Abstract
Background:
The mechanisms of recurrence have been under-studied in rare histologies of invasive epithelial ovarian cancer (EOC) (endometrioid, clear cell, mucinous, and low-grade serous). We hypothesised the existence of an expression signature predictive of outcome in the rarer histologies.
Methods:
In split discovery and validation analysis of 131 Mayo Clinic EOC cases, we used clustering to determine clinically relevant transcriptome classes using microarray gene expression measurements. The signature was validated in 967 EOC tumours (91 rare histological subtypes) with recurrence information.
Results:
We found two validated transcriptome classes associated with progression-free survival (PFS) in the Mayo Clinic EOC cases (P=8.24 × 10−3). This signature was further validated in the public expression data sets involving the rare EOC histologies, where these two classes were also predictive of PFS (P=1.43 × 10−3). In contrast, the signatures were not predictive of PFS in the high-grade serous EOC cases. Moreover, genes upregulated in Class-1 (with better outcome) were showed enrichment in steroid hormone biosynthesis (false discovery rate, FDR=0.005%) and WNT signalling pathway (FDR=1.46%); genes upregulated in Class-2 were enriched in cell cycle (FDR=0.86%) and toll-like receptor pathways (FDR=2.37%).
Conclusions:
These findings provide important biological insights into the rarer EOC histologies that may aid in the development of targeted treatment options for the rarer histologies.
Keywords: epithelial ovarian cancer, prognostic signature, endometrioid carcinomas, clear cell carcinomas, mucinous carcinomas, low-grade serous carcinomas
With an estimated 21 290 new cases in 2015 (Siegel et al, 2015), epithelial ovarian cancer (EOC) is a heterogeneous disease from morphological and molecular perspectives (Kobel et al, 2008; Cancer Genome Atlas Research Network, 2011). As the most common histologic type representing ∼70% of EOC cases, high-grade serous carcinoma (HGSC) has been extensively studied in terms of its molecular profile and genomic landscape in many prognostic studies. For other less common histological types (endometrioid carcinoma (EC), clear cell carcinoma (CCC), mucinous carcinoma (MC), and low-grade serous carcinoma (LGSC)), prognostic studies to understand disease mechanism and find corresponding treatment options are lacking.
Among previous studies investigating rare histological types of EOC, very few focused on molecular associations with patients' prognosis, partially due to relatively small number of cases. In fact, only one published Australian expression array study incorporated LGSC and EC tumours to assess outcome (Tothill et al, 2008), but did not include CCC and MC. Differences between HGSC and other histologies has also been observed at genomic and epigenomic levels (Huang et al, 2012; Cicek et al, 2013), and several inherited susceptibility regions of EOC specific to HGSC (Pharoah et al, 2013; Shen et al, 2013). Compared with HGSC studies with several validated outcome-associated signatures (Cancer Genome Atlas Research Network, 2011; Verhaak et al, 2013; Riester et al, 2014), there is a strong need to better understand the transcriptome of rare histological types from a disease outcome perspective.
In this study, we characterised associations between expression profiling of 131 EC, CCC, MC, and LGSC EOC cases and progression-free survival (PFS). We hypothesised the existence of expression signature predictive of PFS that is shared across EOC of rare histological types, motivated by several studies showing that different cancer types could share similar transcriptome features with therapeutic potential (Cancer Genome Atlas Research Network, 2012; Martinez et al, 2014). We performed a semi-supervised gene expression clustering analysis and revealed two underlying transcriptome classes in the rare EOC histological types, which were associated with differential outcome in both univariate and multivariate analysis. In addition, we conducted validation analyses in public data sets that contained expression measurements on the rare histological types, confirming existence of the two discovered transcriptome classes and significant PFS association. Noticeably, resulting classes were consistently predictive of PFS in rare histological tumours but not in HGSC tumours for the Mayo Clinic and public data sets (for example, ‘Class-1' cases had better outcome compared with ‘Class-2' cases). Through pathway enrichment analysis, we also found that ‘Class-1' tumours have more active metabolic activities producing steroid hormones and also more enriched with WNT signalling pathway, while the ‘Class-2' tumours are associated with the upregulation of cell cycle signalling pathway and toll-like receptor (TLR) signalling pathway.
Materials and Methods
Mayo Clinic study participants and expression profiling
Eligible cases (n=131) were women aged 20 years or above who were ascertained between 1992 and 2009 at the Mayo Clinic with pathologically confirmed rarer histological types of invasive EOC (73 EC, 39 CCC, 14 MC, and 5 LGSC). Initial clinical diagnoses were confirmed by a gynaecologic pathologist (GLK), who verified histology and tumour grade and reviewed each tissue to ensure 70% tumour content prior to RNA extraction. Progression and vital status were obtained from the Mayo Clinic Tumor Registry, electronic medical records, and active patient contact. All cases provided informed consent for use of their tissues and medical records in research; all protocols were approved by the Mayo Clinic Institutional Review Board. Additional details on study participants have been described elsewhere (Cicek et al, 2013).
PFS time was defined as time from the date of diagnosis to the date that second-line therapy was initiated for a clinically-actionable tumour recurrence, accounting for date of study entry (left truncation). Clinical characteristics examined as covariates included histology (EC, CCC, MC, LGSC), stage (I, II, III/IV), grade (low, high), surgical debulking status (no macroscopic disease, others), age (<50, 50–59, 60–69, 70–79, 80+), body mass index at diagnosis, pre-surgical CA125, and ascites (yes, no). Cox regression was used to estimate hazard ratios and 95% confidence intervals, including multivariate stepwise variable selection. Univariate analysis of clinical features revealed that histology, grade, stage, and surgical debulking status significantly associated with PFS with P-value <0.05. Following stepwise variable selection for the multivariate analysis, only stage and surgical debulking status remained significantly associated with PFS. Clinical characteristics, along with univariate and multivariate associations with PFS are shown in Table 1.
Table 1. Clinical characteristics and association with PFS for 131 Mayo Clinic EOC patients with endometrioid, clear cell, mucinous, or low-grade serous tumours.
| N (%) or median (range) | Univariate HRa (95%CI) | Univariate P-value | Multivariate HRb (95%CI) | Multivariate P-value | |
|---|---|---|---|---|---|
| Progression status at last follow-up | |||||
| No progression | 94 (71.8%) | NA | NA | ||
| Progression | 37 (28.2%) | ||||
| Length of follow-up, months | 102.7 (0.4, 198.7) | NA | NA | ||
| Age at diagnosis | Per category trend HR | 0.06 | |||
| <50 | 27 (20.6%) | ||||
| 50–59 | 45 (34.6%) | ||||
| 60–69 | 34 (26.0%) | 1.35 (0.99, 1.83) | |||
| 70–79 | 15 (11.5%) | ||||
| 80+ | 10 (7.6%) | ||||
| Age at diagnosis | 58 (21.0, 88.0) | 1.03 (1.00, 1.06) | 0.06 | ||
| Body mass index (kg m−2) | 28.4 (18.1, 53.3) | 1.03 (0.97, 1.09) | 0.36 | ||
| Log10 pre-surgery CA125 | 2.27 (1.06, 4.57) | 0.99 (0.54, 1.82) | 0.98 | ||
| Stage | |||||
| I | 61 (46.6%) | REF | REF | ||
| II | 18 (13.7%) | 3.63 (1.05, 12.59) | 0.0418 | 2.62 (0.72, 9.52) | 0.1424 |
| III/IV | 52 (39.7%) | 9.38 (3.59, 24.52) | <.0001 | 7.50 (2.78, 20.26) | <.0001 |
| Grade | |||||
| Low | 29 (22.1%) | REF | |||
| High | 102 (77.9%) | 2.88 (0.88, 9.40) | 0.0792 | ||
| Debulking | |||||
| Optimal; no macroscopic disease | 94 (71.8%) | REF | REF | ||
| Others | 37 (28.2%) | 3.69 (1.90, 7.18) | 0.0001 | 2.21 (1.10, 4.44) | 0.0255 |
| Ascites | |||||
| Yes | 44 (41.5%) | REF | 0.24 | ||
| No | 62 (58.5%) | 0.62 (0.28, 1.37) | |||
| Unknown | 25 | ||||
| Histology | |||||
| Endometrioid | 73 (55.7%) | REF | 0.3394 | ||
| Clear cell | 39 (29.8%) | 1.61(0.82, 3.17) | |||
| Mucinous | 14 (10.7%) | 0.00(0.00,—) | |||
| Low-grade serous | 5 (3.8%) | 2.98(0.67, 12.93) |
Abbreviations: CI=confidence interval; EOC=epithelial ovarian cancer; HR=hazards ratio; NA=not applicable; PFS=progression-free survival; REF=reference group.
Unadjusted univariate hazard ratios from Cox proportional hazards regression analysis; bold indicates P<0.05; progression status as of August 28, 2015; length of follow-up calculated among 94 patients without progression; body mass index HR represents risk per-unit increase; HRs for pre-surgery CA125 based on log10(pre-surgery CA125).
Multivariate hazard ratios from Cox proportional hazards regression analysis using significant variables selected from univariate analysis and stepwise model selection.
RNA from fresh frozen tumours of each patient was extracted and assessed using Agilent Whole Human Genome 4 × 44 K Expression Arrays as previously described (Goode et al, 2013; Konecny et al, 2014). Batch effects were corrected to adjust Cy5, Cy3 labelling difference observed among experimental batches, using ‘ComBat', an empirical Bayesian approach (Johnson et al, 2007). TCGA-based HGSC transcriptome subtypes were assigned to each tumour as described previously (Konecny et al, 2014).
Semi-supervised expression clustering
For Mayo Clinic internal discovery set (n=66), a semi-supervised clustering technique implemented in R ‘Superpc' package (Bair and Tibshirani, 2004) was applied to normalised log-ratio expression (Supplementary Figure 1A). The ‘semi-supervised' aspect of the analysis determined a reduced set of features (gene probe sets), expression levels of which were associated with PFS. Using the internal discovery set, Cox models were fit to each of the features separately to examine the association between expression level and PFS. The features were then ranked based on their strength of association with PFS, and the top expression probes were selected for subsequent clustering analysis using principal component analysis. The optimal number of expression probes was selected to be 960 using a 10-fold cross-validation procedure.
As implemented in the R ‘Superpc' package, clustering was done by projecting a probe-by-sample expression data matrix of the selected probes in the first principal component direction, using singular value decomposition. To achieve a discrete group assignment, the median of the first principal component projection was used as a cutoff (Bair and Tibshirani, 2004). On the basis of the predictive projection generated using the discovery set, we further predicted transcriptome class memberships using centroid-based similarity score, as described in following section.
Expression centroid-based class similarity score
Based on derived class-predictive probes from semi-supervised clustering, we summarised expression centroid for Class-1 and Class-2 as the averaged expression vector for assigned samples in Mayo Clinic discovery set. Similar to breast cancer studies using an expression centroid to determine transcriptome class (Tibshirani et al, 2002; Parker et al, 2009), we defined a class similarity score as the Pearson's correlation coefficient between expression centroid and corresponding signature gene expression of one test sample. For simplicity, we defined a differential correlation score for each tumour sample as DiffCorr1vs2=Pearson's Correlation(sample, centroid1)–Pearson's Correlation(sample, centroid2). Therefore, a tumour sample with DiffCorr1vs2>0 will be assigned as ‘Class-1' membership, and <0 to ‘Class-2'.
For Mayo Clinic validation samples (N=65) having the same Agilent 4 × 44 k platform with discovery set, probe-level expressions were used to compute DiffCorr1vs2 and predict class memberships, according to probe-level centroids (Supplementary Table 1). For public data sets with expression measurements from different microarray platforms, gene-level expressions were used, with the overlapped genes between given sample and gene-level centroids (Supplementary Table 2).
Other validation data sets
Several validation sets were also used (Bonome et al, 2008; Tothill et al, 2008; Crijns et al, 2009; Denkert et al, 2009; Mok et al, 2009; Cancer Genome Atlas Research Network, 2011; Mateescu et al, 2011; Bentink et al, 2012; Ferriss et al, 2012; Pils et al, 2012; Yoshihara et al, 2012; Karlan et al, 2014). One Mayo Clinic HGSC data set consisted of additional 372 HGSC cases, in which 174 cases from a previous study for EOC patients in the Mayo Clinic (Konecny et al, 2014). Fifteen public expression data sets were retrieved from a curated ovarian cancer transcriptome database (Ganzfried et al, 2013), with organised clinical information, such as survival/progression time, histology, grade, stage, and debulking status. From the database, two clinical annotations ‘summarygrade' (low-grade/high-grade) and ‘histological_type' were used to determine expression samples of rare histological types. With a note, we used ‘grade' 1 and ‘histological_type' to determine LGSC samples according to previous studies (Ayhan et al, 2009; Vang et al, 2009). Expression samples of tumours with undetermined or other histological type (e.g., borderline) were excluded. Where multiple probe sets mapped a gene, the probe set with the highest mean across all data sets of the sample platform was utilised (Miller et al, 2011).
After using ‘Combat' analysis to eliminate per-study batch effects across 15 data sets with at least 50 eligible samples, total 2460 EOC expression samples were used in the analysis, including 78 EC, 70 LGSC, 27 MC, 24 CCC, and 2261 HGSC. Out of 199 rare histological cases across 9 public data sets, 91 samples had recurrence information, and 57 samples had PFS, stage, and debulking information for univariate and multivariate associations, respectively. Out of 2261 HGSC cases across fifteen public data sets, 967 had recurrence information, and 595 samples had PFS, stage, and debulking information for univariate and multivariate associations, respectively. Information on the 15 studies included in the validation and summary of the clinical covariates are summarised in Supplementary Tables 3 and 4.
Results
Among the 131 Mayo Clinic invasive EOC patients studied, 37 (28.2%) experienced recurrent disease (Table 1). The internal discovery (n=66) and validation (n=65) sets were split randomly with a balance distribution in clinical characteristics (Supplementary Table 5, Supplementary Figure 1A). Two transcriptome classes were derived from the discovery set only (n=66), referred as Class-1 and Class-2. In the discovery set, cases in the Class-1 were associated with longer PFS and fewer recurrence events as compared with the Class-2 (Figure 1A and Table 1). Using a cross-validation procedure, 960 expression probe sets mappable to 705 genes were determined to be used in semi-supervised clustering and predictive of derived tumour classes (Supplementary Tables 1 and 2, Supplementary Figure 1B and C). For classifying unseen samples in internal and public validation sets, we assigned the class membership with more resembled expression centroid (seen in Materials and methods section).
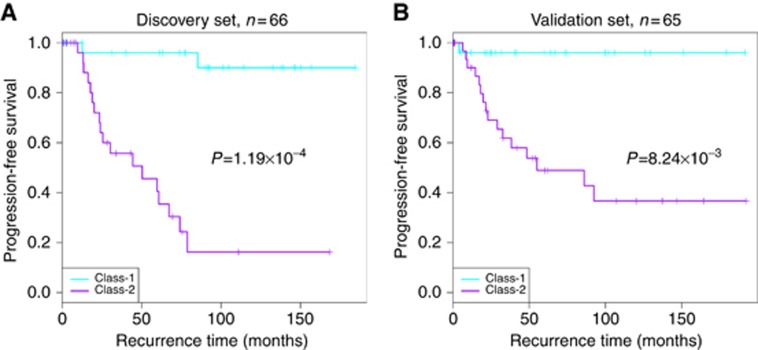
Figure 1.
Rare histological EOC cases with PFS information in Mayo Clinic dataset. (A) For the samples in Mayo Clinic internal discovery set (n=66), Kaplan-Meier plot of progression-free survival demonstrates a clear outcome difference between Class-1 and Class-2, with better outcome in Class-2 group. (B) For the samples in Mayo Clinic internal validation set (n=65), Kaplan–Meier plot of progression-free survival confirms the outcome difference between Class-1 and Class-2. Cyan and purple colours indicate recurrence curves of Class-1 and Class-2 cases, respectively.
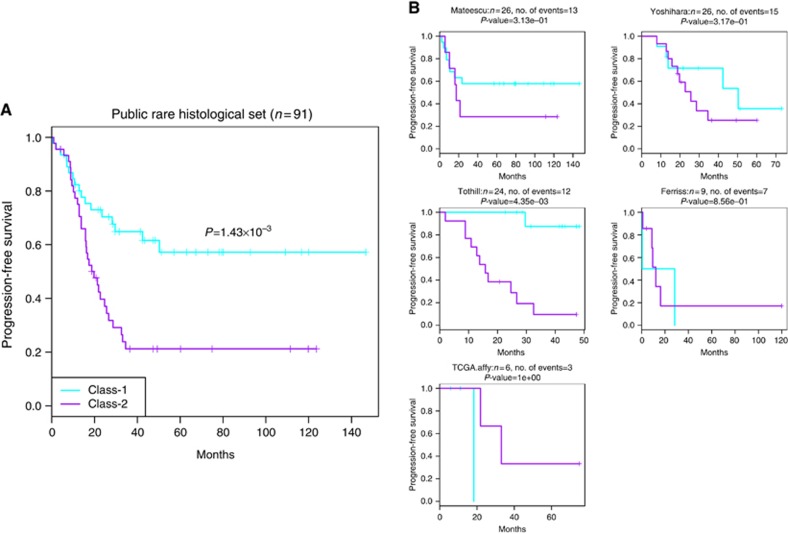
In the Mayo Clinic validation set, Class-1 membership showed an independent contribution to predict better PFS, shown as Figure 1B and Table 2, with univariate analysis P=8.2 × 10−3 and multivariate analysis P=2.8 × 10−2 after adjusting for stage and debulking status. In public validation sets for samples with PFS information (n=91), the Class-1 membership was associated with better prognosis in univariate analysis (P=1.4 × 10−3), shown as Figure 2, and still marginally significant association with multivariate analysis (P=6.8 × 10−2, n=57). Univariate and multivariate analysis details were shown in Table 2. When examining clinical characteristics with predicted classes in internal validation set and public combined validation set, we found several consistent relationships with discovered two classes (Supplementary Table 3): Class-1 tumours were significantly enriched for patients with low-grade, early stage diseases, and had a lower proportion of patients with histology of CCC, and higher proportion of MC.
Table 2. Univariate and multivariate PFS associations in the Mayo Clinic and public data sets of endometrioid, clear cell, mucinous, or low-grade serous EOC patients.
|
The Mayo Clinic data set (n=131) |
Public rarer histological samples (n=91) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Discovery set (n=66) |
Validation set (n=65) |
Public validation set (n=91) |
|||||||
| HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value | |
|
Univariate analysis | |||||||||
| Class-1 vs Class-2 | 0.05 | (0.01–0.24) | 1.2 × 10−4 | 0.07 | (0.01–0.49) | 8.2 × 10−3 | 0.38 | (0.21–0.69) | 1.4 × 10−3 |
| Multivariate analysis |
n=66 |
n=65 |
n=57 |
||||||
| Class-1 vs Class-2 | 0.09 | (0.02–0.42) | 2.4 × 10−3 | 0.08 | (0.009–0.77) | 2.8 × 10−2 | 0.48 | (0.22–1.05) | 6.8 × 10−2 |
| Stage (I vs III and IV) | 0.15 | (0.02–0.95) | 4.4 × 10−2 | 0.35 | (0.093–1.34) | 0.12 | 1.5 × 10−8 | (4.96 × 10−9–4.43 × 10−8) | <2 × 10−16 |
| Stage (II vs III and IV) | 0.35 | (0.11–1.17) | 0.09 | 0.80 | (0.23–2.78) | 0.73 | 1.41 | (0.42–4.74) | 0.58 |
| Debulking statusa | 1.10 | (0.42–2.90) | 0.84 | 3.44 | (1.35–8.76) | 9.4 × 10−3 | 3.29 | (1.76–6.16) | 2.0 × 10−4 |
Abbreviations: CI=confidence interval; EOC=epithelial ovarian cancer; HR=hazards ratio; PFS=progression-free survival.
Mayo: other vs having no macroscopic disease; Public: suboptimal vs optimal.
Figure 2.
For rare histological EOC cases with PFS information across five data sets (n=91). (A) Kaplan–Meier plot of PFS showing the Class-1 and Class-2 outcome association for all the combined samples. (B) Per-study Kaplan–Meier plots. Cyan and purple colours indicate recurrence curves of predicted Class-1 and Class-2 cases, respectively.
In addition, when TCGA expression signatures of four molecular subtypes of HGSC EOC was applied to these samples, Class-1 tumours had higher proportion of patients with the TCGA-defined ‘Differentiated' molecular subtype and lower proportion of ‘Immunoreactive' subtype (Cancer Genome Atlas Research Network, 2011; Verhaak et al, 2013). In order to examine the relationship between expression classes and stromal contamination, we also evaluated association between class membership vs tumour content, which were evaluated by a gynaecologic pathologist (GLK), and found no significant association (P=0.14, Supplementary Table 3).
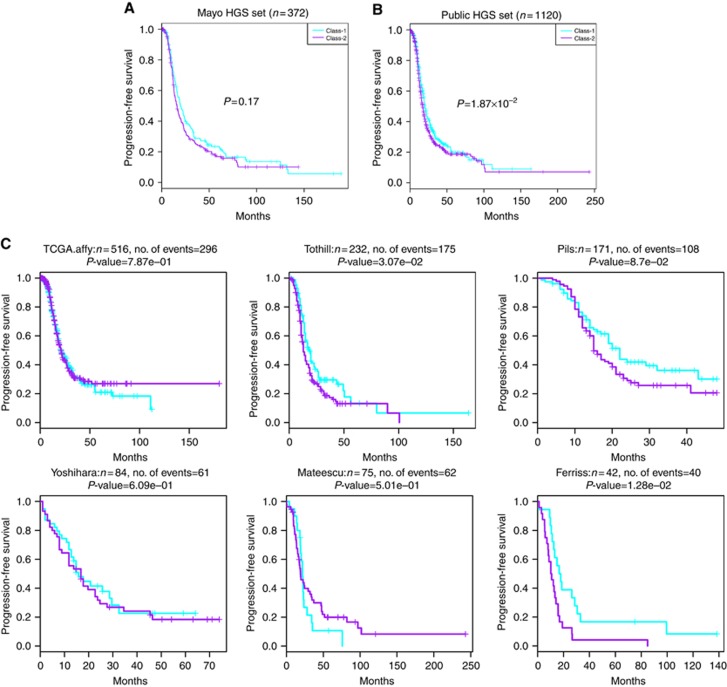
To investigate whether transcriptome classes from rarer histology samples predict PFS in HGSC, we also assigned Class-1/Class-2 memberships in Mayo HGSC data set and across 15 public data sets (Dressman et al, 2007; Wu et al, 2007; Bonome et al, 2008; Tothill et al, 2008; Crijns et al, 2009; Denkert et al, 2009; Mok et al, 2009; Yoshihara et al, 2010; Cancer Genome Atlas Research Network, 2011; Mateescu et al, 2011; Bentink et al, 2012; Ferriss et al, 2012; Pils et al, 2012; Yoshihara et al, 2012; Karlan et al, 2014). In Mayo HGSC data set (n=372), the class membership resulted from the expression signature was not associated with PFS (P=0.17) (Figure 3A). In 15 public data sets involving HGSC cases (n=967), Class-1/-2 membership was significant (P=1.9 × 10−2) in univariate PFS association (Figure 3B and C) but not significant in multivariate association (P=0.15) when adjusting for stage and debulking status (details seen in Table 3).
Figure 3.
Mayo Clinic and public HGS datasets with PFS information. (A) Kaplan–Meier plot of PFS showing the Class-1 and Class-2 outcome association for Mayo Clinic HGSC cases. (B) Kaplan–Meier plot for all the HGSC cases with PFS information in public data sets (n=1120). (C) Per-study Kaplan–Meier plots. Cyan and purple colours indicate recurrence curves of predicted Class-1 and Class-2 cases, respectively.
Table 3. Univariate and multivariate PFS associations in the Mayo Clinic and public data sets of high-grade serous EOC patients.
|
The Mayo Clinic high-grade serous data set (n=372) |
Public high-grade serous data sets (n=967) |
|||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
|
Univariate analysis | ||||||
| Class-1 vs Class-2 | 0.84 | (0.65–1.08) | 0.17 | 0.83 | (0.72–0.97) | 1.9 × 10−2 |
| Multivariate analysis |
n=372 |
n=595 |
||||
| Class-1 vs Class-2 | 0.94 | (0.72–1.23) | 0.66 | 0.89 | (0.75–1.05) | 0.15 |
| Stage (I vs III and IV) | 0.13 | (0.019–0.84) | 3.19 × 10−2 | 0.18 | (0.05–0.56) | 3.5 × 10−3 |
| Stage (II vs III and IV) | 0.46 | (0.22–0.94) | 3.34 × 10−2 | 0.59 | (0.38–0.93) | 2.4 × 10−2 |
| Debulking statusa | 2.06 | (1.59–2.68) | 5.06 × 10−8 | 1.51 | (1.27–1.79) | 3.3 × 10−6 |
Abbreviations: CI=confidence interval; EOC=epithelial ovarian cancer; HR=hazards ratio; PFS=progression-free survival.
Mayo: other vs having no macroscopic disease; Public: suboptimal vs optimal.
To identify pathways differentially enriched between two transcriptome classes in entire Mayo rare histology cohort, we first chose differentially expressed genes based on statistical confidence and substantial fold changes (false discovery rate (FDR) <1% and absolute log2 fold-change>0.5), resulting to 965 and 713 genes upregulated in Class-1 and Class-2, respectively (Supplementary Figure 2 and Supplementary Table 6). Then, we performed KEGG pathway enrichment analysis of these genes using DAVID online annotation tool (http://david.abcc.ncifcrf.gov/) (Huang da et al, 2009a, 2009b). Significantly enriched pathways with FDR <20% are presented in Supplementary Table 7. For upregulated genes in Class-1, the most enriched pathways included ‘hsa00140: Steroid hormone biosynthesis' (FDR=0.005%), which may suggest differential metabolite activities producing hormones between Class-1 and Class-2. Noticeably, Class-2 upregulated genes were highly enriched in cell cycle activities: ‘hsa04110: Cell cycle' (FDR=0.86%), consistent with more rapid recurrence in Class-2 patients. Class-2 upregulated genes were also found enriched in another immune-related ‘hsa04620: Toll-like receptor signalling pathway' (FDR=2.37%). Clinical covariates and predicted membership information for all the public expression samples used in this study (n=2460) are presented in Supplementary Table 8.
Discussion
Historically, EOC has been classified according to patterns of abnormal differentiation and morphology, as serous (fallopian tube-like), endometrioid (endometrium-like), mucinous (endocervical-like), and clear cell (mesonephros-like) (Auersperg et al, 2001), with serous histology samples further classified into low- and high-grade categories (Vang et al, 2009). In addition, some researchers have suggested that EOC histological types could be collapsed as two types: the so-called type-II refers to the HGSC type arising from the fallopian tube, and all the remaining types (i.e., EC, CCC, MC, and LGSC) belonging to type-I (Kurman et al, 2008). Meanwhile, CCC, MC, and EC tumours present with differing clinical characteristics; CCC tumours are usually of advanced stage, and MC tumours are often low grade and diagnosed at an early stage. EC is a very diverse histology type by itself, while lower grade usually associated with better outcome, high-grade EC cases were often reported with TP53 mutations and genome instability, resembling HGSC disease (Prat, 2012).
Despite established histological classification of EOC, huge challenges reside in clinical practices in that very few therapeutic options are available for treating women with EOC, and treatment is not specific to histological type. Therefore, revealing prognostic tumour-based molecular information is critical to understand and potentially find treatment solutions for EOC patients. As the most common EOC histological type, HGSC has received much research attention including by the TCGA (Cancer Genome Atlas Research Network, 2011), which resulted in the discovery of four expression subtypes with different pathways activated, that are prognostic (Konecny et al, 2014), and may lead to different potential therapeutic targets (Liu and Matulonis, 2014; Secord et al, 2014). Inspired by the successful example in HGSC, we investigated the existence of prognostic tumour expression classes in collections of rarer non-HGSC EOC.
In this expression study, we investigated the utility of using transcriptome classes as markers to define EC, MC, CCC, and LGSC EOC tumours with different progression risk. Our semi-supervised clustering analysis on whole-genome gene expression data identified two prognosis classes derived from a discovery set and replicated in a validation set of rare histological EOC tumours seen at the Mayo Clinic. The association with PFS of derived classes was statistically significant even after controlling for the covariates that contribute to PFS in multivariate models (stage and debulking status); as a contrast, histology was not predictive of PFS in multivariate model as shown in Table 1, underscoring the needs to better understand molecular mechanisms. With expression signature derived from Mayo Clinic discovery set, we externally validated the existence of two classes in nine public expression data sets with rare histological samples, and confirmed Class-1 was associated with better PFS in subset of samples with PFS information. Comparing with established clinical factors stage and debulking status, Class-1/Class-2 membership provided additional prognostic value, shown in Table 2. These PFS associations were not suggested in analysis of either Mayo Clinic or public HGSC cohorts after controlling for stage and debulking (Table 3).
According to PFS analysis of Mayo Clinic patients, stage and surgical debulking as established factors affecting recurrence achieve association with high significance (debulking status: univariate P-value=0.0001). This underscores the importance of early detection of EOC. Also, an aggressive surgical effort leading to no remaining macroscopic disease, whenever possible, is critical to reduce the risk of tumour progression. Tumour grade was a factor significantly associated with PFS in univariate analysis but lost its significance in multivariate analysis which can likely be explained by the correlation of grade and stage (65.5% of low-grade rare histological cancers were diagnosed at stage I, Fisher's exact test P=3.9 × 10−2). Similarly, histology alone was predictive of PFS, but did not provide additional prediction towards PFS beyond stage and grade information. In contrast, our rare subtype transcriptome memberships significantly predicted PFS outcome after accounting for stage and grade in Mayo Clinic patients, as well as in the public data sets.
Compared with previous non-HGSC EOC studies (Tothill et al, 2008), our study represents the largest and most comprehensive collection with the greatest number of histological types and external validations. Also, instead of combining HGSC and non-HGSC types (Tothill et al, 2008), we performed semi-supervised clustering only in non-HGSC types, and we investigated resulting class in HGSC and non-HGSC tumours separately. The advantage of this approach is that the predominance of HGSC did not impact clustering. The distinctly different PFS associations for Class-1/Class-2 membership in patients with HGSC and non-HGSC tumours suggest that they should be separately studied in the future.
Through pathway enrichment analysis after differential expression analysis, we also highlighted different biological pathways behind each class of tumours. Class-1 tumours were associated with more active hormone activities, reflected by enrichment of ‘Steroid hormone biosynthesis' pathway and ‘Metabolism of xenobiotics by cytochrome P450'. Progesterone and oestrogen are steroid hormones regulating normal menopause cycle, and have been studied for potential roles in ovarian cancer aetiologies and prognosis (Lukanova and Kaaks, 2005; Sieh et al, 2013). Noticeably, progesterone receptor (PR) was significantly upregulated (Class-1 vs -2 log2 ratio foldchange=2.2, t-test FDR=1.36 × 10−11, Supplementary Table 6). The known protective effects of progesterone may contribute to the less aggressive progression in Class-1 tumours. This observation was also reported by the Ovarian Tumour Tissue Analysis Consortium (Sieh et al, 2013), in which they found an association between high immunohistochemistry based protein expression PR measurements and improved disease-specific survival in EC (log-rank P<0.0001) and HGSC (log-rank P=0.0006). The other enriched pathway in ‘Class-1' upregulated genes is WNT signalling pathway. Although frequent mutations of pathway members were only expected for EC, WNT pathway was found implicated in other ovarian histological types and therefore has been studied for potential target treatments (Arend et al, 2013, 2014).
As a contrast, genes upregulated in Class-2 were associated with noticeably active cell cycle activities with several cell cycle regulator genes, including cyclin E1 (Class-2 vs −1 log2 ratio foldchange=1.0, t-test FDR=3.26 × 10−7), which is a gene found frequently amplified independent of BRCA1/2 mutations, and associated with primary treatment resistance in HGSC (Nakayama et al, 2010; Etemadmoghadam et al, 2013). Another enriched pathway associated with Class-2 is TLR pathway, the signalling pathway of which in tumour cells may result in immunosuppression and thereby furthering tumour growth (Muccioli and Benencia, 2014). With summarised pathway interpretations, we can possibly name ‘Class-1' as ‘hormone-WNT' class and ‘Class-2' as ‘cyclin-TLR' class, call for future studies focusing on class-specific pathway aberrations, and investigate treating rarer histological of ovarian tumours according to discovered classes.
In conclusion, this comprehensive study revealed the existence of two tumour transcriptome classes among EC, MC, CCC, and LGSC EOC and found that transcriptome classes associated with PFS. Results in Mayo Clinic cases were validated in non-HGSC cases of public rarer histological expression samples, but not in either Mayo Clinic HGSC or public HGSC EOC patients, suggesting discovered classes are unique to rarer histological EOC. Pathway enrichment analysis further showed that differentially upregulated genes in Class-1 and Class-2 appeared to be associated with distinct molecular pathways. Future work is needed to validate current findings in even larger non-HGSC EOC collections and to consolidate pathway mechanisms of the revealed transcriptome classes for investigating therapeutic potentials.
Acknowledgments
This work was supported by a Developmental Research Programme Project from the Mayo Clinic Ovarian Cancer SPORE (P50 CA136393), R01 CA122443, Mayo Clinic Cancer Center (P30 CA015083), and the Fred C. and Katherine B. Andersen Foundation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Arend RC, Londono-Joshi AI, Samant RS, Li Y, Conner M, Hidalgo B, Alvarez RD, Landen CN, Straughn JM, Buchsbaum DJ (2014) Inhibition of Wnt/beta-catenin pathway by niclosamide: a therapeutic target for ovarian cancer. Gynecol Oncol 134(1): 112–120. [DOI] [PubMed] [Google Scholar]
- Arend RC, Londono-Joshi AI, Straughn JM Jr, Buchsbaum DJ (2013) The Wnt/beta-catenin pathway in ovarian cancer: a review. Gynecol Oncol 131(3): 772–779. [DOI] [PubMed] [Google Scholar]
- Auersperg N, Wong AS, Choi KC, Kang SK, Leung PC (2001) Ovarian surface epithelium: biology, endocrinology, and pathology. Endocr Rev 22(2): 255–288. [DOI] [PubMed] [Google Scholar]
- Ayhan A, Kurman RJ, Yemelyanova A, Vang R, Logani S, Seidman JD, Shih IeM (2009) Defining the cut point between low-grade and high-grade ovarian serous carcinomas: a clinicopathologic and molecular genetic analysis. Am J Surg Pathol 33(8): 1220–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bair E, Tibshirani R (2004) Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol 2(4): E108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentink S, Haibe-Kains B, Risch T, Fan JB, Hirsch MS, Holton K, Rubio R, April C, Chen J, Wickham-Garcia E, Liu J, Culhane A, Drapkin R, Quackenbush J, Matulonis UA (2012) Angiogenic mRNA and microRNA gene expression signature predicts a novel subtype of serous ovarian cancer. PLoS ONE 7(2): e30269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC, Boyd J, Birrer MJ (2008) A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res 68(13): 5478–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicek MS, Koestler DC, Fridley BL, Kalli KR, Armasu SM, Larson MC, Wang C, Winham SJ, Vierkant RA, Rider DN, Block MS, Klotzle B, Konecny G, Winterhoff BJ, Hamidi H, Shridhar V, Fan JB, Visscher DW, Olson JE, Hartmann LC, Bibikova M, Chien J, Cunningham JM, Goode EL (2013) Epigenome-wide ovarian cancer analysis identifies a methylation profile differentiating clear-cell histology with epigenetic silencing of the HERG K+ channel. Hum Mol Genet 22(15): 3038–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crijns AP, Fehrmann RS, de Jong S, Gerbens F, Meersma GJ, Klip HG, Hollema H, Hofstra RM, te Meerman GJ, de Vries EG, van der Zee AG (2009) Survival-related profile, pathways, and transcription factors in ovarian cancer. PLoS Med 6(2): e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkert C, Budczies J, Darb-Esfahani S, Gyorffy B, Sehouli J, Konsgen D, Zeillinger R, Weichert W, Noske A, Buckendahl AC, Muller BM, Dietel M, Lage H (2009) A prognostic gene expression index in ovarian cancer–validation across different independent data sets. J Pathol 218(2): 273–280. [DOI] [PubMed] [Google Scholar]
- Dressman HK, Berchuck A, Chan G, Zhai J, Bild A, Sayer R, Cragun J, Clarke J, Whitaker RS, Li L, Gray J, Marks J, Ginsburg GS, Potti A, West M, Nevins JR, Lancaster JM (2007) An integrated genomic-based approach to individualized treatment of patients with advanced-stage ovarian cancer. J Clin Oncol 25(5): 517–525. [DOI] [PubMed] [Google Scholar]
- Etemadmoghadam D, Weir BA, Au-Yeung G, Alsop K, Mitchell G, George J Australian Ovarian Cancer Study G Davis S, D'Andrea AD, Simpson K, Hahn WC, Bowtell DD (2013) Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc Natl Acad Sci USA 110(48): 19489–19494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferriss JS, Kim Y, Duska L, Birrer M, Levine DA, Moskaluk C, Theodorescu D, Lee JK (2012) Multi-gene expression predictors of single drug responses to adjuvant chemotherapy in ovarian carcinoma: predicting platinum resistance. PLoS ONE 7(2): e30550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganzfried BF, Riester M, Haibe-Kains B, Risch T, Tyekucheva S, Jazic I, Wang XV, Ahmadifar M, Birrer MJ, Parmigiani G, Huttenhower C, Waldron L (2013) curatedOvarianData: clinically annotated data for the ovarian cancer transcriptome. Database (Oxford) 2013: bat013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode EL, DeRycke M, Kalli KR, Oberg AL, Cunningham JM, Maurer MJ, Fridley BL, Armasu SM, Serie DJ, Ramar P, Goergen K, Vierkant RA, Rider DN, Sicotte H, Wang C, Winterhoff B, Phelan CM, Schildkraut JM, Weber RP, Iversen E, Berchuck A, Sutphen R, Birrer MJ, Hampras S, Preus L, Gayther SA, Ramus SJ, Wentzensen N, Yang HP, Garcia-Closas M, Song H, Tyrer J, Pharoah PP, Konecny G, Sellers TA, Ness RB, Sucheston LE, Odunsi K, Hartmann LC, Moysich KB, Knutson KL (2013) Inherited variants in regulatory T cell genes and outcome of ovarian cancer. PLoS ONE 8(1): e53903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, da W, Sherman BT, Lempicki RA (2009. a) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, da W, Sherman BT, Lempicki RA (2009. b) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1): 44–57. [DOI] [PubMed] [Google Scholar]
- Huang RY, Chen GB, Matsumura N, Lai HC, Mori S, Li J, Wong MK, Konishi I, Thiery JP, Goh L (2012) Histotype-specific copy-number alterations in ovarian cancer. BMC Med Genomics 5: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1): 118–127. [DOI] [PubMed] [Google Scholar]
- Karlan BY, Dering J, Walsh C, Orsulic S, Lester J, Anderson LA, Ginther CL, Fejzo M, Slamon D (2014) POSTN/TGFBI-associated stromal signature predicts poor prognosis in serous epithelial ovarian cancer. Gynecol Oncol 132(2): 334–342. [DOI] [PubMed] [Google Scholar]
- Kobel M, Kalloger SE, Boyd N, McKinney S, Mehl E, Palmer C, Leung S, Bowen NJ, Ionescu DN, Rajput A, Prentice LM, Miller D, Santos J, Swenerton K, Gilks CB, Huntsman D (2008) Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med 5(12): e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecny GE, Wang C, Hamidi H, Winterhoff B, Kalli KR, Dering J, Ginther C, Chen HW, Dowdy S, Cliby W, Gostout B, Podratz KC, Keeney G, Wang HJ, Hartmann LC, Slamon DJ, Goode EL (2014) Prognostic and therapeutic relevance of molecular subtypes in high-grade serous ovarian cancer. J Natl Cancer Inst 106: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurman RJ, Visvanathan K, Roden R, Wu TC, Shih IeM (2008) Early detection and treatment of ovarian cancer: shifting from early stage to minimal volume of disease based on a new model of carcinogenesis. Am J Obstet Gynecol 198(4): 351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Matulonis UA (2014) New strategies in ovarian cancer: translating the molecular complexity of ovarian cancer into treatment advances. Clin Cancer Res 20(20): 5150–5156. [DOI] [PubMed] [Google Scholar]
- Lukanova A, Kaaks R (2005) Endogenous hormones and ovarian cancer: epidemiology and current hypotheses. Cancer Epidemiol Biomarkers Prev 14(1): 98–107. [PubMed] [Google Scholar]
- Martinez E, Yoshihara K, Kim H, Mills GM, Trevino V, Verhaak RG (2014) Comparison of gene expression patterns across 12 tumor types identifies a cancer supercluster characterized by TP53 mutations and cell cycle defects. Oncogene 34(21): 2732–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P, Sastre-Garau X, Mechta-Grigoriou F (2011) miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med 17(12): 1627–1635. [DOI] [PubMed] [Google Scholar]
- Miller JA, Cai C, Langfelder P, Geschwind DH, Kurian SM, Salomon DR, Horvath S (2011) Strategies for aggregating gene expression data: the collapseRows R function. BMC Bioinformatics 12: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok SC, Bonome T, Vathipadiekal V, Bell A, Johnson ME, Wong KK, Park DC, Hao K, Yip DK, Donninger H, Ozbun L, Samimi G, Brady J, Randonovich M, Pise-Masison CA, Barrett JC, Wong WH, Welch WR, Berkowitz RS, Birrer MJ (2009) A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: microfibril-associated glycoprotein 2. Cancer Cell 16(6): 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muccioli M, Benencia F (2014) Toll-like receptors in ovarian cancer as targets for immunotherapies. Front Immunol 5: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N, Nakayama K, Shamima Y, Ishikawa M, Katagiri A, Iida K, Miyazaki K (2010) Gene amplification CCNE1 is related to poor survival and potential therapeutic target in ovarian cancer. Cancer 116(11): 2621–2634. [DOI] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27(8): 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah PD, Tsai YY, Ramus SJ, Phelan CM, Goode EL, Lawrenson K, Buckley M, Fridley BL, Tyrer JP, Shen H, Weber R, Karevan R, Larson MC, Song H, Tessier DC, Bacot F, Vincent D, Cunningham JM, Dennis J, Dicks E Australian Cancer Study, Australian Ovarian Cancer Study Group Aben KK, Anton-Culver H, Antonenkova N, Armasu SM, Baglietto L, Bandera EV, Beckmann MW, Birrer MJ, Bloom G, Bogdanova N, Brenton JD, Brinton LA, Brooks-Wilson A, Brown R, Butzow R, Campbell I, Carney ME, Carvalho RS, Chang-Claude J, Chen YA, Chen Z, Chow WH, Cicek MS, Coetzee G, Cook LS, Cramer DW, Cybulski C, Dansonka-Mieszkowska A, Despierre E, Doherty JA, Dork T, du Bois A, Durst M, Eccles D, Edwards R, Ekici AB, Fasching PA, Fenstermacher D, Flanagan J, Gao YT, Garcia-Closas M, Gentry-Maharaj A, Giles G, Gjyshi A, Gore M, Gronwald J, Guo Q, Halle MK, Harter P, Hein A, Heitz F, Hillemanns P, Hoatlin M, Hogdall E, Hogdall CK, Hosono S, Jakubowska A, Jensen A, Kalli KR, Karlan BY, Kelemen LE, Kiemeney LA, Kjaer SK, Konecny GE, Krakstad C, Kupryjanczyk J, Lambrechts D, Lambrechts S, Le ND, Lee N, Lee J, Leminen A, Lim BK, Lissowska J, Lubinski J, Lundvall L, Lurie G, Massuger LF, Matsuo K, McGuire V, McLaughlin JR, Menon U, Modugno F, Moysich KB, Nakanishi T, Narod SA, Ness RB, Nevanlinna H, Nickels S, Noushmehr H, Odunsi K, Olson S, Orlow I, Paul J, Pejovic T, Pelttari LM, Permuth-Wey J, Pike MC, Poole EM, Qu X, Risch HA, Rodriguez-Rodriguez L, Rossing MA, Rudolph A, Runnebaum I, Rzepecka IK, Salvesen HB, Schwaab I, Severi G, Shridhar V, Shu XO, Sieh W, Southey MC, Spellman P, Tajima K, Teo SH, Terry KL, Thompson PJ, Timorek A, Tworoger SS, van Altena AM, van den Berg D, Vergote I, Vierkant RA, Vitonis AF, Wang-Gohrke S, Wentzensen N, Whittemore AS, Wik E, Winterhoff B, Woo YL, Wu AH, Yang HP, Zheng W, Ziogas A, Zulkifli F, Goodman MT, Hall P, Easton DF, Pearce CL, Berchuck A, Chenevix-Trench G, Iversen E, Monteiro AN, Gayther SA, Schildkraut JM, Sellers TA (2013) GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat Genet 45(4): 362–370, e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pils D, Hager G, Tong D, Aust S, Heinze G, Kohl M, Schuster E, Wolf A, Sehouli J, Braicu I, Vergote I, Cadron I, Mahner S, Hofstetter G, Speiser P, Zeillinger R (2012) Validating the impact of a molecular subtype in ovarian cancer on outcomes: a study of the OVCAD Consortium. Cancer Sci 103(7): 1334–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat J (2012) New insights into ovarian cancer pathology. Ann Oncol 23(Suppl 10): x111–x117. [DOI] [PubMed] [Google Scholar]
- Riester M, Wei W, Waldron L, Culhane AC, Trippa L, Oliva E, Kim SH, Michor F, Huttenhower C, Parmigiani G, Birrer MJ (2014) Risk prediction for late-stage ovarian cancer by meta-analysis of 1525 patient samples. J Natl Cancer Inst 106: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secord AA, Nixon AB, Hurwitz HI (2014) The search for biomarkers to direct antiangiogenic treatment in epithelial ovarian cancer. Gynecol Oncol 135(2): 349–358. [DOI] [PubMed] [Google Scholar]
- Shen H, Fridley BL, Song H, Lawrenson K, Cunningham JM, Ramus SJ, Cicek MS, Tyrer J, Stram D, Larson MC, Kobel M Consortium P Ziogas A, Zheng W, Yang HP, Wu AH, Wozniak EL, Woo YL, Winterhoff B, Wik E, Whittemore AS, Wentzensen N, Weber RP, Vitonis AF, Vincent D, Vierkant RA, Vergote I, Van Den Berg D, Van Altena AM, Tworoger SS, Thompson PJ, Tessier DC, Terry KL, Teo SH, Templeman C, Stram DO, Southey MC, Sieh W, Siddiqui N, Shvetsov YB, Shu XO, Shridhar V, Wang-Gohrke S, Severi G, Schwaab I, Salvesen HB, Rzepecka IK, Runnebaum IB, Rossing MA, Rodriguez-Rodriguez L, Risch HA, Renner SP, Poole EM, Pike MC, Phelan CM, Pelttari LM, Pejovic T, Paul J, Orlow I, Omar SZ, Olson SH, Odunsi K, Nickels S, Nevanlinna H, Ness RB, Narod SA, Nakanishi T, Moysich KB, Monteiro AN, Moes-Sosnowska J, Modugno F, Menon U, McLaughlin JR, McGuire V, Matsuo K, Adenan NA, Massuger LF, Lurie G, Lundvall L, Lubinski J, Lissowska J, Levine DA, Leminen A, Lee AW, Le ND, Lambrechts S, Lambrechts D, Kupryjanczyk J, Krakstad C, Konecny GE, Kjaer SK, Kiemeney LA, Kelemen LE, Keeney GL, Karlan BY, Karevan R, Kalli KR, Kajiyama H, Ji BT, Jensen A, Jakubowska A, Iversen E, Hosono S, Hogdall CK, Hogdall E, Hoatlin M, Hillemanns P, Heitz F, Hein R, Harter P, Halle MK, Hall P, Gronwald J, Gore M, Goodman MT, Giles GG, Gentry-Maharaj A, Garcia-Closas M, Flanagan JM, Fasching PA, Ekici AB, Edwards R, Eccles D, Easton DF, Durst M, du Bois A, Dork T, Doherty JA, Despierre E, Dansonka-Mieszkowska A, Cybulski C, Cramer DW, Cook LS, Chen X, Charbonneau B, Chang-Claude J, Campbell I, Butzow R, Bunker CH, Brueggmann D, Brown R, Brooks-Wilson A, Brinton LA, Bogdanova N, Block MS, Benjamin E, Beesley J, Beckmann MW, Bandera EV, Baglietto L, Bacot F, Armasu SM, Antonenkova N, Anton-Culver H, Aben KK, Liang D, Wu X, Lu K, Hildebrandt MA Australian Ovarian Cancer Study G Australian Cancer S, Schildkraut JM, Sellers TA, Huntsman D, Berchuck A, Chenevix-Trench G, Gayther SA, Pharoah PD, Laird PW, Goode EL, Pearce CL (2013) Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat Commun 4: 1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65(1): 5–29. [DOI] [PubMed] [Google Scholar]
- Sieh W, Kobel M, Longacre TA, Bowtell DD, deFazio A, Goodman MT, Hogdall E, Deen S, Wentzensen N, Moysich KB, Brenton JD, Clarke BA, Menon U, Gilks CB, Kim A, Madore J, Fereday S, George J, Galletta L, Lurie G, Wilkens LR, Carney ME, Thompson PJ, Matsuno RK, Kjaer SK, Jensen A, Hogdall C, Kalli KR, Fridley BL, Keeney GL, Vierkant RA, Cunningham JM, Brinton LA, Yang HP, Sherman ME, Garcia-Closas M, Lissowska J, Odunsi K, Morrison C, Lele S, Bshara W, Sucheston L, Jimenez-Linan M, Driver K, Alsop J, Mack M, McGuire V, Rothstein JH, Rosen BP, Bernardini MQ, Mackay H, Oza A, Wozniak EL, Benjamin E, Gentry-Maharaj A, Gayther SA, Tinker AV, Prentice LM, Chow C, Anglesio MS, Johnatty SE, Chenevix-Trench G, Whittemore AS, Pharoah PD, Goode EL, Huntsman DG, Ramus SJ (2013) Hormone-receptor expression and ovarian cancer survival: an Ovarian Tumour Tissue Analysis consortium study. Lancet Oncol 14(9): 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474(7353): 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418): 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G (2002) Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA 99(10): 6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, Traficante N, Fereday S, Hung JA, Chiew YE, Haviv I Australian Ovarian Cancer Study G Gertig D, DeFazio A, Bowtell DD (2008) Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res 14(16): 5198–5208. [DOI] [PubMed] [Google Scholar]
- Vang R, Shih IeM, Kurman RJ (2009) Ovarian low-grade and high-grade serous carcinoma: pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv Anat Pathol 16(5): 267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, Fereday S, Lawrence M, Carter SL, Mermel CH, Kostic AD, Etemadmoghadam D, Saksena G, Cibulskis K, Duraisamy S, Levanon K, Sougnez C, Tsherniak A, Gomez S, Onofrio R, Gabriel S, Chin L, Zhang N, Spellman PT, Zhang Y, Akbani R, Hoadley KA, Kahn A, Kobel M, Huntsman D, Soslow RA, Defazio A, Birrer MJ, Gray JW, Weinstein JN, Bowtell DD, Drapkin R, Mesirov JP, Getz G, Levine DA, Meyerson M Cancer Genome Atlas Research Network (2013) Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest 123(1): 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Hendrix-Lucas N, Kuick R, Zhai Y, Schwartz DR, Akyol A, Hanash S, Misek DE, Katabuchi H, Williams BO, Fearon ER, Cho KR (2007) Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/beta-catenin and PI3K/Pten signalling pathways. Cancer Cell 11(4): 321–333. [DOI] [PubMed] [Google Scholar]
- Yoshihara K, Tajima A, Yahata T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, Kudo Y, Kotera K, Masuzaki H, Tashiro H, Katabuchi H, Inoue I, Tanaka K (2010) Gene expression profile for predicting survival in advanced-stage serous ovarian cancer across two independent datasets. PLoS ONE 5(3): e9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, Tsunoda T, Shigemizu D, Fujiwara H, Hatae M, Masuzaki H, Katabuchi H, Kawakami Y, Okamoto A, Nogawa T, Matsumura N, Udagawa Y, Saito T, Itamochi H, Takano M, Miyagi E, Sudo T, Ushijima K, Iwase H, Seki H, Terao Y, Enomoto T, Mikami M, Akazawa K, Tsuda H, Moriya T, Tajima A, Inoue I, Tanaka K (2012) High-risk ovarian cancer based on 126-gene expression signature is uniquely characterized by downregulation of antigen presentation pathway. Clin Cancer Res 18(5): 1374–1385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.