Abstract
A safe and effective vaccine against human immunodeficiency virus type 1 (HIV-1) is urgently needed to combat the worldwide AIDS pandemic, but still remains elusive. The fact that uncontrolled replication of an attenuated vaccine can lead to regaining of its virulence creates safety concerns precluding many vaccines from clinical application. We introduce a novel approach to control HIV-1 replication, which entails the manipulation of essential HIV-1 protein biosynthesis through unnatural amino acid (UAA*)-mediated suppression of genome-encoded blank codon. We successfully demonstrate that HIV-1 replication can be precisely turned on and off in vitro.
Keywords: attenuated vaccine, genetic code expansion, HIV-1 vaccine, unnatural amino acid, virus engineering
Genetic code expansion with the evolved orthogonal suppressor tRNA–aminoacyl-tRNA synthetase (aaRS) pair has been widely applied to site-specific incorporation of unnatural amino acids (UAA*s) with unique chemical and physical properties into proteins in living cells to facilitate the study of protein structure and function.[1] Here we report a new application of genetic code expansion, which couples the UAA*-mediated blank codon (codon that does not encode natural proteinogenic amino acids) suppression with the virus assembly process for a potential solution to the development of a live-attenuated HIV-1 vaccine. In this strategy, the replication of HIV-1 is controlled by UAA*s that are not present in nature and constitute unique chemical probes for future immunological studies.
The search for safe and effective HIV-1 preventive vaccines has been ongoing for more than 30 years since HIV-1 was identified as the causative agent for AIDS.[2] Unfortunately, safe and effective HIV-1 vaccines remain elusive.[2b, 3] More than 60 million people worldwide have been infected with HIV-1 and nearly half of these individuals have died. Although progress has been made in preventing new HIV-1 infections and in lowering the annual number of AIDS-related deaths through comprehensive prevention programs and increased access to antiretroviral therapy,[2b] the number of people living with HIV-1, now over 34 million, continues to grow. The development of a safe and effective HIV-1 vaccine would undoubtedly be one of the best solutions for the ultimate control of the worldwide AIDS pandemic.[4] One visible achievement of the past few years was the results of the RV144 trial—the HIV-1 vaccine trial conducted in Thailand—which showed that a poxvirus-protein prime-boost combination provided modest (31%) protection against HIV-1 acquisition.[5] These results represent the first demonstration of any level of efficacy in preventing HIV-1 acquisition in humans by a vaccine. However, the low level of protection elicited by RV144 vaccine makes this vaccine not useful in clinic. Of all the HIV-1 vaccine modalities developed and tested thus far, deletion of nef gene in HIV-1 as a live-attenuated vaccine (Δnef-LAV) has shown the best efficacy against HIV-1 acquisition (Δnef-LAV protection rate is 95% versus 7% for all other types of vaccines) in SIV/rhesus macaque model of HIV-1 infection.[6] Despite its best efficacy, the current version of this live-attenuated HIV-1 vaccine cannot be developed for clinical use due to safety concerns. Therefore, finding a safe and effective HIV-1 vaccine using novel approaches remains a top priority.
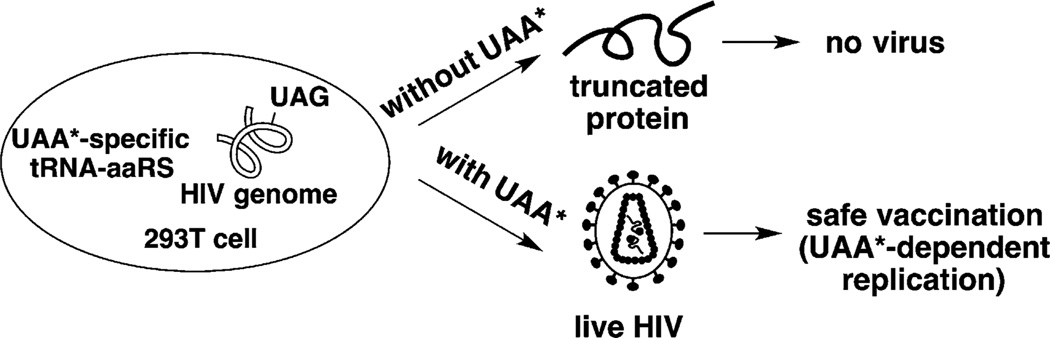
We sought to solve the safety problem by turning virus replication on and off using the UAA*-mediated blank codon suppression system (Figure 1).[1b,c, 7] We envisaged that if we could introduce blank codons at positions within the HIV-1 genome (such as the Δnef-LAV) that encode essential proteins, we would prevent proper translation and assembly of viable HIV-1. On the other hand, live and functional HIV-1 can be assembled in the presence of a special tRNA–aaRS pair that could decode the blank codon. The assembled HIV-1 can be subsequently used as a live-attenuated vaccine. Since the host does not contain the special tRNA–aaRS pair, HIV-1 can only undergo one-cycle infection and is not able to replicate, thus making the vaccine safer. To render the system more stringent, special tRNA–aaRS pairs that only recognize desired UAAs* (the host lacks these UAAs*) are used. The UAA*-mediated blank codon suppression would also allow us to turn on and off multiple replication cycles of HIV-1 in vivo using UAA* as the control element in future vaccine development. Here we report a successful demonstration of controlling HIV-1 replication in vitro.
Figure 1.
Construction of novel live-attenuated HIV vaccine through genetic code expansion. UAA*, unnatural amino acid.
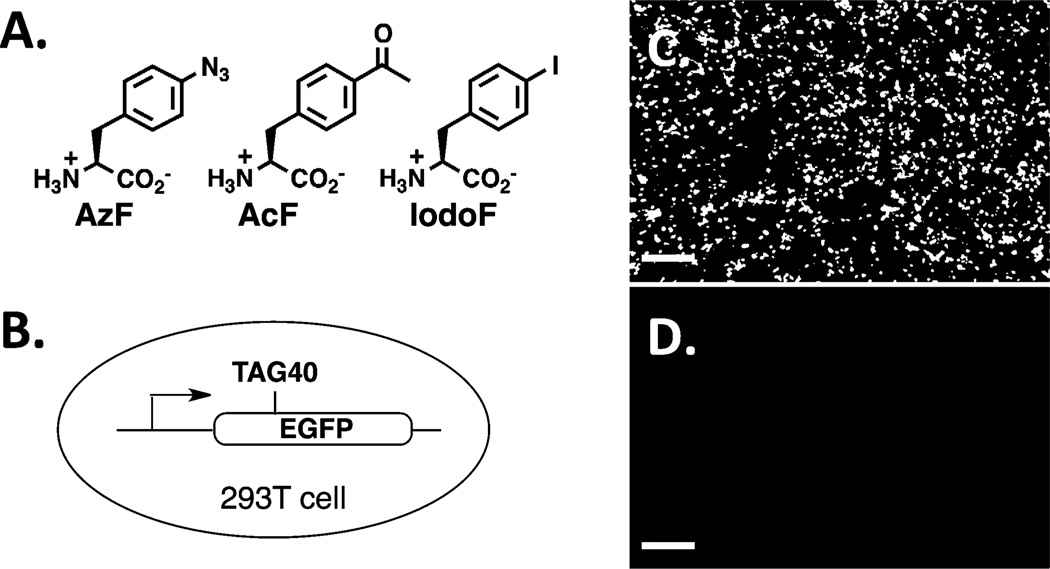
Two systems, E. coli tRNATyr–tyrosyl-tRNA synthetase (TyrRS) pair and archaeal tRNAPyl–pyrrolysyl-tRNA synthetase (PylRS) pair, have been developed to decode either nonsense[8] or frameshift[9] codons in mammalian cells. However, the complicated assembly process of human viruses makes it very challenging to genetically incorporate UAA* into proteins in live human viruses using the above two systems. During our HIV-1 studies, Chen and co-workers reported the first site-specific incorporation of UAA* into surface proteins of hepatitis D virus using the tRNAPyl–PylRS pair.[10] In this work, we focus on the E. coli tRNATyr–TyrRS pair, and specifically on the tRNATyr–4-azidophenylalanyl-tRNA synthetase (AzFRS), tRNATyr–4-acetylphenylalanyl-tRNA synthetase (AcFRS), and tRNATyr–4-iodophenylalanyl-tRNA synthetase (IodoFRS) pairs.[8a] Since these three amino acids have similar size to tyrosine, we envisaged that the incorporation of these three amino acids will unlikely have detrimental effects on the function of HIV-1 proteins. AzF and AcF (Figure 2A) also contain unique functional groups that allow selective modification of viral proteins through bioorthogonal reactions, which is expected to facilitate future studies on viral replications and immunological evaluation of vaccine candidates. The recent development of tRNAPyl–PylRS pair may also allow us to conservatively replace lysine residues with pyrrolysine analogues in future vaccine studies.
Figure 2.
Controlled EGFP expression in 293T cells. A) Chemical structures of 4-azidophenylalanine (AzF), 4-acetylphenylalanine (AcF), and 4-iodophenylalanine (IodoF). B) EGFP with an Amber codon at position 40. C) EGFP expression in the presence of the tRNATyr–AzFRS pair and 1 mm of AzF. D) EGFP expression in the presence of the tRNATyr–AzFRS pair, but without AzF. Scale bars, 200 µm.
To examine the amber suppression efficiency and fidelity of the tRNATyr–AzFRS pair, 293T cells were co-transfected with a plasmid containing tRNATyr under the control of a human U6 promoter and AzFRS (pAzFRS) with a plasmid encoding enhanced green fluorescent protein (EGFP) with an amber mutation at residue 40 (pEGFP-TAG40). Following transfection, cells were cultured in DMEM media (containing 10% fetal bovine serum (FBS) and 2 mm l-glutamine) with or without 1 mm AzF for 12 h before visualization under a fluorescence microscope (Figure 2C,D). Full-length EGFP was detected only in cells supplemented with 1 mm AzF (Figure 2C), while no EGFP was observed otherwise (Figure 2D). The tandem mass spectrometry data (Figure S4 in the Supporting Information) showed no undesirable incorporation of tyrosine or any other natural amino acids. The amber mutation site contained exclusively 4-aminophenylalanine (aminoF), which is the reduction product of AzF. This observation is consistent with previous reports on mass spectrometry analyses of AzF-containing proteins.[8a, 11] The above results confirm an excellent fidelity of AzF incorporation.
We then went on to test the suppression of an amber codon on HIV-1 genome (pSUMA.c/2821, cat#11748, the infectious molecular clone of a founder/transmitter HIV-1 virus from Dr. John Kappes and Dr. Christina Ochsenbauer through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH; abbreviated hereafter as pSUMA). We first chose to mutate the Tyr132 codon on the Gag (group-specific antigen) protein-encoding gene into an amber codon. The resulting pSUMA-Tyr132 was co-transfected into 293T cells with plasmid pAzFRS. After the cells were grown for 48 h in the presence and the absence of 1 mm AzF, viruses were harvested and the titer of HIV-1 was analyzed using anti-p24 antibody. The presence and the strength of a blue color suggest the presence and the level of the capsid protein p24. As shown in Figure 3A (wells 3 and 4), we observed an AzF-dependent p24 synthesis in pSUMA-Tyr132 mutant due to the essential role of Gag (Gag is processed during maturation to p24) in p24 protein synthesis. The p24 assay confirmed a very high fidelity of AzF incorporation (absorbance values of 0.001 versus 0.453 in the absence and the presence of AzF, respectively; Figure 3A, wells 3 and 4). However, only very low level of p24 synthesis was observed compared to the wild-type pSUMA control (Figure 3A, well 1). On the other hand, very strong p24 synthesis was observed with the tRNATyr–AcFRS pair (Figure S1, well 4) and an undetectable p24 synthesis was observed with the tRNATyr–IodoFRS pair (Figure S1, well 7). Despite amber suppression and the synthesis of p24, unfortunately, no viral infection was observed when the harvested viruses were used to infect TZM-bl cells (Figure 3D and Figure S2A,D). (The TZM-bl cell line stably expresses large amounts of CD4 and CCR5 and should be highly sensitive to infections by diverse isolates of HIV-1. The TZM-bl also contains a genome copy of β-galactosidase gene under the control of the HIV-1 promoter. The infection assay is based on the expression level of β-galactosidase.)
Figure 3.
Control HIV-1 replication with Amber suppression. A) p24 assay after transfection of 293T with pSUMA variants. The absorbance values were determined at 450 nm after the colorimetric reactions were stopped by the addition of 1m H2SO4. The well 11 was used as blank for all measurements. 1, wild-type pSUMA only; 2, wild-type pSUMA + tRNATyr–AzFRS pair + 1 mm AzF; 3, pSUMA-Tyr132 + tRNATyr–AzFRS pair, without AzF; 4, pSUMA-Tyr132 + tRNATyr–AzFRS pair, with 1 mm AzF; 5, pSUMA-Tyr132 + tRNATyr–TyrRS pair; 6, pSUMA-Tyr132; 7, pSUMA-Ala119 + tRNATyr–AzFRS pair, without AzF; 8, pSUMA-Ala119 + tRNATyr-AzFRS pair, with 1 mm AzF; 9, pSUMA-Leu365 + tRNATyr-AzFRS pair, without AzF; 10, pSUMA-Leu365+tRNATyr-AzFRS pair, with 1 mm AzF; 11, negative ELISA control, no p24; 12, positive ELISA control, 125 pgmL−1 p24; B)–I) Infection assays with TZM-bl cells: B) infected with virus collected from (A-1); C) infected with virus collected from (A-2); D) infected with virus collected from (A-4); E) infected with virus collected from (A-3); F) infected with virus collected from (A-5); G) infected with virus collected from (A-6); H) infected with virus collected from (A-8); I) infected with virus collected from (A-10). Scale bars, 100 µm.
To explain the lack of live HIV-1 assembly from the above experiments, we first examined whether amber suppression can negatively affect HIV-1 protein syntheses, virion assembly, and/or infection since HIV-1 uses seven amber codons as stop signal. To this end, wild-type pSUMA was co-transfected with plasmid pAzFRS into 293T cells. After 48 h of cultivation in the presence of 1 mm AzF, viruses were harvested and the titer of HIV-1 was analyzed using anti-p24 antibody. We observed no obvious difference in p24 synthesis (Figure 3A, wells 1 and 2) and infection rate (Figure 3B,C) of wild-type pSUMA in the presence or the absence of the amber suppressing tRNATyr–AzFRS pair. To further eliminate the possibility, we constructed a different HIV-1 mutant that was derived from pNL-GI,[12] which contains a copy of GFP inserted at the nef locus. In this new HIV-1 construct, an amber mutation was made at position 40 of GFP (originally encode a tyrosine residue) and there was no amber mutation on the HIV-1 protein-encoding genes. The resulting HIV-1 variant, pNL-GI-Tyr40, was transfected into 293T cells together with plasmid pAzFRS. Strong GFP fluorescence (Figure 4B) was detected when 1 mm of AzF was included in the culture medium. The fluorescence intensity is comparable to the wild-type pNL-GI (Figure 4A), which indicated a very efficient amber suppression with the tRNATyr–AzFRS pair. The produced HIV-1 viruses were then harvested and used to infect TZM-bl cells. Near wild-type infection was observed (Figure 4C,D). Based on the above observations, we concluded that amber suppression does not have detectable detrimental effects on the viability of HIV-1.
Figure 4.
Expression and infection with pNL-GI variants. A) Fluorescence image of pNL-GI. B) Fluorescence image of pNL-GI-Tyr40 + tRNATyr-AzFRS pair, with 1 mm AzF. C) TZM-bl cell infection with virus collected from (A). D) TZM-bl cell infection with virus collected from (B). Scale bars, 200 µm.
We then tested if UAA* incorporation at Tyr132 of Gag protein interfered with either the proper function or the posttranslational processing of the Gag protein. To this end, the pSUMA-Tyr132 mutant was transfected into 293T cells with a plasmid containing tRNATyr–TyrRS pair. The amber suppression led to the incorporation of a tyrosine residue at position 132, which produced the wild-type Gag protein. The p24 (Figure 3A, well 5) and infection (Figure 3F) assays showed that the pSUMA-Tyr132 mutant, after amber suppression, had near wild-type activities. As a control, when pSUMA-Tyr132 was transfected into 293T cells that did not contain the tRNATyr–TyrRS pair, no p24 synthesis was detected (Figure 3A, well 6) and no live viruses were assembled according to the infection assay (Figure 3G). Combining these results and the results from UAA* incorporation, we concluded that incorporation of UAA* at position Tyr132 must have a negative effect on Gag protein expression and/or function.
We next decided to examine other mutation sites and generated two new HIV-1 mutants, with amber mutation at Ala119 of Gag (pSUMA-Ala119) and Leu365 of Pol (pSUMA-Leu365), respectively. We initially focused on the amber suppression with the tRNATyr–AzFRS pair in the presence of AzF. We observed an AzF-dependent p24 synthesis in pSUMA-Ala119 (Figure 3A, wells 7 and 8). The amber suppression at Ala119 position (Figure 3A well 8) was apparently stronger than that at position Tyr132 of Gag (Figure 3A well 4) and has similar good fidelity (absorbance values of 0.000 versus 2.127 in the absence and the presence of AzF, respectively; Figure 3A, wells 7 and 8). On the other hand, the synthesis of p24 in pSUMA-Leu365 was not AzF-dependent (Figure 3A, wells 9 and 10) since the amber mutation is in Pol, which does not affect p24 synthesis. Both mutants showed infection (Figure 3H,I) but with much lower activity than wild-type pSUMA (Figure 3B). As a control, no infection was observed when AzF was not provided in the culture medium of 293T cells after transfection (Figure S3 A,B). We next examined the tRNATyr–AcFRS pair and tRNATyr–IodoFRS pair. As shown in Figure S1, a range from non-detectable to relative strong amber suppression was observed with the two tRNA–aaRS pairs and the different amber mutation sites (Figure S1, wells 5, 6, 8, and 9). However, no infection was detected when tRNATyr–AcFRS pair and tRNATyr–IodoFRS pair were used to generate pSUMA-Ala119 and pSUMA-Leu365 mutants (Figure S2B,C,E,F). Since the amber suppression efficiency did not correlate well with the infection results, we suspected that the structure of UAA*s and the incorporation sites might still partially interfere with the protein function.
An additional mutation site, Tyr59TAG (pSUMA-Tyr59) of HIV-1 protease was then examined. Since Tyr59 has similar structure to AzF and is away from the active site (PDB 1EBZ),[13] mutation of Tyr59 to AzF mightd not cause undesired disturbance of protein structure and function. Indeed, the pSUMA-Tyr59 mutant showed better infection activity (Figure 5B) than pSUMA-Ala119 (Figure 3H) and pSUMA-Leu365 (Figure 3 I). As a control, no live pSUMA-Tyr59 virus was produced in the absence of AzF during viral assembly in 293T cells according to the infection assay (Figure S3C). A comparison between the observed tissue culture infectious dose 50 (TCID50) values in the presence (1.31 × 103) and the absence of AzF (0.00) implicates high fidelity of the tRNATyr–AzFRS pair. But the amber suppression efficiency still needs further improvement in order to obtain better titer of live HIV-1 mutants. Nonetheless, the above results suggest that the UAA*-mediated amber suppression strategy can be used to produce live HIV-1 and the resulting virus is infectious. Since the infected cells do not contain the amber suppression machinery (the special tRNA–aaRS pair and UAA*) that is required for HIV-1 assembly, no new virus can be assembled after the initial infection.
Figure 5.
Infection assays with TZM-bl cells. A) Infected with wild-type pSUMA. B) Infected with pSUMA-Tyr59. C) Infected with pSUMA-Trp36Gln127. Scale bars, 200 µm. The doses used for infection are shown as TCID50 values. The wild-type pSUMA was diluted 4-fold and the pSUMA mutants were concentrated 15-fold in order to adjust the infectious dose of each variant to similar levels.
While the fidelity of UAA* incorporation is very good with a comparable fidelity of protein synthesis in nature, it remains possible that the HIV-1 can regain functional replication by mutating amber codon back to a sense codon since HIV-1 has relatively high genetic variability. Therefore, although we did not observe any case where the amber codon was mutated back to sense codon under the cell culture conditions, such possibility might become greater in a longer time span or in vivo. We envisaged that the introduction of multiple amber codons into the essential genes of HIV-1 could be used to further tighten the control on virus replication. If we take the virus mutation rate as approximately 3 × 10−5 per nucleotide base per cycle of replication[14] and the virus life cycle as one day, the possibility of mutating all amber codons back to sense codons is approximately 1% and 0.01%, respectively, during human life span (100 years) when one and two amber codons are used. Since the amber sites we chose are not in regions with high mutation rate, we expect a very low chance for HIV-1 mutant to regain virulence if two or more amber codons are used. To examine if the suppression of two amber codons on the HIV-1 genome by the tRNATyr–AzFRS pair is efficient enough to produce meaningful amounts of live HIV-1, we constructed an HIV-1 mutant in which two amber mutations, Trp36 and Gln127 of matrix domain (MA) of Gag (pSUMA-Trp36Gln127), were made according to the crystal structure of the MA protein (PDB: 2H3F).[15] The Trp36 and Gln127 are not key residues for MA’s interaction with phosphatidylinositol 4,5-bisphosphate.[15, 16] In addition, these mutation sites are located relative earlier on the HIV-1 genome, which reduces the possibility of interference with the alternative reading frame and posttranslational processing of HIV-1 proteins. Encouragingly, we observed similar infection activity with pSUMA-Trp36Gln127 (Figure 5C) compared to HIV-1 containing single amber mutation (Figure 5B). As a control, no live pSUMA-Trp36Gln127 virus was produced in the absence of AzF during viral assembly in 293T cells according to the infection assay (Figure S3D).
In conclusion, we have demonstrated that precise control of HIV-1 viability in vitro can be achieved by using a UAA*-mediated amber codon suppression strategy. The above work is an important step towards the development of a safe and effective HIV-1 vaccine in order to combat worldwide AIDS pandemic. Our approach can also be used to improve the safety of other live-attenuated or replication-competent vector-based vaccines. In addition, the ability to introduce unique functional groups into HIV-1 proteins enables further modification of those proteins through bioorthogonal reactions, which could facilitate our future and other studies on HIV-1 in order to find a cure for HIV-1 related diseases.
Encouraged by our initial results, we are currently working on improving the efficiency of UAA*-mediated amber codon suppression. We are also constructing HIV-1 mutants that contain genome insertion of either amber- or quadruplet-decoding tRNA-aaRS pair. These mutants will be examined for multi-cycle infection in vitro with the addition and the removal of UAA* as an on and off switch. Such manipulation will later allow us to control HIV-1 replication in humanized mouse for immunological evaluation of the vaccine candidates.[17]
Supplementary Material
Acknowledgments
This work was supported by a grant from the Nebraska Research Initiative and a New Faculty Startup Fund to J.G. from the Chemistry Department of the University of Nebraska–Lincoln.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201402092.
Contributor Information
Nanxi Wang, Department of Chemistry, University of Nebraska–Lincoln, Lincoln, NE 68588 (USA).
Dr. Yue Li, Nebraska Center for Virology & School of Biological Sciences, University of Nebraska–Lincoln, Lincoln, NE 68588 (USA)
Dr. Wei Niu, Department of Chemistry, University of Nebraska–Lincoln, Lincoln, NE 68588 (USA)
Dr. Ming Sun, Nebraska Center for Virology & School of Biological Sciences, University of Nebraska–Lincoln, Lincoln, NE 68588 (USA)
Dr. Ronald Cerny, Department of Chemistry, University of Nebraska–Lincoln, Lincoln, NE 68588 (USA)
Prof. Qingsheng Li, Department of Chemistry, University of Nebraska–Lincoln, Lincoln, NE 68588 (USA)
Prof. Jiantao Guo, Department of Chemistry, University of Nebraska–Lincoln, Lincoln, NE 68588 (USA)
References
- 1.a) Wu X, Schultz PG. J. Am. Chem. Soc. 2009;131:12497–12515. doi: 10.1021/ja9026067. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chin JW. Science. 2012;336:428–429. doi: 10.1126/science.1221761. [DOI] [PubMed] [Google Scholar]; c) Liu CC, Schultz PG. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 2.a) Barré-Sinoussi F, Ross AL, Delfraissy J-F. Nat. Rev. Microbiol. 2013;11:877–883. doi: 10.1038/nrmicro3132. [DOI] [PubMed] [Google Scholar]; b) Fauci AS, Folkers GK, Dieffenbach CW. Nat. Immunol. 2013;14:1104–1107. doi: 10.1038/ni.2735. [DOI] [PubMed] [Google Scholar]
- 3.Virgin HW, Walker BD. Nature. 2010;464:224–231. doi: 10.1038/nature08898. [DOI] [PubMed] [Google Scholar]
- 4.Fauci AS. Nature. 2008;453:289–290. doi: 10.1038/453289a. [DOI] [PubMed] [Google Scholar]
- 5.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaew-kungwal J, Chiu J, Paris R, Premsri N, Namwat C, De SM, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. N. Engl. J. Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 6.Koff WC, Johnson PR, Watkins DI, Burton DR, Lifson JD, Hasenkrug KJ, McDermott AB, Schultz A, Zamb TJ, Boyle R, Desrosiers RC. Nat. Immunol. 2006;7:19–23. doi: 10.1038/ni1296. [DOI] [PubMed] [Google Scholar]
- 7.Hohsaka T, Sisido M. Curr. Opin. Chem. Biol. 2002;6:809–815. doi: 10.1016/s1367-5931(02)00376-9. [DOI] [PubMed] [Google Scholar]
- 8.a) Liu W, Brock A, Chen S, Chen S, Schultz PG. Nat. Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]; b) Chen PR, Groff D, Guo J, Ou W, Cellitti S, Geierstanger BH, Schultz PG. Angew. Chem. 2009;121:4112–4115. doi: 10.1002/anie.200900683. Angew. Chem. Int. Ed. 2009, 48, 4052 – 4055. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee K-F, Slesinger PA, Wang L. Nat. Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Gautier A, Deiters A, Chin JW. J. Am. Chem. Soc. 2011;133:2124–2127. doi: 10.1021/ja1109979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu W, Schultz PG, Guo J. ACS Chem. Biol. 2013;8:1640–1645. doi: 10.1021/cb4001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin S, Yan H, Li L, Yang M, Peng B, Chen S, Li W, Chen PR. Angew. Chem. 2013;125:14220–14224. doi: 10.1002/anie.201305787. Angew. Chem. Int. Ed. 2013, 52, 13970 – 13974. [DOI] [PubMed] [Google Scholar]
- 11.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 12.Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 13.Andersson HO, Fridborg K, Lowgren S, Alterman M, Muhlman A, Bjorsne M, Garg N, Kvarnstrom I, Schaal W, Classon B, Karlen A, Danielsson UH, Ahlsen G, Nillroth U, Vrang L, Oberg B, Samuelsson B, Hallberg A, Unge T. Eur. J. Biochem. 2003;270:1746–1758. doi: 10.1046/j.1432-1033.2003.03533.x. [DOI] [PubMed] [Google Scholar]
- 14.Rambaut A, Posada D, Crandall KA, Holmes EC. Nat. Rev. Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 15.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Proc. Natl. Acad. Sci. USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Vlach J, Saad JS. Proc. Natl. Acad. Sci. USA. 2013;110:3525–3530. doi: 10.1073/pnas.1216655110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chow JYH, Jeffries CM, Kwan AH, Guss JM, Trewhella J. J. Mol. Biol. 2011;413:742. [Google Scholar]
- 17.Lan P, Tonomura N, Shimizu A, Wang S, Yang Y-G. Blood. 2006;108:487–492. doi: 10.1182/blood-2005-11-4388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.