Abstract
We present the design, and analyze the performance of a multi-stage natural language processing system employing named entity recognition, Bayesian statistics, and rule logic to identify and characterize heart disease risk factor events in diabetic patients over time. The system was originally developed for the 2014 i2b2 Challenges in Natural Language in Clinical Data. The system's strengths included a high level of accuracy for identifying named entities associated with heart disease risk factor events. The system's primary weakness was due to inaccuracies when characterizing the attributes of some events. For example, determining the relative time of an event with respect to the record date, whether an event is attributable to the patient's history or the patient's family history, and differentiating between current and prior smoking status. We believe these inaccuracies were due in large part to the lack of an effective approach for integrating context into our event detection model. To address these inaccuracies, we explore the addition of a distributional semantic model for characterizing contextual evidence of heart disease risk factor events. Using this semantic model, we raise our initial 2014 i2b2 Challenges in Natural Language of Clinical data F1 score of 0.838 to 0.890 and increased precision by 10.3% without use of any lexicons that might bias our results.
Keywords: Biomedical text mining, clinical informatics, translational research, natural language processing, named entity recognition, distributional semantic models, heart disease risk factors, diabetes
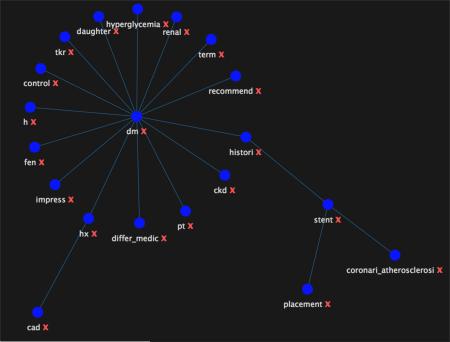
Graphical abstract
Force-directed graph of diabetes mellitus and cad concepts extracted with distributional semantic model.
Introduction
Diabetes Mellitus is a common disease with cardiovascular complications. Complications such as an ST elevation myocardial infarction (STEMI) are associated with mortality, significant morbidity, and healthcare spending. The ability to identify patients likely to have a significant cardiovascular event within 1 to 3 years provides an opportunity for successful intervention. A significant challenge to developing models for predicting cardiac risk involves the identification of temporally related events and measurements in the unstructured text in electronic health records. The 2014 i2b2 Challenges in Natural Language Processing in Clinical Data track for identifying risk factors for heart disease over time was created to facilitate development of natural language processing systems to address this challenge [1]. The details of the i2b2 Natural Language Challenge are documented in the annotation guidelines [2], and are summarized in the following Annotation section. Teams were provided with 521 de-identified medical record note text containing 64,035 risk factors. For evaluation, risk factor instances were rolled up to the document level for a total of 16,167 distinct record/document risk factors.
Accurate identification of risk factors requires proper characterization of time, and whether a risk factor is attributable to the patient or a family member. For example, a mention of hypertension could be a past or current condition, and the condition could be attributable to either the patient or a patient's family member. Capturing this information requires an effective approach for integrating contextual semantics within the event detection model. Distributional semantic models (DSM) can be used to quantify the semantic similarity between linguistic terms based on their distributional properties in large samples of text. The central assumption here is that the context surrounding a given word or phrase provides important information about its meaning [3, 4, 5]. DSMs provide a mechanism for representing terms, concepts, relations, or sentence meaning by using distributional statistics. The semantic properties of terms are captured in a multi-dimensional space by vectors that are constructed from large bodies of text by observing the distributional patterns of co-occurrence with their neighboring words. These vectors can then be used as measures of text similarity between words, phrases, concepts, relations, or snips of arbitrary text. Early work on use of distributional semantic modeling in EHRs (Electronic Health Records) has focused on providing vector-based representations of medical concepts, i.e., SNOMED [6], and for synonym recognition [7].
Annotation Task
Given a set of medical records, the annotation task was to create a set of text annotations that track the progression of heart disease in diabetic patients. Multiple records were annotated for each patient, which provides a general timeline to be created from the set. Annotation tags and attributes were used to indicate the presence and progression of disease (diabetes, heart disease), associated risk factors (hypertension, hyperlipidemia, smoking status, obesity status, and family history), disease-related medications, and the time they were present in the patient's medical history. Each disease and risk factor associated with this task was assigned its own set of indicators that is used to identify whether or not the disease or risk factor is present for that patient, and when it is present. Annotations are summarized in Table 1.
Table 1.
Risk factor tags, attributes, and descriptions
| Risk Factor | Indicator | Descriptions |
|---|---|---|
| Diabetes | Mention | Type 1 or Type 2 diabetes diagnosis, e.g., HXDM. |
| High A1c | A1c > 6.5; E.g., A1c: 6.2 | |
| High glucose | 2 fasting blood glucose measurements > 126; E.g., SMBP 130. | |
| CAD | Mention | Diagnosis or history of CAD. |
| Event | E.g., MI, STEMI, NSTEMI, bypass surgery, CABG, percutaneous, cardiac arrest, ischemic cardiomyopathy. | |
| Test result | Exercise or pharmacologic stress test showing ischemia, abnormal cardiac catheterization showing coronary stenoses (narrowing) | |
| Symptom | Chest pain consistent with angina | |
| Hyperlipidemia / Hypercholesterolemia | Mention | Diagnosis/history of Hyperlipidemia or Hypercholesterolemia |
| High Cholesterol | Total cholesterol of over 240 | |
| High LDL | LDL measurement of over 100 mg/dL | |
| Hypertension | Mention | Diagnosis or preexisting condition of Hypertension. |
| High BP | BP measurement of over 140/90 mm/hg | |
| Obesity | Mention | Description of obesity |
| BMI | > 30 | |
| Waist | Men >= 40”; woman >= 35” | |
| Medications | Diabetes | Metformin, insulin, sulfonylureas, thiazolidinediones, GLP-1 agonists, Meglitinides, DPP-4 inhibitors, Amylin, anti-diabetes medications, combinations. |
| CAD | Aspirin, Thienopyridines, beta blockers, ACE inhibitors, nitrates, calcium-channel blockers, combinations. | |
| Hyperlipidemia | Statins, fibrates, niacins, ezetimibes, combinations. | |
| Hypertension | Beta-blockers, ACE inhibitors ARBs, Thiazide diuretics, calcium-channel blockers, combinations. | |
| Obesity | Orlistat (xenical) or Lorqess (Lorcaserin). | |
| Family History | Present if the patient has a first-degree relative (parents, siblings, or children) who was diagnosed prematurely (< 55 for male relatives, < 65 for female relatives) with CAD. | |
| Smoker | Status: CURRENT, PAST (quit > 1 year ago), EVER (smoked at some point but it is unclear), NEVER (never smoked), or UNKNOWN (not mentioned). | |
Every tag except for SMOKER and FAMILY_HIST has a time attribute that is used to show when the indicator for each medical problem is known to have existed. These reflect when the indicator occurred/was active in relation to the date the medical record was written, i.e., document creation time (DCT): before DCT, during DCT, after DCT, not mentioned.
Methods
We developed our own NLP pipeline for this challenge. The processing pipeline consisted of the following process:
Preprocessing and dimensional indexing for distributional statistics
Risk factor named entity recognition
Attribute and measurement extraction
Contextual measurement via distributional semantic model
Risk factor event classification
Record level aggregation of risk factor classification
Preprocessing and Dimensional Indexing for Distributional Statistics
XML-formatted EHR data and training annotations were first imported into a relational database. Individual patient records are parsed into sentences; and sentences are parsed into words, noun phrases and candidate named entities. An inverted index is constructed using a data warehousing style dimensional data model [8, 9]. We have scaled a variation of this model to several hundred Gigabytes for chemical patent retrieval [10]. The grain of the index is the individual word with attributes for position, part-of-speech, phrase and entity membership. Dimensional indexing facilitates efficient OLAP style SQL queries for aggregating distributional statistics of candidate risk events. Data can be efficiently aggregated by word, phrase, entity, sentence, or document to construct distributional co-occurrence vector representations of words, phrases, entities, or sentences.
Risk Factor Event Recognition
Heart disease risk factor event recognition consisted of training conditional random fields (CRF) based named entity recognition (NER) models [11], and subsequent execution of the NER models on test data to identify candidate instances of risk factor events. A CRF is a conditional sequence model, which represents the probability of a hidden state sequence given some observations. NER models were trained using the extensive set of features developed by Finkel, et al. [12]. Example features include: capitalization, word text, prior word, part of speech, etc.
NER training files were generated for each Risk Factor listed in Table 1 using the i2b2 training data. Since the training data annotation boundaries were not consistent, we could not use the annotations directly to train a NER model without further processing. For example, “NITROGLYCERIN” and “NITROGLYCERIN 1/150 (0.4 MG)” are both annotated as the same nitrate medication from the training data. These annotations are automatically extracted from the training data and written to a text file. The text file is manually reviewed, and annotations like “NITROGLYCERIN 1/150 (0.4 MG)” are edited down to “NITROGLYCERIN.” This manual review process took approximately 1-person 1-day. Sample annotations are provided in Appendix A. A program was written to take these annotation files and convert them into word-classification-part of speech triplets for training NER classifiers. The NER process is listed below. Performance of our NER subsystem is listed in Table 2. A sample NER training file is provided in Appendix B.
Distinct training data annotations are exported into a text files for each Risk Factor tag.
Tag files are manually inspected and edited down to specific noun phrases.
Software was written to generate two named entity training files: one for medications, and one for all other type tags, by recognizing words and phrases within the note text of the training data.
Two conditional random fields-based named entity recognizers are trained: one for medications, one for all other medical events. The named entity recognizers are subsequently used to identify candidate risk factor instances.
Table 2.
Risk factor NER subsystem performance on test partition
| Entity | P | R | F1 | TP | FP | FN |
|---|---|---|---|---|---|---|
| CAD|event | 0.991 | 0.977 | 0.984 | 832 | 8 | 20 |
| CAD|mention | 0.992 | 0.980 | 0.986 | 585 | 5 | 12 |
| CAD|symptom | 0.998 | 0.977 | 0.988 | 596 | 1 | 14 |
| CAD|test | 0.990 | 0.983 | 0.987 | 888 | 9 | 15 |
| DIABETES|A1C | 0.963 | 0.919 | 0.940 | 363 | 14 | 32 |
| DIABETES|glucose | 0.994 | 0.879 | 0.933 | 503 | 3 | 69 |
| DIABETES|mention | 0.979 | 0.932 | 0.955 | 1137 | 25 | 83 |
| FAMILY_HIST|not_present | 0.997 | 0.966 | 0.981 | 648 | 2 | 23 |
| HISTORY|mention | 0.997 | 0.972 | 0.984 | 1346 | 4 | 39 |
| HYPERLIPIDEMIA|O | 1.000 | 1.000 | 1.000 | 11 | 0 | 0 |
| HYPERLIPIDEMIA|high_LDL | 0.951 | 0.936 | 0.944 | 117 | 6 | 8 |
| HYPERLIPIDEMIA|high_chol. | 0.879 | 0.872 | 0.876 | 109 | 15 | 16 |
| HYPERLIPIDEMIA|mention | 0.995 | 0.940 | 0.967 | 758 | 4 | 48 |
| HYPERTENSION|high_bp | 0.976 | 0.940 | 0.958 | 827 | 20 | 53 |
| HYPERTENSION|mention | 0.968 | 0.894 | 0.929 | 755 | 25 | 90 |
| OBESE|BMI | 1.000 | 0.800 | 0.889 | 16 | 0 | 4 |
| OBESE|mention | 0.956 | 0.908 | 0.931 | 197 | 9 | 20 |
| SMOKER|current | 0.792 | 0.815 | 0.803 | 167 | 44 | 38 |
| SMOKER|never | 0.922 | 0.435 | 0.591 | 47 | 4 | 61 |
| SMOKER|past | 0.981 | 0.823 | 0.895 | 102 | 2 | 22 |
| SMOKER|unknown* | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0 |
| Totals | 0.980 | 0.938 | 0.959 | 10004 | 200 | 667 |
Note: Smoking named entity recognition was lower than other named entities due to our system not being designed to recognize smoking UNKNOWN status.
Attribute and Measurement Extraction
Regular expressions were developed to extract specific measurements for A1C, blood glucose, blood pressure, hyperlipidemia, LDL and cholesterol, obesity BMI and waist circumference, and Date. This process consisted of creating a list of training data examples for each risk event annotation that was accompanied by a measurement, and writing regular expressions to extract each measurement. Regular expressions were applied after a candidate event was recognized by the NER-based event recognition system described above. A list of the regular expressions used is provided in Appendix C.
Extracted risk factors for Medication, CAD, Diabetes, Hyperlipidemia, and Hypertension are assigned default times (DCT) based on each risk factor's most likely assignment in the training data. For example, given a CAD mention like ‘MI’ what is the conditional probability that the event happened before, during, or after the record time? In a similar matter, smoker risk factors are assigned the most likely status, and obesity risk factors are assigned the most likely indicator.
Contextual Measurement via Distributional Semantic Model
Popular methods for corpus-based distributional measures of word semantic similarity include pointwise mutual information (PMI), latent semantic analysis (LSA), and higher order tensor models [13]. PMI measures the pointwise mutual information between two objects as the log ratio of the joint probability of two objects co-occurring relative to the probability of those objects occurring independently. PMI using information retrieval (PMI-IR) was suggested by Turney [14] as an unsupervised measure for the evaluation of the semantic similarity of words (Eq. 1).
| (1) |
Multiple evaluations have demonstrated that using PMI within multi-evidence models meets or exceed the performance of LSA and TSM (tensor space models) on semantic similarity benchmarks [15, 16]. Due to its performance and efficiency within a DSM, we focused our efforts on developing distributional semantic similarity measurements using PMI. We captured distributional measurements from the i2b2 training data by running OLAP style SQL queries on our dimensional data model. To illustrate our approach, tables 3 and 4 show the non-normalized PMI of words for the terms Diabetes and CHF. The collection of semantically similar words (stemmed terms) for each disease can be used to infer the underlying concepts Diabetes and CHF respectively.
Table 3.
PMI of words for Diabetes
| Concept | Stem term | PMI |
|---|---|---|
| diabet | mellitu | 4.12 |
| diabet | depend | 3.52 |
| diabet | type | 2.67 |
| diabet | retinopathi | 2.14 |
| diabet | insulin | 2.13 |
| diabet | nephropathi | 2.02 |
| diabet | noninsulin | 1.84 |
| diabet | hyperlipidemia | 1.76 |
| diabet | esrd | 1.54 |
| diabet | adult | 1.52 |
| diabet | glaucoma | 1.42 |
| diabet | hypercholesterolemia | 1.10 |
Table 4.
PMI of words for CHF
| Concept | Stem term | PMI |
|---|---|---|
| chf | exacerb | 2.34 |
| chf | ef | 1.5 |
| chf | drainag | 1.4 |
| chf | leukocytosi | 0.71 |
| chf | lvh | 0.47 |
| chf | treat | 0.34 |
| chf | secondari | 0.33 |
| chf | etiolog | 0.31 |
| chf | cad | 0.29 |
| chf | diuresi | 0.27 |
| chf | evid | 0.25 |
| chf | pleural | 0.21 |
Mihalcea, et al. [16] extended semantic similarity measurements to two arbitrary text segments. Given a measurement for the semantic similarity of two (bag of words) text segments and a measurement for term specificity (IDF), the semantic similarity of two text segments C1 and C2 can be defined using a model that combines the semantic similarities of each text segment in turn with respect to the other text segment. We extended the original bag-of-words text-to-text measurement to include phrases (named entities). Using PMI as the underlying measure of semantic similarity, we developed the following 2nd order PMI-based model for measuring the semantic similarity between concepts C1, C2. (Eq. 2).
| (2) |
We applied our semantic similarity measurement (SemSim) to risk-attribute event instances identified in the training data by aggregating words within a +/− 4 word window risk-attribute start and end positions. This resulted in distributional semantic models for each risk-attribute listed in table 5. For example, models were created for MEDICATION_beforeDCT, MEDICATION_duringDCT, and MEDICATION_afterDCT, or SMOKING_current, SMOKING_never, etc. The highest SemSim measurement terms for smoking status of never, past, and current are provided in tables 6, 7, and 8.
Table 5.
Risk-attribute categories for semantic similarity (SemSim) models
| Risk | Time | Status | Count |
|---|---|---|---|
| CAD | after DCT | 756 | |
| CAD | before DCT | 1738 | |
| CAD | during DCT | 897 | |
| DIABETES | after DCT | 1687 | |
| DIABETES | before DCT | 2057 | |
| DIABETES | during DCT | 1780 | |
| FAMILY_HIST | 2026 | ||
| HYPERLIPIDEMIA | after DCT | 997 | |
| HYPERLIPIDEMIA | before DCT | 1126 | |
| HYPERLIPIDEMIA | during DCT | 1013 | |
| HYPERTENSION | after DCT | 1711 | |
| HYPERTENSION | before DCT | 1782 | |
| HYPERTENSION | during DCT | 2430 | |
| MEDICATION | after DCT | 7907 | |
| MEDICATION | before DCT | 8138 | |
| MEDICATION | during DCT | 7935 | |
| OBESE | after DCT | 330 | |
| OBESE | before DCT | 330 | |
| OBESE | during DCT | 419 | |
| SMOKER | current | 131 | |
| SMOKER | ever | 10 | |
| SMOKER | never | 457 | |
| SMOKER | past | 432 | |
| SMOKER | unknown | 967 |
Table 6.
Smoking Status never
| term2 | idf | semsim |
|---|---|---|
| ex | 0.667 | 5.951 |
| former | 0.477 | 5.582 |
| tob | 0.439 | 4.278 |
| smoker | 0.31 | 4.194 |
| ,ks. | 0.795 | 4.058 |
| technologist | 0.795 | 4.015 |
| retir | 0.344 | 3.424 |
| duluth | 0.795 | 3.266 |
| heavi | 0.457 | 3.170 |
| yr | 0.282 | 2.857 |
| pk | 0.742 | 2.846 |
| girl | 0.795 | 2.775 |
| tobacco | 0.261 | 2.769 |
| hyperglycemia | 0.573 | 2.697 |
Table 7.
Smoking Status past
| term2 | idf | semsim |
|---|---|---|
| li | 0.700 | 5.865 |
| nonsmok | 0.700 | 5.791 |
| smoker | 0.310 | 5.023 |
| oh | 0.700 | 4.929 |
| tob | 0.439 | 4.563 |
| cigarett | 0.477 | 3.924 |
| ma | 0.700 | 3.568 |
| habit | 0.376 | 3.544 |
| mason | 0.700 | 3.391 |
| tobacco | 0.261 | 3.340 |
| et | 0.288 | 3.310 |
| oh | 0.306 | 3.278 |
| sigmoidoscopi | 0.614 | 3.237 |
| never | 0.285 | 3.224 |
Table 8.
Smoking status current
| term2 | idf | semsim |
|---|---|---|
| cig | 0.487 | 3.123 |
| homeless | 0.742 | 2.789 |
| calista | 0.795 | 2.594 |
| svt | 0.795 | 2.594 |
| lgi | 0.795 | 2.594 |
| tob | 0.439 | 2.551 |
| pack | 0.349 | 2.459 |
| postmenopaus | 0.795 | 2.452 |
| corpor | 0.871 | 2.375 |
| smoker | 0.31 | 2.339 |
| niddm | 0.614 | 1.786 |
| oh | 0.306 | 1.709 |
| et | 0.288 | 1.585 |
| ppd | 0.382 | 1.571 |
Classification
Risk factor specific rules were developed to assign risk factors to the patient or to the patient's family history; override the default time, status, or indicator; qualify measurements as hypertensive, hyperlipidemia, high glucose, and high A1C; override smoking status, and determine obesity (Table 1). For example, if an extracted systolic blood pressure measurement was > 140 mmHg, the extracted risk factor was considered a legitimate heart risk factor. Otherwise the extracted risk factor is not tagged in the output data stream. One of the weakest components of the subsystem was determination of smoking status and family history.
Record Level Aggregation of Risk Factor Classification
The final stage of the system consisted of aggregating risk factor instances to the record (document) level, translating highly specific risk factor classifications to more general classifications, e.g., “antianginal” to “nitrate,” and generating risk factor tag specific XML output for evaluation. If more than one smoking status event was detected within an individual record, we assigned the most likely smoking status by calculating the prior likelihood of smoking status from the training data (Table 10).
Table 10.
Prior probability of smoking status (from training data)
| Smoking status | P |
|---|---|
| current | 0.076 |
| ever | 0.009 |
| never | 0.244 |
| past | 0.199 |
| unknown | 0.473 |
Results
Official results from the i2b2 NLP Challenge are shown in Table 11. The results augmented by our SemSim distributional semantic model are shown in Table 12. Most impressive is an increase in precision of 10.3% when using the semantic model to capture distributional context. These results can be contrasted with the top performing result from the official challenge which had an impressive micro- (macro-) precision of 0.8951 (0.8965), recall of 0.9625 (0.9611), and F1-measure of 0.9276 (0.9277) [17]. It is worthy to note that this team added extensive additional annotation information on top of the provided corpus.
Table 11.
i2b2 NLP Challenge results
| (514) | Measure | Macro | Micro (Primary) |
|---|---|---|---|
| Total | Precision | 0.793 | 0.800 |
| Recall | 0.887 | 0.887 | |
| F1 | 0.838 | 0.841 |
Table 12.
i2b2 NLP Challenge results with distributional semantic models
| (514) | Measure | Macro | Micro (Primary) |
|---|---|---|---|
| Total | Precision | 0.875 | 0.877 |
| Recall | 0.906 | 0.907 | |
| F1 | 0.890 | 0.892 |
The devil is of course in the details. We've identified 94 distinct classifications, i.e., medications permute across of type1, type 2, and time attributes. We did not consider type2 classifications for medications (see Table 1). 38% of our initial system's error is due to incorrect smoking status and family history assignment. The remaining classifications (87), contribute on average, 0.7% error each.
As previously discussed, the NER subsystem was very effective at identifying risk factor event candidate instances (Table 2). Exceptions of our initial system included OBESE|BMI(0.889), SMOKER|current (0.803), SMOKER|never (0.591), SMOKER|past (0.895). The system using the distributional semantic model was able to capture the context of risk events to improve risk attribute classification, and clearly improved the overall performance of the system. Precision increased by 10.3%. F1 score improved from 0.838 to 0.890.
The remaining, and most significant source of errors in the system were due to risk factor attribute assignment. To gain a better understanding to the source of these errors, we analyzed statistical distribution of risk factor attributes from the test data (Medications only include the most significant assignment). We find the assignment of time to be relatively ad hoc. For example, why would the most significant assignment of ACE inhibitor be “after DCT, before DCT “(58%), and calcium channel blocker be “after DCT” (0.53) when both drugs are prescribed to treat chronic conditions? Similarly, how could a patient be hypertensive “after DCT”? Getting a handle on these attribute assignments is fundamental to improving the performance of the system.
Conclusions
We presented the design and analysis of a multi-stage natural language processing system employing named entity recognition, Bayesian statistics, and rule logic for identifying heart disease risk factor events in diabetic patients over time. The most significant shortcoming of the system was due to inaccuracies characterizing the attributes of these events. To address these inaccuracies, we introduced a novel distributional semantic model to capture event context. Using our distributional semantic model, we were able to improve our F1 score from 0.838 to 0.890, and increase precision by 10.3% on the 2014 i2b2 Challenges in Natural Language of Clinical dataset without use of any lexicons that might bias our results. We believe there is significant potential for integrating distributional semantics in the form of vector space models for improving named entity and event accuracies in healthcare natural language processing applications. Future plans include construction of a much larger EHR dataset from our clinical data warehouse to gather more robust distributional statistics and to perform a more thorough evaluation.
Highlights.
NLP system to identify heart disease risk factors in diabetic patients over time
Employ named entity recognition, Bayesian statistics, and rule logic
Introduce distributional semantic model for characterizing contextual evidence
Distributional semantic model improves precision by 10.3% without use of lexicons
Acknowledgements
This publication and project was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number 8UL1TR000055. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Appendix A
Sample annotation file
Format: term, classification
ASA, aspirin
Ecotrin, aspirin
Colfarit, aspirin
Acetylsalicylic, aspirin
Acetylsalicylic acid, aspirin
aspirin, aspirin
PLAVIX, thienopyridine
TSH, thienopyridine
hTSH, thienopyridine
thienopyridine, thienopyridine
clopidogrel, thienopyridine
ticlopidine, thienopyridine
Ticlid, thienopyridine
prasugrel, thienopyridine
Effient, thienopyridine
ATENOLOL, beta blocker
AMLODIPINE, beta blocker
Sectral, beta blocker
acebutolol, beta blocker
Zebeta, beta blocker
bisoprolol, beta blocker
Brevibloc, beta blocker
esmolol, beta blocker
Inderal, beta blocker
propranolol, beta blocker
Tenormin, beta blocker
atenolol, beta blocker
Normodyne, beta blocker
Trandate, beta blocker
labetalol, beta blocker
Coreg, beta blocker
carvedilol, beta blocker
Lopressor, beta blocker
Toprol, beta blocker
metoprolol, beta blocker
beta blocker, beta blocker
Appendix B
Sample Named Entity Recognition training file snippet.
| Format: word – class – part of speech | ||||||
| PO | O | NNP | ||||
| QD | O | NNP | ||||
| LIPITOR MEDICATIONS|statin | NNP | |||||
| ATORVASTATIN MEDICATIONS|statin | NNP | |||||
| 10MG | O | NN | ||||
| 1 | O | CD | ||||
| Tablet | O | NNP | ||||
| -LRB- | O | -LRB- | ||||
| s | O | NNS | ||||
| -RRB- | O | -RRB- | ||||
| PO | O | NNP | ||||
| QD | O | NNP | ||||
| HCTZ | MEDICATIONS|diuretic | NNP | ||||
| -LRB- | O | -LRB- | ||||
| HYDROCHLOROTHIAZIDE | MEDICATIONS|diuretic | NNP | ||||
Appendix C
Regular expressions for information extraction
//A1C
a1cRegexStr1 = ” \b(Hgb A1c|HgB
A1c|HgbA1C|HBA1c|hgbaic|HgbA1c|HGBA1C|hgba1c|HBGA1c|A1Cs|A1C|Hgb|HGBA1C|HA1C|HbA1c|hemoglobin
HbA1C|Hgb A1c|A1c|A1C|HgB|HgA1c|hemoglobin A1c|Hemoglobin A1c|HgbA1c|HBAIC|HBA1C|Hem A1C|HgB
A1c|a1c|HbA1C|HgA1C|HgA1c|hemoglobin A1c)\b(:|,|-|=)?”
+ “(\s+(([a-zA-Z]|-)+|([0-9]{1,2}/([0-9]{1,2}/)?[0-9]{2,4})))*”
+ “(:|,|-|=)?”
+ “ \s*” + “ \b(\d+(\.\d+)?)s?\b”;
//Glucose
glucoseRegexStr1 = “ \b(Glucose|GLUCOSE|BS|FINGER BLOOD GLUCOSE|FS|FSs|FS's|GLU-POC|BG|BGs|BG's|glucose|GLU|Fingerstick|fingerstick|blood sugars|blood sugar|FG)\b(:|,|-|=)?”
+ “(\s+((([a-zA-Z\(\)]|-|~)+)|([0-9]{1,2}/([0-9]{1,2}/)?[0-9]{2,4})))*”
+ “(:|,|-|=|~)?”
+ “ \s+” + “ \b(\d+)(-(\d+))?s?\b”;
//Blood pressure
bpRegexStr2 =
“ \b(?i)(Sys|Systolic|SBP|blood pressure|blood pressures|BP|Dias|Diastolic|DBP)\.?(?-i)\b”
+ “(\s+(([a-zA-Z]|-)+|([0-9]{1,2}/([0-9]{1,2}/)?[0-9]{2,4})))*”
+ “(:|,|-|=)?”
+ “ \s*” + “ \b(\d+)(-(\d+))?s?\b”;
bpRegexStr3 = “ \b(\d+)(-(\d+))?s?\b“
+ “ \s+”
+ “ \b(?i)(Sys|sys|Systolic|SBP|blood pressure|blood pressures)(?-i)\b”;
//Hyperlipidemia
hyperlipidemiaLDLRegexStr1 = “ \b(?i)(Cholesterol-LDL|LDL|LDL Cholesterol|LDL-Cholesterol)(?i-)\b(:|,|-|=)?”
+ “(\s+((([a-zA-Z\(\)]|-|~)+)|([0-9]{1,2}/([0-9]{1,2}/)?[0-9]{2,4})))*“
+ “(:|,|-|=|~)?”
+ “ \s+” + “ \b(\d+)(-(\d+))?s?\b”;
//Cholesterol
hyperlipidemiaCholRegexStr1 = “ \b(?i)(?!LDL Chol|LDL-Chol|LDL Cholesterol|LDL-Cholesterol)(Chol|Cholesterol)(?i-)\b(:|,|-|=)?”
+ “(\s+((([a-zA-Z\(\)]|-|~)+)|([0-9]{1,2}/([0-9]{1,2}/)?[0-9]{2,4})))*”
+ “(:|,|-|=|~)?”
+ “\s+” + “ \b(\d+)(-(\d+))?s?\b”;
/BMI
bmiRegexStr1 = “ \b(?i)(BMI)(?i-)\b(:|,|-|=)?”
+ “(\s+((([a-zA-Z\(\)]|-|~)+)|([0-9]{1,2}/([0-9]{1,2}/)?[0-9]{2,4})))*”
+ “(:|,|-|=|~)?”
+ “ \s+” + “ \b((\d+)(\.(\d+)))?\b”;
//Date
dateRegex0 = “([0-9]{4})[/\-]([0-9]{1,2})[/\-]([0-9]{1,2})”; // year/month/day
dateRegex1 = “([0-9]{1,2})[/\-]([0-9]{4})”; // month/year
dateRegex2 = “([0-9]{1,2})[/\-]([0-9]{1,2})”; // month/day
dateRegex3 = “([0-9]{1,2})[/\-]([0-9]{1,2})[/\-,]\s*([0-9]{1,4})”; // month/day/year
dateRegex4 = “([ADFJMNOS]\w*)\s+([0-9]{0,2})(th|TH|nd|ND){0,1},{0,1}\s+([0-9]{4})”;
dateRegex5 = “([ADFJMNOS]\w*)\s+([0-9]{4})”;
dateRegex6 = “([ADFJMNOS]\w*)\s+([0-9]{0,2})(th|TH|nd|ND){0,1},{0,1}”;
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stubbs A, Kotfila C, Uzuner O. Practical applications for NLP in Clinical Research: the 2014 i2b2 shared tasks. 2014 doi: 10.1016/j.jbi.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stubbs A, Uzuner O, Kumar V, Shaw S. Annotation guidelines: Risk factors for Heart Disease in Diabetic Patients. 2014 Apr 1; https://www.i2b2.org/NLP/HeartDisease/
- 3.Church KW, Hanks P. Word Association Norms, Mutual Information and Lexicography. Proceedings of the 27th Annual Conference of the Association of Computational Linguistics. 1989:76–83. [Google Scholar]
- 4.Harris Z. Distributional structure. Word. 1954;10(23):146–162. [Google Scholar]
- 5.Firth JR. A synopsis of linguistic theory 1930-1955. In: Palmer FR, editor. Studies in Linguistic Analysis. Philological Society; Longman; Oxford: London: 1957. 1968. pp. 1–32. Selected Papers of J.R. Firth 1952-1959. [Google Scholar]
- 6.Henriksson A, Conway M, Duneld M, Chapman W. Identifying synonymy between SNOMED clinical terms of varying length using distributional analysis of electronic health records. AMIA Annual Symposium Proceedings. 2013:600. [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell J, Brear H, Scichilone R, White S, Giannangelo K, Carlsen B, Solbrig H, Fung K. Semantic interoperation and electronic health records: context sensitive mapping from SNOMED CT to ICD-10. MedInfo. 2013:603–607. [PubMed] [Google Scholar]
- 8.Gray J, et al. Data Cube: A Relational Aggregation Operator Generalizing Group-By, Cross-Tab, and Sub-Totals. Data Mining and Knowledge Discovery. 1997;1(1) [Google Scholar]
- 9.Urbain J, Frieder O, Goharian N. Passage relevance models for genomics search. BMC Bioinformatics. 2009;10(Suppl 3):S3. doi: 10.1186/1471-2105-10-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urbain J, Frieder O. Advances in Multidisciplinary Retrieval. Springer; Berlin Heidelberg: 2010. Exploring contextual models in chemical patent search. pp. 60–69. [Google Scholar]
- 11.Lafferty J, McCallum A, Pereira F. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. In: Brodley Carla E., Pohoreckyj Danyluk Andrea., editors. Proceedings of the Eighteenth International Conference on Machine Learning (ICML '01) Morgan Kaufmann Publishers Inc.; 2001. pp. 282–289. [Google Scholar]
- 12.Finkel J, Grenager T, Manning C. Incorporating Non-local Information into Information Extraction Systems by Gibbs Sampling. Proceedings of the 43nd Annual Meeting of the Association for Computational Linguistics (ACL 2005) 2005:363–370. 2005. [Google Scholar]
- 13.Turney P, Pantel P. From frequency to meaning: Vector space models of semantics. Journal of artificial intelligence research. 37.1(2010):141–188. [Google Scholar]
- 14.Turney P. Mining the Web for Synonyms: PMI-IR versus LSA on TOEFL. NRC Publications Archive; 2001. [Google Scholar]
- 15.Agirre E, Diab M, Cer D, Gonzalez-Agirre A. Proceedings of the First Joint Conference on Lexical and Computational Semantics-Volume 1: Proceedings of the main conference and the shared task, and Volume 2: Proceedings of the Sixth International Workshop on Semantic Evaluation. Association for Computational Linguistics; 2012. Semeval-2012 task 6: A pilot on semantic textual similarity. [Google Scholar]
- 16.Mihalcea R, Corley C, Strapparava C. Corpus-based and knowledge-based measures of text semantic similarity. Vol. 6. AAAI; 2006. pp. 775–780. [Google Scholar]
- 17.Roberts K, Shooshan S, Rodriguez L, Abhyankar S, Kilicoglu H, Demner-Fushman D. Machine Learning Methods for Detecting Risk Factors for Heart Disease in EHRs. NLP in Clinical Research: the 2014 i2b2 shared tasks workshop proceedings. 2014 [Google Scholar]