Abstract
Vertebrate sex‐determining mechanisms (SDMs) are triggered by the genotype (GSD), by temperature (TSD), or occasionally, by both. The causes and consequences of SDM diversity remain enigmatic. Theory predicts SDM effects on species diversification, and life‐span effects on SDM evolutionary turnover. Yet, evidence is conflicting in clades with labile SDMs, such as reptiles. Here, we investigate whether SDM is associated with diversification in turtles and lizards, and whether alterative factors, such as lifespan's effect on transition rates, could explain the relative prevalence of SDMs in turtles and lizards (including and excluding snakes). We assembled a comprehensive dataset of SDM states for squamates and turtles and leveraged large phylogenies for these two groups. We found no evidence that SDMs affect turtle, squamate, or lizard diversification. However, SDM transition rates differ between groups. In lizards TSD‐to‐GSD surpass GSD‐to‐TSD transitions, explaining the predominance of GSD lizards in nature. SDM transitions are fewer in turtles and the rates are similar to each other (TSD‐to‐GSD equals GSD‐to‐TSD), which, coupled with TSD ancestry, could explain TSD's predominance in turtles. These contrasting patterns can be explained by differences in life history. Namely, our data support the notion that in general, shorter lizard lifespan renders TSD detrimental favoring GSD evolution in squamates, whereas turtle longevity permits TSD retention. Thus, based on the macro‐evolutionary evidence we uncovered, we hypothesize that turtles and lizards followed different evolutionary trajectories with respect to SDM, likely mediated by differences in lifespan. Combined, our findings revealed a complex evolutionary interplay between SDMs and life histories that warrants further research that should make use of expanded datasets on unexamined taxa to enable more conclusive analyses.
Keywords: Evolution and natural selection; life span and longevity, life history; phenotypic plasticity; sex chromosomes; sexual development; speciation, extinction, and net diversification; temperature‐dependent (TSD) and genotypic (GSD) sex determination; turtles, lizards, snakes, and squamate reptiles; vertebrate speciation and extinction
Introduction
Vertebrate sex determination, or the commitment to a male or female developmental fate, can be triggered by an individual's genotype (genotypic sex determination [GSD]) or by environmental factors such as temperature (temperature‐dependent sex determination [TSD]; Bull 1983; Valenzuela and Lance 2004). GSD is found in all mammals, birds, and amphibians, while TSD exists in some fishes and in many reptiles (Bachtrog et al. 2014). Fewer examples of species with mixed mechanisms where GSD systems are overridden by certain temperatures have been documented in reptiles and fish (e.g., Shine et al. 2002; Yamamoto et al. 2014; Holleley et al. 2015). A clear explanation for the evolution of this diversity in sex‐determining mechanisms (SDM) remains elusive as our understanding of the causes and consequences of SDM turnover is inadequate.
Links between the evolution of SDMs and some important traits have been documented. For instance, transitions between SDM correlate with profound evolutionary changes, such as turtle genome reorganization (Valenzuela and Adams 2011), the evolution of viviparity in some marine reptiles (Organ et al. 2009), and adult sex ratio and demography of populations (Pipoly et al. 2015). Life history can also play a role in SDM turnover as theory predicts that lifespan influences whether TSD and GSD is adaptive, maladaptive, or neutral (Bull and Bulmer 1989; Valenzuela 2004a; Schwanz and Proulx 2008; Freedberg and Debenport 2014) and thus the tendency for TSD to be retained or replaced by GSD over evolutionary time. However, large‐scale empirical tests of these predictions are lacking.
Sex determination is expected to influence species diversification because it affects life history parameters (e.g., sexual development) and sex ratios and, consequently, population growth (via the effect of sex ratio on effective population size) that are linked to speciation and extinction (Bessa‐Gomes et al. 2004; Girondot et al. 2004; Valenzuela and Lance 2004; Bachtrog et al. 2014). Furthermore, sex determination strongly affects speciation in species with sex chromosomes as sex chromosomes often show the first signs of reproductive incompatibilities (Haldane 1922; Presgraves 2008; Elgvin et al. 2011). Yet, in groups where sex determination is evolutionarily labile, such as reptiles, evidence that sex determination is associated with diversification remains inconclusive as reports linking SDM and reptile speciation or extinction are conflicting. For instance, because TSD taxa are vulnerable to climate change, sex ratios could be skewed leading to extinction (Janzen 1994; Neuwald and Valenzuela 2011). TSD may thus lower diversification (i.e., speciation minus extinction) rates. However, TSD families appeared to have suffered lower extinction rates than GSD lineages during the climate change of the Cretaceous/Palaeogene transition (Silber et al. 2011; Escobedo‐Galvan and Gonzalez‐Salazar 2012). This observation suggests that TSD taxa may have been better adapted to past climate change than GSD taxa due to their phenotypic plasticity (Kallimanis 2009; Escobedo‐Galvan et al. 2011; Valenzuela et al. 2013). Conversely, transitions to GSD may be an adaptation to climate change to counter highly biased sex ratios in TSD taxa (Valenzuela and Adams 2011). Similarly, the transition to GSD was proposed to explain the adaptive radiation of extinct marine reptiles (Organ et al. 2009). Yet again, family‐level analyses found no relationship between diversification rates and the prevalence of GSD in Sauropsida (reptiles plus birds; Organ and Janes 2008). Thus, whether SDM is a causal driver or whether other correlated factors such as lifespan might be more important for diversification remains obscure.
Here, we take a phylogenetic, species‐level approach to examine the factors that influence the relative prevalence of SDMs in turtles and lizards. These two groups are ideal to address this question as TSD and GSD evolved multiple times independently within both groups (Valenzuela and Lance 2004). TSD is more common in turtles (78%) and GSD in lizards (86%) (Pokorná and Kratochvíl 2009; Valenzuela and Adams 2011). We test whether sex determination is associated with diversification rates, and whether alterative factors, such as lifespan's effect on transition rates, could explain the relative prevalence of SDMs in turtles and lizards. Based on existing data and analytical methods, we generate hypotheses to guide future research.
Methods
Data and phylogenies
An initial reptilian SDM database was obtained from (The Tree of Sex Consortium, 2014) and complemented with an extensive literature search (Ota et al. 1992; Gamble 2010; Pokorná et al. 2011, 2014; Trifonov et al. 2011; Badenhorst et al. 2013; Matsubara et al. 2013, 2014; Gamble et al. 2014, 2015; Koubová et al. 2014; Pokorna et al. 2014; Rovatsos et al. 2014a,b; Schmid et al. 2014; Sulandari et al. 2014). The resulting dataset contains information for 87 turtle and 303 lizard species (Table S1a) that have been studied across families (Table S1b). TSD and GSD were defined following (Valenzuela et al. 2003). To account for species that possess mixed sex‐determining mechanism where GSD and TSD coexist (termed “GSD+EE” by Valenzuela et al. 2003), and for species for which the evidence for TSD is weak (Harlow 2004; Valenzuela 2004b; Table S1a), we ran alternative analyses using one or the other SDM classification to test the robustness of our results. We used a dated phylogeny of 314 turtle species (Valenzuela and Adams 2011; ~96% of all estimated species by van Dijk et al. 2014), and one of 2899 lizard species (Pyron and Burbrink 2014) [~47% of estimated 6176 species (Uetz and Hosek 2015) representing all recognized families and subfamilies]. Of these, all 87 turtle species in the SDM dataset were present in the turtle phylogeny, and 279 lizard species with known SDM were represented in the phylogeny. Snakes, which share a single ZZ/ZW GSD system (Rovatsos et al. 2015), are nested within lizards. We thus analyzed an additional squamate dataset that included both lizards and snakes. Results did not differ from those found in lizards alone. Unless noted otherwise, because most of the methods that we used (see details below) do not account for unknown character states, the trees were pruned to include only taxa with known SDM.
Diversification analyses
Three different approaches, ranging from nonparametric, semiparametric, and fully parametric, were applied to test for SDM effect on lineages diversification rates. To assess statistical significance, each analysis was complemented with a parametric bootstrap approach to obtain the null distribution expected under a scenario depicting those of the empirical datasets.
First, we used the MacroCAIC method (Agapow and Isaac 2002) implemented in R package “caper” (Orme et al., 2013), to test for a correlation between SDM and species richness under the phylogenetically independent contrast paradigm (Felsenstein 1985). The method produces contrasts across a clade, including only contrasts with a minimal number of species (MNS), and linear regressions are then fitted to these contrasts (e.g., using MNS = 20, the mean size and richness value of all contrasts in the clade that have at least 20 species are calculated). We thus applied the method to turtles and lizards using MNS cutoffs of 10, 20, 30, and 40.
To control for potentially inflated false‐positive rate of MacroCAIC (Freckleton et al. 2008), we used a parametric bootstrapping approach to obtain the null distribution of the F‐statistic inferred by MacroCAIC expected for a neutral character with no effect on diversification patterns. Specifically, we simulated 1000 random distributions of neutral characters (assuming no effect on diversification) on the same empirically derived phylogenies of turtles and lizards. To obtain the simulated parameter values, we first estimated the two transition rates (GSD to TSD and TSD to GSD) according to a MK2 model (using make.mk2 function within the package diversitree [FitzJohn 2012]) and the root state set to TSD (which was inferred as the root state, see Results). We then simulated a binary trait along the tree according to the inferred transition rates (using sim.character function within the package diversitree [FitzJohn 2012]), starting, again, with the root state set to TSD. We then applied MacroCAIC on each simulated set and recorded the F‐statistic values. Finally, the empirically derived F‐statistic value was compared to the corresponding simulated distributions to obtain a P value according to the proportion of simulated values that are equal or greater than the observed value.
Second, we used STRAPP (“STructured RAte Permutations on Phylogenies”), a recently developed semiparametric test for detecting trait‐dependent diversification (Rabosky and Huang 2015). STRAPP first divides the input phylogeny into distinct diversification regimes, without considering the analyzed trait, as estimated by BAMM (Rabosky et al. 2013). It then treats these regimes as distinct data points to test for a trait effect on diversification. Unlike BiSSE (Maddison et al. 2007), which was recently shown to exhibit an elevated Type I error rate in the estimation of diversification rates (FitzJohn 2012; Rabosky and Goldberg 2015), STRAPP does not reconstruct character changes and diversification simultaneously. Consequently, STRAPP was shown to have low Type I error rates that are robust to combinations of character state frequencies and evolutionary rates, as well as to missing data, such that it is proposed for use even when character state data are available for a small fraction of the species in a phylogeny (Rabosky and Huang 2015). We note, however, that the improved lower rate of Type I errors in STRAPP is possibly accompanied by reduced sensitivity (Rabosky and Huang 2015). Thus, results using STRAPP would be conservative. We ran BAMM on each phylogeny, for 2,000,000 generations, keeping event data every 1000 steps. We then removed the first 10% steps as burn‐in and applied STRAPP using 10,000 permutations.
Finally, we conducted a third analysis of diversification as a function of SDM using the BiSSE modeling framework (Maddison et al. 2007; Data S1) and alleviated the potential problem of false‐positives with extensive simulations to test the robustness of the results following (FitzJohn 2012; Rabosky and Goldberg 2015). As detailed above, we simulated random distributions of neutral characters that have no effect on diversification on the same phylogenies and then tested whether the log‐likelihood difference for the competing models is more extreme for the real datasets than what could be expected by chance for neutral simulated traits (Rabosky and Goldberg 2015). For the BiSSE analyses, the full phylogenies containing 314 turtles, 2899 lizards, and 4161 squamates were used, but we also provide the results obtained using the pruned trees (Data S1).
Transition rates analyses
We tested whether the transition rate from GSD to TSD (q GT) is different than from TSD to GSD (q TG), using the MK2 model within the R package diversitree (FitzJohn 2012). We used maximum‐likelihood (ML) analysis to compare two nested models, one in which q GT is different from q TG, and one where the rates are equal. The LRT was then used to choose the best‐fit model.
As a second method, we tested for differences in transitions rates using a Markov chain Monte Carlo (MCMC) sampling approach (FitzJohn et al. 2009) to estimate the posterior probability distributions of the two transition rate parameters. We used an exponential prior distribution with mean set to 0.1. MCMC chains were run for 2000 steps with the first 25% discarded as burn‐in. To test whether transition rates differ between SDMs, we calculated the proportion of MCMC steps (i.e., the posterior probability, PP) in which q TG was higher than q GT. PP value above 0.975 or below 0.025 indicates a significant difference between the two rates. To examine whether the estimation of transition rates is affected by accounting for trait‐dependent diversification, we applied the BiSSE framework (Maddison et al. 2007) as implemented in diversitree version 0.9.7 (FitzJohn 2012). We used the “skeletal” tree approach (FitzJohn et al. 2009), which accounts for the sampling fraction of species in the phylogeny out of the total number of species in the clade (assuming an equal sampling fraction for both TSD and GSD). This method was used to estimate the speciation rates of lineages in states GSD and TSD and extinction rates, in addition to the transition rate parameters. Similar to the ML analysis of the MK2 model, we compared two nested models, one in which q GT is different from q TG, and one where the rates are equal (while speciation and extinction rates are constrained to be equal), using LRT. We note that BiSSE inference could be biased due to several characteristics of our data, including small sample size (turtles), high tip ratio bias (overprevalence of one observed character state), and incomplete sampling (lizards; Davis et al. 2013). Parametric bootstrap was used to test the false‐positive rate of each approach; namely, we simulated character states using the sim.character function (R package diversitree [FitzJohn 2012]), with the root state set to TSD. Here, we applied an equal transition rate model. The rate parameters used in the simulations were identical to those estimated from the real data using the constrained MK2 model. We then compared the empirically derived statistics (∆LL in the ML and PP in the MCMC analysis) to the corresponding simulated distributions to obtain a P value according to the proportion of simulated values that are equal or greater than the observed value.
We also tested the effect of missing data on the estimation of the transition rates in BiSSE, which could afflict the lizard dataset more strongly. For this, we simulated random trees with 1000 tips with equal speciation rates (λ = 0.1), no extinction, and varying transition rates (q01 = 0.1, q10 = 0.1, 0.05, 0.025) and carried out 100 simulations for each parameter combination (Data S1). In each simulation, the data were analyzed by BiSSE with 100, 25, or 5% of the state data while the rest were converted to missing data (Fig. S3).
Ancestral state reconstruction
We used a Markov model of trait change (the MK2 model within the R package diversitree (FitzJohn 2012)) to reconstruct the ancestral state at the root with the asr.marginal function within the R package diversitree (FitzJohn 2012). Because diversification could bias the inference of ancestral state reconstruction (King and Lee 2015), we also reconstructed the ancestral state using a BiSSE model (again, with the asr.marginal function), assuming equal speciation and extinction rates of GSD and TSD states (as the alternative model of unequal diversification rates was not supported; see Results).
Estimating shifts in lifespan in association with SDM
We examined the possible correlation of lifespan with SDM evolution using OUwie (Beaulieu et al. 2012). Lifespan data were obtained from (Tacutu et al. 2013; Scharf et al. 2015) and were log‐transformed. First, we assessed whether the rate of lifespan evolution differs between TSD and GSD lineages by comparing the fit of single‐ and two‐rate models of Brownian motion (BM) evolution. The best‐fit model was chosen using the LRT. The two‐parameter model requires partitioning of the tree into distinct regimes (i.e., a reconstructed phylogenetic history of GSD and TSD lineages, which was performed again with the asr.marginal function). Second, we used OUwie to test whether TSD and GSD lineages differ in their evolutionary trajectory (optimum value) for lifespan. Two nested models were compared using the LRT. In the first (OU1), there is a common optimum for GSD and TSD lineages while the second model (OUM) allows each SDM state to have a distinct optimum. We used AIC to compare the four models (BM1, BM2, OU1, and OUM).
Results
Diversification analyses
We explored whether differential diversification explains the contrasting abundance of SDMs in turtles and lizards in order to illuminate the causes and consequences of SDM evolution in these two lineages. Results were robust to the inclusion of snakes along with lizards in a squamate dataset during analyses, as they did not differ from the results obtained with lizards alone.
In turtles, standard MAcroCAIC procedures using the F‐statistic predicted a marginal statistical support for higher species richness in TSD clades than in GSD clades (P = 0.046, in MNS = 30 and 40; P > 0.25 for MNS = 10 and 20; Table 1). However, results from the parametric bootstrap showed that the observed differences are not significantly different than what was expected by chance in any group (P > 0.12, Table 1). In lizards and squamates, no significant difference between SDMs was observed under both statistical measures (P > 0.5 in all MNS values, Table 1). BAMM predicted 1, 7, and 16 rate shifts in diversification for turtles, lizards, and squamates, respectively (Table S3). The STRAPP method (Rabosky and Huang 2015) consistently detected no significant association between diversification rate estimates and SDM (P > 0.5 in all cases) (Table 2). Results were robust to using alternative SDM assignment for species with mixed or equivocal SDM (Tables 2 and S2).
Table 1.
MacroCAIC results
| Group | MNSa | r 2 | Slope | P value | Simulation P value |
|---|---|---|---|---|---|
| Turtles | 10 | −0.05 | 3.56 | 0.717 | 0.759 |
| 20 | 0.06 | 33.30 | 0.276 | 0.316 | |
| 30 | 0.71 | 66.90 | 0.046 | 0.148 | |
| 40 | 0.71 | 66.90 | 0.046 | 0.135 | |
| Lizards | 10 | −0.01 | −2.79 | 0.734 | 0.699 |
| 20 | −0.02 | −4.79 | 0.819 | 0.808 | |
| 30 | −0.03 | −3.48 | 0.879 | 0.875 | |
| 40 | −0.04 | −1.05 | 0.971 | 0.971 | |
| Squamates | 10 | −0.01 | −2.43 | 0.75 | 0.74 |
| 20 | −0.02 | −5.08 | 0.80 | 0.86 | |
| 30 | −0.02 | −3.84 | 0.86 | 0.85 | |
| 40 | −0.03 | −5.98 | 0.80 | 0.83 |
MNS, minimal number of species included for computing contrasts.
Table 2.
STRAPP results
| Group | P | P (alternative SDM assignment) |
|---|---|---|
| Turtles | 0.95 | 0.96 |
| Lizards | 0.67 | 0.73 |
| Squamates | 0.56 | 0.65 |
Consistent with these results, although initial analysis using the BiSSE approach (Maddison et al. 2007) identified differences by SDM in diversification rates in turtles, lizards, and squamates (Data S1), the results from our parametric bootstrapping procedure using neutral binary traits showed that, in all groups, the inferred differences using BiSSE are not significantly different than expected by chance (Data S1).
Transition rates analyses
Transitions from TSD to GSD were significantly more frequent than transitions from GSD to TSD in lizards and squamates, while in turtles transition rates were not significantly different, regardless of the analysis used (ML or Bayesian; Table 3). Unlike the diversification analyses, here the null model was rejected also when applying the parametric bootstrap approach (Table 3). Results were robust to accounting for diversification (using a BiSSE model) and to using alternative SDM assignment for species with mixed or equivocal SDM (Table S3). Our simulations also indicated that while greater sampling diminishes the variance in the transition rate estimates, the estimates are generally unbiased with the average transition rate estimate centered around to true value also when a large proportion of the tips do not contain trait data (Fig. S3).
Table 3.
Summary of transition rate parameters estimates using the MK2 model with both maximum‐likelihood and Bayesian (MCMC) methodologies and BiSSE for the turtles, lizards, and squamate datasets
| Group | Analysis | qGT | qTG | Significancea | Simulation P value |
|---|---|---|---|---|---|
| Turtles | Maximum likelihood | 8.6 × 10−07 | 0.0017 | 0.10 | 0.11 |
| MCMC | 0.0016 | 0.0021 | 0.73 | 0.09 | |
| BiSSE | 6.5 × 10−06 | 0.0018 | 0.10 | 0.14 | |
| Lizards | Maximum likelihood | 5.7 × 10−04 | 0.0119 | 2.6 × 10 −05 | <0.001 |
| MCMC | 9.5 × 10−04 | 0.0120 | 1 | <0.001 | |
| BiSSE | 5.9 × 10−04 | 0.0119 | 2.5 × 10−05 | <0.001 | |
| Squamates | Maximum likelihood | 3.0 × 10−04 | 0.0121 | 2.7 × 10 −07 | <0.001 |
| MCMC | 5.0 × 10−04 | 0.0124 | 1 | <0.001 | |
| BiSSE | 3.0 × 10−04 | 0.0122 | 2.7 × 10−07 | <0.001 |
Significance is estimated with likelihood ratio test for the maximum‐likelihood and BiSSE analyses; Significance of the MCMC analyses is estimated by calculating the proportion of MCMC steps (i.e., the posterior probability, PP) in which q TG was higher than q GT. PP value above 0.975 or below 0.025 indicates a significant difference between the two rates. Significant p‐values are denoted in bold.
Ancestral state reconstruction
Ancestral state reconstruction revealed that TSD is ancestral for both turtles and lizards (Fig. 1). Results were robust to the inclusion of snakes in the analyses (Fig. S1). The inference of ancestral TSD state was not affected when BiSSE was applied to account for species diversification or when using the alternative SDM classification for taxa with either mixed SDM or with weakly supported TSD (Fig. S1). Our results agree with previous reconstructions obtained with smaller datasets using ML in turtles (Valenzuela and Adams 2011) and maximum parsimony in squamates (Pokorná and Kratochvíl 2009). Altogether, our results suggests that the ancestral TSD state in both clades, combined with the asymmetry in lizard (but not turtle) transition rates (TSD‐to‐GSD surpass GSD‐to‐TSD), explains the observed prevalence of TSD turtles (via TSD retention) and GSD lizards (via TSD‐to‐GSD transitions) observed in nature.
Figure 1.
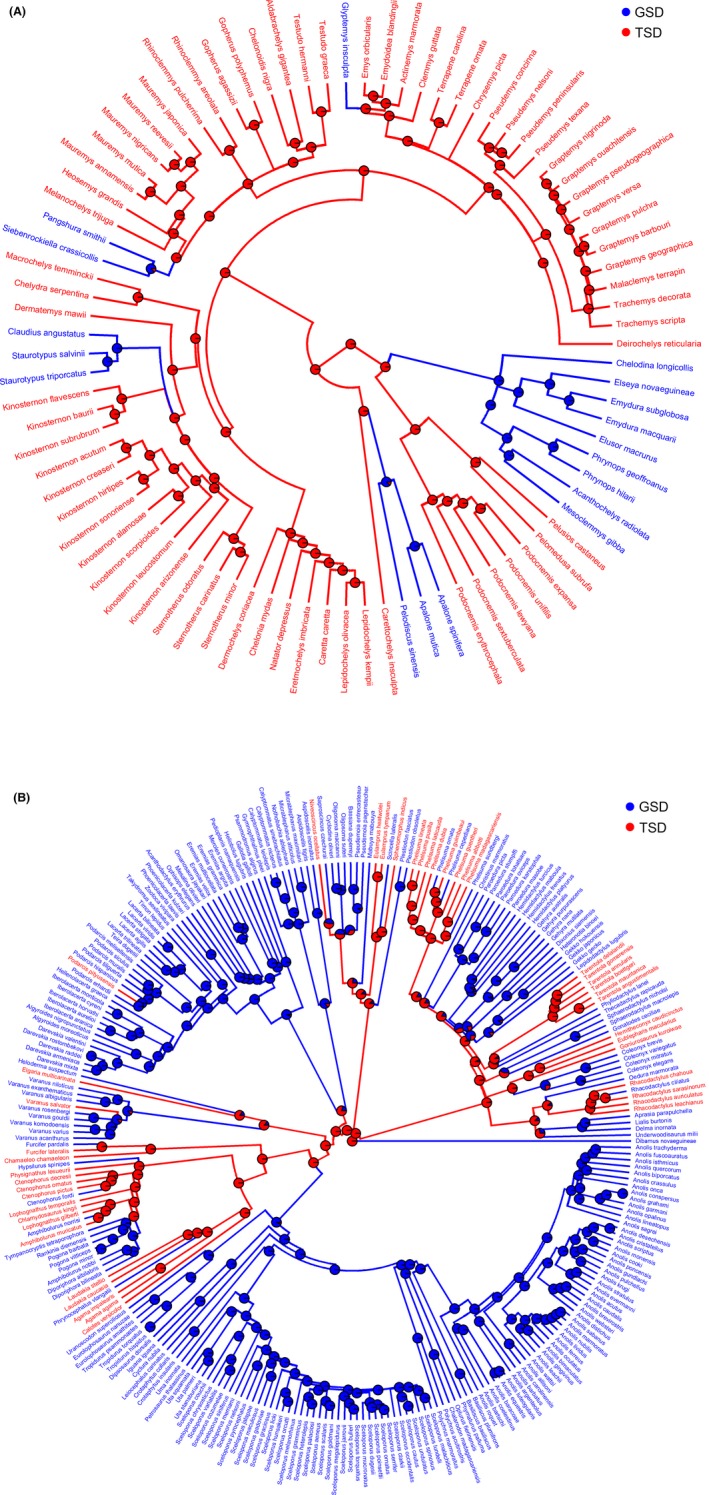
ML ancestral reconstruction of sex‐determining mechanisms in (A) turtles and (B) lizards. Pie charts denote the state probabilities at ancestral nodes.
Coevolution of SDM and longevity
To examine the possibility that differences in longevity might have influenced the evolutionary patterns of lineages in both groups, we modeled the evolution of lifespan with respect to SDM. The BM analysis showed that TSD turtles underwent greater lifespan evolution than their GSD counterparts, whereas the opposite was true in lizards where GSD lineages experienced greater lifespan evolution than TSD lizards (no differences were detected in squamates). In turtles, this greater evolutionary rate resulted in contrasting lifespan optima by SDM, whereas no differences were detected in lizards or squamates. Namely, we found that TSD turtles evolved toward greater longevity than GSD turtles (35.9 and 22.6 years, respectively; P = 0.018), whereas in lizards (and squamates), lifespan did not differ significantly between SDMs (Table 4; Fig. 2). When we compared the four models (BM1, BM2, OU1, and OUM) together, we found that in all datasets the OU models fit the data significantly better. Results were robust to using alternative SDM assignment for species with mixed or equivocal SDM (Table S3).
Table 4.
Log‐likelihood differences (∆LL) obtained between the single (BM1)‐ and two (BM2)‐rate Brownian motion models of evolution, and between the single (OU1) and two (OU2) optimums, as estimated for lifespan in turtles, lizards, and squamates. , , optimumGSD, and optimumTSD: estimated parameters for GSD and TSD lineages. Significant p‐values are denoted in bold
| Group | LogLiks BM1 | LogLiks BM2 | BM P‐valuea |
|
|
LogLiks OU1 | LogLiks OU2 | OU P‐valueb | OptimumGSD | OptimumTSD | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Turtles | −90.7 | −84.7 | 0.0005 | 0.361 | 2.8368 | −72.9 | −70.1 | 0.0181 | 22.6204 | 35.8875 | ||
| Lizards | −192.9 | −189.9 | 0.0135 | 3.3016 | 1.6455 | −163.8 | −162.8 | 0.1496 | 7.9104 | 10.0869 | ||
| Squamates | −246.4 | −245.2 | 0.1126 | 2.5472 | 1.6852 | −220.7 | −220.7 | 0.7911 | 9.5098 | 10.019 |
P‐value comparing the fit of a single‐ and two‐rate BM models based on the likelihood ratio test.
P‐value comparing the fit of a single and two OU models based on the likelihood ratio test.
Figure 2.
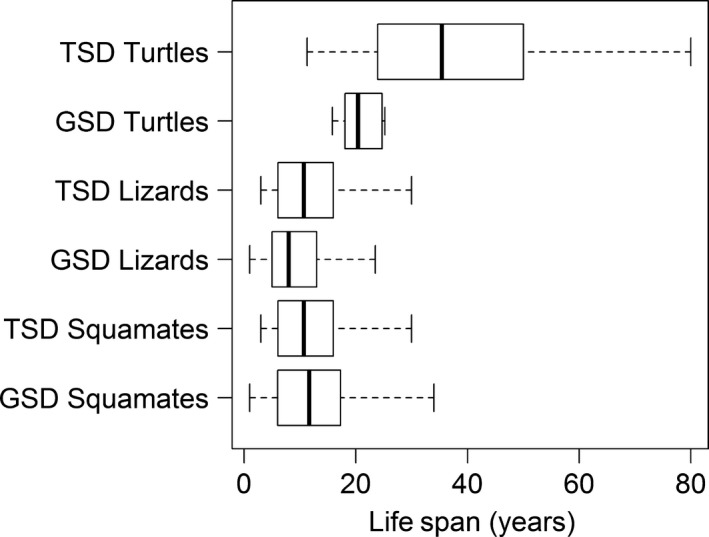
Box plots depicting longevity values for TSD and GSD turtles, lizards, and squamates. A significant difference between TSD and GSD lineages was detected in turtle longevity using the phylogenetic Ornstein–Uhlenbeck model (see text for details).
Discussion
SDM and diversification
Here, we examined the association of sex determination and species richness in reptiles by examining possible differences in diversification rates between TSD and GSD lineages, together with the transition rates between SDM states, in the two reptilian groups with labile sex determination – turtles and lizards (with and without the inclusion of snakes). We did not detect a significant influence of SDMs on turtle and lizard diversification using three alternative methods. Our findings were not affected by the inclusion of snakes in the analyses, nor by the few taxa with documented mixed SDM or by those with equivocal SDM assignment. Instead, the inferred transition rates between SDMs (which differed between the groups examined), coupled with TSD ancestry, could explain the predominance of GSD in lizards and TSD in turtles, without a significant difference between diversification rates.
This result is surprising because SDMs affect demographic and reproductive traits, and consequently, it is expected that SDM should influence species diversification and the prevalence of TSD or GSD found in nature. Indeed, SDMs affects population sex ratios, which in turn affect the effective population size, and, ultimately, population growth and the rate of loss of genetic variation (Hartl and Clark 1989) – all of which are factors underlying adaptation, speciation rates, and extinction probabilities (Bessa‐Gomes et al. 2004; Girondot et al. 2004). Our findings agree with previous family‐level analysis of reptiles and birds, which detected no association between speciation rates and SDMs (Organ and Janes 2008), although it should be noted that their results could also be due to the lower power of family‐level analyses (Organ and Janes 2008) and the fact that their study combined a family‐level tree with a model that assumes complete sampling (Organ and Janes 2008). The lack of evidence of an effect of SDM on diversification contradicts the expectation that TSD species should be more vulnerable to extinction as climate change can drastically bias TSD sex ratios (Janzen 1994; Neuwald and Valenzuela 2011), as well as counter reports that TSD reptilian families suffered lower extinction rates than GSD families during the Cretaceous/Palaeogene transition (Silber et al. 2011; Escobedo‐Galvan and Gonzalez‐Salazar 2012).
The observed lack of support for a relationship between SDM and diversification could also be due to the sparsity of the data (Table S1b) or the limitations of the methods (Freckleton et al. 2008; Maddison and FitzJohn 2015; Rabosky and Goldberg 2015). However, it should be noted that the same independence between SDM and diversification was detected here with three alternative methods (MacroCAIC, STRAPP, and the permutation analyses to test BiSSE results). Yet, all methods employed intrinsically assume a homogenous evolutionary process for both transition and diversification rates. That is, the model is time homogenous and similar across different clades of the phylogeny. This assumption is rather questionable for the large clades analyzed here. However, the sparsity of the data did not allow us to explore more sophisticated models that require a larger number of parameters. Taken together, while our results indicate that asymmetries in transition rates rather than diversification rates lead to the differential SDM diversity observed in squamates, the jury awaits for improved and well‐vetted analytical methods plus the collection of additional information on sex determination in reptiles. Fortunately, SDM data are growing at an accelerated pace thanks to the use of a variety of molecular techniques to complement classic incubation experiments (Ota et al. 1992; Gamble 2010; Pokorná et al. 2011, 2014; Trifonov et al. 2011; Badenhorst et al. 2013; Matsubara et al. 2013, 2014; Mu et al. 2013; Gamble et al. 2014, 2015; Koubová et al. 2014; Pokorna et al. 2014; Rovatsos et al. 2014a,b; Schmid et al. 2014; Sulandari et al. 2014; Valenzuela et al. 2014; Montiel et al. 2016). Extensive research over 50 years has uncovered SDM information in all turtle families (except Platysternidae) for at least 1 species per family, whereas 9 of 37 lizard families remain unstudied. The coverage varies across families, from <10–68% for turtle families that are not monotypic, compared to 1–50% in lizards (Table S1b). Notably, the existing data and methods permit some important insights into why GSD is more prevalent than TSD in squamates while TSD turtles abound over GSD turtles, and these working hypotheses should foster even further research in this area.
SDM and species richness
The contrasting relative prevalence of TSD and GSD in turtles and lizards (as well as squamates) can be explained by their differences in the transition rates between SDMs; namely, in turtles, transition rates between TSD and GSD were similar, such that the higher abundance of TSD turtles derives from the greater retention of the ancestral TSD condition. In contrast, lizards (including and excluding snakes) shifted from TSD into GSD much more often than from GSD to TSD, resulting in greater abundance of GSD lizards overall (and all snakes retain a ZZ/ZW GSD system that evolved at their split from lizards (Rovatsos et al. 2015). In general, a lack of difference between transition rates in turtles could be due to a relatively small number of transitions overall or to an overall short time lineages have been in the GSD state (i.e., “lower rate” vs. “lower opportunity” to transition).
Longevity, sex determination, and diversification
Why did lizards give up TSD so readily while turtles thrived with TSD? We hypothesize that differences in individual longevity between these two clades are key, because lifespan influences whether TSD is adaptive, maladaptive, or neutral (Bull and Bulmer 1989; Valenzuela and Lance 2004). Turtles live, on average, over three times as long (~30 years) as lizards (~9 years) (Tacutu et al. 2013; Scharf et al. 2015; Fig. 3). This may even be an underestimate because lizard data include an overrepresentation of large species that tend to be longer‐lived (Tacutu et al. 2013). These differences in lifespan are relevant because TSD populations are vulnerable to producing drastically biased sex ratios in any given season, which may imperil population survival of short‐lived species, while longevity lessens this detrimental effect by providing reproductive assurance (Girondot and Pieau 1996; Valenzuela and Lance 2004; Grayson et al. 2014). Namely, in theory, a population of an annual species that produces a single sex during a drastically cold or warm year could go extinct in a single generation, while long‐lived TSD taxa would more effectively average sex ratios over multiple years (Girondot and Pieau 1996) because by the time individuals reach maturity, and during their multiple reproductive years, potential mates would have been recruited into the population. Additionally, TSD may lead to the accumulation of deleterious mutations particularly under shorter lifespan (because biased sex ratios produced by TSD reduce effective population sizes; Freedberg and Debenport 2014), and in combination with environmental fluctuation regimes (Schwanz and Proulx 2008). GSD would thus be expected to persist more frequently in short‐lived lineages, while TSD would be expected to persist more readily in longer‐lived lineages. Under such circumstances, heritable genetic variation underlying sexual development of TSD species (Sarre et al. 2011; Valenzuela et al. 2013) might enable the persistence and adaptation of long‐lived taxa during changing climatic conditions. This could explain the persistence of TSD turtles in ways that may have been precluded for many shorter‐lived lizards. The hypothesis that longevity mediates TSD retention was supported when we tested whether TSD and GSD lineages differ in their evolutionary trajectory for lifespan and found that lifespan of TSD turtles evolved toward greater values (and are consequently longer‐lived) than their GSD counterparts (lizards showed a similar tendency but these differences were not significant [Table 4]). Concordant with this notion, the other reptilian lineages that have only TSD are also long‐lived (Fig. 3), that is, crocodilians and the rhynchocephalian (tuatara). We note that if the pace of climate change is too rapid – as occurs today (Diffenbaugh and Field 2013) – adaptive responses such as those inferred here may be limited, particularly for the many TSD taxa that are already endangered and suffer from small population sizes and drastic habitat degradation (van Dijk et al. 2014).
Figure 3.
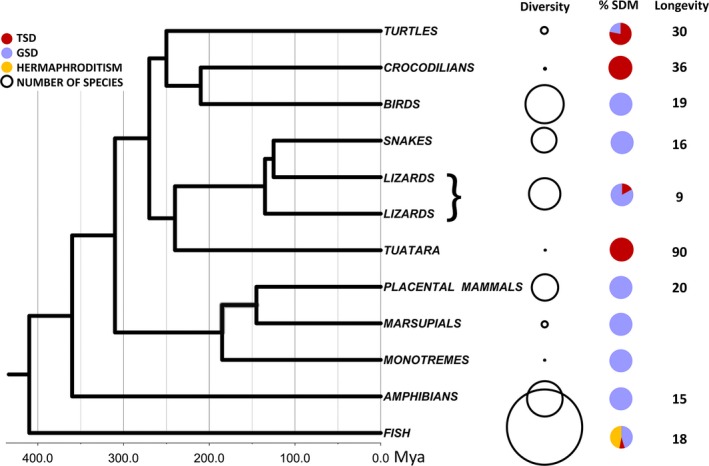
Species diversity, sex determination and longevity of extant vertebrates. Species numbers per lineage vary from 1 (tuatara) to >33,000 (fish) (Eschmeyer and Fong 2014; Frost 2014; Hay et al. 2010; Uetz and Hosek 2015; van Dijk et al. 2011). Sex determination from sources cited in the text. Divergence times as per Chiari et al. (2012) and Jones et al. (2013). Average longevity from (Tacutu et al. 2013) in years (Data S1). Open circle size is proportional to species number per clade. Values are presented for lizards overall, despite the paraphyly with snakes falling within the lizard clade.
Then, how could TSD evolve or persist in short‐lived taxa, such as some lizards and fish? TSD must be much more adaptive than GSD in short‐lived taxa to compensate for the costs associated with fluctuating sex ratios (e.g., sensu Charnov and Bull 1977). Under the Charnov–Bull model, TSD is adaptive and would be favored over GSD, if (1) the environment is patchy (in space or time) and unpredictable by the developing offspring or their parents, (2) the temperature (or a correlated variable) experienced during early development confers differential lifetime fitness to each sex, and (3) individuals mate at random among patches (Charnov and Bull 1977). This model was elegantly demonstrated to apply for Amphibolurus muricatus lizards (Warner and Shine 2008), and for Menidia menidia fish (Conover and Heins 1987). In both these short‐lived vertebrates, spring/summer temperature (when sexual development occurs) is positively correlated with the time available for growth before winter, which determines adult size. In both cases, female fitness (via fecundity gains) increases with body size more than in males (Conover and Heins 1987; Warner and Shine 2008). Females of both species develop at colder temperatures naturally experienced early in the reproductive season, grow for a longer time and attain larger adult sizes, while males develop at warmer temperatures experienced later in the year, experience a shorter growing season and, consequently, attain smaller adult sizes (Conover and Heins 1987; Warner and Shine 2008), in close accord with the Charnov–Bull model. Thus, given the right conditions, TSD can evolve or persist in shorter‐lived taxa. Similar advantages may also exist in TSD turtles (Shine 1999; Valenzuela and Lance 2004), and such adaptive significance would only reinforce the persistence of TSD in chelonians.
In summary, our data support the hypothesis that diversification was not affected by SDM and that the high transition rates from TSD to GSD in lizards accounts for the high abundance of GSD lineages in this group, while TSD prevalence in turtles seems to reflect the retention of the ancestral character state. We hypothesize that turtle longevity helps them cope with fluctuating sex ratios. In contrast, we propose that the general shorter lifespan of lizards hinder TSD persistence (except under strong selection) favoring the transitions to GSD and contributing to their overall prevalence we observe in nature. Thus, our results underscore that turtles and lizards appear to have followed different evolutionary trajectories with respect to SDM likely mediated by differences in life‐history traits. An urgent need remains to expand the existing SDM and life‐history information of the many reptiles that remained unstudied, so as to enable more conclusive analyses. Our work contributes to ongoing efforts to study phenotypic macroevolution in a comprehensive manner to illuminate the relative success and demise of distinct branches of the tree of life, their causes and consequences, and their potential to adapt to a changing world.
Conflict of Interest
None declared.
Supporting information
Figure S1. ML ancestral reconstruction of sex‐determining mechanisms in (A) squamates, and using the alternative SDM classification in (B) squamates and (C) lizards.
Table S1A. Dataset used in this study.
Table S1B. Taxonomic coverage of turtle and squamate families used in this study.
Data S1. Results using alternative SDM assignment for species with mixed or equivocal SDM as listed in Table S1:
Table S2. MacroCAIC results using alternative SDM assignment.
Table S3. BAMM estimation for the number of rate shifts in diversification. The number of rates shifts with the highest probability in each group is marked in bold.
Table S4. Summary of transition rate parameters estimates using the MK2 model with both Maximum Likelihood and Bayesian (MCMC) methodologies and BiSSE for the turtles, lizards, and squamate data sets using the alternative SDM assignment.
Table S5. Log likelihood differences (ΔLL) obtained between the single (BM1) and two rate (BM2) Brownian motion models of evolution, and between the single (OU1) and two (OU2) optimums, as estimated for lifepan in turtles, lizards and squamates. σ2GSD and σ2TSD, OptimumGSD, and OptimumTSD: estimated parameters for GSD and TSD lineages using the alternative SDM assignment.
Data S2. BiSSE Analyses.
Acknowledgments
We thank members of the Tree of Sex Working Group, and Sally Otto and Emma Goldberg, in particular, as well as members of the Valenzuela, Adams and Serb labs at Iowa State University, for comments. Funding was provided in part by National Evolutionary Synthesis Center to the Tree of Sex Consortium Working Group, Marie Curie Reintegration Grant 2011‐293878 to I. Mayrose, Postdoctoral fellowship from the Edmond J. Safra Center for Bioinformatics to N. Sabath, and USA National Science Foundation grant MCB 1244355 to N. Valenzuela.
References
- Agapow, P. M. , and Isaac N. J. B.. 2002. MacroCAIC: revealing correlates of species richness by comparative analysis. Divers. Distrib. 8:41–43. [Google Scholar]
- Bachtrog, D. , Mank J. E., Peichel C. L., Kirkpatrick M., Otto S. P., Ashman T.‐L., et al. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12:e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst, D. , Stanyon R., Engstrom T., and Valenzuela N.. 2013. A ZZ/ZW microchromosome system in the spiny softshell turtle, Apalone spinifera, reveals an intriguing sex chromosome conservation in Trionychidae. Chromosome Res. 21:137–147. [DOI] [PubMed] [Google Scholar]
- Beaulieu, J. M. , Jhwueng D.‐C., Boettiger C., and O'Meara B. C.. 2012. Modeling stabilizing selection: expanding the Ornstein–Uhlenbeck model of adaptive evolution. Evolution 66:2369–2383. [DOI] [PubMed] [Google Scholar]
- Bessa‐Gomes, C. , Legendre S., and Clobert J.. 2004. Allee effects, mating systems and the extinction risk in populations with two sexes. Ecol. Lett. 7:802–812. [Google Scholar]
- Bull, J. J. 1983. Evolution of sex determining mechanisms. Benjamin/Cummings, Menlo Park, CA. [Google Scholar]
- Bull, J. J. , and Bulmer M. G.. 1989. Longevity enhances selection of environmental sex determination. Heredity 63:315–320. [DOI] [PubMed] [Google Scholar]
- Charnov, E. L. , and Bull J. J.. 1977. When is sex environmentally determined? Nature 266:828–830. [DOI] [PubMed] [Google Scholar]
- Chiari, Y. , Cahais V., Galtier N., and Delsuc F.. 2012. Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria). BMC Biol. 10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover, D. O. , and Heins S. W.. 1987. Adaptive variation in environmental and genetic sex determination in a fish. Nature 326:497–498. [DOI] [PubMed] [Google Scholar]
- Davis, M. P. , Midford P. E., and Maddison W.. 2013. Exploring power and parameter estimation of the BiSSE method for analyzing species diversification. BMC Evol. Biol. 13Available at http://bmcevolbiol.biomedcentral.com/articles/10.1186/1471-2148-13-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffenbaugh, N. S. , and Field C. B.. 2013. Changes in ecologically critical terrestrial climate conditions. Science 341:486–492. [DOI] [PubMed] [Google Scholar]
- Elgvin, T. O. , Hermansen J. S., Fijarczyk A., Bonnet T., Borge T., Saether S. A., et al. 2011. Hybrid speciation in sparrows II: a role for sex chromosomes? Mol. Ecol. 20:3823–3837. [DOI] [PubMed] [Google Scholar]
- Eschmeyer, W. N. , and Fong J. D.. 2014. Catalog of fishes. California Academy of Sciences; Available at http://research.calacademy.org/redirect?url=http://researcharchive.calacademy.org/research/Ichthyology/catalog/fishcatmain.asp (accessed 17 January 2014). [Google Scholar]
- Escobedo‐Galvan, A. H. , and Gonzalez‐Salazar C.. 2012. Survival and extinction of sex‐determining mechanisms in Cretaceous tetrapods. Cretac. Res. 36:116–118. [Google Scholar]
- Escobedo‐Galvan, A. H. , Gonzalez‐Salazar C., Lopez‐Alcaide S., Baruch Arroyo‐Pena V., and Martinez‐Meyer E.. 2011. Will all species with temperature‐dependent sex determination respond the same way to climate change? A reply to Kallimanis (2010). Oikos 120:795–799. [Google Scholar]
- Felsenstein, J. 1985. Phylogenies and the comparative method. Am. Nat. 125:1–15. [Google Scholar]
- FitzJohn, R. G. 2012. Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 3:1084–1092. [Google Scholar]
- FitzJohn, R. G. , Maddison W. P., and Otto S. P.. 2009. Estimating trait‐dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58:595–611. [DOI] [PubMed] [Google Scholar]
- Freckleton, R. P. , Phillimore A. B., and Pagel M.. 2008. Relating traits to diversification: a simple test. Am. Nat. 172:102–115. [DOI] [PubMed] [Google Scholar]
- Freedberg, S. , and Debenport S. J.. 2014. Weakened purifying selection leads to elevated mutation load under environmental sex determination. J. Evol. Biol. 27:643–652. [DOI] [PubMed] [Google Scholar]
- Frost, D. R. 2014. Amphibian species of the world: an online reference. Version 6.0, American Museum of Natural History, New York, NY: Available at http://research.amnh.org/herpetology/amphibia/index.html (accessed January 2014). [Google Scholar]
- Gamble, T. 2010. A review of sex determining mechanisms in geckos (Gekkota: Squamata). Sex. Dev. 4:88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble, T. , Geneva A. J., Glor R. E., and Zarkower D.. 2014. Anolis sex chromosomes are derived from a single ancestral pair. Evolution 68:1027–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble, T. , Coryell J., Ezaz T., Lynch J., Scantlebury D. P., and Zarkower D.. 2015. Restriction site‐associated DNA sequencing (RAD‐seq) reveals an extraordinary number of transitions among gecko sex‐determining systems. Mol. Biol. Evol. 32:1296–1309. [DOI] [PubMed] [Google Scholar]
- Girondot, M. , and Pieau C.. 1996. On the limits of age‐structured models for the maintenance of environmental sex determination in crocodilians. Annal. Sci. Nat. 17:85–97. [Google Scholar]
- Girondot, M. , Delmas V., Rivalan P., Courchamp F., Prévot‐Julliard A. C., and Godfrey M. H.. 2004. Implications of temperature‐dependent sex determination for population dynamics Pp. 148–155 in Valenzuela N. and Lance V. A., eds. Temperature dependent sex determination in vertebrates. Smithsonian Books, Washington, DC. [Google Scholar]
- Grayson, K. L. , Mitchell N. J., Monks J. M., Keall S. N., Wilson J. N., and Nelson N. J.. 2014. Sex ratio bias and extinction risk in an isolated population of tuatara (Sphenodon punctatus). PLoS ONE 9:e94214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane, J. B. S. 1922. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 12:101–109. [Google Scholar]
- Harlow, P. S. 2004. Temperature‐dependent sex determination in lizards Pp. 42–52 in Valenzuelaand N. and Lance V. A., eds. Temperature dependent sex determination in vertebrates. Smithsonian Books, Washington, DC. [Google Scholar]
- Hartl, D. L. , and Clark A. G.. 1989. Principles of population genetics. 2nd ed Sinauer Associates Inc, Sunderland, MA. [Google Scholar]
- Hay, J. M. , Sarre S. D., Lambert D. M., Allendorf F. W., and Daugherty C. H.. 2010. Genetic diversity and taxonomy: a reassessment of species designation in tuatara (Sphenodon: Reptilia). Conserv. Genet. 11:1063–1081. [Google Scholar]
- Holleley, C. E. , O'Meally D., Sarre S. D., Marshall Graves J. A., Ezaz T., Matsubara K., et al. 2015. Sex reversal triggers the rapid transition from genetic to temperature‐dependent sex. Nature 523:79–82. [DOI] [PubMed] [Google Scholar]
- Janzen, F. J. 1994. Climate change and temperature‐dependent sex determination in reptiles. Proc. Natl Acad. Sci. USA 91:7487–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M. E. H. , Anderson C. L., Hipsley C. A., Mueller J., Evans S. E., and Schoch R. R.. 2013. Integration of molecules and new fossils supports a Triassic origin for Lepidosauria (lizards, snakes, and tuatara). BMC Evol. Biol. 13:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallimanis, A. S. 2009. Temperature dependent sex determination and climate change. Oikos 119:197–200. [Google Scholar]
- King, B. , and Lee M. S.. 2015. Ancestral state reconstruction, rate heterogeneity, and the evolution of reptile viviparity. Syst. Biol. 64:532–544. [DOI] [PubMed] [Google Scholar]
- Koubová, M. , Pokorná M., Rovatsos M., Farkačová K., Altmanová M., and Kratochvíl L.. 2014. Sex determination in Madagascar geckos of the genus Paroedura (Squamata: Gekkonidae): are differentiated sex chromosomes indeed so evolutionary stable? Chromosome Res. 22:441–452. [DOI] [PubMed] [Google Scholar]
- Maddison, W. P. , and FitzJohn R. G.. 2015. The unsolved challenge to phylogenetic correlation tests for categorical characters. Syst. Biol. 64:127–136. [DOI] [PubMed] [Google Scholar]
- Maddison, W. P. , Midford P. E., and Otto S. P.. 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol. 56:701–710. [DOI] [PubMed] [Google Scholar]
- Matsubara, K. , Knopp T., Sarre S. D., Georges A., and Ezaz T.. 2013. Karyotypic analysis and FISH mapping of microsatellite motifs reveal highly differentiated XX/XY sex chromosomes in the pink‐tailed worm‐lizard (Aprasia parapulchella, Pygopodidae, Squamata). Mol. Cytogenet. 6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, K. , Sarre S. D., Georges A., Matsuda Y., Graves J. A. M., and Ezaz T.. 2014. Highly differentiated ZW sex microchromosomes in the australian Varanus species evolved through rapid amplification of repetitive sequences. PLoS ONE 9:e95226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel, E. E. , Badenhorst D., Tamplin J., Burke R., and Valenzuela N.. 2016. Discovery of youngest sex chromosomes reveals first case of convergent co‐option of ancestral autosomes in turtles. Chromosoma. doi:10.1007/s00412‐016‐0576‐7. [e‐pub ahead of print] [DOI] [PubMed] [Google Scholar]
- Mu, X. , Wen J., Guo M., Wang J., Li G., Wang Z., et al. 2013. Retinoic acid derived from the fetal ovary initiates meiosis in mouse germ cells. J. Cell. Physiol. 228:627–639. [DOI] [PubMed] [Google Scholar]
- Neuwald, J. L. , and Valenzuela N.. 2011. The lesser known challenge of climate change: thermal variance and sex‐reversal in vertebrates with temperature‐dependent sex determination. PLoS ONE 6:e18117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme, D. 2013. The caper package: comparative analysis of phylogenetics and evolution in R. R package version 5.
- Organ, C. L. , and Janes D. E.. 2008. Evolution of sex chromosomes in Sauropsida. Integr. Comp. Biol. 48:512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ, C. L. , Janes D. E., Meade A., and Pagel M.. 2009. Genotypic sex determination enabled adaptive radiations of extinct marine reptiles. Nature 461:389–392. [DOI] [PubMed] [Google Scholar]
- Ota, H. , Hikida T., Matsui M., and Mori A.. 1992. Karyotypes of two species of the genus Cyrtodactylus (Squamata: Gekkonidae) from Sarawak, Malaysia. Caryologia 45:43–49. [Google Scholar]
- Pipoly, I. , Bokony V., Kirkpatrick M., Donald P. F., Szekely T., and Liker A.. 2015. The genetic sex‐determination system predicts adult sex ratios in tetrapods. Nature 527:91–94. [DOI] [PubMed] [Google Scholar]
- Pokorná, M. , and Kratochvíl L.. 2009. Phylogeny of sex‐determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool. J. Linn. Soc. 156:168–183. [Google Scholar]
- Pokorna, M. , Rens W., Rovatsos M., and Kratochvil L.. 2014. A ZZ/ZW sex chromosome system in the thick‐tailed gecko (Underwoodisaurus milii; Squamata: Gekkota: Carphodactylidae), a member of the ancient gecko lineage. Cytogenet. Genome Res. 142:190–196. [DOI] [PubMed] [Google Scholar]
- Pokorná, M. , Giovannotti M., Kratochvíl L., Kasai F., Trifonov V. A., O'Brien P. C. M., et al. 2011. Strong conservation of the bird Z chromosome in reptilian genomes is revealed by comparative painting despite 275 million years divergence. Chromosoma 120:455–468. [DOI] [PubMed] [Google Scholar]
- Pokorná, M. J. , Rovatsos M., and Kratochvíl L.. 2014. Sex chromosomes and karyotype of the (nearly) mythical creature, the gila monster, Heloderma suspectum (squamata: Helodermatidae). PLoS ONE 9:e104716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C. 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron, R. A. , and Burbrink F. T.. 2014. Early origin of viviparity and multiple reversions to oviparity in squamate reptiles. Ecol. Lett. 17:13–21. [DOI] [PubMed] [Google Scholar]
- Rabosky, D. L. , and Goldberg E. E.. 2015. Model inadequacy and mistaken inferences of trait‐dependent speciation. Syst. Biol. 64:340–355. [DOI] [PubMed] [Google Scholar]
- Rabosky, D. L. , and Huang H.. 2015. A robust semi‐parametric test for detecting trait‐dependent diversification. Syst. Biol. 65:181–193. [DOI] [PubMed] [Google Scholar]
- Rabosky, Daniel L. , et al. 2013. “Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation.” Nature communications 4. [DOI] [PubMed] [Google Scholar]
- Rovatsos, M. , Altmanova M., Pokorna M., and Kratochvil L.. 2014a. Conserved sex chromosomes across adaptively radiated Anolis lizards. Evolution 68:2079–2085. [DOI] [PubMed] [Google Scholar]
- Rovatsos, M. , Pokorná M., Altmanová M., and Kratochvíl L.. 2014b. Cretaceous park of sex determination: sex chromosomes are conserved across iguanas. Biol. Lett. 10:20131093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovatsos, M. , Vukić J., Lymberakis P., and Kratochvíl L.. 2015. Evolutionary stability of sex chromosomes in snakes. Proc. R. Soc. B Biol. Sci. 282:20151992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarre, S. D. , Ezaz T., and Georges A.. 2011. Transitions between sex‐determining systems in reptiles and amphibians. Annu. Rev. Genomics Hum. Genet. 12:391–406. [DOI] [PubMed] [Google Scholar]
- Scharf, I. , Feldman A., Novosolov M., Pincheira‐Donoso D., Das I., Bohm M., et al. 2015. Late bloomers and baby boomers: an analysis of ecological drivers of squamate longevity. Glob. Ecol. Biogeogr. 24:396–405. [Google Scholar]
- Schmid, M. , Steinlein C., Haaf T., and Mijares‐Urrutia A.. 2014. Nascent ZW sex chromosomes in Thecadactylus rapicauda (Reptilia, Squamata, Phyllodactylidae). Cytogenet. Genome Res. 143:259–267. [DOI] [PubMed] [Google Scholar]
- Schwanz, L. E. , and Proulx S. R.. 2008. Mutual information reveals variation in temperature‐dependent sex determination in response to environmental fluctuation, lifespan and selection. Proc. R. Soc. B Biol. Sci. 275:2441–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine, R. 1999. Why is sex determined by nest temperature in many reptiles? Trends Ecol. Evol. 14:186–189. [DOI] [PubMed] [Google Scholar]
- Shine, R. , Elphick M. J., and Donnellan S.. 2002. Co‐occurrence of multiple, supposedly incompatible modes of sex determination in a lizard population. Ecol. Lett. 5:486–489. [Google Scholar]
- Silber, S. , Geisler J. H., and Bolortsetseg M.. 2011. Unexpected resilience of species with temperature‐dependent sex determination at the Cretaceous‐Palaeogene boundary. Biol. Lett. 7:295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulandari, S. , Zein M. S. A., Arida E. A., and Hamidy A.. 2014. Molecular sex determination of captive Komodo dragons (Varanus komodoensis) at Gembira Loka zoo, Surabaya zoo, and Ragunan zoo, Indonesia. Hayati J. Biosci. 21:65–75. [Google Scholar]
- Tacutu, R. , Craig T., Budovsky A., Wuttke D., Lehmann G., Taranukha D., et al. 2013. Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 41:D1027–D1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tree of Sex Consortium . 2014. Tree of Sex: a datbase of sexual systems. Sci. Data 1:140015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov, V. A. , Giovannotti M., O'Brien P. C. M., Wallduck M., Lovell F., Rens W., et al. 2011. Chromosomal evolution in Gekkonidae. I. Chromosome painting between Gekko and Hemidactylus species reveals phylogenetic relationships within the group. Chromosome Res. 19:843–855. [DOI] [PubMed] [Google Scholar]
- Uetz, P. , and Hosek J.. 2015. The Reptile Database. Available at http://www.reptile-database.org/ (accessed 20 March 2016).
- Valenzuela, N. 2004a. Evolution and maintenance of temperature‐dependent sex determination Pp. 131–147 in Valenzuela N. and Lance V. A., eds. Temperature dependent sex determination in vertebrates. Smithsonian Books, Washington, DC. [Google Scholar]
- Valenzuela, N. 2004b. Temperature‐dependent sex determination Pp. 211–227 in Deeming D. C., ed. Reptilian incubation: environment & behaviour. Nottingham University Press, Nottingham, U.K. [Google Scholar]
- Valenzuela, N. , and Adams D. C.. 2011. Chromosome number and sex determination co‐evolve in turtles. Evolution 65:1808–1813. [DOI] [PubMed] [Google Scholar]
- Valenzuela N., and Lance V. A., eds. 2004. Temperature dependent sex determination in vertebrates. Smithsonian Books, Washington, DC. [Google Scholar]
- Valenzuela, N. , Adams D. C., and Janzen F. J.. 2003. Pattern does not equal process: exactly when is sex environmentally determined? Am. Nat. 161:676–683. [DOI] [PubMed] [Google Scholar]
- Valenzuela, N. , Neuwald J. L., and Literman R.. 2013. Transcriptional evolution underlying vertebrate sexual development. Dev. Dyn. 242:307–319. [DOI] [PubMed] [Google Scholar]
- Valenzuela, N. , Badenhorst D., Montiel Jiménez E. E., and Literman R.. 2014. Molecular cytogenetic search for cryptic sex chromosomes in painted turtles Chrysemys picta . Cytogenet. Genome Res. 144:39–46. [DOI] [PubMed] [Google Scholar]
- van Dijk, P. P. , Iverson J. B., Shaffer H. B., Bour R., Rhodin A. G. J., and Turtle Taxonomy Working Group . 2011. Turtles of the world, 2011 update: annotated checklist of taxonomy, synonymy, distribution, and conservation status Pp. 000.165–000.241 in Rhodin A. G. J., Pritchard P. C. H., van Dijk P. P., Saumure R. A., Buhlmann K. A., Iverson J. B. and Mittermeier R. A., eds. Conservation biology of freshwater turtles and tortoises: a compilation project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group. Chelonian Research Monographs, Lunenburg, MA. [Google Scholar]
- van Dijk, P. P. , Iverson J. B., Rhodin A. G. J., Shaffer H. B., and Baur B.. 2014. Turtles of the world Pp. 329–479. 7th ed Vol. 5 Annotated checklist of taxonomy, synonymy, distribution with maps, and conservation status. Chelonian Research Monographs, Lunenburg, MA. [Google Scholar]
- Warner, D. A. , and Shine R.. 2008. The adaptive significance of temperature‐dependent sex determination in a reptile. Nature 451:566–568. [DOI] [PubMed] [Google Scholar]
- Yamamoto, Y. , Zhang Y., Sarida M., Hattori R. S., and Struessmann C. A.. 2014. Coexistence of genotypic and temperature‐dependent sex determination in pejerrey Odontesthes bonariensis . PLoS ONE 9:e102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. ML ancestral reconstruction of sex‐determining mechanisms in (A) squamates, and using the alternative SDM classification in (B) squamates and (C) lizards.
Table S1A. Dataset used in this study.
Table S1B. Taxonomic coverage of turtle and squamate families used in this study.
Data S1. Results using alternative SDM assignment for species with mixed or equivocal SDM as listed in Table S1:
Table S2. MacroCAIC results using alternative SDM assignment.
Table S3. BAMM estimation for the number of rate shifts in diversification. The number of rates shifts with the highest probability in each group is marked in bold.
Table S4. Summary of transition rate parameters estimates using the MK2 model with both Maximum Likelihood and Bayesian (MCMC) methodologies and BiSSE for the turtles, lizards, and squamate data sets using the alternative SDM assignment.
Table S5. Log likelihood differences (ΔLL) obtained between the single (BM1) and two rate (BM2) Brownian motion models of evolution, and between the single (OU1) and two (OU2) optimums, as estimated for lifepan in turtles, lizards and squamates. σ2GSD and σ2TSD, OptimumGSD, and OptimumTSD: estimated parameters for GSD and TSD lineages using the alternative SDM assignment.
Data S2. BiSSE Analyses.


