ABSTRACT
Streptococcus pneumoniae is able to integrate exogenous DNA into its genome by natural genetic transformation. Transient accumulation of high levels of the only S. pneumoniae alternative σ factor is insufficient for development of full competence without expression of a second competence-specific protein, ComW. The ΔcomW mutant is 104-fold deficient in the yield of recombinants, 10-fold deficient in the amount of σX activity, and 10-fold deficient in the amount of σX protein. The critical role of ComW during transformation can be partially obviated by σA mutations clustered on surfaces controlling affinity for core RNA polymerase (RNAP). While strains harboring σA mutations in the comW mutant background were transforming at higher rates, the mechanism of transformation restoration was not clear. To investigate the mechanism of transformation restoration, we measured late gene expression in σA* suppressor strains. Restoration of late gene expression was observed in ΔcomW σA* mutants, indicating that a consequence of the σA* mutations is, at least, to restore σX activity. Competence kinetics were normal in ΔcomW σA* strains, indicating that strains with restored competence exhibit the same pattern of transience as wild-type (WT) strains. We also identified a direct interaction between ComW and σX using the yeast two-hybrid (Y2H) assay. Taken together, these data are consistent with the idea that ComW increases σX access to core RNAP, pointing to a direct role of ComW in σ factor exchange during genetic transformation. However, the lack of late gene shutoff in ΔcomW mutants also points to a potential new role for ComW in competence shutoff.
IMPORTANCE The sole alternative sigma factor of the streptococci, SigX, regulates development of competence for genetic transformation, a widespread mechanism of adaptation by horizontal gene transfer in this genus. The transient appearance of this sigma factor is strictly controlled at the levels of transcription and stability. This report shows that it is also controlled at the point of its substitution for SigA by a second transient competence-specific protein, ComW.
INTRODUCTION
Streptococcus pneumoniae, a Gram-positive opportunistic pathogen found in the human nasopharynx, causes diseases such as pneumonia and meningitis. S. pneumoniae's natural competence, or ability to integrate exogenous DNA into its chromosome, provides a major mechanism of rapidly overcoming selective pressure (1, 2). The ability to take up and exchange DNA depends on development of the competent state, which is prompted by quorum sensing (QS) mediated by a small peptide, via a signal transduction pathway that is incompletely understood.
Cells in the exponential growth phase are first primed for transformation by a QS mechanism encoded by two genetic loci initially transcribed at a basal level by core RNA polymerase (RNAP) bound to the primary sigma factor, σA (3). The loci are comAB (4, 5) and comCDE (3). ComC is a propeptide cleaved and exported by ComAB, as the mature peptide called CSP (competence-stimulating peptide) (6). CSP is sensed by the histidine kinase receptor ComD, which then phosphorylates its cognate response regulator, ComE (3, 5, 7, 8). Phosphorylated ComE (ComE∼P) binds at a direct repeat upstream of the promoters of comAB, comCDE, and six additional operons that are transcribed only at competence (3, 9, 10), called the early genes. Induction of the early genes establishes a positive-feedback loop that increases the level of CSP produced.
Another early gene, comX, encodes the single known streptococcal alternative sigma factor, σX (11). In S. pneumoniae and other streptococci, σX forms a holoenzyme with RNA polymerase (RNAP) to recognize noncanonical promoter sequences, termed cin (competence-induced)-boxes, upstream of late competence genes (12–15). Organized in ∼12 principal operons, the so-called late genes encode a core streptococcal set of 28 competence-specific proteins, which create a DNA processing pathway for efficient transport, preservation, and recombination of extracellular genome fragments. The most prominent of these core competence-specific proteins is SsbB, a nonessential paralogue of the essential single-stranded DNA (ssDNA)-binding protein, SsbA. In competent cells, SsbB coats internalized ssDNA fragments in amounts that can exceed half a full genome equivalent. The promoter of ssbB contains the highly conserved −10 octamer element (TACGAATA) that is typical of cin-box promoters.
The ComABCDE/CSP system of competence induction is maintained in the mitis and anginosus groups of streptococci, whereas other streptococcal groups utilize a different set of QS genes, comR and comS, to coordinate population-wide induction of σX genes (16). Strict regulation of σX is imperative in the streptococci as some competence genes are responsible for fratricidal proteins and associated immunity proteins (17). In addition to transcriptional regulation by QS, σX stability and activity rely on a small protein, ComW (18, 19), which is found only in S. pneumoniae and 10 other species of streptococci. As σX is conserved in every Streptococcus species (20), it appears that the remaining species without ComW have other mechanisms of regulation of σX that do not require ComW.
comW was first identified as a gene whose expression was CSP inducible and required for transformation (21). ComW was subsequently implicated in the full σX response during transformation (18), as ectopic coexpression of σX and ComW was sufficient to restore the yield of transformants to 80% of wild-type (WT) levels. ComW was also described as necessary for σX activity (19) and for the protection of σX from degradation by the ATP-dependent ClpEP protease. However, ΔcomW ΔclpP strains were unable to transform despite producing the native amounts of σX (19).
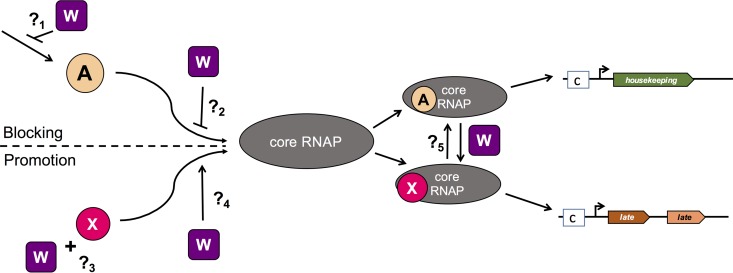
In an effort to pinpoint the critical role of ComW in competence development, a suppressor screen revealed that mutations in the primary σ factor could partially restore transformation yields in the ΔcomW background. That screen took advantage of the strong (10,000-fold) transformation-negative phenotype of comW mutants to enrich for suppressor mutations by means of three successive large-scale transformation steps, which were carried out independently in 15 parallel enrichment series, yielding a total of 27 independent suppressor alleles. All 27 independent spontaneous suppressor mutations mapped to regions of σA that control its affinity to core RNAP and mimic alterations known to reduce σA affinity for core RNAP (22). This suggested that ComW might act at the sigma substitution step to promote substitution of σA for σX; however, there are several possible ways in which ComW could affect the switching of sigma factors (Fig. 1). For example, ComW could act either to block σA association with core RNAP or to promote σX binding to core RNAP. Further, for either mechanism there are multiple steps where ComW could act. We hypothesized that ComW functions as an adapter, binding to σX to promote its access to core RNAP. To test this interpretation, we have asked whether late gene expression is restored in ΔcomW σA* mutants.
FIG 1.
Hypothetical functions of ComW as a σ factor adapter or in sequestration of σA during competence in S. pneumoniae. Two main mechanisms are suggested for ComW's mode of action during transformation, blocking σA association with E (core RNA polymerase) or promoting σX association with core RNAP. However, many steps exist with either mechanism, and ComW could act at any of these steps. For example, if the role of ComW is to prevent σA association with core RNAP, it might act by sequestering σA (?1) or by preventing σA from accessing core RNAP (?2). If ComW acts to increase σX access to core RNAP, it might act by directly binding σX (?3) or by promoting the loading of σX onto core RNAP (?4). It could also act to promote switching between the two sigma factors (?5). Purple squares, ComW; beige circles, σA; magenta circles, σX; gray ovals, core RNA polymerase; green pentagon, housekeeping genes; brown and orange pentagons, late genes; ?, hypothetical locations of ComW action; boxed c's, cin-boxes.
We now show that late gene expression is restored in ΔcomW σA* strains, that competence kinetics in these strains is similar to that of the WT, and that ComW can interact directly with σX. Taken together, these data indicate that one role of ComW is to increase σX access to core RNAP. Finally, we observed persistence of late gene expression in all ΔcomW mutants, suggesting a new function for ComW in late gene shutoff.
MATERIALS AND METHODS
Bacterial strains and culture media.
The strains used in this study are listed in Table 1. CP2137, a Δcps ΔcomA ΔssbB::pEVP3::ssbB derivative of strain Rx1 (11, 22, 23), was used as the wild-type (WT) standard for transformation assays. It produces no CSP, being deficient in the CSP maturation protease and secretion exporter subunit, ComA. Isolation of σA* suppressor mutations and creation of isogenic suppressor strains were described previously (22). All pneumococcal strains were cultured in CAT medium, supplemented as needed with 1.5% agar. CAT medium was prepared from 5 g of tryptone (Difco Laboratories), 10 g of enzymatic casein hydrolysate (ICN Nutritional Biochemicals), 1 g of yeast extract (Difco), and 5 g of NaCl in 1 liter of H2O; sterilized for 40 min at 121°C; and then supplemented to 0.2% glucose and 0.016 M K2HPO4 before use. Antibiotics were used at the concentrations described previously (22). CSP1 (24) was obtained from NeoBioSci (Cambridge, MA) as a custom synthetic peptide with the sequence EMRLSKFFRDFILQRKK at 80% purity and stored as a sterile 0.025% solution in water at −20°C.
TABLE 1.
Strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| S. pneumoniae | ||
| CP2137 | hex malM511 str-1 bgl-1; low α-galactosidase background; ΔcomA Δcps ΔssbB::pEVP3::ssbB SsbB+ Smr Cmr; LacZ reporter | 23 |
| CP2451 | CP2137 rpoD-L363F Smr Cmr | 22 |
| CP2452 | CP2137 rpoD-A171V Smr Cmr | 22 |
| CP2453 | CP2137 rpoD-R355H Smr Cmr | 22 |
| CP2454 | CP2137 rpoD-R316H Smr Cmr | 22 |
| CP2455 | CP2451 ΔcomW::kan Smr Cmr Knr | 22 |
| CP2456 | CP2452 ΔcomW::kan Smr Cmr Knr | 22 |
| CP2457 | CP2453 ΔcomW::kan Smr Cmr Knr | 22 |
| CP2458 | CP2454 ΔcomW::kan Smr Cmr Knr | 22 |
| CP2463 | CP2137 ΔcomW::kan Smr Cmr Knr | 22 |
| E. coli DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Invitrogen |
| S. cerevisiae | ||
| GlodY2H | MATa trp1-901 leu2-3,112 gal4Δ gal80Δ mel1 | Clontech |
| Y187 | MATα trp1-901 leu2-3,112 gal4Δ gal80Δ MEL1 | Clontech |
Transformation assays.
The standard assay for transformation was done essentially as previously described (19). A log-phase culture at an optical density at 550 nm (OD550) of 0.05 at 37°C was incubated with 0.1 μg/ml DNA, 250 ng/ml CSP, 0.5 mM CaCl2, and 0.04% bovine serum albumin (BSA) for 80 min at 37°C. Portions of the culture were then embedded (in 1.5 ml of CAT medium mixed with 1.5 ml of CAT agar) in sandwich plates and overlaid with the relevant antibiotic, as previously described (19). After 15 h at 37°C, colonies were counted. The transformation efficiency is expressed as CFU per milliliter of cells transformed at an OD550 of 0.05.
β-Galactosidase assay.
To measure β-galactosidase activity, a culture was induced to competence at an OD550 of 0.1 by the addition of 0.1 μg/ml DNA, 250 ng/ml CSP, 0.5 mM CaCl2, and 0.04% BSA. At 10 min intervals, 0.5-ml samples were removed, mixed with 0.125 ml lysis buffer (300 mM Na2HPO4, 200 mM NaH2PO4, 50 mM KCl, 5 mM MgSO4, 0.5% Triton X-100, and 250 mM β-mercaptoethanol), held at 37°C for 10 min, and then put on ice. An 0.1-ml portion of each mixture was loaded into a well in a 96-well plate, in duplicate. An 0.05-ml amount of o-nitrophenyl-β-d-galactopyranoside solution was added to each well (4 mg/ml o-nitrophenyl-β-d-galactopyranoside, 60 mM Na2HPO4, 40 mM NaH2PO4). The plate was incubated at 37°C with shaking, and absorbance at 420 nm was read every 10 min for 100 min on the BioTek Synergy 2 reader. The slope of the absorbance curve was used to calculate LacZ activity, which is reported in Miller units.
Competence kinetics assays.
To measure the amount of transformants at several time points after CSP addition, transforming cells were incubated with DNA for 5 min and then exposed to DNase I. DNase I (Sigma-Aldrich) was prepared as a 5-mg/ml solution in 0.15 M NaCl with 50% glycerol and stored at −20°C. A log-phase culture at an OD550 of 0.05 was brought to 250 ng/ml CSP, 0.5 mM CaCl2, and 0.04% BSA and incubated at 37°C. Every 10 min post-CSP addition, 0.1 ng/ml DNA was added to 1 ml cells and incubated at 37°C for 5 min. DNase I was added to a final concentration of 20 μg/ml, and the cells were incubated for another 75 min to allow integration and expression of introduced genes. Cells were then diluted and plated using the method described above.
Yeast two-hybrid (Y2H) strains and culture media.
The Saccharomyces cerevisiae strains used in this study were Y2HGold (MATa trp1-901 leu2-3,112 gal4Δ gal80Δ mel1) and Y187 (MATα trp1-901 leu2-3,112 gal4Δ gal80Δ MEL1) (Clontech, Mountain View, CA). Yeast strains were grown in YPAD medium (1% [wt/vol] Bacto yeast extract, 2% [wt/vol] Bacto peptone, 80 mg liter−1 adenine hemisulfate) or selective minimal (SD) medium that included a carbon and nitrogen source. Essential amino acids were added to SD medium to create various selection media, as previously described (25).
Plasmid construction.
pAD, pBD, and other control plasmids were obtained from Clontech (Mountain View, CA). The recombinant plasmids used for the Y2H assay, listed in Table 2, were constructed by restriction enzyme digestion and ligation of insert DNAs into shuttle vectors. The region of the S. pneumoniae R6 chromosome encoding the protein of interest was amplified by PCR. pAD-comX was created by ligating the comX amplicon generated by PCR into pAD after digestion of both molecules by BamHI and XhoI (New England BioLabs, Ipswich, MA). Similarly, pBD-comW was created by ligating comW into pBD after digestion of both molecules by BamHI and SalI (New England BioLabs, Ipswich, MA). The primers used in this study are listed in Table 2. Primers JB17 and JB20 were used to amplify the full length of comX, and JB37 and JB38 were used for amplification of the full length of comW. The full length of rpoD was amplified by primers JB51 and JB52, digested with BamHI and XhoI, and cloned into pAD. pBD-comX was created by ligating comX generated by PCR into pBD digested by BamHI and SalI. pAD-comW was created by ligating comW to pAD digested by BamHI and XhoI. Recombinant plasmids were first transformed into Escherichia coli (DH5α) with selection on ampicillin (50 μg/ml) or kanamycin (50 μg/ml) plates. DNA from selected single colonies was purified with the Zymo Miniprep kit, and correct cloning in frame was confirmed by sequencing the junction area of DNA by Sanger sequencing (University of Illinois at Chicago Research Resources Center [UIC RRC]).
TABLE 2.
Plasmids and primers used in this study
| Plasmid or primer name | Plasmid description or primer sequencea |
|---|---|
| Plasmids | |
| pAD | pGADT, HA tag, Ampr leu2 |
| pBD | pGBKT, c-Myc epitope tag, Kanr trp1 |
| pAD-comX | pGADT-comX, HA tag, Ampr leu2 |
| pBD-comX | pGBKT-comX, c-Myc epitope tag, Kanr trp1 |
| pBD-W | pGBKT-comW, c-Myc epitope tag, Kanr trp1 |
| pAD-W | pGADT-comW, HA tag, Ampr leu2 |
| pAD-rpoD | pGADT-rpoD, HA tag, Ampr leu2 |
| pAD-T | pGADT-antigen T, HA tag, Ampr leu2 |
| pBD-53 | pGBKT-murine p53, HA tag, Kanr trp1 |
| pBD-lam | pGBKT-lamin, HA tag, Kanr trp1 |
| Primers | |
| JB17 | GTACGGATCCAGGGGAAAATTATGATTAAAGAATTGTAT |
| JB20 | GCATCTCGAGCTAATGGGTACGGATAGTAAACTC |
| JB37 | GTCAGGATCCTTATGTTACAAAAAATTTATGAGCAGATG |
| JB38 | GCATGTCGACTACTAAAATTACCTCAACAAGAAATAAAC |
| JB41 | GTCAGGATCCTTATGTTACAAAAAATTTATGAGCAGATG |
| JB42 | GCATCTCGAGTACTAAAATTACCTCAACAAGAAATAAAC |
| JB45 | GTACGGATCCAGGGGAAAATTATGATTAAAGAATTGTAT |
| JB48 | GTACGGATCCAGGGGAAAATTATGATTAAAGAATTGTAT |
| JB51 | GTACGGATCCCAGCCCTAGAAGAATTGGAACG |
| JB52 | GCATCTCGAGTCAATTTGCTCTTCTGTATAAG |
Underlining indicates restriction enzyme recognition sites. HA, hemagglutinin.
Y2H assay.
Recombinant plasmids were transformed into competent yeast of α or a mating type by the lithium acetate (LiAc)-mediated method (26), with selection on SD-Leu or SD-Trp medium (30°C for 72 h). Single clones of the haploid transformants were verified by sequencing plasmid junctions and stored at −80°C with 15% glycerol. By cross-mating between different mating types of yeast haploids, diploid strains were obtained on SD-Leu-Trp. Five diploid transformants were picked up and saved from each cross. Small identical volumes of five individual frozen strains were mixed by resuspension with double-distilled water (ddH2O) and diluted to inoculate about 105 cells onto patches of SD-Leu-Trp agar with 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-Gal). Diploids on SD-Leu-Trp–X-α-Gal were incubated at 30°C for 5 days before determining interacting partners by production of blue product.
RESULTS
Late gene expression is restored in ΔcomW σA* suppressor strains.
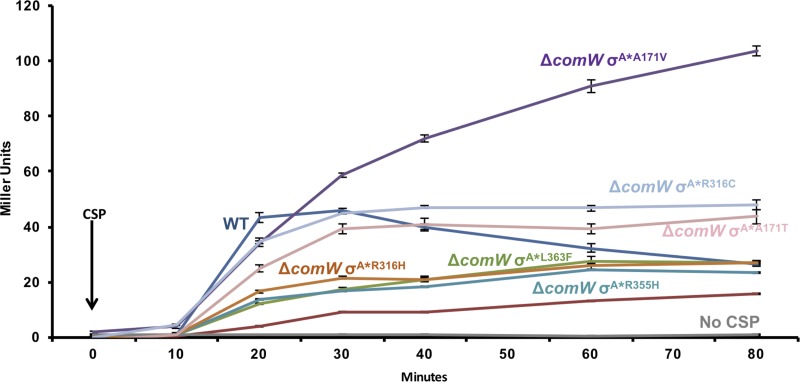
Transformation requires the cooperation of dozens of proteins, many of which are expressed specifically during the development of competence. In comW mutants, expression of the σX-dependent late genes is severely reduced, and the yield of transformants is only 0.01% of that in the WT (19). In ΔcomW σA* suppressor strains, the yield of transformants is restored by 10- to 1,000-fold, reaching as high as 10% of the WT level (22), but the mechanism of this suppression of the strong comW competence phenotype is unclear. Because transformation depends on initiation of transcription of dozens of late genes by σX polymerase, one possibility is that the σA* mutations restore late gene expression sufficiently to allow production of functional levels of effector proteins. To test whether σX polymerase activity is in fact increased by σA* suppressor mutations, we determined expression from a representative late gene promoter after CSP stimulation in ΔcomW σA* suppressor strains. The strains examined had different suppressor mutations in σA, the primary σ factor, causing one of six amino acid replacements (A171T, A171V, R316C, R316H, R355H, and L363F), representing the range of the amino acid replacements found in the previous suppressor screen, and all transformed at 10% of the WT level in the comW mutant background (22).
To monitor late gene expression in ΔcomW σA* strains, we measured the activity of β-galactosidase produced by a lacZ transcriptional fusion to the promoter of ssbB (11), a late gene responsible for protecting the newly imported DNA (27). ssbB is transcribed from a cin-box recognized by σX-RNAP (14, 15). Since cin-boxes are highly conserved among the late genes (14, 28), ssbB expression can be taken as generally representative of late gene expression level (29).
ssbB expression in six σA* comW strains was measured over a period of 80 min after addition of CSP to CSP-deficient cultures (Fig. 2). In the WT, ssbB expression was undetectable at the time of addition of CSP (Fig. 2), but by 20 to 30 min later, maximal expression had been reached, followed by a decrease of LacZ activity after 30 min. The slow decrease of LacZ activity reflects the shutoff of late gene expression, proteolytic decay of σX, loss of competence, and dilution of the stable LacZ protein by cell growth. In the comW mutant, LacZ accumulated much more slowly but continued beyond 80 min at a reduced rate. In contrast, in every ΔcomW σA* strain, there was a significant increase of ssbB expression relative to the ΔcomW control at 20 min post-CSP induction, indicating a partial restoration of ssbB expression. In strains with the suppressors σA*R316H, σA*L363F, and σA*R355H, expression was increased to roughly 20% of that of the WT, whereas in those with σA*A171V, σA*A171T, and σA*R316C, it reached 70 to 80% of WT expression at 20 min. No strain exhibited late gene expression without CSP induction, indicating the absolute need for σX activity to allow late gene expression and indicating that the suppressor alleles of σA do not themselves allow recognition of late gene promoters. We conclude that σA* suppressors not only greatly increase recombination yields but also significantly restore σX-RNAP activity in vivo.
FIG 2.
Late gene expression is restored in ΔcomW σA* strains. Late gene expression patterns were measured in WT and ΔcomW strains and six ΔcomW σA* mutant (σA*A171T, σA*A171V, σA*R316C, σA*R316H, σA*R355H, and σA*L363F) strains. PssbB-driven transcription of lacZ was measured as β-galactosidase activity (Miller units) in samples harvested at indicated times after CSP addition. Each strain is indicated by its σA mutation. The WT and ΔcomW strains have the WT copy of σA. Late gene expression was not detected without CSP treatment. Error bars, standard deviations from three biological replicates. Arrow, addition of CSP.
Competence is transient in σA* mutants.
A characteristic feature of the temporal profile of competence development in S. pneumoniae, mediated by the negative feedback regulator DprA (3, 30) and by degradation of σX by the ATP-dependent ClpP/ClpE protease (13, 31, 32), is the rapid appearance and subsequent shutoff of competence after sudden exposure of naive cells to CSP.
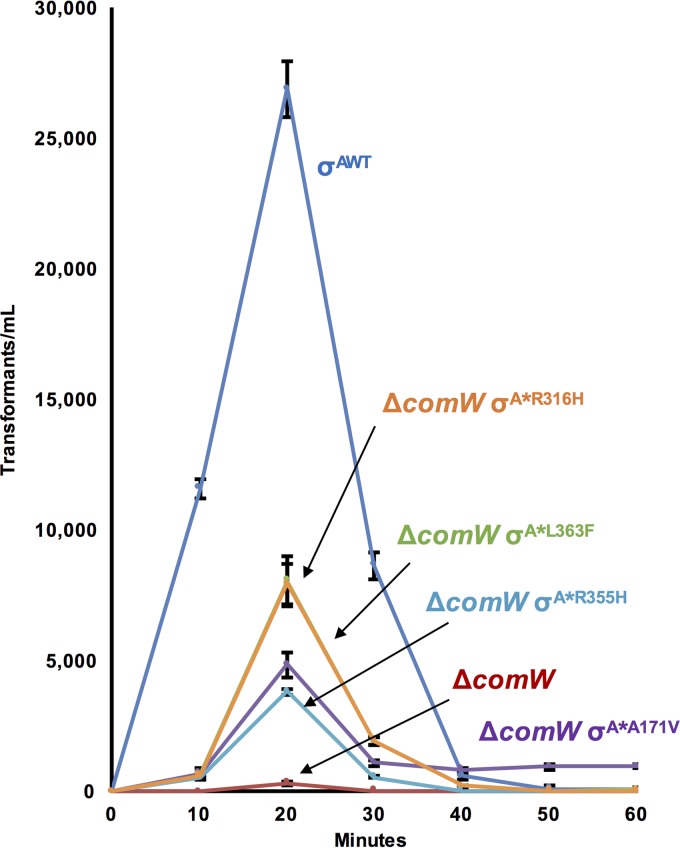
To learn whether the partially restored competence in suppressor strains was also subject to such regulation, we monitored transformation over time during CSP-induced competence development in ΔcomW σA* mutant strains, evaluating the yield of transformants produced by 5-min DNA exposures at various times after CSP addition (Fig. 3). Similarly to the temporal path of ssbB expression, transformation reached peak yields at 20 min after CSP induction; by 30 min, the yield of transformants dropped off sharply, soon becoming undetectable for all strains, except the ΔcomW σA*A171V strain. The latter strain still transformed past 60 min but with 5-fold-lower yields than at the time of peak late gene expression. Interestingly, this is the same strain in which late gene expression did not shut off completely, and it was higher than that for any other ΔcomW σA* suppressor strain (Fig. 2 and Table 3). Presumably, the persistent residual σX activity allows maintenance of sufficient levels of all late proteins for continued DNA-processing capacity. We conclude that the partially restored competence in suppressor strains exhibits the same pattern of transient expression of competence as that in WT strains.
FIG 3.
Competence kinetics are normal in ΔcomW σA* strains. The number of transformants/ml after a 5-min exposure to DNA was measured in the WT (σAWT) and ΔcomW (σAWT) strains and four ΔcomW σA* mutant (σA*A171V, σA*R316H, σA*R355H, and σA*L363F) strains after CSP induction and a 5-min exposure to 5 ng/ml DNA at each time point. Each strain is indicated by its σA mutation. The WT and ΔcomW strains have the WT copy of σA. Transformants were not detected at any time without CSP treatment. Error bars, standard deviations from experiments performed in triplicate. CSP was added at time zero.
TABLE 3.
Persistent late gene expression in strains lacking ComW
| σA allelea | ComW present |
ComW absent |
||
|---|---|---|---|---|
| Inductionb | Shutoffc | Inductionb | Shutoffc | |
| WT | 4.2 | −0.4 | 0.4 | 0.2 |
| A171V | 2.9 | −0.4 | 3.0 | 1.0 |
| R316H | 5.0 | −0.5 | 1.6 | 0.2 |
| R355H | 4.1 | −0.5 | 1.3 | 0.3 |
| L363F | 5.2 | −0.5 | 1.1 | 0.3 |
Allele of σA in strain tested.
Rate of change in Miller units during induction phase, measured as slope of expression between 10 and 20 min.
Rate of change in Miller units during shutoff phase, measured as slope of expression between 40 and 60 min.
Late gene expression kinetics in suppressor strains indicate a role of ComW in determining reversal of σX activation.
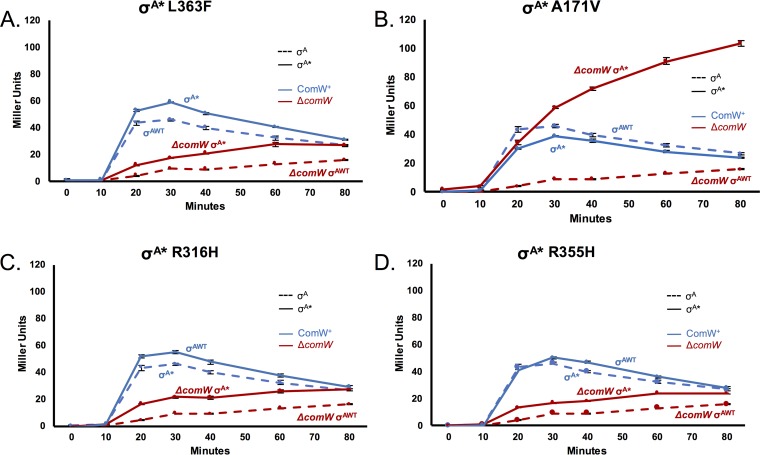
A perplexing feature of the ssbB expression profiles following CSP treatment of σA* suppressor strains (Fig. 2) was that an initial burst of ssbB expression at 10 to 20 min was followed by lower but persistent expression of ssbB in the 20- to 60-min period, as evidenced by continued accumulation of LacZ, in contrast to the growth-dependent dilution of LacZ activity seen in WT strains. Although this persistent but low level of expression of late genes was generally not accompanied by continued competence for DNA uptake (Fig. 3), it nevertheless indicates an apparent partial failure of the escape from competence by inactivation of ComE (by DprA) (30) and/or proteolysis of σX and late gene products (by ClpEP) (13, 33). A simple explanation of this contribution would be suboptimal DprA levels in the suppressor comW strains. However, this explanation is not entirely satisfactory, because the suppressor (σA*A171V) with the highest expression of ssbB, and by extension the largest amounts of DprA, was also the one with the most conspicuous failure of shutoff.
To trace the origin of this failure to either of the new components of the suppressor ΔcomW strains—mutation in σA or lack of ComW—we monitored late gene expression in strains carrying σA suppressor mutations but also carrying wild-type ComW. Comparing the expression profiles of these two sets of strains would indicate whether the lack of ComW was responsible for the persistent pattern of late gene expression or whether the pattern was due to the mutations in σA. The temporal patterns of turn-on and shutoff of ssbB expression in ComW+ strains (Fig. 4; summarized in Table 3) revealed, in all cases, a return to the wild-type temporal profile, suggesting that ComW somehow contributes to the completion of shutoff rather than that the altered σA itself interferes with shutoff. This result implies that ComW is partly responsible for the proper regulation and shutoff of σX activity, because late gene expression is normal regardless of the σA* mutation if ComW is present in this strain, and strains lacking comW all exhibited persistent ssbB expression. We conclude that ComW is responsible for the induction and shutoff of ssbB expression, pointing to another role for ComW in the regulation of σX activity. This suggests that ComW acts as an adapter to promote σX degradation after competence ends.
FIG 4.
Late gene expression is persistent in σA* strains lacking ComW. Late gene expression patterns after CSP induction using a lacZ reporter at gene ssbB. β-Galactosidase activity (Miller units) in samples harvested every 10 min after CSP induction was measured in WT (σAWT), ΔcomW (σAWT), and σA* strains in the presence or absence of ComW. Each panel examines a different single amino acid substitution in σA, denoted as the WT amino acid, the position of the change, and the amino acid at that position: σA*L363F (A), σA*A171V (B), σA*R316H (C), and σA*R355H (D). Dashed lines, strains with WT σA; solid lines, strains with σA* suppressor mutations; red, strains with comW deletions; blue, strains with WT comW. Late gene expression was not detected without CSP induction of σX activity. Error bars, standard deviations from experiments performed in triplicate.
σX interacts with ComW in yeast two-hybrid assays.
Considering the known mechanism of σ factor regulation in bacteria, two classes of action of ComW that might tip the σA/σX balance in favor of σX in competent cells can be considered. Acting negatively, ComW might decrease the amount of σA or decrease its access to polymerase core. Acting positively, ComW might increase the amount of σX or increase its access to polymerase core. Available evidence argues against two of these mechanisms. First, σA remains essentially constant in amount in competent cells, indicating that nothing is directing its destruction (18). Second, increasing the amount of the labile σX protein in ΔcomW strains by inactivation of the ClpP protease does not suppress the ComW transformation-deficient phenotype (18), indicating that the critical consequence of the absence of ComW is not the decreased amount of σX.
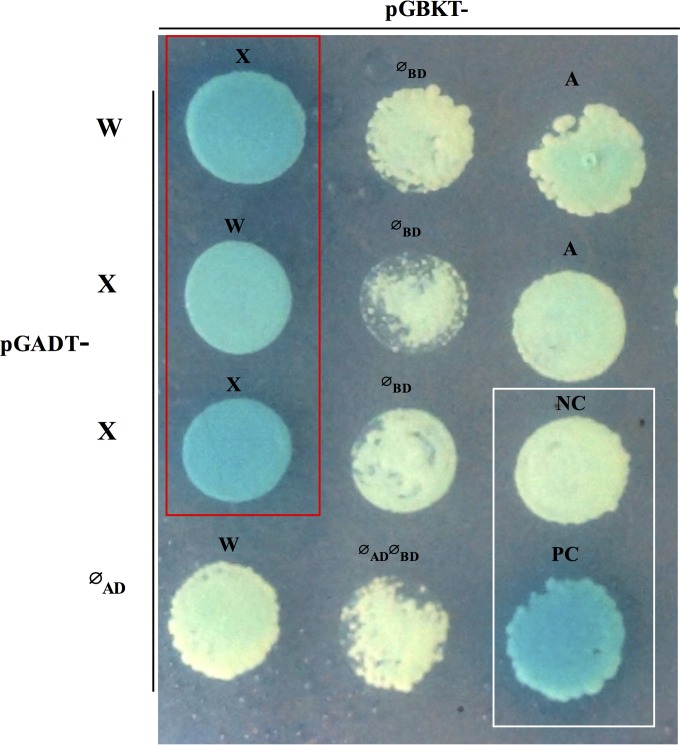
To distinguish whether ComW may function to promote σX access to RNA polymerase directly by binding σX or indirectly by sequestration of σA to weaken its competition with σX for access to RNA polymerase, we turned to a Y2H assay that could detect interacting partners. When the fusion proteins interact with each other, a functional Gal4 transcription factor is detected by expression of the Gal4-responsive mel1 gene, which is monitored by formation of blue colonies on X-α-Gal plates. We observed σX and ComW interaction in both pairings (BD-σX/AD-W and BD-W/AD-σX) indicated by blue colonies, but no interaction of either fusion with empty vector products (Fig. 5, red rectangle). Similarly, σX exhibited self-interaction (BD-σX/AD-σX). Diploids containing the following pairs of fusion proteins formed white colonies on X-α-Gal indicator plates: ØBD/AD-W, ØBD/AD-σX, ØAD/BD-σX, ØBD/ØAD, AD-σX/BD-σA, and BD-W/ØAD. Besides, ComW and σA did not show interactions, as indicated by white colonies on SD-Leu-Trp–X-α-Gal plates. Control partners used in the assay were pGBKT7-53 and pGADT7-T as positive controls (PCs) and pGBKT7-lam and pGADT7 as negative controls (NCs) (Fig. 5, white rectangle).
FIG 5.
Interactions of ComW with σX and σA evaluated by yeast two-hybrid assay. Diploids containing fusion proteins were plated on SD-Leu-Trp–X-α-Gal plates. Interacting proteins that activate α-galactosidase allow colonies to appear blue. Plates were incubated at 30°C for 5 days. Proteins fused to pGADT are indicated at the left; proteins fused to pGBKT are indicated directly above each diploid patch. Ø, empty vector; NC, negative control showing no interaction between lamin and large-T antigen; PC, positive control showing interaction between lamin and a large-T antigen. Empty vectors ØAD and ØBD paired with tested proteins were examined, as well as ØAD and ØBD only. X, σX; A, σA; W, ComW.
The Y2H results are the first indication that σX could interact with ComW and that σX could interact with itself but that ComW does not interact with σA. As the Y2H results indicate an interaction of ComW with σX but not with σA, we are led to the hypothesis that ComW acts to promote σX assembly into polymerase core rather than to reduce access to σA with core.
DISCUSSION
RNA polymerase (RNAP) can be directed to specific noncanonical promoters via different alternative σ factors, which are important mediators of the bacterial response to stress. Bacteria contain at least one σ factor, the housekeeping σA, which is responsible for the transcription of genes required for metabolism and growth, but may also carry alternative σ factors, which can direct RNAP to specific noncanonical promoter sequences, allowing them to quickly respond to their environment.
Despite their ability to rapidly direct the cell to respond to stressors, alternative σ factors are not always actively present. In fact, because alternative σ expression can result in global transcriptional changes in the cell, most alternative σ factors are regulated by occlusion, proteolysis, or other mechanisms. In Streptococcus thermophilus, for example, σX is negatively regulated by degradation through the MecA adapter protein and the ClpP protease complex (34). σ factors can be sequestered by anti-sigma factors, preventing σ factor association with RNAP, as in Bacillus subtilis, where the anti-σ factor RsiW sequesters σW until YluC cleaves RsiW, eventually allowing the release of σW (35). However, few alternative σ factors are known to be regulated directly at the σ-RNAP holoenzyme assembly step. Data from this and our recent study suggest that ComW acts as a σ switching factor that assists in the switch from σA to σX during development of competence for genetic transformation.
ComW has been described as having at least two effects, increasing both σX activity and σX stability (19) (Fig. 6). ComW is also suggested to improve σX affinity for core RNAP (22). A new function proposed for ComW in this study is a potential role in the shutoff of late gene expression. Every comW mutant strain, regardless of whether it had the WT or mutant σA, had unusually persistent residual late gene expression. While this may present an additional function for ComW, it is possible that all the functions of ComW reflect a single role with different effects. For example, if ComW is a labile σ factor activator that assists with loading of σX into core RNAP, this could explain all the separate phenotypes observed. ComW binds to σX to load it onto core RNAP for a brief period during transformation, allowing the transcription of late genes, as shown by its role in late gene transcription. Loading of σX onto core could also allow decreased σX proteolysis because of protection by core RNAP. This reduced proteolysis would explain ComW's role in increasing σX stability. Later, although it is unclear how ComW is involved, the correct shutoff mechanism is ineffective without ComW. Alternatively, it is possible that ComW acts as an adapter, allowing improved σX binding to core RNAP, but also improves its recognition by proteases. The absence of ComW in mutant strains could allow persistence of late gene expression because of the decreased recognition by proteases. However, it is not clear how this promotion of degradation of RNAP activity late in transformation is related to the apparent protection from degradation early in the transformation process (Fig. 6).
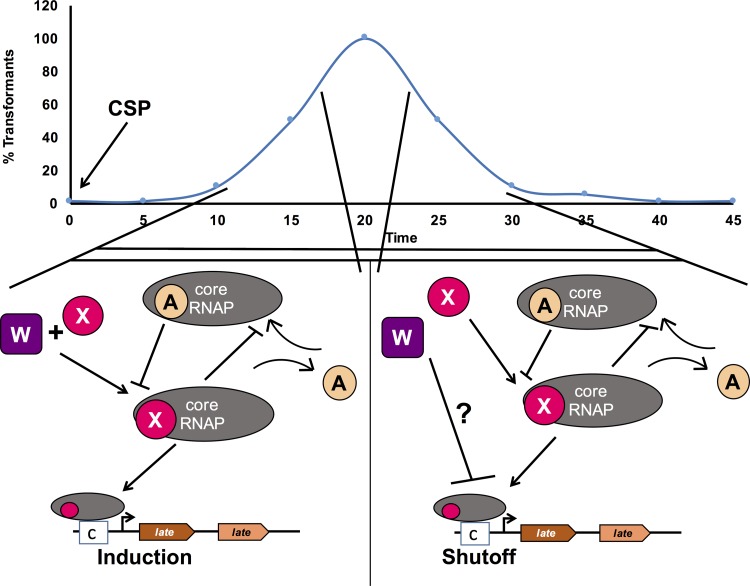
FIG 6.
Model of the potential action of ComW during transformation in S. pneumoniae. From CSP induction until the time of peak σX activity (20 min), ComW functions to promote σX access to E RNA polymerase to allow transcription of the late genes necessary to complete transformation. After peak σX activity, ComW may function to assist in the shutoff of late gene expression. The question mark points to the unknown effect of ComW on the shutoff of late gene expression. The lack of shutoff of late gene expression in ΔcomW mutants points to ComW having a function in controlling both the upregulation and downregulation of late genes during transformation. CSP, competence-stimulating peptide addition; purple squares, ComW; magenta circles, σX; beige circles, σA; gray ovals, E RNAP; boxed c's, cin-boxes; brown and orange pentagons, late genes; bent arrow, promoter.
While many alternative σ factors are negatively regulated by proteolysis and sequestration, the idea of a σ factor activator is not entirely novel (36). The small protein Crl, discovered in E. coli, acts to promote σS binding to core RNAP (37, 38). However, Crl has been identified only in Gram-negative bacteria, even though it is found in many genera such as Escherichia, Salmonella, and Pseudomonas (39, 40). ComW is found only in the streptococci, where it is restricted to the mitis and anginosus groups of species. If ComW is, in fact, a σ factor activator, its function would be novel in Gram-positive bacteria. The current studies on the function of ComW all point to a role as a protein responsible for promoting σ factor exchange during transformation (Fig. 6). Taken together, the data presented in this study suggest that ComW functions as a novel σ factor activator in S. pneumoniae during transformation.
ACKNOWLEDGMENTS
We thank Nicole Inniss, Anna Do, Om Bhetuwal, and Paola Sarmiento for assistance with experiments.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Chaguza C, Cornick JE, Everett DB. 2015. Mechanisms and impact of genetic recombination in the evolution of Streptococcus pneumoniae. Comput Struct Biotechnol J 13:241–247. doi: 10.1016/j.csbj.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston C, Campo N, Bergé MJ, Polard P, Claverys J-P. 2014. Streptococcus pneumoniae, le transformiste. Trends Microbiol 22:113–119. doi: 10.1016/j.tim.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Martin B, Soulet A-L, Mirouze N, Prudhomme M, Mortier-Barrière I, Granadel C, Noirot-Gros M-F, Noirot P, Polard P, Claverys J-P. 2013. ComE/ComE∼P interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol Microbiol 87:394–411. doi: 10.1111/mmi.12104. [DOI] [PubMed] [Google Scholar]
- 4.Hui FM, Morrison DA. 1991. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J Bacteriol 173:372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pestova EV, Håvarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol 21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 6.Havarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Håvarstein LS, Gaustad P, Nes IF, Morrison DA. 1996. Identification of the streptococcal competence-pheromone receptor. Mol Microbiol 21:863–869. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- 8.Ween O, Gaustad P, Havarstein LS. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol Microbiol 33:817–827. doi: 10.1046/j.1365-2958.1999.01528.x. [DOI] [PubMed] [Google Scholar]
- 9.Martin B, Granadel C, Campo N, Hénard V, Prudhomme M, Claverys J-P. 2010. Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol Microbiol 75:1513–1528. doi: 10.1111/j.1365-2958.2010.07071.x. [DOI] [PubMed] [Google Scholar]
- 10.Oggioni M, Morrison D. 2008. Cooperative regulation of competence development in Streptococcus pneumoniae: cell-to-cell signaling via a peptide pheromone and an alternative sigma factor, p 345–362. In Winans SC, Bassler B (ed), Chemical communications among bacteria. ASM Press, Washington, DC. [Google Scholar]
- 11.Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol 181:5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo P, Li H, Morrison DA. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol Microbiol 50:623–633. doi: 10.1046/j.1365-2958.2003.03714.x. [DOI] [PubMed] [Google Scholar]
- 13.Luo P, Morrison DA. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J Bacteriol 185:349–358. doi: 10.1128/JB.185.1.349-358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol 51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 15.Campbell EA, Choi SY, Masure HR. 1998. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol 27:929–939. doi: 10.1046/j.1365-2958.1998.00737.x. [DOI] [PubMed] [Google Scholar]
- 16.Håvarstein LS. 2010. Increasing competence in the genus Streptococcus. Mol Microbiol 78:541–544. doi: 10.1111/j.1365-2958.2010.07380.x. [DOI] [PubMed] [Google Scholar]
- 17.Eldholm V, Johnsborg O, Haugen K, Ohnstad HS, Håvarstein LS. 2009. Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology 155:2223–2234. doi: 10.1099/mic.0.026328-0. [DOI] [PubMed] [Google Scholar]
- 18.Luo P, Li H, Morrison DA. 2004. Identification of ComW as a new component in the regulation of genetic transformation in Streptococcus pneumoniae. Mol Microbiol 54:172–183. doi: 10.1111/j.1365-2958.2004.04254.x. [DOI] [PubMed] [Google Scholar]
- 19.Sung CK, Morrison DA. 2005. Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae. J Bacteriol 187:3052–3061. doi: 10.1128/JB.187.9.3052-3061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston C, Martin B, Fichant G, Polard P, Claverys J-P. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 21.Bartilson M, Marra A, Christine J, Asundi JS, Schneider WP, Hromockyj AE. 2001. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide. Mol Microbiol 39:126–135. doi: 10.1046/j.1365-2958.2001.02218.x. [DOI] [PubMed] [Google Scholar]
- 22.Tovpeko Y, Morrison DA. 2014. Competence for genetic transformation in Streptococcus pneumoniae: mutations in σA bypass the comW requirement. J Bacteriol 196:3724–3734. doi: 10.1128/JB.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng L, Biswas I, Morrison DA. 2009. A self-deleting Cre-lox-ermAM cassette, Cheshire, for marker-less gene deletion in Streptococcus pneumoniae. J Microbiol Methods 79:353–357. doi: 10.1016/j.mimet.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein LS, Piccoli L, Simon D, Morrison DA. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J Bacteriol 178:6087–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendelsohn AR, Brent R. 1994. Applications of interaction traps/two-hybrid systems to biotechnology research. Curr Opin Biotechnol 5:482–486. doi: 10.1016/0958-1669(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 26.Fritz CC, Green MR. 1992. Fishing for partners. Curr Biol 2:403–405. doi: 10.1016/0960-9822(92)90315-2. [DOI] [PubMed] [Google Scholar]
- 27.Attaiech L, Olivier A, Mortier-Barrière I, Soulet A-L, Granadel C, Martin B, Polard P, Claverys J-P. 2011. Role of the single-stranded DNA-binding protein SsbB in pneumococcal transformation: maintenance of a reservoir for genetic plasticity. PLoS Genet 7:e1002156. doi: 10.1371/journal.pgen.1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson S, Cline RT, Tettelin H, Sharov V, Morrison DA. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J Bacteriol 182:6192–6202. doi: 10.1128/JB.182.21.6192-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan R, Rukke HV, Høvik H, Åmdal HA, Chen T, Morrison DA, Petersen FC. 2016. Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core streptococcal competence genes. mSystems 1:e00038-15. doi: 10.1128/mSystems.00038-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirouze N, Bergé MA, Soulet A-L, Mortier-Barrière I, Quentin Y, Fichant G, Granadel C, Noirot-Gros M-F, Noirot P, Polard P, Martin B, Claverys J-P. 2013. Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc Natl Acad Sci U S A 110:E1035–E1044. doi: 10.1073/pnas.1219868110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson GT, Ng W-L, Foley J, Gilmour R, Winkler ME. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J Bacteriol 184:3508–3520. doi: 10.1128/JB.184.13.3508-3520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chastanet A, Prudhomme M, Claverys JP, Msadek T. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J Bacteriol 183:7295–7307. doi: 10.1128/JB.183.24.7295-7307.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desai BV, Morrison DA. 2006. An unstable competence-induced protein, CoiA, promotes processing of donor DNA after uptake during genetic transformation in Streptococcus pneumoniae. J Bacteriol 188:5177–5186. doi: 10.1128/JB.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boutry C, Wahl A, Delplace B, Clippe A, Fontaine L, Hols P. 2012. Adaptor protein MecA is a negative regulator of the expression of late competence genes in Streptococcus thermophilus. J Bacteriol 194:1777–1788. doi: 10.1128/JB.06800-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schöbel S, Zellmeier S, Schumann W, Wiegert T. 2004. The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol Microbiol 52:1091–1105. doi: 10.1111/j.1365-2958.2004.04031.x. [DOI] [PubMed] [Google Scholar]
- 36.Österberg S, del Peso-Santos T, Shingler V. 2011. Regulation of alternative sigma factor use. Annu Rev Microbiol 65:37–55. doi: 10.1146/annurev.micro.112408.134219. [DOI] [PubMed] [Google Scholar]
- 37.Gaal T, Mandel MJ, Silhavy TJ, Gourse RL. 2006. Crl facilitates RNA polymerase holoenzyme formation. J Bacteriol 188:7966–7970. doi: 10.1128/JB.01266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banta AB, Chumanov RS, Yuan AH, Lin H, Campbell EA, Burgess RR, Gourse RL. 2013. Key features of σS required for specific recognition by Crl, a transcription factor promoting assembly of RNA polymerase holoenzyme. Proc Natl Acad Sci U S A 110:15955–15960. doi: 10.1073/pnas.1311642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong T, Schellhorn HE. 2010. Role of RpoS in virulence of pathogens. Infect Immun 78:887–897. doi: 10.1128/IAI.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robbe-Saule V, Lopes MD, Kolb A, Norel F. 2007. Physiological effects of Crl in Salmonella are modulated by sigmaS level and promoter specificity. J Bacteriol 189:2976–2987. doi: 10.1128/JB.01919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]