ABSTRACT
The unique Escherichia coli GTPase Der (double Era-like GTPase), which contains tandemly repeated GTP-binding domains, has been shown to play an essential role in 50S ribosomal subunit biogenesis. The depletion of Der results in the accumulation of precursors of 50S ribosomal subunits that are structurally unstable at low Mg2+ concentrations. Der homologs are ubiquitously found in eubacteria. Conversely, very few are conserved in eukaryotes, and none is conserved in archaea. In the present study, to verify their conserved role in bacterial 50S ribosomal subunit biogenesis, we cloned Der homologs from two gammaproteobacteria, Klebsiella pneumoniae and Salmonella enterica serovar Typhimurium; two pathogenic bacteria, Staphylococcus aureus and Neisseria gonorrhoeae; and the extremophile Deinococcus radiodurans and then evaluated whether they could functionally complement the E. coli der-null phenotype. Only K. pneumoniae and S. Typhimurium Der proteins enabled the E. coli der-null strain to grow under nonpermissive conditions. Sucrose density gradient experiments revealed that the expression of K. pneumoniae and S. Typhimurium Der proteins rescued the structural instability of 50S ribosomal subunits, which was caused by E. coli Der depletion. To determine what allows their complementation, we constructed Der chimeras. We found that only Der chimeras harboring both the linker and long C-terminal regions could reverse the growth defects of the der-null strain. Our findings suggest that ubiquitously conserved essential GTPase Der is involved in 50S ribosomal subunit biosynthesis in various bacteria and that the linker and C-terminal regions may participate in species-specific recognition or interaction with the 50S ribosomal subunit.
IMPORTANCE In Escherichia coli, Der (double Era-like GTPase) is an essential GTPase that is important for the production of mature 50S ribosomal subunits. However, to date, its precise role in ribosome biogenesis has not been clarified. In this study, we used five Der homologs from gammaproteobacteria, pathogenic bacteria, and an extremophile to elucidate their conserved function in 50S ribosomal subunit biogenesis. Among them, Klebsiella pneumoniae and Salmonella enterica serovar Typhimurium Der homologs implicated the participation of Der in ribosome assembly in E. coli. Our results show that the linker and C-terminal regions of Der homologs are correlated with its functional complementation in E. coli der mutants, suggesting that they are involved in species-specific recognition or interaction with 50S ribosomal subunits.
INTRODUCTION
GTPases are one of the most widespread protein families in all kingdoms of life (1). A number of GTPases play key roles in eukaryotes, prokaryotes, and archaea by regulating various cellular processes, such as proliferation, cell growth, protein translation, protein translocation, and signal transduction (1–4). To regulate these cellular functions tightly, GTPase proteins undergo three different conformation states: a nucleotide-free empty state, a GTP-bound active state, and a GDP-bound inactive state (1, 2, 5, 6). Escherichia coli contains numerous GTPase proteins, most of which play pivotal roles in ribosome function (either translation or biogenesis). Among these GTPases, Era and RsgA are involved in the maturation of 30S ribosomal subunits, whereas Der (double Era-like GTPase) and ObgE participate in the completion of 50S ribosomal subunits (7–13).
Der, also known as EngA (essential neisserial GTPase) or YphC, is an essential bacterial GTPase (14, 15). The subfamily of Der GTPases belongs to the TrmE-Era-EngA-YihA-Septin-like superfamily of the TRAFAC (translation-factor-related) class (16, 17) and to the HAS-GTPase class (hydrophobic amino acid substituted for catalytic glutamine GTPases) (18). Der is unique among all GTPases as it bears two consecutive GTP-binding domains, G domain 1 (GD1) and G domain 2 (GD2), at the N-terminal region, followed by a unique C-terminal KH domain (14). Based on the crystal structure of Der of Thermotoga maritima, it was postulated that the KH domain participates in protein-protein and/or protein-nucleic acid interactions (19).
Genetic screening revealed that the overexpression of either of two genes encoding the universally conserved bacterial GTPases, ObgE and Der, restored the growth defect and abnormality of the ribosome profile in the ΔrrmJ strain (HB23 strain) (20). RrmJ is a methyltransferase that methylates the U2552 residue in the A loop of 23S rRNA in intact 50S ribosomal subunits (20, 21). Although rrmJ deletion is not lethal, it causes serious defects in cell growth. This growth impairment is caused by the accumulation of 50S and 30S ribosomal subunits in the polysome profiles at the expense of 70S ribosomes and polysomes. Suppression by Der was found to be mediated without methylation at U2552, suggesting that Der plays a novel role in 50S ribosomal subunit assembly (20).
When lysates of wild-type E. coli were fractionated on sucrose gradients, endogenous Der was specifically cofractionated with 50S ribosomal subunits in a GTP-dependent manner (11, 22, 23). In addition, the polysome profiles of Der-depleted cells revealed decreased levels of 70S ribosomes and the accumulation of both 30S and 50S ribosomal subunits compared with the features of wild-type cells (11, 22, 23). In 50S ribosomal subunits isolated from Der-depleted cells, ribosomal proteins (r-proteins) L9 and L18 were significantly diminished, and r-proteins L2 and L6 were marginally reduced (11). All the r-proteins affected by Der depletion belong to the late assembly proteins in 50S ribosomal subunit biogenesis, among which L9 is necessary to structurally stabilize 50S ribosomal subunits when the GTPase activity of Der is low (24). The depletion of Der also causes the accumulation of the rRNA precursors pre-23S and pre-16S rRNA, which are unprocessed at both the 5′ and 3′ ends (11). This evidence demonstrates that Der participates in 50S ribosomal subunit maturation at a later biogenesis step. Interestingly, both GTP-binding domains (GD1 and GD2) were essential for cell growth. Moreover, the two GTP-binding domains function cooperatively, suggesting that GTP-induced conformational changes and GTPase activities are indispensable for cell viability and function (11, 22). Der-depleted cells also exhibited uncharacterized phenotypes, such as morphological filamentation and aberrant chromosomal segregation (14, 25). Although Der interacts with 50S ribosomal subunits and is important for the production of fully mature 50S ribosomal subunits, its precise role in ribosome biogenesis is unclear.
Der homologs from various bacteria, such as Neisseria gonorrhoeae, Bacillus subtilis, and Staphylococcus aureus, were previously revealed to be essential for cell viability (15, 25–27). In addition, der was presumed to be essential for Mycoplasma genitalium, which has one of the smallest genomes, indicating that it belongs to a minimal gene set in bacteria (28, 29). A unique GTPase subfamily of Der is conserved in eubacteria as well as a few plants, and interestingly, Der proteins in these plants are targeted to parts of chloroplasts, such as stroma and the thylakoid membrane (30). Arabidopsis thaliana der-null mutants exhibited an embryonic lethal phenotype, and virus-induced gene silencing of der in Nicotiana benthamiana resulted in defective rRNA processing and ribosome biogenesis in chloroplasts. These findings indicate that plant Der homologs are also involved in ribosome biogenesis in chloroplasts. Thus, there are emerging evidences that bacterial and plant Der proteins comprise a novel 50S ribosomal subunit maturation (or stabilization) GTPase.
In this study, we heterologously expressed Deinococcus radiodurans, Klebsiella pneumoniae, N. gonorrhoeae, Staphylococcus aureus, and Salmonella enterica serovar Typhimurium Der homologs (DrDer, KpDer, NgDer, SaDer, and StDer, respectively) in an E. coli der-null strain and examined their functional complementations to elucidate the conserved function of bacterial Der homologs in 50S ribosomal subunit biogenesis and stabilization. In addition, we constructed chimeric Der proteins to identify critical regions in Der that enable complementation to the phenotype of the der-null strain.
MATERIALS AND METHODS
Construction of the der-null strain.
For linear DNA transformation, the der::kan fragment (der replaced with a kanamycin resistance cassette) was amplified from pUCDerK by PCR (14). Strain BW25113 harboring pKD46 (containing λ Red recombinase and a temperature-sensitive origin [orits]) (31) and the helper plasmid pINEcDer (der+ [wild type] lppp lacpo cat) was subjected to linear DNA transformation. Transformants resistant to chloramphenicol and kanamycin at 30°C were isolated and confirmed for gene disruption by PCR. The resultant strain was used to prepare for P1 transduction (32). The P1 lysate was transduced into strain MG1655 harboring pIEEcttg (14), a helper plasmid containing der, an ampicillin-resistant gene, and a temperature-sensitive replication origin. The transductant resistant to ampicillin and kanamycin at 30°C was tested for temperature sensitivity to 43°C, yielding strain MK (der-null strain) (Table 1). The der deletion in strain MK was confirmed by PCR.
TABLE 1.
E. coli strains used in this study and their genotypes
| Strain | Description | Source or reference |
|---|---|---|
| MG1655 | F− λ− ilvG rfb-50 rph-1 | 52 |
| BW25113 | lacIp4000(lacIq) rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | 31 |
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| MK | MG1655 der::kan(pIEEcttg) | This study |
| HB23 | MG1655 zgi-203::Tn10 Tetr rrmJ(ftsJ)Δ567 | 53 |
| ESC08 | MG1655 der::kan(pBAD33EcDer) | This study |
Plasmids.
DNA fragments of DrDer, KpDer, SaDer, and StDer were amplified by PCR of the genomic DNA in each bacterium using primer sets designed such that the PCR amplicon carried an NdeI site at the 5′ end and a BamHI site at the 3′ end (see Table S1 in the supplemental material). Primer sets to amplify the DNA fragment of NgDer contained an NdeI site at the 5′ end and an EcoRI site at the 3′ end. Subsequently, each PCR fragment was subjected to blunt-end ligation into the SmaI site of pUC19 (see Table S2 in the supplemental material), and these subclones were digested with the appropriate restriction enzymes. Then, each insert was ligated into pIN vectors, yielding pINDrDer, pINKpDer, pINNgDer, pINSaDer, and pINStDer (Table 2). In this manner, pMAL-DrDer, pMAL-KpDer, pMAL-NgDer, pMAL-SaDer, and pMAL-StDer were constructed using pMAL-c5x (New England BioLabs). To construct plant der expression vectors, plasmids T;AtDer, T;NbDer, and T;T1458 (see Table S2) were digested by NdeI and BamHI, after which inserts were cloned into pIN vectors, yielding pINAtDer, pINNbDer, and pINT1458.
TABLE 2.
Plasmids used in this study and their genotypes
| Plasmid | Description | Source or reference |
|---|---|---|
| pIN | lppp lacpo cat | 54 |
| pINEcDer | der+ in pIN | 14 |
| pINDrDer | Deinococcus radiodurans der+ in pIN | This study |
| pINKpDer | Klebsiella pneumoniae der+ in pIN | This study |
| pINNgDer | Neisseria gonorrhoeae der+ in pIN | This study |
| pINSaDer | Staphylococcus aureus der+ in pIN | This study |
| pINStDer | Salmonella enterica serovar Typhimurium der+ in pIN | This study |
| pINAtDer | pIN expressing N-terminally truncated Der from Arabidopsis thaliana | This study |
| pINNbDer | pIN expressing N-terminally truncated Der from Nicotiana benthamiana | This study |
| pINT1458 | Nicotiana benthamiana der+ in pIN | This study |
| pET28a | His-tagged expression vector | Novagen |
| pET28EcDer | der+ in pET28a | 45 |
| pMAL-c5x | MBP fusion expression vector | New England Biolabs |
| pMAL-EcDer | der+ in pMAL-c5x | This study |
| pMAL-DrDer | Deinococcus radiodurans der+ in pMAL-c5x | This study |
| pMAL-KpDer | Klebsiella pneumoniae der+ in pMAL-c5x | This study |
| pMAL-NgDer | Neisseria gonorrhoeae der+ in pMAL-c5x | This study |
| pMAL-SaDer | Staphylococcus aureus der+ in pMAL-c5x | This study |
| pMAL-StDer | Salmonella enterica serovar Typhimurium der+ in pMAL-c5x | This study |
| pBAD33 | Expression vector, Para promoter, cat | 55 |
| pBAD33EcDer | der+ in pBAD33 | This study |
| pBAD33DrC1 | Chimeric D. radiodurans der in pBAD33 | This study |
| pBAD33DrC2 | Chimeric D. radiodurans der in pBAD33 | This study |
| pBAD33DrC3 | Chimeric D. radiodurans der in pBAD33 | This study |
| pBAD33NgC1 | Chimeric N. gonorrhoeae der in pBAD33 | This study |
| pBAD33NgC2 | Chimeric N. gonorrhoeae der in pBAD33 | This study |
| pBAD33NgC3 | Chimeric N. gonorrhoeae der in pBAD33 | This study |
| pKD46 | γ β exo (λ Red recombinase; bla orits) | 31 |
| pIEEcttg | der+ bla orits | 14 |
The chimeric der genes were constructed using an overlapping PCR method, as shown in Fig. S1 in the supplemental material. Primers P1 + P2 and P3 + P4 were used to construct pBAD33DrC1 and pBAD33NgC1, respectively, both of which contain an E. coli linker. The E. coli C-terminal region was amplified using primers P6 and P7, and this PCR fragment was fused to the PCR product amplified by primers P1 + P5, resulting in pBAD33DrC2 and pBAD33NgC2. Plasmids pBAD33DrC3 and pBAD33NgC3 were constructed with primers P1 + P5 and P6 + P7 using pBAD33DrC1 and pBAD33NgC1 as the templates, respectively. Each PCR fragment was digested with NdeI-EcoRI or NdeI-BamHI and ligated into pBAD33 vectors (Table 2). To construct pBAD33EcDer, pINEcDer was digested with NdeI and EcoRI, and the insert was ligated into pBAD33.
Bacterial strains and growth conditions.
Strain MK (der-null strain) was routinely used for the heterologous expression of Der homologs. Transformants resistant to ampicillin, chloramphenicol, and kanamycin at 30°C were isolated and precultivated at 30°C in Luria-Bertani (LB) medium containing the same antibiotics for 4 h. Portions of cultures were centrifuged to remove antibiotics and then resuspended in prewarmed LB medium containing kanamycin and chloramphenicol. The cultures were shifted to 43°C and then repeatedly diluted (1:3 to 1:5 ratio) into a fresh prewarmed medium during culture at this temperature. HB23 strains were grown in LB medium, as described previously (33), after which they were diluted five times, and their optical density at 600 nm (OD600) values were measured to develop a growth curve. Each antibiotic was applied at a concentration of 50 μg/ml.
Protein overexpression and purification.
All pMAL-c5x expression vectors were introduced to the E. coli BL21(DE3) strain, after which transformants were grown to an OD600 of 0.5 to 0.6 in 4 liters of LB medium containing ampicillin. Cells were then induced with 0.3 mM isopropyl β-d-thiogalactopyranoside (IPTG) and further grown at 37°C for 2 to 4 h. The induced cells were subsequently harvested by centrifugation at 4°C, after which the cell pellets were washed with 10 mM Tris HCl (pH 6.8) and subjected to a second centrifugation. The cell pellets were then resuspended in 20 ml of buffer A (20 mM Tris HCl [pH 7.5], 200 mM KCl, 1 mM EDTA), lysed by sonication, and centrifuged at 10,000 × g for 25 min at 4°C. The resulting supernatant was subjected to ultracentrifugation at 70,000 × g for 1 h at 4°C in a Beckman 70Ti rotor to remove the membrane and insoluble fractions. Next, the soluble fractions were diluted with six volumes of buffer A, and the diluted soluble fraction was applied to amylose resin (New England BioLabs) that had been equilibrated with buffer A. The column was subsequently washed with buffer A, after which maltose-binding protein (MBP) fusion proteins were eluted with buffer A containing 10 mM maltose. Fractions containing MBP fusion proteins were pooled and dialyzed twice against 2 liters of buffer A without EDTA.
To purify His-tagged proteins, pET28EcDer was introduced into E. coli BL21(DE3), and transformants were grown to an OD600 of 0.5 to 0.6 in 1 liter of LB medium containing kanamycin. Next, cells were induced with 0.5 mM IPTG and grown at 37°C for 1.5 h, after which they were harvested by centrifugation at 4°C. Cell pellets were washed with 10 mM Tris HCl (pH 6.8), followed by a second centrifugation. The cell pellets were subsequently resuspended in 20 ml of buffer B (50 mM Tris HCl [pH 7.5], 200 mM KCl, 10 mM imidazole), lysed by sonication, and centrifuged at 10,000 × g for 25 min to remove cell debris and unbroken cells. The membrane fraction was removed, as described previously. The final soluble fractions were applied to a nickel-nitrilotriacetic acid-agarose resin (Qiagen) column that had been equilibrated with buffer B. The column was washed with buffer C (50 mM Tris HCl [pH 7.5], 200 mM KCl, 20 mM imidazole), after which His-tagged proteins were eluted with buffer D (50 mM Tris HCl [pH 7.5], 200 mM KCl, 250 mM imidazole). Fractions containing His-tagged proteins were pooled and dialyzed twice against 2 liters of buffer E (20 mM Tris HCl [pH 7.5], 200 mM KCl, 5 mM β-mercaptoethanol, and 5% glycerol).
For further purification, dialyzed proteins were loaded onto a Hi-Load 16/60 Superdex 200 prep-grade column (GE Healthcare) and eluted with buffer A without EDTA at a flow rate of 1 ml/min. Fractions (3 ml per fraction) were collected and monitored using a UV monitor.
SDS-PAGE and Western blot.
SDS-PAGE and Western blot analyses were conducted as described previously (14). The immunoassay part of Western blot analyses was conducted using polyclonal anti-Der antiserum generated by Pocono Rabbit Farm and Laboratory (Canadensis, PA).
Sucrose density gradient sedimentation.
Polysomes were prepared and resolved as described previously, with minor modifications (34). In brief, strain MK was grown at 30°C for 3 h in 100 ml of LB medium containing kanamycin and ampicillin, and MK transformants harboring pINEcDer, pINKpDer, or pINStDer were grown at 30°C for 4 h in 100 ml of LB medium containing ampicillin, kanamycin, and chloramphenicol. Portions of cultures were centrifuged to remove antibiotics and then resuspended in prewarmed LB medium containing 1 mM IPTG, after which the cultures were shifted to 43°C. During incubation at 43°C, the cultures were repeatedly diluted (1:10 for strain MK or 1:3 to 1:5 for MK transformants) using fresh prewarmed medium. After 6 h of incubation at 43°C, polysomes were trapped by the addition of chloramphenicol to the culture at a final concentration of 0.1 mg/ml. Following an additional 3 min of incubation, cells were harvested by centrifugation. The cell pellet was then resuspended in 0.5 ml of buffer BP (20 mM Tris HCl [pH 7.5], 10 mM MgCl2, 100 mM NH4Cl, and 5 mM β-mercaptoethanol). For immunoassay analysis of colocalization, the cell pellet was resuspended in 0.5 ml of buffer BP containing 0.1 mM GMP-PNP (a nonhydrolyzable analog of GTP) or GDP and placed in a Beckman ultracentrifuge tube. Cells were then lysed by immersing the tube into a liquid nitrogen bath for 1 min, followed by thawing in cold water until no traces of ice remained. This freeze-thaw cycle was repeated twice, and the lysate was subsequently subjected to centrifugation at 17,000 × g for 30 min at 4°C. Polysomes and subunits were resolved as described previously (11). HB23 strains were grown at 30°C in 100 ml of LB medium until reaching an OD600 of approximately 0.6, and their polysome and subunit profiles were analyzed as described previously.
Isolation of ribosomal subunits.
The ribosomal subunits from the wild-type strain (MG1655) were separated using a 5% to 25% linear sucrose density gradient containing 1 mM MgCl2. The fractions containing 50S and 30S ribosomal subunits were combined, and sucrose in the combined fraction was diluted by adding buffer BP containing 1 mM MgCl2. Isolated subunits were concentrated by ultrafiltration (10-kDa molecular-weight cutoff; Vivaspin). The quantity of ribosomal subunits in buffer BP containing 1 mM MgCl2 was determined by absorbance at 260 nm (35).
GTPase assay.
Enzymatic assays were performed as described previously, with minor modifications (14). The release of free inorganic phosphate was measured using the malachite green phosphate assay kit (BioAssay Systems), according to the manufacturer's instructions. The reaction was conducted in a 100-μl reaction mixture of 50 mM Tris HCl (pH 7.5), 400 mM KCl, 5 mM MgCl2, 128 μM GTP, and 0.2 μM purified Der proteins at 37°C for 20 min. The reaction was stopped by adding working reagent prepared as described in the kit, after which the reaction mixture was incubated for 30 min at room temperature for color development. The absorbance was then measured at 620 nm using an Epoch microplate spectrophotometer (Biotek). Each value was averaged from at least two independent experiments.
RESULTS
Restoration of growth of E. coli Der-depleted cells via the expression of Der homologs.
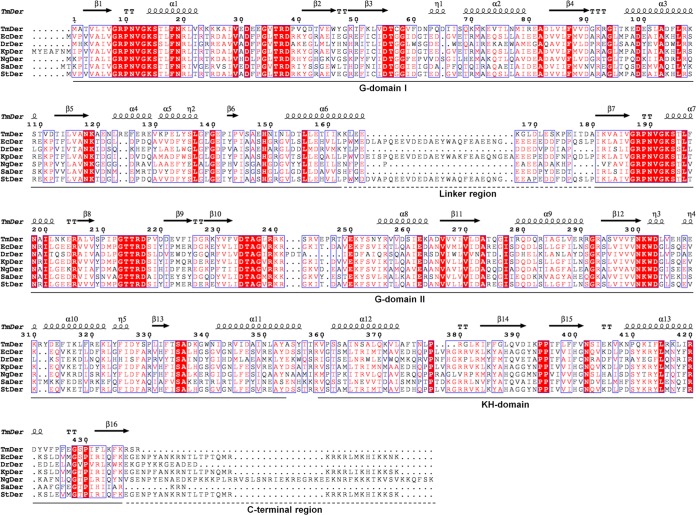
Our goal in this study was to elucidate the conserved and essential role of Der GTPase from various bacteria; thus, we compared and analyzed the primary sequences through multiple-sequence alignment of Der homologs prior to functional characterization of Der GTPases (Fig. 1). The alignment revealed that both G domains were highly similar and well conserved, whereas the linker region between GD1 and GD2 and the C-terminal region varied in length and amino acid composition. Among these, StDer and KpDer displayed the highest homology with E. coli Der (EcDer). In the case of DrDer and SaDer, the linker and C-terminal regions were shorter than those of EcDer. Interestingly, NgDer has a shorter linker region but a longer C-terminal region than EcDer. In addition, we examined the similarity and identity scores of each domain of the Der homologs using BLAST. Comparison with the full amino acid sequence of EcDer revealed that StDer had the highest similarity and identity (99% and 97%, respectively), followed by KpDer (96% and 92%, respectively) (see Table S3 in the supplemental material). The similarities of DrDer, NgDer, and SaDer were 55%, 67%, and 61%, respectively, and the identities of these Der homologs were less than 50%. Among the Der homologs, DrDer displayed the lowest similarity and identity. We also examined the similarity and identity scores of each domain of the Der homologs by analyzing their partial sequences. KpDer and StDer, which are most similar to EcDer, exhibited similarity scores of 96% to 100% in GD1, 98% to 99% in GD2, and 96% to 98% in the KH domain. The remaining Der homologs, namely, DrDer, NgDer, and SaDer, had similarity scores of 59% to 67% in GD1, 64% to 80% in GD2, and 51% to 68% in the KH domain, which were lower than the scores of StDer and KpDer. As the lengths of the linker and C-terminal regions of Der homologs varied, some homologs could not be compared with EcDer. With regard to the linker region, only KpDer and StDer could be analyzed. The similarity scores of these homologs were 90% and 95%, respectively. For the C-terminal region, KpDer, StDer, and NgDer were subjected to multiple-sequence alignment, and they had similarity scores of 96%, 100%, and 71%, respectively. These findings indicate that GD2 is the most highly conserved region in Der homologs, followed by GD1 and the KH domain. Overall, comparison of the primary sequences indicated that KpDer and StDer can likely serve as substitutes for EcDer in E. coli.
FIG 1.
Multiple-sequence alignment of Der homologs. Sequence alignment was conducted using ClustalW and ESPript (50, 51). G domain 1, G domain 2, and the KH domain are shown, and high conservation is noted within these domains. In contrast, there are significant variations between the linker (first dashed line) and C-terminal regions (second dashed line). The numbers correspond to the residues. The GenBank accession numbers for the Der homologs are as follows: TmDer, AHD18661.1; EcDer, AAC75564.2; DrDer, AAF11852.1; KpDer, CCN30978.1; NgDer, AAC63508; SaDer, BAF78346.1; and StDer, AAL21413.1.
To determine whether Der homologs in other bacteria inherently function in 50S ribosomal subunit assembly and can complement E. coli der deletion, we constructed a der-null strain (strain MK). In this strain, the open reading frame of der on the chromosome was replaced with a kanamycin resistance cassette. In addition, strain MK harbors pIEEcttg, a helper plasmid containing E. coli der with its own promoter and a temperature-sensitive replication origin (14). We cloned Der homolog genes in the IPTG-inducible expression vector pIN, yielding pINDrDer, pINKpDer, pINNgDer, pINSaDer, and pINStDer, as described in Materials and Methods.
These plasmids were transformed into strain MK, overnight cultures of each transformant were diluted 105-fold in LB medium, and a 50-μl aliquot of the diluted culture was spread onto LB agar plates. Because the temperature-sensitive helper plasmid pIEEcttg can replicate at 30°C, colonies must be formed on LB agar plates containing ampicillin, kanamycin, and chloramphenicol (LB + AKC) at 30°C. Conversely, pIEEcttg does not replicate at 43°C, and it is diluted out of the cells after proliferation. If Der homologs of other bacteria were functionally complementary, colonies could be formed at 43°C on LB agar plates containing kanamycin and chloramphenicol (LB + KC). All transformants harboring the aforementioned plasmids formed colonies at 30°C on LB + AKC plates (Fig. 2A). Among these, only MK/pINStDer (MK strain harboring pINStDer) formed colonies on LB + KC plates even in the absence of IPTG, whereas MK/pINKpDer could form colonies on LB + KC plates only in the presence of 1 mM IPTG at 43°C. To confirm the absence of a helper plasmid, the diluted culture was spread onto LB agar plates containing ampicillin and kanamycin (LB + AK) at 43°C. As shown in Fig. 2A, colonies were not formed on LB + AK plates, indicating that all transformants lost a helper plasmid at 43°C and the functional complementation by StDer and KpDer was solely due to expression from pINStDer and pINKpDer.
FIG 2.
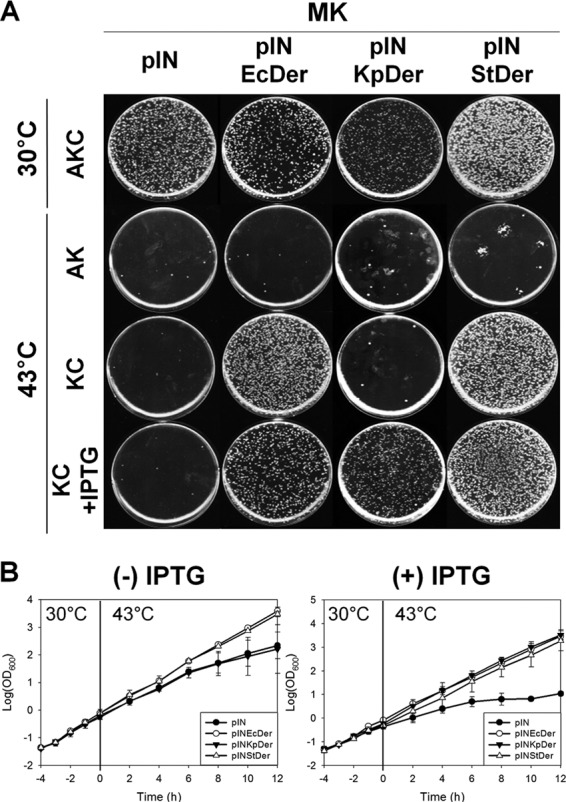
Growth of the der-null strain expressing E. coli Der or its homologs. (A) Complementation of strain MK by Der homologs found in other bacteria. First, 50-μl aliquots of diluted overnight cultures were spread onto LB plates containing the indicated antibiotics with or without 1 mM IPTG, after which the plates were incubated at 30°C or 43°C overnight. A, C, and K indicate ampicillin, chloramphenicol, and kanamycin, respectively. (B) Growth curves of the der-null strain producing E. coli Der or Der homologs. Transformants were incubated in LB medium with or without 1 mM IPTG. Growth of transformants was measured as described in Materials and Methods.
To confirm these results in liquid medium, we monitored the growth of strain MK expressing Der homologs for 12 h at 43°C. After a temperature shift from 30°C to 43°C, only MK/pINStDer exhibited similar growth to the pINEcDer transformant in LB medium without IPTG (Fig. 2B). MK/pINKpDer displayed nearly identical growth to strain MK harboring an empty vector. However, when cells were cultured in LB medium containing 1 mM IPTG, both pINStDer and pINKpDer transformants grew similarly to the isogenic wild-type pINEcDer transformant. MK/pINDrDer and MK/pINNgDer grew similarly to strain MK harboring pIN, even in the presence of IPTG (see Fig. S2 in the supplemental material). We also examined whether plant Der could complement E. coli der deletion, and neither Nicotiana benthamiana Der (NbDer) nor Arabidopsis thaliana Der (AtDer) could rescue the der-null phenotype (see Fig. S3A in the supplemental material). These results suggest that the Der homologs of gammaproteobacteria StDer and KpDer were able to complement the der-null phenotype, which is likely to be due to the similarity between primary sequences.
Expression of Der homologs rescues ribosome defects of E. coli Der-depleted cells.
Bacillus subtilis YphC and E. coli Der interact with the 50S ribosomal subunit in a GMP-PNP-dependent manner, whereas Der-depleted cells disintegrate 50S ribosomal subunits accumulating aberrant particles under low Mg2+ conditions (11, 23). In the next experiment, we conducted sucrose gradient sedimentation analysis and tested the colocalization of Der homologs with E. coli 50S ribosomal subunits to determine whether Der homologs could bind to the E. coli 50S ribosomal subunit and restore the abnormal ribosome profile of the der-null strain. First, we checked whether anti-EcDer antiserum cross-reacted with other Der homologs. To accomplish this, wild-type E. coli cells were transformed with an empty vector, pINEcDer, pINDrDer, pINKpDer, pINNgDer, pINSaDer, or pINStDer. Cell lysates of each transformant were then subjected to 12.5% SDS-PAGE, followed by Western blot analysis using anti-EcDer antiserum (see Fig. S4A in the supplemental material). The results revealed that anti-EcDer antiserum cross-reacts as strongly with KpDer and StDer as with EcDer; however, it did not interact with the remaining Der homologs. The result of Coomassie blue staining showed that all Der homologs were expressed at similar levels, suggesting that cross-reactivity with Der homologs is dependent on antiserum specificity but not on the expression level (see Fig. S4B in the supplemental material).
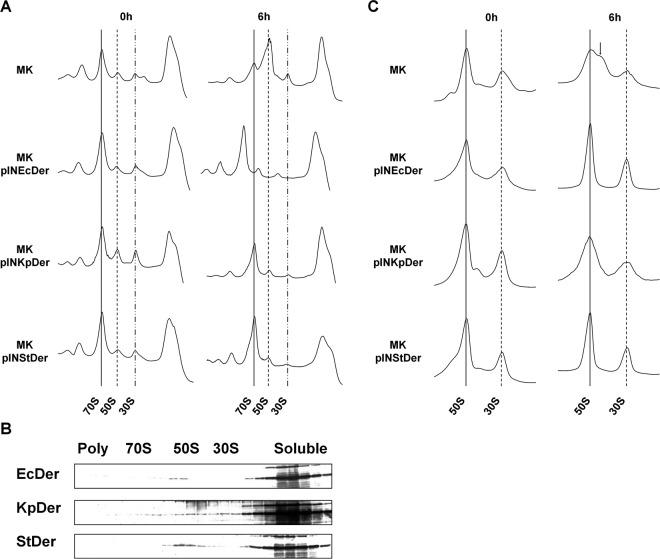
We next investigated whether the expression of Der homologs could recover the ribosome profile of Der-depleted cells. Strain MK cells harboring Der homolog expression vectors were cultured in medium containing 1 mM IPTG, and cell cultures from 0 and 6 h after temperature upshift were withdrawn for polysome profile analysis. Cleared cell lysates were then subjected to 5% to 40% sucrose density gradient fractionation. As shown in Fig. 3A, strain MK had dramatically reduced amounts of 70S ribosomes with a concomitant increase in free 50S and 30S ribosomal subunits at 6 h. However, this abnormal ribosome profile was not observed in MK/pINEcDer. Lysates obtained from MK/pINKpDer or MK/pINStDer exhibited a normal ribosome profile that closely resembled that of MK/pINEcDer. These results suggest that KpDer and StDer are capable of replacing EcDer during ribosome assembly. Western blot analyses of the fractions collected upon polysome analysis were performed using anti-EcDer antiserum. The results revealed that the exogenously expressed EcDer, KpDer, and StDer specifically comigrated with 50S ribosomal subunits in the presence of GMP-PNP but not with 70S monosomes or 30S ribosomal subunits (Fig. 3B). These results demonstrate that KpDer or StDer specifically interacts with the 50S ribosomal subunit of E. coli and restores ribosome assembly in der-null strains. Accordingly, these findings are consistent with the idea that KpDer or StDer participates in 50S ribosomal subunit biogenesis in the respective bacterium.
FIG 3.
The phenotypes of heterologously complemented der deletion and Western blot analysis of Der homologs. (A) Polysome profiles of der-null strains complemented with Der homologs. Samples were collected at 0 and 6 h after temperature shift. Cell lysate for polysome analysis was prepared and separated as described in Materials and Methods. (B) Cofractionation of Der with 50S ribosomal subunits. Samples collected at 6 h after temperature shift were mixed with GMP-PNP at a final concentration of 100 μM, and cell lysates were prepared and separated as in panel A. Fractions containing trisomes to free RNA were subjected to 12.5% SDS-PAGE, followed by Western blotting. (C) Subunit profiles of der-null mutants complemented with Der homologs. Cell pellets were resuspended in buffer BP containing 1 mM MgCl2. Arrow indicates approximately 40S particles.
Under low levels of Mg2+, Der depletion in E. coli causes 50S ribosomal subunits to be destabilized with a concomitant accumulation of approximately 40S subunits (11). To examine whether KpDer or StDer expression in strain MK rescues this instability of 50S ribosomal subunits, ribosomal subunit profiles were analyzed via a 5% to 25% sucrose gradient sedimentation experiment. As shown in Fig. 3C, the amount of 50S ribosomal subunits in strain MK was decreased at 6 h, whereas that of the approximately 40S particle was increased significantly. Nevertheless, the amount and stability of 30S ribosomal subunits remained unaffected. The integrity of 50S ribosomal subunits was maintained in strain MK cells exogenously expressing EcDer or StDer. It should be noted that the expression of KpDer gave rise to a small 40S peak at 30°C and a wider 50S peak compared to EcDer after 6 h of shift to higher temperature, suggesting partial complementation by KpDer. These results implied that the conserved role of Der homologs contributes to the conformational stability of 50S ribosomal subunits.
Suppression of rrmJ deletion by E. coli Der or Der homologs.
RrmJ participates in 50S ribosomal subunit biogenesis by methylating 23S rRNA and generating the highly conserved 2-O-methyluridine at position 2552 in 23S rRNA (20, 21). The rrmJ-null strain has substantial defects in growth and ribosome assembly, but the phenotypes can be restored via the overexpression of either Der or ObgE (20). In this study, strain HB23 cells were transformed with Der homolog expression vectors to investigate whether Der homologs could also restore the growth defects of strain HB23.
The 50-μl aliquot of overnight cultures was diluted 105-fold in LB medium and spread onto LB agar plates containing chloramphenicol (LB + Cm), after which the plates were incubated at 30°C. HB23/pINStDer (strain HB23 harboring pINStDer) grew as normally as HB23/pINRrmJ or HB23/pINEcDer, even without IPTG (Fig. 4A). HB23/pINKpDer exhibited a similar colony size to transformants containing pIN, whereas HB23/pINDrDer, HB23/pINNgDer, and HB23/pINSaDer grew very slowly (see Fig. S5A in the supplemental material). When 1 mM IPTG was added to LB + Cm plates, consistent results were obtained, indicating that both EcDer and StDer suppress the null phenotype of rrmJ.
FIG 4.
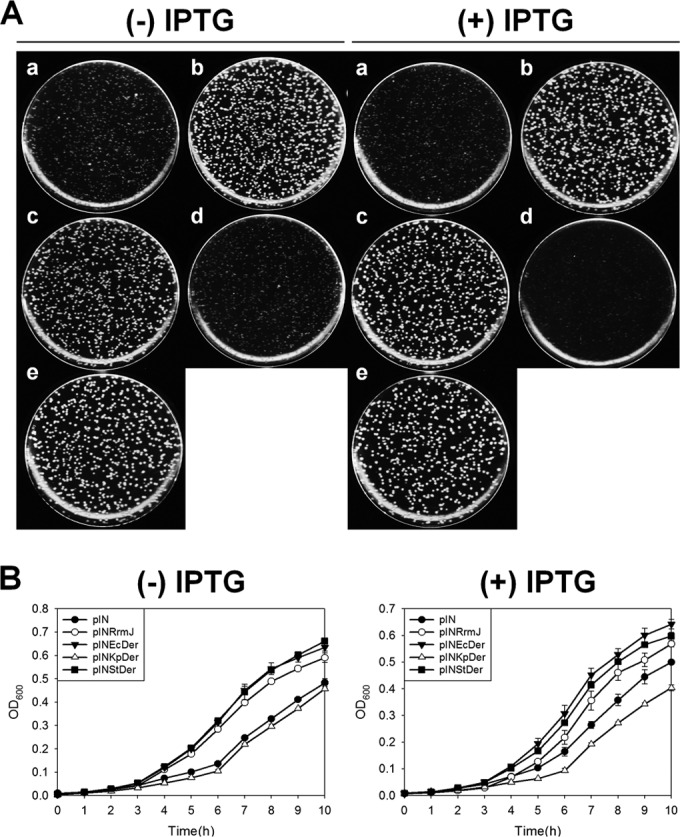
Suppression of the slow growth of rrmJ-null strains by E. coli Der or Der homologs. (A) Suppression of rrmJ deletion by the overexpression of Der homologs. HB23 cells were transformed with the following plasmids: pIN (a), pINRrmJ (b), pINEcDer (c), pINKpDer (d), or pINStDer (e). pIN was used as a negative control, and pINRrmJ was used as a positive control. (B) Growth curve of strain HB23 harboring E. coli Der or other Der homolog expression vectors. Cells were cultured in LB medium containing chloramphenicol or chloramphenicol and 1 mM IPTG at 30°C, after which cultures were diluted five times before measurement of the optical density at 600 nm.
Comparison of the growth curves revealed that the growth defects of the rrmJ-null strain were suppressed only by the overexpression of EcDer or StDer; conversely, the other homologs could not restore the phenotype, and they exhibited a similar growth rate to HB23/pIN (Fig. 4B). Unlike the results obtained from the der deletion complementation experiment, the overexpression of KpDer could not suppress the growth defect of the rrmJ-null strain, consistent with the idea that 50S ribosomal subunits containing unmethylated U2552 were not recognized by KpDer. Furthermore, HB23 transformants expressing DrDer, NgDer, SaDer, NbDer, or AtDer grew more slowly than HB23 cells harboring pIN vectors (see Fig. S3B and S5B in the supplemental material). It is possible that this hampered growth resulted from the toxicity of overexpressed foreign proteins, which was also observed in Fig. S2 in the supplemental material. We cannot exclude the possibility that DrDer, NgDer, and SaDer bind to 50S ribosome subunits in E. coli, preventing the formation of functional 70S ribosomes.
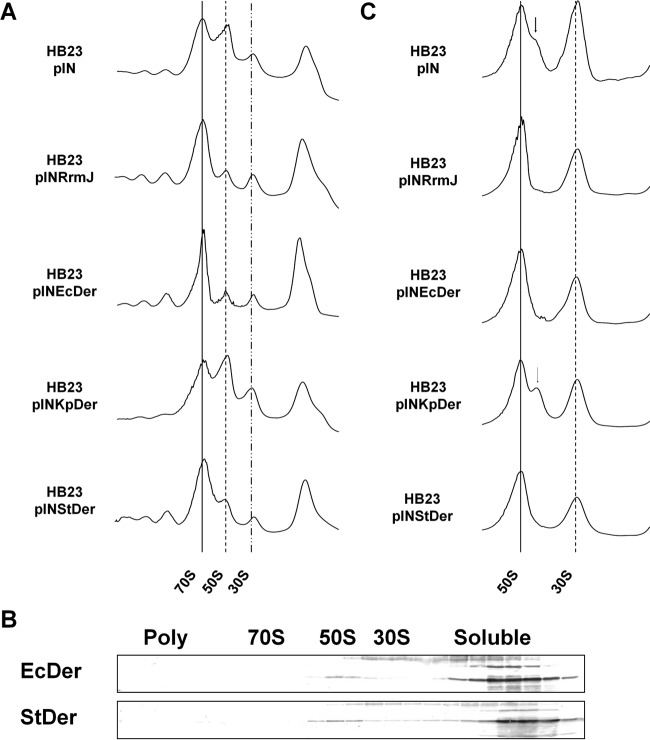
The rrmJ suppression effect was further investigated by analyzing the polysome and ribosomal subunits. HB23 cells harboring each plasmid were cultured in LB medium containing chloramphenicol and 1 mM IPTG until the midexponential growth phase. Cell cultures were then centrifuged, after which cleared cell lysates were prepared, as described previously. Polysome profiles showed that EcDer and StDer restored impairment of the ribosome profile of strain HB23 (Fig. 5A). Ribosomal subunit profiles also revealed that StDer alleviated the accumulation of destabilized 50S particles in strain HB23 (Fig. 5C). To determine whether restoration of the abnormal ribosome profile was caused by the binding of StDer to the 50S ribosomal subunit, Western blot analyses were performed using anti-EcDer antiserum. StDer comigrated with 50S ribosomal subunits but not with 70S monosomes or 30S ribosomal subunits in the presence of 0.1 mM GMP-PNP (Fig. 5B). These results demonstrate that overexpression of StDer led to the normal assembly of 50S ribosomal subunits in strain HB23.
FIG 5.
Phenotypes of rrmJ-null strains expressing Der homologs. (A) Polysome profiles of rrmJ-null strain expressing Der homologs. Cell lysate for polysome analysis was prepared and separated as described in Materials and Methods. (B) Cofractionation of Der with 50S ribosomal subunits in ΔrrmJ. Samples were mixed with GMP-PNP at a final concentration of 100 μM, and cell lysate was prepared and separated as described in Fig. 3B. Fractions containing trisomes to free RNA were subjected to 12.5% SDS-PAGE followed by Western blotting. (C) Ribosomal subunit profiles of the rrmJ-null strain expressing Der homologs. The cell pellets were resuspended with a buffer BP (1 mM Mg2+), and cleared lysates were layered onto a 5% to 25% sucrose gradient (1 mM Mg2+). Arrows indicate approximately 40S particles.
GTPase activities of Der homologs.
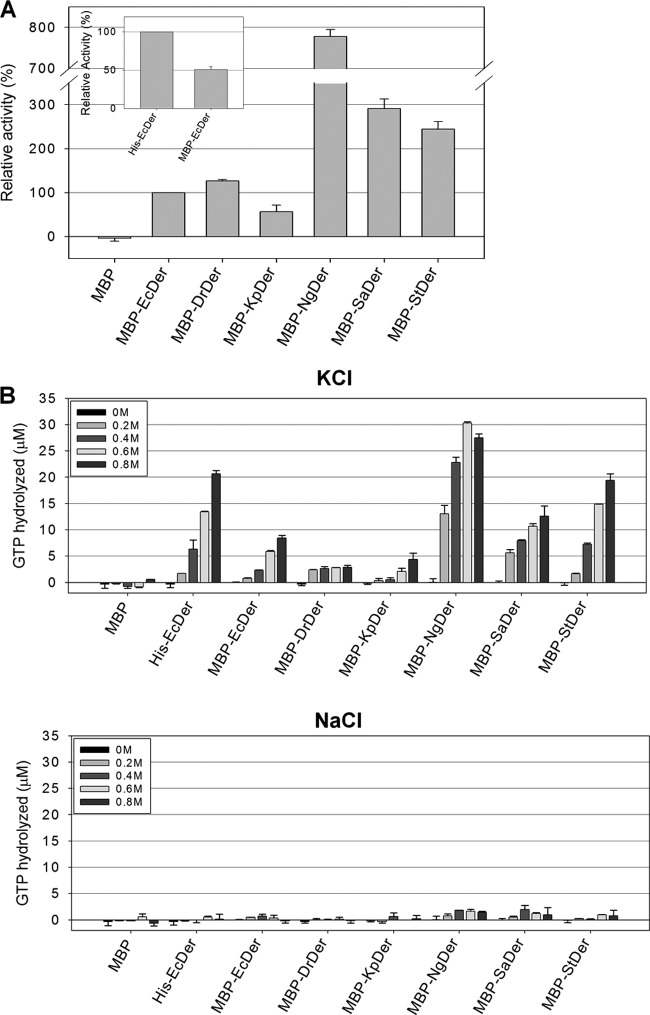
Our results demonstrated that only two Der homologs of gammaproteobacteria rescue the der-null phenotype. As mentioned previously, because the GTPase activity of Der is essential to cellular viability, we compared the GTP hydrolysis activities of EcDer, DrDer, KpDer, NgDer, SaDer, and StDer. Initially, we tried to purify His-tagged Der homologs; however, His-tagged KpDer and StDer began to aggregate within a short time. In order to resolve this problem, the open reading frames of Der homologs were cloned into pMAL-c5x, yielding pMAL-EcDer, pMAL-DrDer, pMAL-KpDer, pMAL-NgDer, pMAL-SaDer, and pMAL-StDer. These plasmids express N-terminal MBP fusion proteins. Each vector was transformed into BL21(DE3), and the proteins were purified as described in Materials and Methods. A GTPase assay was performed in a 100-μl reaction mixture of 20 mM Tris HCl (pH 8.0) containing 5 mM MgCl2, 400 mM KCl, 0.2 μM protein, and 128 μM GTP for 20 min at 37°C. The results revealed that all Der homologs had intrinsic GTPase activity, with MBP-NgDer displaying the greatest activity, followed by MBP-SaDer, MBP-StDer, MBP-DrDer, MBP-EcDer, and MBP-KpDer (Fig. 6A). The GTPase activities of MBP-NgDer, MBP-SaDer, and MBP-StDer were significantly higher than that of MBP-EcDer (777%, 286%, and 243%, respectively). Among the homologs, the GTPase activity of MBP-DrDer was the most similar to that of MBP-EcDer (124%). Only MBP-KpDer had lower activity than MBP-EcDer.
FIG 6.
Comparison of enzymatic properties of Der homologs. (A) GTPase activities of Der homologs. The GTPase assay was conducted in a 100-μl reaction mixture containing 50 mM Tris HCl (pH 7.5), 400 mM KCl, 5 mM MgCl2, 128 μM GTP, and 0.2 μM protein for 20 min at 37°C. (B) Effects of salt on the GTPase activities of Der homologs. The GTPase assay was performed in the same reaction mixture described previously but containing different concentrations (0 to 0.8 M) of KCl or NaCl for 20 min at 37°C.
The MBP tag is bulky, and it may therefore affect the biochemical behavior of the target protein. Therefore, to examine the effects of the MBP tag on the GTPase activity of EcDer, the open reading frame of EcDer was cloned into pET28a to generate pET28EcDer, which was transformed into BL21(DE3). We then expressed and purified His-EcDer, as described previously. MBP-EcDer exhibited lower GTPase activity (approximately 51%) than His-EcDer. Investigation of Thermotoga maritima Der demonstrated that the GTPase activity of GD1 is approximately 2-fold higher than that of GD2, and the substrate-binding site of GD1 faces the C-terminal KH-like domain, which negatively modulates nucleotide binding and/or dissociation (19, 36, 37). As GTPase undergoes a large conformational change upon nucleotide binding and hydrolysis, it is assumed that the decreased GTPase activity of MBP-EcDer is due to the bulky MBP tag, which may interfere with conformational changes and the nucleotide association of Der, or at least GD1. Nevertheless, MBP-EcDer, MBP-KpDer, and MBP-StDer could complement the growth phenotype of strain MK (see Fig. S6 in the supplemental material).
A previous study showed that the GTPase activity of Thermotoga maritima Der was preferentially stimulated by KCl (14). Therefore, a GTPase assay was performed in a 100-μl reaction mixture under the same conditions as those described previously to examine the salt dependence of Der homologs using various concentrations of KCl or NaCl. Interestingly, the GTPase activities of most Der homologs tended to increase with increasing concentrations of KCl (Fig. 6B). Maximal activity of His-EcDer and MBP-EcDer was observed in the presence of 0.8 M KCl. Similarly, MBP-KpDer, MBP-SaDer, and MBP-StDer exhibited maximal GTP hydrolysis in response to 0.8 M KCl, whereas the highest activity of MBP-NgDer was obtained in the presence of 0.6 M KCl. However, at 1 or 1.2 M KCl, the GTPase activities of all homologs were slightly reduced (see Fig. S7 in the supplemental material). MBP-EcDer and MBP-StDer were strongly stimulated by KCl relative to other Der homologs; however, levels of KCl greater than 0.2 M did not further stimulate MBP-DrDer activity. It should be noted that stimulation by NaCl was not observed in all Der homologs. In addition, most ribosome-associating GTPases are activated upon interaction with ribosomal subunits (36, 38–40); therefore, we examined whether the 50S ribosomal subunit of E. coli could stimulate the GTPase activities of Der homologs. As shown in Fig. S8 in the supplemental material, the GTPase activities of all Der homologs were not stimulated by 50S ribosomal subunits. Although significant enhancement of the GTP hydrolysis rate was not observed in the presence of 50S ribosomal subunits, it is possible that the precursor 50S particle is indeed a native stimulator of Der GTPase activity.
Overall, the finding that the GTPase activity of MBP-NgDer was significantly higher than those of MBP-StDer and MBP-KpDer indicates that GTP hydrolysis function is not correlated with the complementation phenotype.
Complementation of der deletion by Der chimeras.
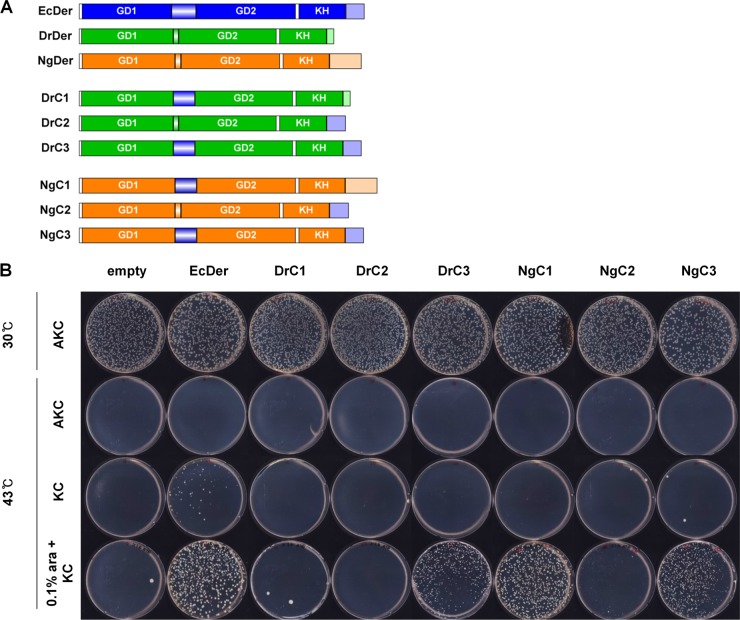
Previously, we observed that truncation of the last 30 amino acid residues of EcDer causes it to lose its specific association with 50S ribosome subunits (33). In addition, the linker of YphC (Bacillus subtilis Der) contributes to the rotation of GTP-binding domain upon nucleotide binding, consequently exposing the ribosome-binding surface (37). Thus, we rationalized that, because noncomplementing Der homologs have very short linker and C-terminal regions (Fig. 1), key factors for a species-specific interaction between Der and 50S ribosomes may reside in the linker and C-terminal regions. To test this hypothesis, we constructed six Der chimeras by incorporating the linker or C-terminal region of EcDer into DrDer or NgDer, as described in Materials and Methods (Fig. 7A; see also Fig. S1 in the supplemental material). To examine complementation ability, strain MK was transformed with chimeric plasmids, and an overnight culture of each transformant was diluted 105-fold in LB medium. Then, a 50-μl aliquot of the diluted culture was spread on LB agar plates. Among six Der chimeras, only DrC3, NgC1, and NgC3 complemented the null phenotype of strain MK in the presence of arabinose (Fig. 7B). All successfully complementing chimeras harbored both linker and long C-terminal regions. Interestingly, although NgC1 possessed a C-terminal region with low homology to EcDer, it could be substituted for EcDer. These results suggested that linker and C-terminal regions are required for Der homologs to replace EcDer. However, due to the abundance of positively charged amino acids in the C-terminal region, we cannot exclude the possibility that the sequence of the C-terminal region is important for complementation.
FIG 7.
Complementation of the der-null strain by Der chimera. (A) Schematic representation of chimeric Der proteins. Six chimeric der clones were constructed from Ecder and Drder or Ngder. Either the linker or C-terminal region of DrDer or NgDer was replaced with that of EcDer, as described in Materials and Methods. (B) MK cells were transformed with pBAD33, pBAD33EcDer, pBAD33DrC1, pBAD33DrC2, pBAD33DrC3, pBAD33NgC1, pBAD33NgC2, or pBAD33NgC3. The transformants were grown at 30°C overnight in LB + AKC medium, and cultures were then diluted 105-fold in LB medium. Next, 50 μl of diluted cultures was spread onto LB plates containing the indicated antibiotics with or without 0.1% arabinose and incubated at 30°C or 43°C overnight.
DISCUSSION
In this study, we found that six Der homologs have intrinsic GTP hydrolysis activities and two homologs (K. pneumoniae and S. enterica serovar Typhimurium Der proteins) can substitute the essential function of E. coli Der in 50S ribosomal subunit assembly. In both solid and liquid cultures, the expression of KpDer or StDer enabled strain MK cells to grow under nonpermissive conditions (Fig. 2). Polysome analysis and cosedimentation experiments revealed that KpDer and StDer can synthesize structurally stable 50S ribosomal subunits in der-null strains (Fig. 3). Therefore, it is more likely that KpDer and StDer are engaged in late steps of 50S ribosomal subunit biosynthesis in the respective bacterium. Interestingly, KpDer was able to complement the der-null phenotype only when IPTG was added. When Der homologs were expressed in MG1655 strains, the expression level of KpDer was lower than that of EcDer or StDer (see Fig. S4B in the supplemental material). Thus, the low expression level of KpDer in MG1655 strains could explain why KpDer complemented der deletion in the presence of IPTG (Fig. 2).
Interestingly, although the der-null phenotype was complemented by the overexpression of either StDer or KpDer, the rrmJ null-phenotype was suppressed by StDer but not by KpDer. U2552 methylated by RrmJ is located in the A loop of 23S rRNA, which is a required modification of the ribosome peptidyltransferase center that forms a noncanonical base pair with C2556 (41, 42). A structural study of fragmented rRNA demonstrated that methylation at U2552 substantially influenced the conformational features of loop residues, including U2552, U2555, and C2556, which mediate tertiary interactions of the A loop within the ribosome structure (43). Although U2552 was universally conserved in the 23S rRNA sequences of all bacteria used in this study, the conformation around U2552 may be differently adopted in each bacterium, as S. aureus does not possess a homolog of RrmJ. Therefore, it is likely that not all Der homologs expressed in E. coli can recognize the local arrangements formed by unmethylated U2552 of the 50S subunit in the ΔrrmJ strain. Apparently, the KH domain of E. coli Der is surrounded by a P loop (nucleotides G2246 to C2258) and A loop (nucleotides G2545 to U2563) and interacts with them (36). Analysis of P-loop or A-loop sequences in 23S rRNA from bacteria used in this study revealed that the P loop was highly conserved among the six bacteria; however, there was extensive variation of A-loop sequences. The A loops of K. pneumoniae, S. enterica serovar Typhimurium, and N. gonorrhoeae were identical to that of E. coli, but the A loops of D. radiodurans and S. aureus were different. Therefore, it appears that complementation by Der homologs is more correlated with A-loop sequences than with P-loop sequences.
Given that noncomplementing Der homologs did have comparably high GTPase activity and that they were expressed well, these factors do not appear to be crucial for functional complementation. Rather, complementing Der homologs do have similar linker and C-terminal regions to EcDer. Among chimeric Der proteins, chimeras having linker and long C-terminal regions restored the growth of strain MK (Fig. 7). The linker likely contributes to the flexibility between two G domains upon nucleotide binding, after which the KH domain is positioned to recognize its binding site on 50S ribosomal subunits (33). Although ribosomes are well-conserved macromolecular structures, rRNA, ribosomal proteins, and mechanisms of ribosome assembly differ notably among organisms. It is thus likely that these differences prevented DrDer, NgDer, and SaDer from restoring the defects of growth and ribosome assembly and that functional complementation effects were mostly due to the structural recognition.
Previously, it was observed that, similar to our complementation results, Rickettsia rickettsii and Rickettsia typhi SecA homologs could not reverse the growth defect of the E. coli secAts mutant (44). R. rickettsii and R. typhi are distantly related to E. coli, and the alignment of amino acid sequences revealed that the N-terminal ATPase domain is conserved, whereas the C-terminal regions are variable. The chimeric SecA combining the N-terminal region of R. rickettsii SecA with the C-terminal region of E. coli SecA recovered the growth defect of the E. coli secAts mutant, suggesting that the C terminus provides species-specific characteristics to SecA for its preprotein translocation activity (44). The parallels to Der are intriguing, because the KH domain and C-terminal region of Der protein may confer species specificity to the protein.
None of the Der homologs was stimulated by either of the ribosomal subunits (see Fig. S8 in the supplemental material). We previously demonstrated that immature 50S ribosomal subunits lacking L9 and L18 are accumulated in Der-depleted cells (11). The overexpression of YihI, which stimulates the GTPase activity of Der, also promoted the accumulation of aberrant 50S ribosomal subunits lacking L9, L18, and L25 (45). Accordingly, it is possible that the fully processed mature 50S subunit does not interact with EcDer or influence GTPase activity. Nevertheless, it is apparent that KCl acts as a GTPase-activating element for Der proteins. Similar potassium-dependent activation was observed in TrmE (also known as MnmE), which is a tRNA modification GTP-binding protein (46). The presence of KCl triggered dimerization of the N-terminal α/β domain of TrmE, resulting in juxtaposed alignment of the nucleotide-binding sites of the respective GTP-binding domains (47). Size-exclusion chromatography and yeast two-hybrid experiments revealed that EcDer can dimerize (45). Therefore, it is necessary to determine the structural organization of Der in the presence of KCl to address whether potassium ion affects dimerization of Der and to determine the mechanism by which it functions as a GTPase activating cofactor.
Despite the importance of Der GTPase in ribosome biogenesis, little is known about the enzymatic regulation and detailed mechanism of Der in the ribosome maturation process. Our findings shed light on the conserved role of Der GTPases in ribosome assembly and the enzymatic characteristics of Der homologs. More importantly, we uncovered that the linker and C-terminal region are pivotal to 50S ribosome association. These key elements of Der appear to be attractive targets to develop novel antimicrobial agents against Der or Der-50S ribosome association, as attempted previously (48, 49).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Research Fund Program of the Research Institute for Basic Sciences, Pusan National University, Republic of Korea, 2012, project no. RIBS-PNU-201211260001, and by the Research Fund Program of the Genetic Engineering Institute, Pusan National University, Republic of Korea, 2013. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (NRF-2015R1A2A2A01005881).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00384-16.
REFERENCES
- 1.Bourne HR, Sanders DA, McCormick F. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 2.Bourne HR, Sanders DA, McCormick F. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 3.Wittinghofer A, Pal EF. 1991. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem Sci 16:382–387. doi: 10.1016/0968-0004(91)90156-P. [DOI] [PubMed] [Google Scholar]
- 4.Vetter IR, Wittinghofer A. 2001. The guanine nucleotide-binding switch in three dimensions. Science 294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 5.Sprang SR. 1997. G protein mechanisms: insights from structural analysis. Annu Rev Biochem 66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 6.Mohr D, Wintermeyer W, Rodnina MV. 2002. GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry 41:12520–12528. doi: 10.1021/bi026301y. [DOI] [PubMed] [Google Scholar]
- 7.Britton RA. 2009. Role of GTPases in bacterial ribosome assembly. Annu Rev Microbiol 63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- 8.Tu C, Zhou X, Tropea JE, Austin BP, Waugh DS, Court DL, Ji X. 2009. Structure of ERA in complex with the 3′ end of 16S rRNA: implications for ribosome biogenesis. Proc Natl Acad Sci U S A 106:14843–14848. doi: 10.1073/pnas.0904032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goto S, Kato S, Kimura T, Muto A, Himeno H. 2011. RsgA releases RbfA from 30S ribosome during a late stage of ribosome biosynthesis. EMBO J 30:104–114. doi: 10.1038/emboj.2010.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Q, Yuan Y, Xu Y, Feng B, Liu L, Chen K, Sun M, Yang Z, Lei J, Gao N. 2011. Structural basis for the function of a small GTPase RsgA on the 30S ribosomal subunit maturation revealed by cryoelectron microscopy. Proc Natl Acad Sci U S A 108:13100–13105. doi: 10.1073/pnas.1104645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang J, Inouye M. 2006. The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli. Mol Microbiol 61:1660–1672. doi: 10.1111/j.1365-2958.2006.05348.x. [DOI] [PubMed] [Google Scholar]
- 12.Jiang M, Datta K, Walker A, Strahler J, Bagamasbad P, Andrews PC, Maddock JR. 2006. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J Bacteriol 188:6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato A, Kobayashi G, Hayashi H, Yoshida H, Wada A, Maeda M, Hiraga S, Takeyasu K, Wada C. 2005. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells 10:393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- 14.Hwang J, Inouye M. 2001. An essential GTPase, Der, containing double GTP-binding domains from Escherichia coli and Thermotoga maritima. J Biol Chem 276:31415–31421. doi: 10.1074/jbc.M104455200. [DOI] [PubMed] [Google Scholar]
- 15.Mehr IJ, Long CD, Serkin CD, Seifert HS. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leipe DD, Wolf YI, Koonin EV, Aravind L. 2002. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 17.Mittenhuber G. 2001. Comparative genomics of prokaryotic GTP-binding proteins (the Era, Obg, EngA, ThdF [TrmE], YchF and YihA families) and their relationship to eukaryotic GTP binding proteins (the DRG, ARF, RAB, RAN, RAS and RHO families). J Mol Microbiol Biotechnol 3:21–35. [PubMed] [Google Scholar]
- 18.Mishra R, Gara SK, Mishra S, Prakash B. 2005. Analysis of GTPases carrying hydrophobic amino acid substitutions in lieu of the catalytic glutamine: implications for GTP hydrolysis. Proteins 59:332–338. doi: 10.1002/prot.20413. [DOI] [PubMed] [Google Scholar]
- 19.Robinson VL, Hwang J, Fox E, Inouye M, Stock AM. 2002. Domain arrangement of Der, a switch protein containing two GTPase domains. Structure 10:1649–1658. doi: 10.1016/S0969-2126(02)00905-X. [DOI] [PubMed] [Google Scholar]
- 20.Tan J, Jakob U, Bardwell JC. 2002. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J Bacteriol 184:2692–2698. doi: 10.1128/JB.184.10.2692-2698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caldas T, Binet E, Bouloc P, Richarme G. 2000. Translational defects of Escherichia coli mutants deficient in the Um 2552 23S ribosomal RNA methyltransferase RrmJ/FtsJ. Biochem Biophys Res Commun 271:714–718. doi: 10.1006/bbrc.2000.2702. [DOI] [PubMed] [Google Scholar]
- 22.Bharat A, Jiang M, Sullivan SM, Maddock JR, Brown ED. 2006. Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J Bacteriol 188:7992–7996. doi: 10.1128/JB.00959-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaefer L, Uicker WC, Wicker-Planquart C, Foucher AE, Jault JM, Britton RA. 2006. Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis. J Bacteriol 188:8252–8258. doi: 10.1128/JB.01213-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naganathan A, Moore SD. 2013. Crippling the essential GTPase Der causes dependence on ribosomal protein L9. J Bacteriol 195:3682–3691. doi: 10.1128/JB.00464-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morimoto T, Loh PC, Hirai T, Asai K, Kobayashi K, Moriya S, Ogasawara N. 2002. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148:3539–3552. doi: 10.1099/00221287-148-11-3539. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi G, Moriya S, Wada C. 2001. Deficiency of essential GTP-binding protein ObgE in Escherichia coli inhibits chromosome partition. Mol Microbiol 41:1037–1051. [DOI] [PubMed] [Google Scholar]
- 27.Forsyth R, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 28.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, Fritchman RD, Weidman JF, Small KV, Sandusky M, Fuhrmann J, Nguyen D, Utterback TR, Saudek DM, Phillips CA, Merrick JM, Tomb JF, Dougherty BA, Bott KF, Hu PC, Lucier TS, Peterson SN, Smith HO, Hutchison CA III, Venter JC. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 29.Gil R, Silva FJ, Pereto J, Moya A. 2004. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev 68:518–537. doi: 10.1128/MMBR.68.3.518-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeon Y, Ahn CS, Jung HJ, Kang H, Park GT, Choi Y, Hwang J, Pai HS. 2014. DER containing two consecutive GTP-binding domains plays an essential role in chloroplast ribosomal RNA processing and ribosome biogenesis in higher plants. J Exp Bot 65:117–130. doi: 10.1093/jxb/ert360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller J. 1993. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Trends Biochem Sci-Library Compendium 18:193. [Google Scholar]
- 33.Hwang J, Inouye M. 2010. Interaction of an essential Escherichia coli GTPase, Der, with the 50S ribosome via the KH-like domain. J Bacteriol 192:2277–2283. doi: 10.1128/JB.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ron EZ, Kohler RE, Davis BD. 1966. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science 153:1119–1120. doi: 10.1126/science.153.3740.1119. [DOI] [PubMed] [Google Scholar]
- 35.Daigle DM, Brown ED. 2004. Studies of the interaction of Escherichia coli YjeQ with the ribosome in vitro. J Bacteriol 186:1381–1387. doi: 10.1128/JB.186.5.1381-1387.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Yan K, Zhang Y, Li N, Ma C, Li Z, Zhang Y, Feng B, Liu J, Sun Y, Xu Y, Lei J, Gao N. 2014. Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly. Nucleic Acids Res 42:13430–13439. doi: 10.1093/nar/gku1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muench SP, Xu L, Sedelnikova SE, Rice DW. 2006. The essential GTPase YphC displays a major domain rearrangement associated with nucleotide binding. Proc Natl Acad Sci U S A 103:12359–12364. doi: 10.1073/pnas.0602585103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng B, Mandava CS, Guo Q, Wang J, Cao W, Li N, Zhang Y, Zhang Y, Wang Z, Wu J. 2014. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol 12:e1001866. doi: 10.1371/journal.pbio.1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Himeno H, Hanawa-Suetsugu K, Kimura T, Takagi K, Sugiyama W, Shirata S, Mikami T, Odagiri F, Osanai Y, Watanabe D, Goto S, Kalachnyuk L, Ushida C, Muto A. 2004. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res 32:5303–5309. doi: 10.1093/nar/gkh861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agirrezabala X, Frank J. 2009. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q Rev Biophys 42:159–200. doi: 10.1017/S0033583509990060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim DF, Green R. 1999. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell 4:859–864. doi: 10.1016/S1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- 42.Samaha RR, Green R, Noller HF. 1995. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature 377:309–314. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- 43.Blanchard SC, Puglisi JD. 2001. Solution structure of the A loop of 23S ribosomal RNA. Proc Natl Acad Sci U S A 98:3720–3725. doi: 10.1073/pnas.051608498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahman MS, Simser JA, Macaluso KR, Azad AF. 2005. Functional analysis of secA homologues from Rickettsiae. Microbiology 151:589–596. doi: 10.1099/mic.0.27556-0. [DOI] [PubMed] [Google Scholar]
- 45.Hwang J, Inouye M. 2010. A bacterial GAP-like protein, YihI, regulating the GTPase of Der, an essential GTP-binding protein in Escherichia coli. J Mol Biol 399:759–772. doi: 10.1016/j.jmb.2010.04.040. [DOI] [PubMed] [Google Scholar]
- 46.Armengod M, Moukadiri I, Prado S, Ruiz-Partida R, Benítez-Páez A, Villarroya M, Lomas R, Garzón MJ, Martínez-Zamora A, Meseguer S. 2012. Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie 94:1510–1520. doi: 10.1016/j.biochi.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 47.Scrima A, Vetter IR, Armengod ME, Wittinghofer A. 2005. The structure of the TrmE GTP-binding protein and its implications for tRNA modification. EMBO J 24:23–33. doi: 10.1038/sj.emboj.7600507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hwang J, Tseitin V, Ramnarayan K, Shenderovich MD, Inouye M. 2012. Structure-based design and screening of inhibitors for an essential bacterial GTPase, Der. J Antibiot 65:237–243. doi: 10.1038/ja.2012.9. [DOI] [PubMed] [Google Scholar]
- 49.Bharat A, Blanchard JE, Brown ED. 2013. A high-throughput screen of the GTPase activity of Escherichia coli EngA to find an inhibitor of bacterial ribosome biogenesis. J Biomol Screen 18:830–836. doi: 10.1177/1087057113486001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 52.Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 53.Bügl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U. 2000. RNA methylation under heat shock control. Mol Cell 6:349–360. doi: 10.1016/S1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- 54.Ghrayeb J, Kimura H, Takahara M, Hsiung H, Masui Y, Inouye M. 1984. Secretion cloning vectors in Escherichia coli. EMBO J 3:2437–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.