneTech Inc., Carlisle, MA, USA). All primary antibodies used in RPPA have been previously tested by Western Blotting to assess their specificity for the protein of interest.
Raw data were normalised using Normacurve (Troncale et al, 2012), which normalises for fluorescent background per spot, a total protein stain and potential spatial bias on the slide. Next, each RPPA slide was median centred and scaled (divided by median absolute deviation). We then corrected for remaining sample loadings effects individually for each array by correcting the dependency of the data for individual arrays on the median value of each sample over all arrays using a linear regression.
Statistical analysis
The distributions of target mRNA levels were characterised by their median values and ranges. Relationships between mRNA levels of the different target genes, and between mRNA levels and clinical parameters, were identified using nonparametric tests, namely the χ2-test (relation between two qualitative parameters), the Mann–Whitney's U-test (relation between one qualitative parameter and one quantitative parameter) and the Spearman's rank correlation test (relation between two quantitative parameters). Differences were considered significant at confidence levels >95% (P<0.05).
To visualise the efficacy of a molecular marker (MALAT1 level) to discriminate two populations (patients that developed/did not develop metastases) in the absence of an arbitrary cut-off value, data were summarised in an receiver operating characteristic curve (Hanley and McNeil, 1982). The AUC (area under curve) was calculated as a single measure for discriminate efficacy. Metastasis-free survival (MFS) was determined as the interval between initial diagnosis and detection of the first metastasis. Survival distributions were estimated by the Kaplan–Meier method, and the significance of differences between survival rates were ascertained with the log-rank test. The Cox-proportional hazards regression model was used to assess prognostic significance and the results are presented as hazard ratios and 95% confidence intervals.
Results
MALAT1 expression in breast tumours and relationship with classical clinico-pathological parameters and patient outcome
In order to determine the prognostic significance of MALAT1 expression pattern in human breast tumours, we analysed MALAT1 mRNA levels in a large series of 446 primary breast tumours from patients with known clinical/pathological status and long-term outcome (Supplementary Table 1).
Among the 446 breast tumour RNA samples tested, 63 (14.1%) tumours showed MALAT1 mRNA overexpression (NMALAT1 from 3.02 to 13.4), and only 13 (2.9%) tumours showed MALAT1 mRNA underexpression (NMALAT1 from 0.15 to 0.32), as compared with normal breast tissues. We sought links between MALAT1 mRNA level status and standard clinico-pathological and biological factors in breast cancer (Supplementary Table 3). Significant positive associations were observed between the tumour group showing MALAT1 overexpression and ERα-positive (P=0.000015), PR-positive (P=0.00079) and molecular subtypes (P=0.00000075). It is noteworthy that majority (9/13) of the MALAT1 underexpressed tumours was of triple-negative (HR−/ERBB2−) subtype.
We also examined PIK3CA mutation status, and expressions of EGFR and MKI67 (which encodes the proliferation-related antigen Ki-67). None of these three markers showed significant link with MALAT1 expression.
To further investigate whether MALAT1 mRNA expression could be of prognostic relevance, the log-rank test was used to identify relations between MFS and MALAT1 mRNA expression. Results showed that MFS was not significantly influenced by MALAT1 overexpression status (P=0.23; data not shown).
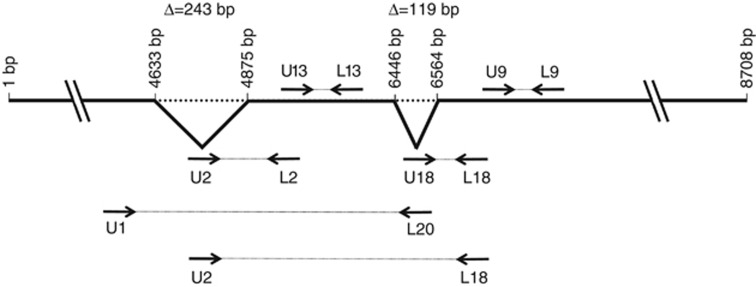
Results of MALAT1 mRNA levels shown in Supplementary Table 3 were obtained by using a primer pair (U13/L13) that encompass region 4891–4975 of published MALAT1 cDNA sequence (GenBank #NR_002819.2; Figure 1). Similar results (frequency of MALAT1 overexpression links with classical clinico-pathological parameters and patient outcome) were obtained with a second primer pair (U9/L9) localised in MALAT1 gene at region 6565–6658 (Figure 1).
Figure 1.
Location of primers used for MALAT1 mRNA expression analysis.Alternative splicing variants were amplified using primer pairs with one of the two primers placed at the junction of the two spliced regions: U2/L2 for the 243 bp alternatively spliced MALAT1 transcript (Δ2sv-MALAT1) and U18/L18 for the 119 bp alternatively spliced MALAT1 transcript (Δ1sv-MALAT1).
Relationship between MALAT1 mRNA level and HOTAIR and ANRIL mRNA levels
We tested possible relation between MALAT1 and HOTAIR and ANRIL (the two most documented lncRNAs that also interact with Polycomb repressive complexes) mRNA levels. We did not observe any association between MALAT1 and these two Polycomb complexes associated lncRNAs (r=+0.069, P=0.14 for HOTAIR; r=+0.014, P=0.76 for ANRIL; Spearman's rank correlation test).
Localisation of MALAT1 transcript in epithelial tumour cells
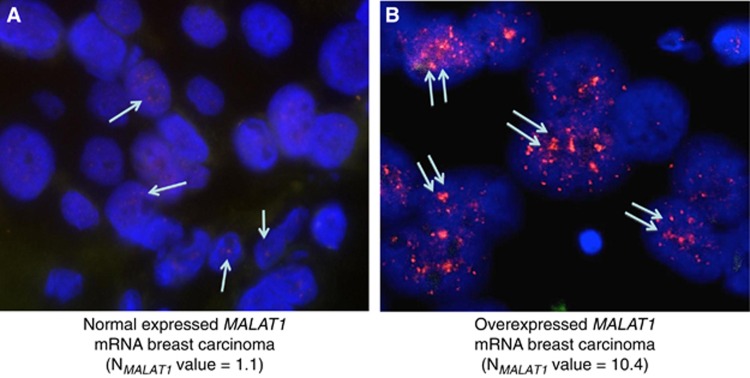
We detected specific MALAT1 RNA in epithelial and stromal cells of all ten tumour samples studied by in situ hybridisation (ISH). MALAT1 mRNA was found exclusively in the nucleus of both stromal and tumour epithelial cells. We detected strong specific MALAT1 RNA level in epithelial cells of the five tumours, which overexpressed MALAT1 mRNA (using qRT–PCR analysis) and low specific MALAT1 RNA level in the five tumours which did not overexpress MALAT1 mRNA. We thus obtained a perfect match between MALAT1 mRNA expression by using qRT–PCR and ISH analysis. MALAT1-overexpressed tumour epithelial cells showed marked diffuse nuclear signals and numerous nuclear speckles of variable size and shape as compared with MALAT1 normal-expressed tumour epithelial cells (Figure 2).
Figure 2.
ISH of MALAT1 RNA in breast tumours.(A) Example of normal-expressed MALAT1 mRNA breast tumour (NMALAT1 value=1.1, as determined by qRT–PCR analysis). Weak signals represented by small speckles (speckles in red, arrow) of equivalent size and shape, regularly distributed within nuclei of tumour cells (nuclei in blue) × 400. (B) Example of overexpressed MALAT1 mRNA breast tumour (NMALAT1 value=10.4). Marked diffuse signals and numerous and frequently huge nuclear speckles (speckles in red, two arrows) of variable size and shape within nuclei of tumour cells (nuclei in blue) × 600.
Identification of a major alternatively spliced MALAT1 transcript in breast tumours, and relationships with classical clinico-pathological parameters and patient outcome
Screening of the dbEST database with the MALAT1 cDNA led to identification of two major groups of alternatively spliced MALAT1 ESTs. The first major alternatively spliced MALAT1 transcript (named Δ1sv-MALAT1) had a 119-bp deletion (from 6446 to 6564, from the NR_002819.2 sequence), resulting from alternative splicing of MALAT1 mRNA, whereas the second alternatively spliced MALAT1 transcript (named Δ2sv-MALAT1) had a 243-bp deletion (from 4633 to 4875, from the NR_002819.2 sequence). The deleted nucleotide sequences show consensus sequences of donor/acceptor splice sites.
To verify presence and quantify mRNA levels of these alternative splicing variants in our breast cancer series, we carried out non-quantitative (classical) RT–PCR using primer pairs with one of the two primers placed at the junction of the two spliced regions: U2/L2 for the 243-bp alternatively spliced MALAT1 transcript (Δ2sv-MALAT1) and U18/L18 for the 119-bp alternatively spliced MALAT1 transcript (Δ1sv-MALAT1; Figure 1). However, additional qualitative analyses using primer couples U1/L20 or U2/L18 (Figure 1 and Supplementary Figure 1) showed that these two splices were always associated together. This unique transcript, showing both 119-bp and 243-bp deletions, is named Δsv-MALAT1 for the remaining part of the manuscript, and FL-MALAT1 for the FL transcript.
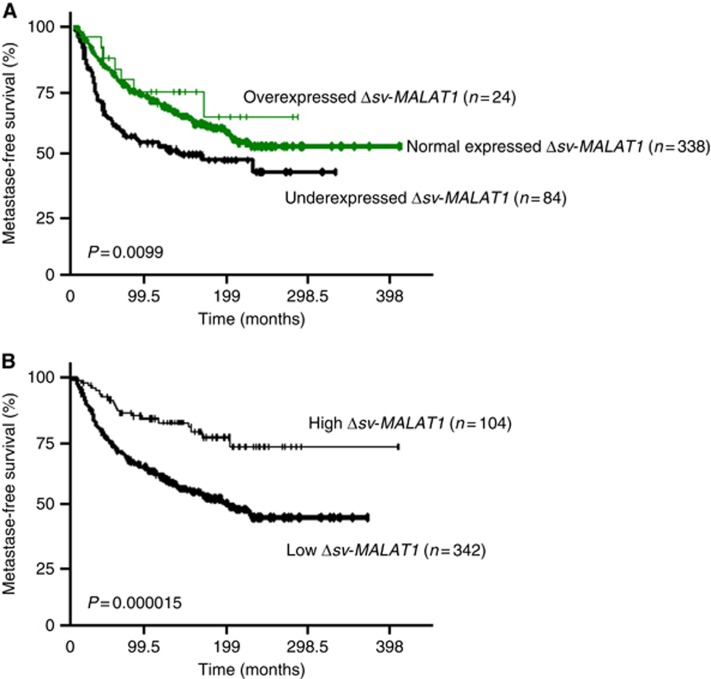
All the 446 breast tumour RNA samples tested showed a marked presence of Δsv-MALAT1 transcript. We observed a highly positive correlation between Δsv-MALAT1 and FL-MALAT1 expressions (Supplementary Figure 2A). The relative expression of Δsv-MALAT1 and FL-MALAT1 for each individual sample listed as dot/box plots is indicated in Supplementary Figure 2B. As compared with normal breast tissues, 24 (5.4%) tumours showed Δsv-MALAT1 mRNA overexpression (NΔsv-MALAT1 from 3.16 to 8.4), and surprisingly 84 (18.8%) tumours showed Δsv-MALAT1 mRNA underexpression (NΔsv-MALAT1 from 0.05 to 0.32). Marked significant positive associations were observed between the tumour group showing Δsv-MALAT1 underexpression and large macroscopic tumour size (P=0.0023), ERα-negative (P=0.000062), PR-negative (P=0.0000051) and molecular subtypes (P=0.00074; Table 1). We observed the same associations between Δsv-MALAT1 mRNA levels and molecular subtypes in a series of 21 breast cell lines, including 4 non-cancerous cell lines, 8 triple-negative cell lines, 4 ERBB2 cell lines and 5 RE+ cell lines (Supplementary Table 4). In particular, we observed a marked Δsv-MALAT1 underexpression in the triple-negative subtype. Δsv-MALAT1 underexpression was also highly associated with MKI67 mRNA levels (P=0.00033). MFS was significantly influenced by Δsv-MALAT1 (overexpression vs normal expression vs underexpression) expression status (P=0.0099; Figure 3A). AUC analyses was then performed to identify a putative cut-point to divide the cohort into two relevant Δsv-MALAT1 expression subgroups. Results confirmed that MFS of patients with low Δsv-MALAT1-expressing tumours (5-year RFS 70.3±2.5% 10-year RFS 58.5±2.8% 15-year RFS 51.4±3.0%) was shorter than that of patients whose tumours highly expressed Δsv-MALAT1 (5-year RFS 85.9±3.5% 10-year RFS 82.3±3.9% 15-year RFS 76.0±5.1% P=0.000015; Figure 3B).
Table 1. Relationship between Δsv-MALAT1-spliced transcript levels and classical clinical biological parameters in a series of 446 breast cancer.
|
Number of patients (%) |
|||||
|---|---|---|---|---|---|
| Total population (%) | Δsv-MALAT1 underexpression | Δsv-MALAT1 normal expression | Δsv-MALAT1 overexpression | P-valuea | |
| Total | 446 (100.0) | 84 (18.8) | 338 (75.8) | 24 (5.4) | |
|
Age (years) | |||||
| ⩽50 | 94 (21.1) | 25 (26.6) | 67 (71.3) | 2 (2.1) | 0.038 |
| >50 | 352 (78.9) | 59 (16.8) | 271 (77.0) | 22 (6.3) | |
|
SBR histological gradeb,c | |||||
| I | 57 (13.0) | 4 (7.0) | 49 (86.0) | 4 (7.0) | 0.043 |
| II+III | 380 (87.0) | 79 (20.8) | 282 (74.2) | 19 (5.0) | |
|
Lymph node statusd | |||||
| 0 | 117 (26.3) | 17 (14.5) | 90 (76.9) | 10 (8.5) | 0.043 |
| 1–3 | 231 (51.9) | 39 (16.9) | 182 (78.8) | 10 (4.3) | |
| >3 | 97 (21.8) | 27 (27.8) | 66 (68.0) | 4 (4.1) | |
|
Macroscopic tumour sizee | |||||
| ⩽25 mm | 218 (49.8) | 28 (12.8) | 174 (79.8) | 16 (7.3) | 0.0023 |
| >25 mm | 220 (50.2) | 55 (25.0) | 157 (71.4) | 8 (3.6) | |
|
ERα status | |||||
| Negative | 115 (25.8) | 37 (32.2) | 76 (66.1) | 2 (1.7) | 0.000062 |
| Positive | 331 (74.2) | 47 (14.2) | 262 (79.2) | 22 (6.6) | |
|
PR status | |||||
| Negative | 191 (42.8) | 55 (28.8) | 132 (69.1) | 4 (2.1) | 0.0000051 |
| Positive | 255 (57.2) | 29 (11.4) | 206 (80.8) | 20 (7.8) | |
|
ERBB2 status | |||||
| Negative | 353 (79.1) | 60 (17.0) | 273 (77.3) | 20 (5.7) | 0.15 (NS) |
| Positive | 93 (20.9) | 24 (25.8) | 65 (70.0) | 4 (4.3) | |
|
Molecular subtypes | |||||
| RH− ERBB2− | 68 (15.2) | 22 (32.4) | 46 (67.6) | 0 (0) | 0.00074 |
| RH− ERBB2+ | 42 (9.4) | 14 (33.3) | 26 (61.9) | 2 (4.8) | |
| RH+ ERBB2− | 285 (63.9) | 38 (13.3) | 227 (79.6) | 20 (7.0) | |
| RH+ ERBB2+ | 51 (11.4) | 10 (19.6) | 39 (76.5) | 2 (3.9) | |
|
PIK3CA mutation status | |||||
| Wild type | 299 (67.0) | 69 (23.1) | 215 (71.9) | 15 (5.0) | 0.0049 |
| Mutated | 147 (33.0) | 15 (10.2) | 123 (83.7) | 9 (6.1) | |
|
MKI67 mRNA expression | |||||
| Median | 12.5 (0.80–117) | 17.6 (0.86–117) | 11.8 (0.80–94.5) | 11.2 (0.93–47.1) | 0.00033f |
|
EGFR mRNA expression | |||||
| Median | 0.22 (0.00–106) | 0.13 (0.02–3.20) | 0.23 (0.00–106) | 0.20 (0.03–1.51) | 0.019f |
Abbreviations: ERα=oestrogen receptor-α ERBB2=human epidermal growth factor receptor 2; HR=hormone receptor; NS=not significant; PR=progesterone receptor.
The bold values are statistically significant (P-value<0.05).
χ2-test.
Scarff Bloom Richardson classification.
Information available for 437 patients.
Information available for 445 patients.
Information available for 438 patients.
Kruskal–Wallis's H test.
Figure 3.
MFS curves of patient groups according to Δsv-MALAT1 mRNA expression level in the series of 446 breast tumours.(A) MFS curves of three patients groups with under, normal and overexpressed Δsv-MALAT1 tumours, as compared with normal breast tissues. (B) MFS curves for patients with high-Δsv-MALAT1-expressing and low-MALAT1-expressing tumours, using an optimal cut-off value.
Results of Δsv-MALAT1 mRNA levels shown in Table 1 were obtained by using a primer pair (U18/L18) that encompasses the 119 bp deleted region. Similar results (frequency of Δsv-MALAT1 overexpression, links with classical clinico-pathological parameters and patient outcome) were obtained with a second primer pair (U2/L2) that encompass the 243 bp deleted region (Figure 1; Supplementary Table 5).
Finally, the prognostic significance of the five parameters identified in univariate analysis, including histopathological grade, lymph node status, macroscopic tumour size, PR status (Supplementary Table 1) and Δsv-MALAT1 expression status (Figure 3B) persisted (except for lymph node and PR status) in Cox multivariate regression analysis of MFS (Supplementary Table 6).
Relationship between Δsv-MALAT1 mRNA levels and Hsa-miR-125b expression status
As Hsa-miR-125b suppresses bladder cancer development by downregulating MALAT1 (Han et al, 2013), we tested the possible negative correlation between Δsv-MALAT1 and Hsa-miR-125b mRNA level in breast cancer. Hsa-miR-125b levels were analysed in 20 low-Δsv-MALAT1-expressing (median mRNA value: 0.36) and 20 high-Δsv-MALAT1-expressing breast tumours (median mRNA value: 2.13). We found no link between Δsv-MALAT1 and Hsa-miR-125b expression status: the median Hsa-miR-125b value was 0.13 in low-Δsv-MALAT1-expressing breast tumours and 0.11 in high-Δsv-MALAT1-expressing breast tumours. Similar results were obtained with 20 low-FL-MALAT1-expressing and 20 high-FL-MALAT1-expressing breast tumours.
Relationship between Δsv-MALAT1 mRNA levels and YAP protein level
As YAP protein regulates transcription of MALAT1 gene in liver cancer (Wang et al, 2014), we tested the possible positive correlation between YAP protein and Δsv-MALAT1 mRNA levels in breast cancer. YAP protein levels were analysed by using RPPA assay in 143 samples from our series of 446 breast tumours. We found a significant positive link with Δsv-MALAT1 mRNA level (r=+0.303, P=0.00032; Spearman's rank correlation test) but no link between YAP protein and FL-MALAT1 mRNA levels of expression. As Δsv-MALAT1 low level is associated with a poor outcome and with low-YAP-protein level, we tested if YAP protein level could be also of prognostic relevance. We did not observe in our smaller series of 143 samples, any statistical correlation between low-YAP-protein expression and poor outcome.
Relationship between Δsv-MALAT1 mRNA levels and a large panel of selected genes involving in various signalling pathways
To obtain further insight into MALAT1 dysregulated pathways in breast cancer, we evaluated by qRT–PCR mRNA expression of a large number of selected genes in 20 low-Δsv-MALAT1-expressing and 20 high-Δsv-MALAT1-expressing breast tumours. We assessed expression level of 48 genes involved in various cellular and molecular phenomena associated with carcinogenesis. These genes encode proteins involved in cell cycle control (n=7), cell migration (n=5), polycomb repressive complexes (PRC1) (n=3) and PRC2 (n=5), EMT (n=7), apoptosis (n=6) and DNA repair (Yang et al, 2011). We also focused on expression of well-known regulators (DGCR8; AGO2) and interactors (SRSF1, SRSF2, SRSF3, UHMK1) of MALAT1 (Gutschner et al, 2013a), as well as transcriptional dysregulated genes after MALAT1 depletion in A549 lung adenoma cell line (ROBO1, MCAM; Gutschner et al, 2013b) and HeLa cells (IFI44; Miyagawa et al, 2012).
Expression of 19 (39.5%) of these 48 genes was significantly positively associated with Δsv-MALAT1 expression (Table 2). Genes significantly associated to Δsv-MALAT1 were mainly involved in cell migration (RHOB, PLAU/UPA and MMP14), Polycomb repressive complex PRC2 (EED, SUZ12, JARID2, TUG1), apoptosis (BIRC6), DNA repair (ATM, MSH2, XRCC1) and regulators (DGCR8) and interactors (SRSF1, SRSF3, UHMK1) of MALAT1. Genes involved in cell cycle control and EMT, as well as putative MALAT1-inducible genes (identified by MALAT1 depletion in cell lines) were not linked to the MALAT1 in breast cancer.
Table 2. Relationship between Δsv-MALAT1-mRNA and target-gene expression.
| Genes | Normal breast tissues (n=10) | Breast tumours with low level of Δsv-MALA(n=20) | Breast tumours with high level of Δsv-MALAT1(n=20) | P-valuea | ROC–AUC |
|---|---|---|---|---|---|
|
Cell cycle control (n=7) | |||||
| MKI67 | 1.0 (0.00–4.74)b | 12.11 (1.88–25.62)b | 13.01 (6.15–81.52)b | 0.42 (NS) | 0.575 |
| AURKA | 1.0 (0.17–2.73) | 8.07 (2.79–27.11) | 7.0 (4.28–123.17) | 0.45 (NS) | 0.570 |
| FOXM1 | 1.0 (0.00–12.25) | 10.91 (2.87–30.02) | 13.74 (4.1–101.38) | 0.40 (NS) | 0.577 |
| PCNA | 1.0 (0.46–3.22) | 2.72 (0.65–8.31) | 3.59 (0.45–6.88) | 0.24 (NS) | 0.626 |
| CCNE1 | 1.0 (0.00–6.82) | 3.34 (0.90–26.50) | 3.18 (1.14–52.07) | 0.96 (NS) | 0.505 |
| E2F1 | 1.0 (0.44–3.08) | 4.50 (1.00–8.60) | 5.05 (1.76–51.10) | 0.73 (NS) | 0.532 |
| AURKB | 1.0 (0.00–16.86) | 23.59 (2.90–37.43) | 15.38 (6.77–160.48) | 0.75 (NS) | 0.47 |
|
Cell migration (n=5) | |||||
| RHOB | 1.0 (0.48–5.75) | 0.88 (0.19–1.36) | 1.13 (0.36–65.33) | 0.015 | 0.724 |
| PLAU | 1.0 (0.42–2.29) | 1.76 (0.31–12.59) | 2.86 (0.84–33.28) | 0.017 | 0.72 |
| MMP14 | 1.0 (0.69–1.52) | 1.31 (0.05–6.21) | 2.52 (0.30–39.17) | 0.0087 | 0.743 |
| RHOA | 1.0 (0.42–3.34) | 1.36 (0.22–3.85) | 1.59 (0.22–6.39) | 0.38 (NS) | 0.581 |
| MMP13 | 1.0 (0.00–4.69) | 45.38 (1.33–688.61) | 38.70 (2.62–250.99) | 0.83 (NS) | 0.48 |
|
Polycomb repressive complex 1 (n=3) | |||||
| CBX7 | 1.0 (0.39–1.62) | 0.36 (0.05–0.95) | 0.43 (0.15–2.43) | 0.045 | 0.685 |
| CBX4 | 1.0 (0.41–3.80) | 1.46 (0.79–2.98) | 1.55 (0.00–3.10) | 0.91 (NS) | 0.501 |
| BMI1 | 1.0 (0.59–1.47) | 1.42 (0.17–4.91) | 1.98 (0.66–6.43) | 0.014 | 0.728 |
|
Polycomb repressive complex 2 (n=5) | |||||
| SUZ12 | 1.0 (0.56–1.25) | 1.19 (0.53–1.93) | 1.63 (0.92–2.99) | 0.00084 | 0.793 |
| JARID2 | 1.0 (0.83–1.46) | 1.19 (0.71–1.83) | 1.89 (0.51–4.13) | 0.0015 | 0.794 |
| EED | 1.0 (0.58–1.84) | 1.02 (0.18–2.61) | 1.36 (0.19–3.13) | 0.0036 | 0.769 |
| TUG1 | 1.0 (0.79–3.06) | 0.73 (0.35–2.01) | 1.16 (0.34–1.98) | 0.044 | 0.686 |
| EZH2 | 1.0 (0.48–2.32) | 4.58 (1.36–12.70) | 5.99 (2.76–14.64) | 0.15 (NS) | 0.632 |
|
EMT (n=7) | |||||
| CDH1 | 1.0 (0.33–1.79) | 0.91 (0.06–2.74) | 1.28 (0.23–5.55) | 0.051 (NS) | 0.68 |
| VIM | 1.0 (0.42–3.11) | 0.22 (0.09–0.70) | 0.31 (0.10–2.16) | 0.22 (NS) | 0.614 |
| ZEB2 | 1.0 (0.44–5.58) | 0.30 (0.12–0.64) | 0.32 (0.14–2.35) | 0.60 (NS) | 0.549 |
| ZEB1 | 1.0 (0.43–4.08) | 0.31 (0.19–1.47) | 0.44 (0.16–1.09) | 0.34 (NS) | 0.587 |
| SNAI2 | 1.0 (0.43–1.50) | 0.23 (0.05–0.82) | 0.24 (0.11–1.41) | 0.52 (NS) | 0.56 |
| TWIST1 | 1.0 (0.53–3.62) | 0.30 (0.08–1.16) | 0.32 (0.07–3.83) | 0.63 (NS) | 0.545 |
| SNAI1 | 1.0 (0.34–7.88) | 0.83 (0.06–2.66) | 0.89 (0.20–6.03) | 0.80 (NS) | 0.524 |
|
Apoptosis (n=6) | |||||
| BIRC6 | 1.0 (0.74–3.10) | 0.85 (0.47–1.23) | 1.13 (0.52–1.75) | 0.00017 | 0.841 |
| BAX | 1.0 (0.41–8.88) | 1.38 (0.62–3.68) | 1.88 (0.80–8.26) | 0.02 | 0.715 |
| BIRC2 | 1.0 (0.73–3.61) | 0.62 (0.30–1.18) | 0.82 (0.26–1.99) | 0.044 | 0.686 |
| BIRC4c | 0.0 (0.00–1.16) | 1.09 (0.00–6.88) | 2.16 (0.00–8.59) | 0.12 (NS) | 0.645 |
| BCL2L1 | 1.0 (0.48–3.76) | 1.19 (0.65–2.19) | 1.47 (0.52–4.69) | 0.22 (NS) | 0.612 |
| BCL2 | 1.0 (0.34–6.13) | 0.85 (0.08–3.75) | 0.78 (0.24–2.43) | 0.82 (NS) | 0.521 |
|
DNA repair (n=6) | |||||
| ATM | 1.0 (0.67–1.66) | 0.60 (0.40–1.87) | 1.36 (0.55–2.24) | 0.00007 | 0.868 |
| MSH2 | 1.0 (0.66–1.55) | 0.98 (0.60–1.57) | 1.40 (0.83–2.95) | 0.0003 | 0.834 |
| BRCA1 | 1.0 (0.00–2.56) | 2.23 (0.05–13.56) | 3.33 (0.00–8.85) | 0.11 (NS) | 0.649 |
| BRCA2 | 1.0 (0.27–2.49) | 3.12 (0.86–7.82) | 3.87 (0.00–9.57) | 0.19 (NS) | 0.631 |
| RAD51 | 1.0 (0.0–2.00) | 7.55 (1.62–18.9) | 5.29 (1.35–21.6) | 0.37 (NS) | 0.417 |
| XRCC1 | 1.0 (0.70–1.30) | 0.78 (0.53–1.34) | 1.41 (0.78–2.84) | 0.00001 | 0.908 |
|
Regulators of MALAT1 (n=6) | |||||
| DGCR8 | 1.0 (0.70–2.59) | 0.80 (0.40–1.71) | 1.12 (0.80–5.05) | 0.000083 | 0.864 |
| AGO2 | 1.0 (0.57–4.50) | 0.8 (0.36–2.64) | 0.89 (0.33–5.87) | 0.26 (NS) | 0.604 |
| SFRS1 | 1.0 (0.66–4.32) | 1.13 (0.87–1.68) | 1.51(0.98–2.80) | 0.00097 | 0.805 |
| SFRS3 | 1.0 (0.65–3.93) | 1.06 (0.69–1.87) | 1.30 (0.78–3.05) | 0.036 | 0.694 |
| SFRS2 | 1.0 (0.60–2.76) | 1.52 (0.88–2.24) | 1.89 (0.81–4.04) | 0.18 (NS) | 0.624 |
| UHMK1 | 1.0 (0.59–5.09) | 1.48 (0.93–2.74) | 2.04 (0.95–3.88) | 0.0094 | 0.74 |
|
MALAT1-inducible genes (n=3) | |||||
| ROBO1 | 1.0 (0.54–1.54) | 0.36 (0.13–4.14) | 0.52 (0.14–1.33) | 0.14 (NS) | 0.635 |
| MCAM | 1.0 (0.57–33.8) | 0.52 (0.22–0.81) | 0.61 (0.22–10.9) | 0.21 (NS) | 0.616 |
| IFI44 | 1.0 (0.34–9.38) | 1.92 (0.06–29.23) | 2.63 (0.11–30.15) | 0.43 (NS) | 0.572 |
Abbreviations: AUC=area under the curve; NS=not significant; ROC=receiver operating curves. Values in bold correspond to significant P-values (P<0.05).
Kruskal–Wallis H-test.
Median (range) of gene mRNA levels ; the mRNA values of the samples were normalised such that the median of the 10 normal breast tissues mRNA values was 1.
The mRNA values of the samples were normalised such that a Ct value of 35 was 1.
Relationship between levels of Δsv-MALAT1 mRNA and RTK/MAPK/PI3K proteins
As several studies recently suggested that MALAT1 promotes proliferation and metastasis of various cancers by activating the RTK/MAPK/PI3K pathways (Wu et al, 2014; Dong et al, 2015; Xu et al, 2015), we tested possible correlation between Δsv-MALAT1 and various proteins involved in these signalling pathways.
Twenty-eight protein (non-phosporylated or/and phosphorylated) levels were analysed using RPPA assays in 143 samples from our series of 446 breast tumours. These selected proteins are involved in TKR (n=9), MAPK (n=4) and PI3K/AKT (n=15) pathways (Table 3). Low-Δsv-MALAT1 mRNA level were associated to high levels of 4 among the 15 proteins involved in the PI3K/AKT pathway (i.e., FOXO1, p70 S6 Kinase total protein and phosphorylated in Threonine 389, S6 ribosomal protein phosphorylated in Ser240/Ser244), but to none of the two others signalling pathways. Low-FL-MALAT1 mRNA level was exclusively associated to high level of FOXO1 (Table 3).
Table 3. Relationship between levels of Δsv-MALAT1 and FL-MALAT1 and a panel of RTK/MAPK/PI3K proteins in a panel of 143 breast tumours.
| Δsv-MALAT1 | FL-MALAT1 | |||
|---|---|---|---|---|
| ra | P-valuea | ra | P-valuea | |
|
RTK proteins (n=9) | ||||
| EGFR | 0.068 | NS | 0.033 | NS |
| p.EGFR.Thr669 | 0.008 | NS | −0.047 | NS |
| p.EGFR.Tyr1173 | 0.036 | NS | 0.052 | NS |
| Her2/ErbB2 | 0.091 | NS | −0.032 | NS |
| Her3/ErbB3 | −0.128 | NS | 0.000 | NS |
| p.Her3/Erbb3.Tyr1289 | −0.060 | NS | −0.020 | NS |
| p.Her4.Tyr1162 | 0.108 | NS | 0.018 | NS |
| Met | −0.025 | NS | −0.029 | NS |
| p.Met.Tyr1234.1235 | −0.064 | NS | −0.026 | NS |
|
MAPK pathway proteins (n=4) | ||||
| MEK1/2 | −0.029 | NS | −0.073 | NS |
| p.MEK1/2.Ser217/221 | 0.113 | NS | −0.047 | NS |
| p44.42.MAPK | 0.116 | NS | 0.150 | NS |
| p.p44.42.MAPK.Thr202.Tyr204 | 0.127 | NS | 0.030 | NS |
|
PI3K/AKT pathway proteins (n=15) | ||||
| PTEN | 0.101 | NS | 0.050 | NS |
| p.PTEN.Ser380.Thr382.383 | −0.018 | NS | −0.026 | NS |
| INPP4b | 0.001 | NS | 0.042 | NS |
| Akt1 | 0.030 | NS | −0.017 | NS |
| p.Akt1.Ser473 | 0.119 | NS | −0.032 | NS |
| Akt2 | −0.102 | NS | 0.028 | NS |
| mTor | −0.155 | NS | −0.066 | NS |
| p.mTor.Ser2448 | −0.077 | NS | 0.008 | NS |
| FOXO1 | −0.194 | 0.0210 | −0.196 | 0.0180 |
| TSC2 | −0.075 | NS | 0.003 | NS |
| p70.S6.Kinase | −0.172 | 0.0400 | 0.099 | NS |
| p.p70.S6.Kinase.Thr389 | −0.225 | 0.0070 | 0.073 | NS |
| S6.Ribosomal.protein | −0.034 | NS | 0.075 | NS |
| p.S6.Ribosomal.protein.Ser235.236 | −0.095 | NS | 0.006 | NS |
| p.S6.Ribosomal.protein.Ser240.244 | −0.201 | 0.0160 | −0.024 | NS |
Abbreviations: Akt=protein kinase B, also known as Akt; EGFR=epidermal growth factor receptor; MAPK=mitogen-activated protein kinase; mTor=mechanistic target of rapamycin; NS=not significant; PI3K=phosphoinositide 3-kinase; PTEN=phosphatase and tensin homologue.
Spearman's rank correlation test.
Discussion
Recent studies have demonstrated the importance of non-protein-coding part of human genome in carcinogenesis. Among numerous kinds of non-protein-coding RNAs, lncRNAs have a key regulatory role in cancer biology. LncRNAs are dysregulated in different types of cancer and the expression levels of certain lncRNAs are associated with metastasis and prognosis of cancer. Overexpression of certain lncRNAs, behaving like oncogenes, can promote tumour growth and cancer cell invasion (Cheetham et al, 2013).
In this study, we focused on the lncRNA MALAT1 that has been shown dysregulated in various cancer types (Zhang et al, 2015), but poorly studied in breast cancer. One study, using deep-sequencing technology, identified MALAT1 as one of the highly expressed lncRNAs in breast tumours (Guffanti et al, 2009).
We tested 10 normal breast tissue RNAs and 446 unilateral invasive primary breast tumour RNAs, using qRT–PCR method. MALAT1 mRNA was detected in all breast tumour samples and also in all normal breast tissues.
Overexpression of MALAT1 mRNA was detected in 14% (63/446) of breast tumours, confirming the oncogenic role of MALAT1. Indeed, MALAT1 is overexpressed in several cancer types, including lung, colon and hepatocarcinoma, and overexpression of MALAT1 in various cell lines enhanced cell proliferation, whereas in nude mice, increased levels of MALAT1 promoted tumour formation (Ji et al, 2003; Guo et al, 2010; Gupta et al, 2010; Gibb et al, 2011; Schmidt et al, 2011; Lai et al, 2012). Additional studies have also demonstrated that depletion of MALAT1 impaired proliferative and invasiveproperties of cancer cells (Guo et al, 2010, Schmidt et al, 2011, Gutschner et al, 2013b).
By using ISH, we showed that MALAT1 transcripts were predominantly localised in nuclear speckles. Nucleus of the MALAT1-overexpressed tumour epithelial cells showed marked diffuse nuclear signals and numerous huge nuclear speckles.
No significant links were observed between MALAT1 mRNA overexpression and markers of aggressiveness, including histopathological grade, lymph node status and macroscopic tumour size, suggesting that overexpression of MALAT1 does not have a major role in aggressiveness of breast carcinomas. Moreover, we observed a link between MALAT1 mRNA overexpression and HR-positive tumours (a marker of good prognostic), suggesting that MALAT1 could be an ER-induced gene in breast cancer. Finally, survival analysis did not reveal that patients with MALAT1-overexpressed tumour had shorter MFS.
Alternative mRNA splicing is a common mechanism for regulating gene expression in higher eukaryotes, and there are many examples of development-specific, tissue-specific and tumour-specific differences in splicing events. In the GENCODE v7 catalogue of human lncRNAs, >25% of lncRNA genes show evidence of alternative splicing with at least two different transcript isoforms per gene locus (Derrien et al, 2012). The vast majority of alternatively spliced lncRNA introns are flanked by canonical splice sites (GT/AG), with no differences in splicing signal compared with the protein-coding genes (Derrien et al, 2012). In the present study, by screening the dbEST database with the FL-MALAT1 cDNA (named FL-MALAT1), we identified a major alternatively spliced MALAT1 transcript (named Δsv-MALAT1) with two concomitant deleted regions of 119 bp and 243 bp. As expected, these alternatively spliced sequences showed consensus sequences of donor/acceptor splice sites. In our cohort, Δsv-MALAT1 showed a very different expression pattern, as compared with FL-MALAT1. Indeed, Δsv-MALAT1 expression varied widely in tumour tissues, being both underexpressed (18.8%) and overexpressed (5.4%). Surprisingly, a significant link was observed between Δsv-MALAT1 underexpression and tumours with large macroscopic size, negative for HRs and expressing high MKI67 mRNA levels, suggesting that underexpression of Δsv-MALAT1 has a role in aggressiveness of breast tumours. In this regard, in contrast to the FL-MALAT1 expression, survival analysis revealed that patients with low-Δsv-MALAT1-expressed tumours had shorter MFS. Moreover, multivariate analysis showed that Δsv-MALAT1 expression status was an independent prognostic marker for MFS. This alternatively spliced MALAT1 transcript isoform could act as decoys, sequestering biomolecules that fixed on the FL-MALAT1 transcript and thus dysregulating its function. Taken together, these results suggest that this alternatively spliced Δsv-MALAT1 transcript isoform has a significant contribution to overall MALAT1 function and breast carcinogenesis.
Further studies are necessary to elucidate the genetic (or epigenetic) mechanisms responsible for the observed underexpression of Δsv-MALAT1, in breast cancer. It is unlikely that gene amplification is one of the mechanisms for MALAT1 overexpression because MALAT1 is located in a chromosomal region (11q13.1) non-recurrently amplified in breast cancer (Curtis et al, 2012; Zack et al, 2013). Mutations in the MALAT1 gene, recently discovered in human cancers, are rare in breast (1.1%) as compared with other cancer types such as bladder cancer (15.3%) (Kandoth et al, 2013). MALAT1 epigenetic dysregulation mediated by CpG island methylation is not reported till date. In the present study, post-transcriptional regulation of MALAT1 by Hsa-miR-125b as described in bladder cancer (Han et al, 2013) was not observed in our breast tumour series. Conversely, our data suggested a transcriptional regulation of the Δsv-MALAT1 (but not of the FL-MALAT1) by the transcriptional co-activator YAP, as described in liver cancer (Wang et al, 2014). Further studies are necessary, using functional assays, to confirm this association. In the downstream MALAT1 pathway, our results suggested that Δsv-MALAT1 (but not FL-MALAT1) could activate the PI3K/AKT pathway, in partial agreement with previous data (Wu et al, 2014; Dong et al, 2015; Xu et al, 2015).
We also assessed the expression levels of gene panel putatively involved in various cellular and molecular phenomena associated with carcinogenesis via dysregulation of Δsv-MALAT1. These genes encode proteins involved in cell cycle control, cell migration, polycomb repressive complexes (PRC1 and 2), EMT, apoptosis and DNA repair, as well as regulators and interactors of Δsv-MALAT1, or known MALAT1-induced genes (Miyagawa et al, 2012; Gutschner et al, 2013b). We identified a strong positive link between Δsv-MALAT1 overexpression and DGCR8 expression, suggesting that Drosha-DGCR8 complex (Microprocessor) controlled the abundance of Δsv-MALAT1 in breast cancer as in HEK 293T cells (Macias et al, 2012). No such link was observed with Ago2, a second putative major regulator of MALAT1 (Weinmann et al, 2009). We observed a positive link between Δsv-MALAT1 overexpression and several interactors of MALAT1, in particular SFRS1 and SFRS3, confirming the involvement of MALAT1 in the regulation of alternative splicing of pre-mRNA in nuclear speckle domains (Tripathi et al, 2010). We also identified a link between Δsv-MALAT1 overexpression and several genes involved in DNA repair (ATM, MSH2, XRCC1). Only one study has recently suggested that MALAT1 depletion could dysregulate ATM-CHK2 pathway in oesophageal squamous cell carcinoma (Hu et al, 2015). More interesting, we observed a link between Δsv-MALAT1 expression and the major members (except EZH2) of Polycomb repressive complex PRC2, including SUZ12, EED and JARID2, but no (or little) link with the members of the Polycomb repressive complex PRC1 (i.e., CBX4, CBX7 and BMI1), as well as the genes regulated by this complex: PCNA and CCNE1 (Yang et al, 2011). In this regard, Yang et al (2011) showed a major role for MALAT1 in the relocation of transcription units by the PRC2 complex in the three-dimensional space of the nucleus, to coordinate the gene-expression programs.
In conclusion, this study suggests that the lncRNA MALAT1 (as the well-known lncRNA HOTAIR) is involved in breast cancer. These data revealed a complex expression pattern of various MALAT1 transcript variants, and suggest that this pattern of expression should be taken into account when evaluating antitumoural drugs designed to target this lncRNA. Further studies are also necessary to elucidate roles of these different MALAT1 transcript variants in breast tumourigenesis and their genetic (or epigenetic) dysregulation molecular mechanisms in this cancer.
Acknowledgments
We thank the staff of Institut Curie–Hospital René Huguenin for their assistance in specimen collection and patient care. We thank Aurélie Barbet, Floriane Bard and Caroline Lecerf for performing RPPA experiments. The RPPA platform is supported by Cancéropôle Ile-de-France. This work was supported by the Comité départemental des Hauts-de-Seine de la Ligue Nationale Contre le Cancer, the Association pour la recherche en cancérologie de Saint-Cloud (ARCS), and by Grant INCa-DGOS-4654.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Bieche I, Onody P, Laurendeau I, Olivi M, Vidaud D, Lidereau R, Vidaud M (1999) Real-time reverse transcription-PCR assay for future management of ERBB2-based clinical applications. Clin Chem 45: 1148–1156. [PubMed] [Google Scholar]
- Bieche I, Parfait B, Laurendeau I, Girault I, Vidaud M, Lidereau R (2001) Quantification of estrogen receptor alpha and beta expression in sporadic breast cancer. Oncogene 20: 8109–8115. [DOI] [PubMed] [Google Scholar]
- Cheetham SW, Gruhl F, Mattick JS, Dinger ME (2013) Long noncoding RNAs and the genetics of cancer. Br J Cancer 108: 2419–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S METABRIC Group Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale AL, Brenton JD, Tavaré S, Caldas C, Aparicio S (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi M, Fletcher JA, Fisher DE (2003) Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci USA 100: 6051–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X (2015) MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol 36: 1477–1486. [DOI] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, Ng S, Lin L, Crowder R, Snider J, Ballman K, Weber J, Chen K, Koboldt DC, Kandoth C, Schierding WS, McMichael JF, Miller CA, Lu C, Harris CC, McLellan MD, Wendl MC, DeSchryver K, Allred DC, Esserman L, Unzeitig G, Margenthaler J, Babiera GV, Marcom PK, Guenther JM, Leitch M, Hunt K, Olson J, Tao Y, Maher CA, Fulton LL, Fulton RS, Harrison M, Oberkfell B, Du F, Demeter R, Vickery TL, Elhammali A, Piwnica-Worms H, McDonald S, Watson M, Dooling DJ, Ota D, Chang LW, Bose R, Ley TJ, Piwnica-Worms D, Stuart JM, Wilson RK, Mardis ER (2012) Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S, Brown CJ, Lam WL (2011) Human cancer long non-coding RNA transcriptomes. PLoS One 6: e25915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti A, Iacono M, Pelucchi P, Kim N, Soldà G, Croft LJ, Taft RJ, Rizzi E, Askarian-Amiri M, Bonnal RJ, Callari M, Mignone F, Pesole G, Bertalot G, Bernardi LR, Albertini A, Lee C, Mattick JS, Zucchi I, De Bellis G (2009) A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics 10: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Li Y, Liu Y, Wang J, Li Y, Li G (2010) Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin 42: 224–229. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hämmerle M, Diederichs S (2013. a) MALAT1–a paradigm for long noncoding RNA function in cancer. J Mol Med 91: 791–801. [DOI] [PubMed] [Google Scholar]
- Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zörnig M, MacLeod AR, Spector DL, Diederichs S (2013. b) The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 73: 1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Liu Y, Zhang H, Wang T, Diao R, Jiang Z, Gui Y, Cai Z (2013) Hsa-miR-125b suppresses bladder cancer development by down-regulating oncogene SIRT7 and oncogenic long noncoding RNA MALAT1. FEBS Lett 587: 3875–3882. [PubMed] [Google Scholar]
- Hanley JA, McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143: 29–36. [DOI] [PubMed] [Google Scholar]
- Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K (2015) Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res 34: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22: 8031–8041. [DOI] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MD, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L (2013) Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS (2012) Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol 29: 1810–1816. [DOI] [PubMed] [Google Scholar]
- Macias S, Plass M, Stajuda A, Michlewski G, Eyras E, Cáceres JF (2012) DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol 19: 760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews J, Duncavage EJ, Pfeifer JD (2013) Characterization of translocations in mesenchymalhamartoma and undifferentiated embryonal sarcoma of the liver. Exp Mol Pathol 95: 319–324. [DOI] [PubMed] [Google Scholar]
- Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T, Akimitsu N (2012) Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 18: 738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C (2011) The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 6: 1984–1992. [DOI] [PubMed] [Google Scholar]
- Spizzo R, Almeida MI, Colombatti A, Calin GA (2012) Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 31: 4577–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV (2010) The nuclearretained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncale S, Barbet A, Coulibaly L, Henry E, He B, Barillot E, Dubois T, Hupé P, de Koning L (2012) NormaCurve: a SuperCurve-based method that simultaneously quantifies and normalizes reverse phase protein array data. PLoS One 7: e38686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang H, Zhang Y, Zhen N, Zhang L, Qiao Y, Weng W, Liu X, Ma L, Xiao W, Yu W, Chu Q, Pan Q, Sun F (2014) Mutual inhibition between YAP and SRSF1 maintains long non-coding RNA, Malat1-induced tumourigenesis in liver cancer. Cell Signal 26: 1048–1059. [DOI] [PubMed] [Google Scholar]
- Weinmann L, Höck J, Ivacevic T, Ohrt T, Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G (2009) Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136: 496–507. [DOI] [PubMed] [Google Scholar]
- Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L, Cao Y, Bao RF, Mu JS, Tan ZJ, Tao F, Liu YB (2014) MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther 15: 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Sui S, Zhang J, Bai N, Shi Q, Zhang G, Gao S, You Z, Zhan C, Liu F, Pang D (2015) Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int J Clin Exp Pathol 8: 4881–4891. [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, LiuW, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG (2011) ncRNA- and Pc2 methylationdependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–78838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH, Sougnez C, Gabriel SB, Hernandez B, Shen H, Laird PW, Getz G, Meyerson M, Beroukhim R (2013) Pan-cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang B, Wang T, Wang H (2015) LncRNA MALAT1 overexpression is an unfavorable prognostic factor in human cancer: evidence from a meta-analysis. Int J Clin Exp Med 8: 5499–5505. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.