Abstract
Background:
A better understanding of the molecular profile of anal squamous cell carcinomas (ASCCs) is necessary to consider new therapeutic approaches, and the identification of prognostic and predictive factors for response to treatment.
Methods:
We retrospectively analysed tumours from ASCC patients for mutational analysis of KRAS, NRAS, HRAS, BRAF, PIK3CA, MET, TP53 and FBXW7 genes by HRM and Sanger sequencing analysis.
Results:
Specimens from 148 patients were analysed: 96 treatment-naive tumours and 52 recurrences after initial radiotherapy (RT) or chemoradiotherapy (CRT). Mutations of KRAS, PIK3CA, FBXW7 and TP53 genes were present in 3 (2.0%), 30 (20.3%), 9 (6.1%) and 7 tumours (4.7%), respectively. The distribution of the mutations was similar between treatment-naive tumours and recurrences, except for TP53 mutations being more frequent in recurrences (P=0.0005). In patients treated with abdominoperineal resection (APR) after relapse (n=38, median follow-up of 18.2 years), overall survival (OS) was significantly correlated with HPV16 status (P=0.048), gender (P=0.045) and PIK3CA mutation (P=0.037). The PIK3CA status retained its prognostic significance in Cox multivariate regression analysis (P=0.025).
Conclusions:
Our study identified PIK3CA mutation as an independent prognostic factor in patients who underwent APR for ASCC recurrence, suggesting a potential benefit from adjuvant treatment and the evaluation of targeted therapies with PI3K/Akt/mTor inhibitors in PIK3CA-mutated patients.
Keywords: anal squamous cell carcinoma, mutation status, PIK3CA, prognostic factor, abdominoperineal resection, therapeutic target
Anal squamous cell carcinoma (ASCC) is a rare tumour that accounts for <5% of all lower gastrointestinal tract malignancies in Europe (Glynne-Jones et al, 2014). The incidence of ASCC has increased steadily in the past decades, particularly in women (Forman et al, 2012) and in men who have sex with men and those with HIV infection (Silverberg et al, 2012). Infection from human papilloma virus (HPV) is the main aetiologic factor in the development of ASCC and >90% of patients are HPV positive (mainly HPV16 and 18) (Frisch et al, 1997; Abramowitz et al, 2011). Recent results of high-sensitivity HPV genotyping in a large series of ASCC patients showed a positivity rate of >95%. This supports the development of multivalent HPV vaccination for prevention (Baricevic et al, 2015). Concomitant chemoradiotherapy (CRT) is the standard of care for locally advanced tumours (Flam et al, 1996; Bartelink et al, 1997; Cacheux et al, 2012). So far, no predictive factor (to CRT) has been identified, excepted p16 expression, HPV status and TP53 mutations (Gilbert et al, 2013; Koerber et al, 2014; Serup-Hansen et al, 2014; Baricevic et al, 2015; Mai et al, 2015; Meulendijks et al, 2015; Rödel et al, 2015). Salvage abdominoperineal resection (APR) is the standard treatment for local failure or recurrence after CRT, but 30 to 60% of operated patients will experience a locoregional and/or metastatic recurrence (Mullen et al, 2007; Mariani et al, 2008; Lefèvre et al, 2012; Correa et al, 2013). For these patients with an inoperable locally advanced or metastatic disease, very few treatments are available and their effectiveness is limited. New therapeutic approaches and predictive factors of outcome are required in this context. A better understanding of molecular markers involved in anal carcinogenesis might lead to the identification of new therapeutic targets as well as prognostic and predictive biomarkers. Recently, the potential effectiveness of anti-epidermal growth factor receptor (EGFR) monoclonal antibodies in advanced ASCC has been suggested by case reports (Lukan et al, 2009; Barmetter et al, 2012) that may be explained by both a high frequency of EGFR overexpression (80–90%) and the rarity of KRAS mutations in these tumours (Van Damme et al, 2010; Paliga et al, 2012, Smaglo et al, 2015). The incidence of other major gene alterations, especially those implicated in the EGFR pathway, has been rarely studied in ASCC. In the present study, we examined the mutation status of RAS (KRAS, NRAS and HRAS), BRAF, MET, FBXW7, TP53 and PIK3CA genes in a large series of 148 ASCC patients and correlated mutation status with clinicopathological characteristics and patient survival.
Materials and methods
Patient population
We retrospectively analysed tumours from ASCC patients consecutively treated from 1992 to 2015 at the Institut Curie Hospital. We included all consecutive patients for whom formalin-fixed, paraffin-embedded (FFPE) tumour tissue was available, and collected clinicopathological data and outcomes. This retrospective study was reviewed and approved by the Ethics Committee of the Institut Curie (No. A10-024). According to French regulations, patients were informed of research performed with the biological specimens obtained during their treatment and did not express opposition. Staging of the disease was based on the 7th revised edition (2010) of the AJCC Anus Cancer.
DNA extraction
Six tissue sections of 6 μm thickness were obtained from FFPE tissues and a seventh tissue section stained with HE staining. The tumour-rich areas were microdissected using a single-use blade and the samples underwent proteinase K digestion in a rotating incubator at 56 °C for 3 days. DNA was extracted with the NucleoSpin kit (Macherey-Nalgen, Hoerdt, France) according to the supplier recommendations in two separate aliquots that were analysed in parallel.
Gene mutation screening
The primer sequences used both for HRM and Sanger sequencing are shown in Supplementary Table 1. The majority of the HRM primers were designed to span the entire exons with product sizes under 200 bp. Primers were designed for KRAS (exons 2–4), HRAS (exons 2 and 3), NRAS, (exons 2 and 3), BRAF (exon 15), FBXW7 (exons 9 and 10), PIK3CA (exons 9 and 20), MET (exons 18 and 19) and TP53 genes (exons 4–8) (Supplementary Table 1). The PCR for HRM and Sanger sequencing analysis was performed on a 384-well plate in the presence of the fluorescent DNA intercalating dye, LC green (Idaho Technology, Salt Lake City, UT, USA) in a LightCycler480 (Roche Diagnosis, Meylan, France). The reaction mixture in a 15 μl final volume contained LC green, UDP Glycosylase (Roche) and Roche Master Mix (Roche). The cycling and melting conditions were as follows: with an initial cycle of 10 min at 40 °C, one cycle of 95 °C for 10 min; 50 cycles of 95 °C for 10 s, 55–65 °C for 10 s, 72 °C for 30 s; one cycle of 97 °C for 1 min and a melt from 70 °C to 95 °C rising 0.2 °C per s. Depending on the melting temperature, a touchdown approach was done for some primers. All samples were tested in duplicate. The HRM data were analysed using the Genescan software (Roche). All samples including the wild-type exons were plotted according to their melting profiles on the differential plot graph. Any difference of the horizon line based on the wild-type sample was sequenced with Sanger sequencing.
Sanger sequencing
The reaction mixture in a total of 50 μl was made using 1 μl of PCR products without first purification followed by a sequencing reaction with Big Dye Terminator v3.1 (Thermofisher, Courtaboeuf, France) according to the manufacturer's protocol. The sequencing products were purified with a Sephadex gel (GE Healthcare, Velizy-Villacoublay, France) before running on a 3500 Genetic Analyser (Applied Biosystems, Foster City, CA, USA). The sequencing data were visualised using Finch TV (Geospiza, Inc., Seattle, WA, USA) with detection sensibility of 10% mutated cells.
HPV detection
From 1998 to 2013, all samples were analysed by PCR using specific primers to identify HPV16, 18, 33, 45, 6 and 11 types and using GP5+/GP6+ primers to detect HPV L1 DNA as previously described (Lombard et al, 1998). After 2013, real-time PCR using Sybr Green (Roche Diagnostics, Mannheim, Germany) and specific primers for HPV16, 18 and 33 and the human GAPDH gene was performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems). HPV L1 amplicons from HPV16-, 18- and 33-negative samples were sequenced by Sanger method with GP6+ primer and HPV type identification was performed by alignment of the sequence with HPV sequence references, using the nucleotide blast program from NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical analysis
The statistical analysis plan was predefined jointly by the authors. Overall survival (OS) was defined as the period from the first day of radiotherapy (RT) or CRT to death from any cause. Data on patients who were alive at the end of follow-up (November 2015) were regarded as censored. Progression-free survival (PFS) was defined as the period from the first day of RT or RCT to the date of first disease progression or death from any cause. Cox univariate and multivariate regression was used for survival analysis and Fisher's exact test was used for the analysis of contingency tables. All statistical tests were two sided and P-values of <0.05 were considered statistically significant. All analyses have been implemented in R version 3.2.1 R Development Core Team, 2013. The applicable R code can be found in the Supplementary Information.
Results
Tumour and patient characteristics
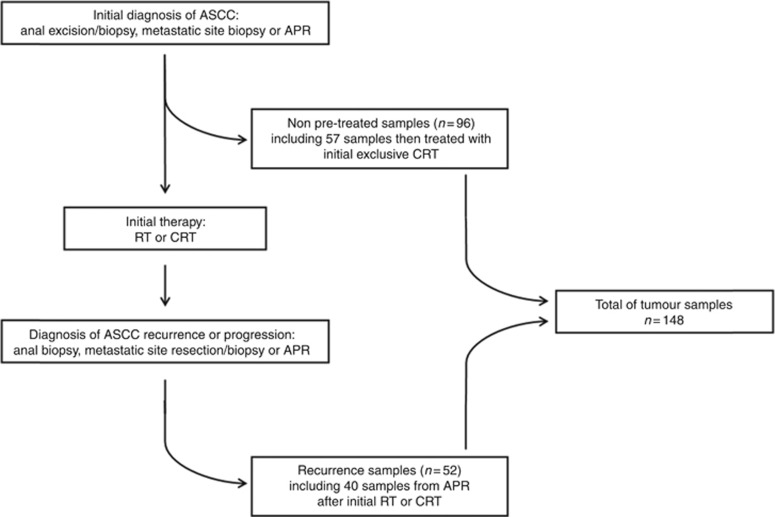
A total of 148 ASCC samples from patients treated in our institution were included and analysed in our gene mutation screening as summarised in the consort diagram (Figure 1): 96 tumours were treatment naive and 52 were samples from recurrence after initial RT or CRT. In total, 142 tumours (95.9%) were HPV positive among which 131 tumours (88.5%) had HPV16 infection. Only 16 patients (10.8%) had HIV infection. In the HIV+ population (n=16), all patients had concomitant HPV infection: 11 HPV16, 2 HPV18, 2 HPV6–11 and 1 HPV33. In the HIV− population (n=132), 6 patients had no HPV infection and 126 had concomitant HPV infection: 120 HPV16, 1 HPV6–11, 1 HPV33, 2 HPV35, 1 HPV59 and 1 HPV67. Tumour characteristics according to the treatment-naive or recurrence status of samples are summarised in Table 1.
Figure 1.
Consort diagram of the study.
Table 1. Clinicopathological features of treatment-naive and recurrence tumour samples with subsequent treatment received (n=148).
| Treatment-naive tumour | Tumour recurrence | |
|---|---|---|
| Total | n=96 | n=52 |
|
Gender | ||
| Female | 77 | 37 |
| Male | 19 | 15 |
|
Site of tumour samples | ||
| Anus | 91 | 42 |
| Lymph node | 5 | 5 |
| Liver | — | 3 |
| Other site | — | 2 |
|
Concomitant HIV infection | ||
| Yes | 7 | 9 |
| No | 89 | 43 |
|
Tumour differentiation | ||
| Poor | 8 | 11 |
| Moderate/well | 88 | 41 |
|
HPV status | ||
| HPV positive | 95 | 47 |
| Genotype 16 | 90 | 41 |
| Other genotypes (6–11/33/35/67) | 5 (1/1/2/1) | 6 (2/2/1/1) |
| HPV negative | 1 | 5 |
|
Prognostic groups (AJCC 2010) | ||
| I | 7 | 7 |
| II | 35 | 18 |
| IIIA | 21 | 6 |
| IIIB | 24 | 13 |
| IV (liver/lymph node) | 5 (4/1) | 3 (2/1) |
| ND | 4 | 5 |
|
Initial therapya | ||
| Surgery (local excision/APR): | 25 | — |
| alone | 5 | — |
| followed by RT | 11 | — |
| followed by CRT | 9 | — |
| Radiation | 11 | 21 |
| Chemoradiation: | 57 | 31 |
| with concomitant 5FU-CDDP | 47 | 26 |
| with concomitant 5FU-MMC | 5 | 3 |
| with other concomitant CT | 5 | 2 |
| Chemotherapy | 1 | — |
| No treatment | 2 | — |
Abbreviations: AJCC=American Joint Committee on Cancer; APR=abdominoperineal resection; CRT=chemoradiotherapy; CT=chemotherapy; HIV=human immunodeficiency virus; HPV=human papilloma virus; ND=not determined; RT=radiotherapy; 5FU-CDDP=5-fluorouracil-cisplatin; 5FU-MMC=5fluorouracil-mitomycin C.
Treatment received after diagnostic tumour samples for treatment-naive tumours and initial treatment received for tumour recurrence samples.
There were 114 females and 34 males. The median age at the diagnosis was 61 years (range: 37–96 years). Twenty-five patients were treated by initial surgery: exclusive surgery (n=5) and surgery followed by RT (n=11) or RCT (n=19). Thirty-two patients were treated by initial RT and 88 by initial CRT. One was treated by chemotherapy for an initial metastatic disease and 2 were not treated after the initial diagnosis. Only 16 patients (10.8%) had HIV infection. Forty-five patients underwent APR: 40 for local recurrence after RT or RCT, 3 at diagnosis and 2 for suspicion of local recurrence with complete histological response on surgical specimens. The median follow-up of 148 patients was 3.3 years (range: 0.2–39.6 years).
Gene mutation screening
Of the 148 tumours, 3 (2.0%) showed a KRAS exon 2 mutation, 30 (20.3%) a PIK3CA mutation, 9 (6.1%) a FBXW7 mutation and 7 (4.7%) a TP53 mutation (Table 2A and Supplementary Table 2). Five tumours (3.4%) had 2 synchronous mutations concerning these previous genes (PIK3CA/FBXW7 mutations in 3 tumours, KRAS/TP53 in 1 tumour and FBXW7/TP53 in 1 tumour). All tumours were wild type for HRAS, NRAS, BRAF and MET genes. In 15 ASCC patients, we analysed several available samples obtained at different therapeutic times or in different sites. We observed a total concordance of the Sanger analysis in 12 patients but the mutational profile was different between samples for 3 patients (Table 2B).
Table 2. (A) Prevalence of identified mutations in the 148 ASCC samples and distribution among treatment-naive tumours and tumour recurrences. (B) Heterogeneity of mutational profiles in different tumour samples from the same patient (n=3).
|
(A) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Genes | KRAS | HRAS | NRAS | BRAF | PIK3CA | MET | FBXW7 | TP53 |
| Total (%) (n=148) | 3 (2.0%) | 0 | 0 | 0 | 30 (20.3%) | 0 | 9 (6.1%) | 7 (4.7%) |
| Exons | Exon 2: 3 | — | — | — | Exon 9: 27 | — | Exon 9: 1 | Exon 4: 1 |
| Exon 20: 3 | Exon 10: 8 | Exon 6: 1 | ||||||
| Exon 7: 2 | ||||||||
| Exon 8: 3 | ||||||||
| Treatment-naive samples (%) (n=96) | 1 (1.1%) | — | — | — | 19 (19.8%) | — | 4 (4.2%) | 0 (0%) |
| Samples from recurrence (%) (n=52) | 2 (3.8%) | — | — | — | 11 (21.2%) | — | 5 (9.6%) | 7 (13.5%) |
| P (Fisher's test) | NS | — | — | — | NS | — | NS | 0.0005 |
|
(B) |
|||
|---|---|---|---|
| KRAS | PIK3CA | FBXW7 | |
|
Patient 1 |
|||
| Recurrence (lymph node metastasis) | — | — | — |
| Recurrence (peritoneal metastasis) | — | exon9:c.G1624A;p.E542K | exon10:c.1436G>A:p.R479Q |
|
Patient 2 |
|||
| Treatment-naive anal tumour | — | exon9:c.G1633A:p.E545K | — |
| Anal recurrence | exon2:c.G34C:p.G12R | — | — |
|
Patient 3 |
|||
| Treatment-naive anal tumour | — | exon9: c.G1624A;p.E542K | — |
| Treatment-naive liver metastasis | exon2:c.G34C:p.G12R | exon9:.G1624A;p.E542K | — |
Abbreviations: ASCC=anal squamous cell carcinoma; NS=not significant.
Correlation between gene mutations and clinicopathological features and prognostic value
The distribution of the mutations was similar between treatment-naive tumours and tumour recurrences, except for TP53 mutations (Table 2A). We found that TP53 mutations were restricted to recurrence samples: 7 of 52 (13.5%) tumour recurrences vs 0 of 96 (0%) treatment-naive tumours (Fisher's test, P=0.0005). Moreover, we observed that TP53 mutations were more frequently associated with HPV16negative samples: 3 of 131 (2.3%) HPV16-positive tumours vs 4 of 17 (23.5%) HPV16-negative tumours (Fisher's test, P=0.003).
As the site and therapeutic status of tumour samples were heterogeneous in this large retrospective cohort of ASCC patients, we focussed our tumour analysis on homogenous groups of patients to study the association between mutational status and clinicopathological characteristics of the patients, and the impact of these parameters on OS. We also excluded nontreated tumours, tumours with ongoing treatment and those without sufficient follow-up (<6 months) in our prognostic analysis.
We identified a first group of treatment-naive tumours from 57 ASCC patients treated by initial exclusive CRT with a median follow-up of 3.1 years (range: 0.3–14 years) (Supplementary Table 3). Overall, recurrence rate was 24.6% (n=14 of 57). All tumours were HPV positive and 52 of 57 (91,2%) had HPV type 16. Only 1 (1.7%) KRAS, 3 (5.3%) FBXW7 and 1 (1.7%) TP53 mutations were identified in this group, whereas PIK3CA mutations were identified in 10 (17.5%) of them (8 in exon 9 and 2 in exon 20). No association was found between PIK3CA mutations and clinicopathological characteristics of patients (data not shown). Moreover, no correlation was found between PI3KCA mutation and PFS or OS (Supplementary Table 4).
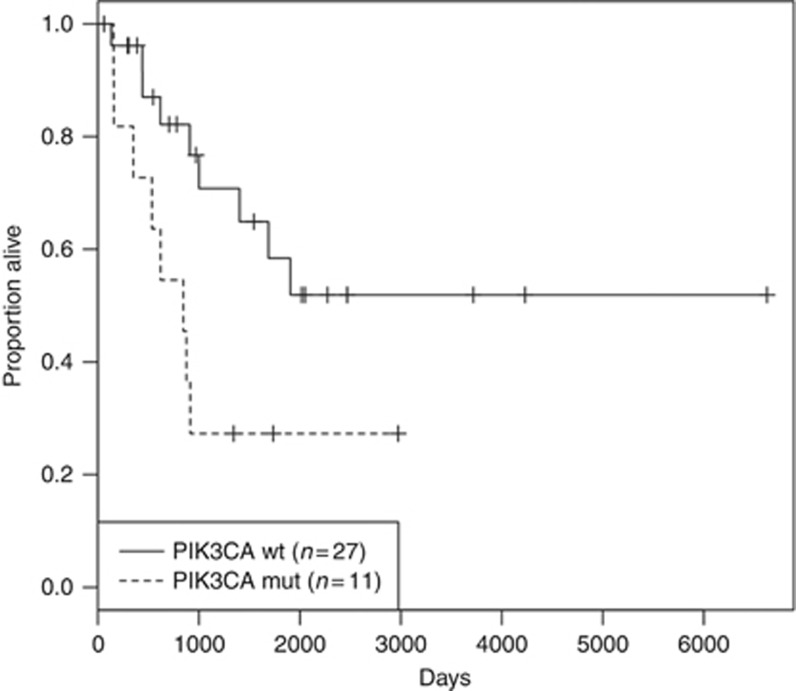
We also selected a second group of 40 recurrent tumour samples from ASCC patients who underwent APR for local recurrence after initial RT or CRT. We excluded 2 samples from patients who died early after APR from postoperative complications (at day 6 and 10 respectively). We obtained a final cohort of 38 ASCC samples with a median follow-up of 18.2 years (range: 0.82–39.6 years). Overall, recurrence rate was 57.9% (n=22 of 38). Clinicopathological characteristics of this group of patients are summarised in Table 3. PIK3CA, FBXW7 and TP53 mutations were identified in 11 (28.9%), 5 (13.2%) and 4 (10.5%) recurrent tumours out of 38 respectively. No association was found between PIK3CA mutations and clinicopathological characteristics (Supplementary Table 5). A significant correlation by univariate Cox regression analysis was found between OS and gender (P=0.045), HPV16 status (P=0.048) and PIK3CA mutation (P=0.037) (Table 4 and Figure 2). Multivariate Cox analysis showed that HPV16 status (P=0.004), HIV status (P=0.032) and PIK3CA mutation (P=0.025) were independent prognostic factors (Table 4).
Table 3. Clinicopathological features of the 38 tumour relapse samples from APR after initial RT/CRT.
| Total | n=38 |
|---|---|
|
Gender | |
| Female | 29 |
| Male | 9 |
|
Concomitant HIV infection | |
| Yes | 7 |
| No | 31 |
|
HPV status | |
| HPV positive | 34 |
| genotype 16 | 29 |
| other genotypes (6–11/18/33/59) | 5 (1/2/1/1) |
| HPV negative | 4 |
|
Initial therapy before APR | |
| Radiation | 14 |
| Chemoradiation with 5FU-CDDP | 20 |
| Chemoradiation with 5FU-MMC | 3 |
| Chemoradiation with other CT | 1 |
|
Pathological results | |
| Tumour differentiation | |
| Poor | 6 |
| Moderate/well | 32 |
| ypT stage | |
| T0 | 1 |
| T1 | 5 |
| T2 | 19 |
| T3 | 9 |
| T4 | 3 |
| ND | 1 |
| ypN stage | |
| N− | 31 |
| N+ | 7 |
| Vascular emboli | |
| Yes | 8 |
| No | 30 |
| Lymphatic invasion | |
| Yes | 9 |
| No | 29 |
| Perineural invasion | |
| Yes | 10 |
| No | 28 |
| R1 resection | |
| Yes | 8 |
| No | 30 |
|
PIK3CA mutation | |
| Yes | 11 |
| Exon 9 | 10 |
| c.1624G>A;p.E542K | 5 |
| c.1633G>A:p.E545K | 5 |
| Exon 20 | 1 |
| No | 27 |
|
FBXW7 mutation | |
| Yes | 5 |
| No | 33 |
|
TP53 mutation | |
| Yes | 4 |
| No | 34 |
Abbreviations: APR=abdominoperineal resection; CRT=chemoradiotherapy; CT=chemotherapy; HIV=human immunodeficiency virus; HPV=human papilloma virus; ND=not determined; RT=radiotherapy; 5FU-CDDP=5-fluorouracil-cisplatin; 5FU-MMC=5fluorouracil-mitomycin C.
Table 4. Overall survival according to clinicopathological and mutational characteristics of the 38 patients who underwent APR for tumour recurrence after RT/CRT.
|
Univariate analysis |
Multivariate analysis |
|||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% Interval | P | HR | 95% Interval | P | |||
| Gender (male) | 3.034 | 1.026 | 8.968 | 0.045 | — | — | — | NSa |
| ypT stage (T3–T4 vs T1–T2) | 1.362 | 0.770 | 2.411 | 0.288 | ||||
| ypN+ stage | 2.302 | 0.792 | 6.700 | 0.126 | 3.108 | 0.861 | 11.211 | 0.083 |
| R1 resection | 2.398 | 0.770 | 7.463 | 0.131 | — | — | — | NSa |
| Moderate/well tumoural differentiation | 0.611 | 0.198 | 1.885 | 0.391 | ||||
| Vascular emboli | 1.392 | 0.450 | 4.309 | 0.566 | ||||
| Lymphatic invasion | 1.031 | 0.334 | 3.181 | 0.958 | ||||
| Perineural invasion | 1.063 | 0.373 | 3.034 | 0.909 | ||||
| HPV16-positive status | 0.344 | 0.119 | 0.990 | 0.048 | 0.155 | 0.043 | 0.558 | 0.004 |
| HIV-positive status | 2.700 | 0.866 | 8.390 | 0.087 | 4.259 | 1.130 | 16.054 | 0.032 |
| Initial therapy (CRT vs RT) | 1.430 | 0.525 | 3.895 | 0.482 | ||||
| PIK3CA mutation | 2.808 | 1.066 | 7.394 | 0.037 | 3.729 | 1.180 | 11.781 | 0.025 |
| FBXW7 mutation | 0.668 | 0.151 | 2.950 | 0.595 | ||||
| TP53 mutation | 0.590 | 0.133 | 2.609 | 0.486 | ||||
Abbreviations: APR=abdominoperineal resection; CRT=chemoradiotherapy; HIV=human immunodeficiency virus; HPV=human papilloma virus; HR=hazard ratio; NS=not significant; RT=radiotherapy.
Univariate and multivariate Cox regression. Bold entries are used for significative P data values which are <0.05 (as explained in the statistical analysis).
Contribution to the model was not significant after stepwise reduction.
Figure 2.
Overall survival depending on the PIK3CA mutation in the 38 relapse APR patients after initial RT/RCT (P=0.025).
Discussion
ASCC is known to be a very well radiosensitive tumour but 20% of patients failed to CRT, and no predictive markers of response have been prospectively validated. Moreover, in case of recurrence after RT/CRT, APR is the treatment of choice without any prognostic factor identified or any adjuvant treatment recommendation, although at least 50% of patients experience recurrence after this surgery (Mullen et al, 2007; Mariani et al, 2008; Lefèvre et al, 2012; Correa et al, 2013). In this context, a better biological and molecular characterisation of anal carcinogenesis is needed to improve the medical care of ASCC patients by identifying new therapeutic targets or prognostic biomarkers.
In the present study, which is the largest retrospective cohort of ASCC samples analysed by sequencing for multiple genes with complete clinicobiological data and long-term patient outcome available, we found frequent PIK3CA mutations (20.3%), as observed in previous smaller studies identifying PIK3CA mutation in 22% (11 out of 53, by pyrosequencing) and 32.5% (28 out of 86, by next-generation sequencing) of tumours (Casadei Gardini et al, 2014; Smaglo et al, 2015). The high level of PIK3CA mutation in ASCC provides a rationale to evaluate specific inhibitors of the PIK3CA/Akt/mTor pathway as demonstrated in preclinical models (Stelzer et al, 2010; Sun et al, 2013).
We identified very few KRAS exon 2 mutations (2.3%), in line with previous studies reporting low rates (Van Damme et al, 2010; Martin et al, 2014; Smaglo et al, 2015) or the absence of KRAS mutations (Paliga et al, 2012; Gilbert et al, 2013; Casadei Gardini et al, 2014) that could explain the effectiveness of EGFR monoclonal antibodies observed in ASCC patients (Lukan et al, 2009). The TP53 mutations were also rarely described in the literature, although a high frequency of TP53 protein expression is reported (Patel et al, 2007).
In our series, we confirm the low frequency of TP53 mutations (5.3%) but we found they were restricted to recurrence samples and more frequently associated to HPV16-negative samples. As observed in our cohort, it was recently reported that TP53 mutations were correlated with HPV16-negative status, predictive of resistance to RT/CRT and correlated with a poor prognosis in ASCC patients (Meulendijks et al, 2015). The same correlation between HPV status and TP53 mutations was previously described in head and neck cancer (Westra et al, 2008).
We also focussed our gene screening on FBXW7 gene. Mutations of FBXW7 gene have been frequently reported in not only various squamous cell carcinomas (Agrawal et al, 2011; Gao et al, 2014; Ojesina et al, 2014) but also adenocarcinoma (The Cancer Genome Atlas Network, 2012; Laforest et al, 2014; The Cancer Genome Atlas Research Network, 2014) and melanoma where a protein inactivation was found (Aydin et al, 2014). For the first time, we report FBXW7 mutations in 6% of ASCC. FBWX7 is known to be a key regulator of the cell cycle involved in the maintenance of normal stem cells and cancer-initiating cells (Takeishi and Nakayama, 2014). It could act as a critical tumour suppressor gene by targeting the NOTCH1 oncoprotein and therefore be an effective biomarker for the evaluation of Notch inhibitors in ASCC (Aydin et al, 2014). Our study finally shows that HRAS, NRAS, BRAF and MET genes are not mutated in ASCC.
Of note, we observed a different gene mutational status in 3 out of 15 patients for whom several tumour samples were available. For one of them, this could be the consequence of CRT on tumour DNA as the different mutational profiles were obtained from anal treatment-naive and pretreated samples respectively. For the two remaining patients, we can make the assumption of tumour heterogeneity as the different mutational profiles were observed in samples from two different tumour sites.
Our study is the first one assessing the relation between clinicopathological characteristics and mutational status of several genes in a large series of ASCC patients with details on treatments received, allowing an exploratory assessment of gene mutation predictive and prognostic value. In the large biomarker analysis of 199 ASCCs recently reported by Smaglo et al (2015), only part of the tumours were the subject of a gene sequencing (8 to 86 according to the gene analysed) and almost no clinicopathological information was available, therefore avoiding any correlation to be performed. In another series of 103 ASCC patients, paraffin-embedded tumour tissue was sufficient to perform analysis of KRAS, BRAF and PIK3CA gene mutation in only 50 patients (Casadei Gardini et al, 2014). In our study, none of the gene mutations identified was associated with clinicopathological characteristics. Casadei Gardini et al (2014) also found no association between PIK3CA mutations (found in 22% of cases) and clinical characteristics.
Several studies have reported on potential prognostic and predictive biomarkers of response to RT/CRT in ASCC (Lampejo et al, 2010; Myklebust et al, 2012; Fraunholz et al, 2013) but none of them was a gene mutation and none has been sufficiently validated to be used in clinical practice. In our large cohort, we selected 2 homogenous groups of patients regarding the treatments received to analyse: (1) the prognostic and predictive value on response to treatment of identified mutations in treatment-naive samples of patients exclusively treated by CRT (n=57) and (2) the prognostic value of identified mutations after APR for local recurrence following RT/CRT (n=38).
In the naive tumours treated by CRT, PIK3CA mutation identified on pretreatment samples was not found prognostic or predictive of response to CRT. This result is concordant with the study of Casadei Gardini et al (2014) in which PI3KCA mutation was not associated with PFS or OS of patients treated by CRT. The predictive impact of this mutation on tumour response to CRT was not explored in this study (Casadei Gardini et al, 2014). We could not study the prognostic or predictive value of KRAS, FBXW7 and TP53 mutations given their low frequency in our study.
To our knowledge, this is the first study assessing gene mutations as potential prognostic biomarkers in ASCC patients who underwent APR for local recurrence after RT or RCT. After multivariate Cox analysis we identified three independent factors associated with worse survival: a negative HPV16 status (P=0.004) and a positive HIV infection (P=0.032), which has already been reported (Wexler et al, 2008; Yhim et al, 2011), and also the presence of PIK3CA gene mutation (P=0.025) that is identified for the first time as a new independent prognostic marker in this setting. Of course, the prognosis value of PIK3CA mutations we report need to be validated in an independent and larger prospective cohort of ASCC, considering the relatively small sample size of our series. These PIK3CA mutations have been previously reported to be associated with poor prognostic in colorectal cancer (Barault et al, 2008; Ogino et al, 2009) but data in cervical squamous cell carcinoma are more divergent with both an association with better OS in early tumour stages (McIntyre et al, 2013) and a poor response following standard CRT in more advanced stages (de la Rochefordiere et al, 2015). Finally, gynecological cancer patients with PIK3CA mutations are more responsive to PI3K/Akt/mTor inhibitors than nonmutated patients (Husseinzadeh and Husseinzadeh, 2014). These results, together with our findings, suggest that PIK3CA mutations might play a major role in HPV-related squamous cell carcinoma, including anal carcinogenesis, especially in mechanisms of resistance to RT or CRT. They provide a rationale for the use of PI3K/Akt/mTor pathway inhibitors in radioresistant tumours, particularly in adjuvant setting after APR. Aspirin therapy, recently shown to be of particular efficacy in adjuvant treatment of PIK3CA-mutated colorectal cancer, could be another therapeutic option in this setting (Liao et al, 2012). In addition, there are recent data suggesting that the host immune reaction mediates response (Gilbert et al, 2016) via tumour-infiltrating lymphocytes. These data suggest the evaluation of immunotherapy in anal cancer, whose efficiency might be enhanced by cyclooxygenase inhibitors such as aspirin (Zelaney et al, 2015).
Acknowledgments
We thank Emmanuelle Lappartient, Didier Meseure and Carine Perennou for technical help and Intidhar Labidi-Galy for helpful discussions. This work was supported by the CEST (Comité d'Evaluation et de Suivi des projets de recherche de Transfert) from the Institut Curie.
Author contributions
All authors contributed to the conception and design of the study, acquisition of data, analysis and interpretation of data, drafting the article or revising it critically for important intellectual content and final approval of the version to be published.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Abramowitz L, Jacquard AC, Jaroud F, Haesebaert J, Siproudhis L, Pradat P, Aynaud O, Leocmach Y, Soubeyrand B, Dachez R, Riethmuller D, Mougin C, Pretet JL, Denis F (2011) Human papillomavirus genotype distribution in anal cancer in France: the EDiTH V study. Int J Cancer 129: 433–439. [DOI] [PubMed] [Google Scholar]
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Treviño L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN (2011) Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333: 1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin IT, Melamed RD, Adams SJ, Castillo-Martin M, Demir A, Bryk D, Brunner G, Cordon-Cardo C, Osman I, Rabadan R, Celebi JT (2014) FBXW7 mutations in melanoma and a new therapeutic paradigm. J Natl Cancer Inst 106: dju107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barmettler H, Komminoth P, Schmid M, Duerr D (2012) Efficacy of cetuximab in combination with FOLFIRI in a patient with KRAS wild-type metastatic anal cancer. Case Rep Oncol 5: 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barault L, Veyrie N, Jooste V, Lecorre D, Chapusot C, Ferraz JM, Lièvre A, Cortet M, Bouvier AM, Rat P, Roignot P, Faivre J, Laurent-Puig P, Piard F (2008) Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer 122: 2255–2259. [DOI] [PubMed] [Google Scholar]
- Baricevic I, He X, Chakrabarty B, Oliver AW, Bailey C, Summers J, Hampson L, Hampson I, Gilbert DC, Renehan AG (2015) High-sensitivity human papilloma virus genotyping reveals near universal positivity in anal squamous cell carcinoma: different implications for vaccine prevention and prognosis. Eur J Cancer 51: 776–785. [DOI] [PubMed] [Google Scholar]
- Bartelink H, Roelofsen F, Eschwege F, Rougier P, Bosset JF, Gonzalez DG, Peiffert D, van Glabbeke M, Pierart M (1997) Concomitant radiotherapy and chemotherapy is superior to radiotherapy alone in the treatment of locally advanced anal cancer: results of a phase III randomized trial of the European Organization for Research and Treatment of Cancer Radiotherapy and Gastrointestinal Cooperative Groups. J Clin Oncol 15: 2040–2049. [DOI] [PubMed] [Google Scholar]
- Cacheux W, Lievre A, De La Rochefordiere A, Dieumegard B, Cvitkovic F, Labib A, Mitry E, Buecher B (2012) Chemotherapy in the treatment of anal canal carcinoma. Dig Liver Dis 44: 803–811. [DOI] [PubMed] [Google Scholar]
- Casadei Gardini A, Capelli L, Ulivi P, Giannini M, Freier E, Tamberi S, Scarpi E, Passardi A, Zoli W, Ragazzini A, Amadori D, Frassineti GL (2014) KRAS, BRAF and PIK3CA status in squamous cell anal carcinoma (SCAC). PLoS One 9: e92071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa JH, Castro LS, Kesley R, Dias JA, Jesus JP, Olivatto LO, Martins IO, Lopasso FP (2013) Salvage abdominoperineal resection for anal cancer following chemoradiation: a proposed scoring system for predicting postoperative survival. J Surg Oncol 107: 486–492. [DOI] [PubMed] [Google Scholar]
- de la Rochefordiere A, Kamal M, Floquet A, Thomas L, Petrow P, Petit T, Pop M, Fabbro M, Kerr C, Joly F, Sevin E, Maillard S, Curé H, Weber B, Brunaud C, Minsat M, Gonzague L, Berton-Rigaud D, Aumont M, Gladieff L, Peignaux K, Bernard V, Leroy Q, Bieche I, Margogne A, Nadan A, Fourchotte V, Diallo A, Asselain B, Plancher C, Armanet S, Beuzeboc P, Scholl SM (2015) PIK3CA pathway mutations predictive of poor response following standard radiochemotherapy±cetuximab in cervical cancer patients. Clin Cancer Res 21: 2530–2537. [DOI] [PubMed] [Google Scholar]
- Flam M, John M, Pajak TF, Petrelli N, Myerson R, Doggett S, Quivey J, Rotman M, Kerman H, Coia L, Murray K (1996) Role of mitomycin in combination with fluorouracil and radiotherapy, and of salvage chemoradiation in the definitive nonsurgical treatment of epidermoid carcinoma of the anal canal: results of a phase III randomized intergroup study. J Clin Oncol 14: 2527–2539. [DOI] [PubMed] [Google Scholar]
- Forman D, de Martel C, Lacey CJ, Soerjomataram I, Lortet-Tieulent J, Bruni L, Vignat J, Ferlay J, Bray F, Plummer M, Franceschi S (2012) Global burden of human papillomavirus and related diseases. Vaccine 30: 12–23. [DOI] [PubMed] [Google Scholar]
- Fraunholz I, Rödel F, Kohler D, Diallo-Georgiopoulou M, Distel L, Falk S, Rödel C (2013) Epidermal growth factor receptor expression as prognostic marker in patients with anal carcinoma treated with concurrent chemoradiation therapy. Int J Radiat Oncol Biol Phys 86: 901–907. [DOI] [PubMed] [Google Scholar]
- Frisch M, Glimelius B, van den Brule AJ, Wohlfahrt J, Meijer CJ, Walboomers JM, Goldman S, Svensson C, Adami HO, Melbye M (1997) Sexually transmitted infection as a cause of anal cancer. N Engl J Med 337: 1350–1358. [DOI] [PubMed] [Google Scholar]
- Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, Zhao YD, Sun J, Zhou CC, Yao R, Wang SY, Wang P, Sun N, Zhang BH, Dong JS, Yu Y, Luo M, Feng XL, Shi SS, Zhou F, Tan FW, Qiu B, Li N, Shao K, Zhang LJ, Zhang LJ, Xue Q, Gao SG, He J (2014) Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 46: 1097–1102. [DOI] [PubMed] [Google Scholar]
- Gilbert DC, Williams A, Allan K, Stokoe J, Jackson T, Linsdall S, Bailey CM, Summers J (2013) p16INK4A, p53, EGFR expression and KRAS mutation status in squamous cell cancers of the anus: correlation with outcomes following chemo-radiotherapy. Radiother Oncol 109: 146–151. [DOI] [PubMed] [Google Scholar]
- Gilbert DC, Serup-Hansen E, Linnemann D, Høgdall E, Bailey C, Summers J, Havsteen H, Thomas GJ (2016) Tumour-infiltrating lymphocyte scores effectively stratify outcomes over and above p16 post chemo-radiotherapy in anal cancer. Br J Cancer 114: 134–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynne-Jones R, Nilsson PJ, Aschele C, Goh V, Peiffert D, Cervantes A, Arnold D (2014) Anal cancer: ESMO-ESSO-ESTRO clinical practice guidelines for diagnosis, treatment and follow-up. Eur J Surg Oncol 40: 1165–1176. [DOI] [PubMed] [Google Scholar]
- Husseinzadeh N, Husseinzadeh HD (2014) mTOR inhibitors and their clinical application in cervical, endometrial and ovarian cancers: a critical review. Gynecol Oncol 133: 375–381. [DOI] [PubMed] [Google Scholar]
- Koerber SA, Schoneweg C, Slynko A, Krug D, Haefner MF, Herfarth K, Debus J, Sterzing F, von Knebel Doeberitz M, Prigge ES, Reuschenbach M (2014) Influence of human papillomavirus and p16(INK4a) on treatment outcome of patients with anal cancer. Radiother Oncol 113: 331–336. [DOI] [PubMed] [Google Scholar]
- Laforest A, Aparicio T, Zaanan A, Silva FP, Didelot A, Desbeaux A, Le Corre D, Benhaim L, Pallier K, Aust D, Pistorius S, Blons H, Svrcek M, Laurent-Puig P (2014) ERBB2 gene as a potential therapeutic target in small bowel adenocarcinoma. Eur J Cancer 50: 1740–1746. [DOI] [PubMed] [Google Scholar]
- Lampejo T, Kavanagh D, Clark J, Goldin R, Osborn M, Ziprin P, Cleator S (2010) Prognostic biomarkers in squamous cell carcinoma of the anus: a systematic review. Br J Cancer 103: 1858–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre JH, Corte H, Tiret E, Boccara D, Chaouat M, Touboul E, Svrcek M, Lefrancois M, Shields C, Parc Y (2012) Abdominoperineal resection for squamous cell anal carcinoma: survival and risk factors for recurrence. Ann Surg Oncol 19: 4186–4192. [DOI] [PubMed] [Google Scholar]
- Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, Sun R, Nosho K, Meyerhardt JA, Giovannucci E, Fuchs CS, Chan AT, Ogino S (2012) Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med 367: 1596–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard I, Vincent-Salomon A, Validire P, Zafrani B, de la Rochefordière A, Clough K, Favre M, Pouillart P, Sastre-Garau X (1998) Human papillomavirus genotype as a major determinant of the course of cervical cancer. J Clin Oncol 16: 2613–2619. [DOI] [PubMed] [Google Scholar]
- Lukan N, Ströbel P, Willer A, Kripp M, Dinter D, Mai S, Hochhaus A, Hofheinz RD (2009) Cetuximab-based treatment of metastatic anal cancer: correlation of response with KRAS mutational status. Oncology 77: 293–299. [DOI] [PubMed] [Google Scholar]
- Mai S, Welzel G, Ottstadt M, Lohr F, Severa S, Prigge ES, Wentzensen N, Trunk MJ, Wenz F, von Knebel-Doeberitz M, Reuschenbach M (2015) Prognostic relevance of HPV infection and p16 overexpression in squamous cell anal cancer. Int J Radiat Oncol Biol Phys 93: 819–827. [DOI] [PubMed] [Google Scholar]
- Mariani P, Ghanneme A, De la Rochefordière A, Girodet J, Falcou MC, Salmon RJ (2008) Abdominoperineal resection for anal cancer. Dis Colon Rectum 51: 1495–1501. [DOI] [PubMed] [Google Scholar]
- Martin V, Zanellato E, Franzetti-Pellanda A, Molinari F, Movilia A, Paganotti A, Deantonio L, De Dosso S, Assi A, Crippa S, Boldorini R, Mazzucchelli L, Saletti P, Frattini M (2014) EGFR, KRAS, BRAF, and PIK3CA characterization in squamous cell anal cancer. Histol Histopathol 29: 513–521. [DOI] [PubMed] [Google Scholar]
- McIntyre JB, Wu JS, Craighead PS, Phan T, Köbel M, Lees-Miller SP, Ghatage P, Magliocco AM, Doll CM (2013) PIK3CA mutational status and overall survival in patients with cervical cancer treated with radical chemoradiotherapy. Gynecol Oncol 128: 409–414. [DOI] [PubMed] [Google Scholar]
- Meulendijks D, Tomasoa NB, Dewit L, Smits PH, Bakker R, van Velthuysen ML, Rosenberg EH, Beijnen JH, Schellens JH, Cats A (2015) HPV-negative squamous cell carcinoma of the anal canal is unresponsive to standard treatment and frequently carries disruptive mutations in TP53. Br J Cancer 112: 1358–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JT, Rodriguez-Bigas MA, Chang GJ, Barcenas CH, Crane CH, Skibber JM, Feig BW (2007) Results of surgical salvage after failed chemoradiation therapy for epidermoid carcinoma of the anal canal. Ann Surg Oncol 14: 478–483. [DOI] [PubMed] [Google Scholar]
- Myklebust MP, Fluge Ø, Immervoll H, Skarstein A, Balteskard L, Bruland O, Dahl O (2012) Expression of DSG1 and DSC1 are prognostic markers in anal carcinoma patients. Br J Cancer 106: 756–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, Kure S, Chan AT, Engelman JA, Kraft P, Cantley LC, Giovannucci EL, Fuchs CS (2009) PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol 27: 1477–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio L, Cibulskis K, Bertelsen B, Romero-Cordoba S, Treviño V, Vazquez-Santillan K, Guadarrama AS, Wright AA, Rosenberg MW, Duke F, Kaplan B, Wang R, Nickerson E, Walline HM, Lawrence MS, Stewart C, Carter SL, McKenna A, Rodriguez-Sanchez IP, Espinosa-Castilla M, Woie K, Bjorge L, Wik E, Halle MK, Hoivik EA, Krakstad C, Gabiño NB, Gómez-Macías GS, Valdez-Chapa LD, Garza-Rodríguez ML, Maytorena G, Vazquez J, Rodea C, Cravioto A, Cortes ML, Greulich H, Crum CP, Neuberg DS, Hidalgo-Miranda A, Escareno CR, Akslen LA, Carey TE, Vintermyr OK, Gabriel SB, Barrera-Saldaña HA, Melendez-Zajgla J, Getz G, Salvesen HB, Meyerson M (2014) Landscape of genomic alterations in cervical carcinomas. Nature 506: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliga A, Onerheim R, Gologan A, Chong G, Spatz A, Niazi T, Garant A, Macheto D, Alcindor T, Vuong T (2012) EGFR and K-ras gene mutation status in squamous cell anal carcinoma: a role for concurrent radiation and EGFR inhibitors? Br J Cancer 107: 1864–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H, Polanco-Echeverry G, Segditsas S, Volikos E, McCart A, Lai C, Guenther T, Zaitoun A, Sieber O, Ilyas M, Northover J, Silver A (2007) Activation of AKT and nuclear accumulation of wild type TP53 and MDM2 in anal squamous cell carcinoma. Int J Cancer 121: 2668–2673. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Rödel F, Wieland U, Fraunholz I, Kitz J, Rave-Fränk M, Wolff HA, Weiss C, Wirtz R, Balermpas P, Fokas E, Rödel C (2015) Human papillomavirus DNA load and p16INK4a expression predict for local control in patients with anal squamous cell carcinoma treated with chemoradiotherapy. Int J Cancer 136: 278–288. [DOI] [PubMed] [Google Scholar]
- Serup-Hansen E, Linnemann D, Skovrider-Ruminski W, Høgdall E, Geertsen PF, Havsteen H (2014) Human papillomavirus genotyping and p16 expression as prognostic factors for patients with American Joint Committee on Cancer stages I to III carcinoma of the anal canal. J Clin Oncol 32: 1812–1817. [DOI] [PubMed] [Google Scholar]
- Silverberg MJ, Lau B, Justice AC, Engels E, Gill MJ, Goedert JJ, Kirk GD, D'Souza G, Bosch RJ, Brooks JT, Napravnik S, Hessol NA, Jacobson LP, Kitahata MM, Klein MB, Moore RD, Rodriguez B, Rourke SB, Saag MS, Sterling TR, Gebo KA, Press N, Martin JN, Dubrow R North American AIDS Cohort Collaboration on Research and Design (NA-ACCORD) of IeDEA (2012) Risk of anal cancer in HIV-infected and HIV-uninfected individuals in North America. Clin Infect Dis 54: 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaglo BG, Tesfaye A, Halfdanarson TR, Meyer JE, Wang J, Gatalica Z, Reddy S, Arguello D, Boland PM (2015) Comprehensive multiplatform biomarker analysis of 199 anal squamous cell carcinomas. Oncotarget 6: 43594–43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer MK, Pitot HC, Liem A, Lee D, Kennedy GD, Lambert PF (2010) Rapamycin inhibits anal carcinogenesis in two preclinical animal models. Cancer Prev Res 3: 1542–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZJ, Zhang L, Zhang W, Hall B, Bian Y, Kulkarni AB (2013) Inhibition of mTOR reduces anal carcinogenesis in transgenic mouse model. PLoS One 8: e74888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeishi S, Nakayama KI (2014) Role of Fbxw7 in the maintenance of normal stem cells and cancer-initiating cells. Br J Cancer 111: 1054–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network (2014) Comprehensive molecular characterization of gastric cancer. Nature 513: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme N, Deron P, Van Roy N, Demetter P, Bols A, Van Dorpe J, Baert F, Van Laethem JL, Speleman F, Pauwels P, Peeters M (2010) Epidermal growth factor receptor and K-RAS status in two cohorts of squamous cell carcinomas. BMC Cancer 10: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra WH, Taube JM, Poeta ML, Begum S, Sidransky D, Koch WM (2008) Inverse relationship between human papillomavirus-16 infection and disruptive p53 gene mutations in squamous cell carcinoma of the head and neck. Clin Cancer Res 14: 366–369. [DOI] [PubMed] [Google Scholar]
- Wexler A, Berson AM, Goldstone SE, Waltzman R, Penzer J, Maisonet OG, McDermott B, Rescigno J (2008) Invasive anal squamous-cell carcinoma in the HIV-positive patient: outcome in the era of highly active antiretroviral therapy. Dis Colon Rectum 51: 73–81. [DOI] [PubMed] [Google Scholar]
- Yhim HY, Lee NR, Song EK, Kwak JY, Lee ST, Kim JH, Kim JS, Park HS, Chung IJ, Shim HJ, Hwang JE, Kim HR, Nam TK, Park MR, Shim H, Park HS, Kim HS, Yim CY (2011) The prognostic significance of tumor human papillomavirus status for patients with anal squamous cell carcinoma treated with combined chemoradiotherapy. Int J Cancer 129: 1752–1760. [DOI] [PubMed] [Google Scholar]
- Zelenay S, van der Veen AG, Böttcher JP, Snelgrove KJ, Rogers N, Acton SE, Chakravarty P, Girotti MR, Marais R, Quezada SA, Sahai E, Reis e Sousa C (2015) Cyclooxygenase-dependent tumor growth through evasion of immunity. Cell 162: 1257–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.