Abstract
Emergence of antimicrobial resistance mediated through New Delhi metallo-β-lactamases (NDMs) is a serious therapeutic challenge. Till date, 16 different NDMs have been described. In this study, we report the molecular and structural characteristics of NDM-5 isolated from an Escherichia coli isolate (KOEC3) of bovine origin. Using PCR amplification, cloning and sequencing of full blaNDM gene, we identified the NDM type as NDM-5. Cloning of full gene in E. coli DH5α and subsequent assessment of antibiotic susceptibility of the transformed cells indicated possible role of native promoter in expression blaNDM-5. Translated amino acid sequence had two substitutions (Val88Leu and Met154Leu) compared to NDM-1. Theoretically deduced isoelectric pH of NDM-5 was 5.88 and instability index was 36.99, indicating a stable protein. From the amino acids sequence, a 3D model of the protein was computed. Analysis of the protein structure elucidated zinc coordination and also revealed a large binding cleft and flexible nature of the protein, which might be the reason for broad substrate range. Docking experiments revealed plausible binding poses for five carbapenem drugs in the vicinity of metal ions. In conclusion, results provided possible explanation for wide range of antibiotics catalyzed by NDM-5 and likely interaction modes with five carbapenem drugs.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-016-0569-5) contains supplementary material, which is available to authorized users.
Keywords: Metallo-β-lactamase, blaNDM-5, Structure, Carbapenem, Cleft, Docking
Introduction
Emergence of antimicrobial resistance is an evolving public health crisis [1]. Among many available antimicrobials, β-lactam antibiotics had been the preferred choice for clinical application due to their efficacy, low cost and minimal toxicity [2]. β-lactamases constitute most important resistance mechanisms for inactivating the β-lactam antibiotics by hydrolysis of the β-lactam ring [3]. Of the two classes of β-lactamases, serine-β-lactamases and metallo-β-lactamases, the latter poses the stiffest clinical challenge by inactivating almost all β-lactams, except for monobactams [3, 4]. New Delhi metallo-β-lactamase (NDM) is the most recent addition in the list of these difficult enzymes and is associated with extensive resistance in Gram negative bacteria [5, 6]. Since first report of NDM in 2009, 16 variants of NDM enzymes (NDM-1 to -16) have been discovered and assigned [7].
Occurrences of NDM have been reported from many countries including Asian countries such as China, Bangladesh, India, Vietnam [3, 8–16]. Among many variants, NDM-5 has been reported to possess increased resistance to carbapenems and broad-spectrum cephalosporins and was first reported in 2011 in an Escherichia coli isolate from United Kingdom [10]. Since then, an increasing number of human infections with organisms harbouring NDM-5 have been reported from many parts of the world [3] including Spain [17], China [9], Algeria [18], Japan [19], South Korea [20] and India [11]. Evidences of animal infections with NDM-5 are rare, though infections with organisms carrying NDM-1 have been reported in companion animals [21].
Structural characterization (such as modelling, docking) of protein molecules, provides insight into molecular functions and biological interactions, thus providing a platform for identification of plausible antibacterial agent [22]. However, unlike NDM-1, till date, there have been no reports on the structural characteristics of the NDM-5. In a previous report, we reported the presence of blaNDM, from an E. coli isolate (KOEC3) of bovine origin [23]. Since NDM-5 is known to possess more resistance to carbapenems, we intended to investigate the molecular and structural basis of carbapenem inactivation by NDM-5 through a combined wet lab and in silico approach.
Materials and Methods
Full Gene Amplification, Cloning and Characterization of Transformed Cells
Full gene of blaNDM from KOEC3 isolate was amplified by PCR using the reported primers [10] at 58 °C for annealing and 72 °C for 45 s for extension. Amplification was checked by agarose gel (1.5 %) electrophoresis. The PCR product was purified by QiaQuick PCR purification kit (Qiagen, USA) as per manufacturer’s instruction. The purified PCR product was then ligated into a pTZ57R/T vector using InsTAclone PCR cloning kit (Thermo Scientific, Waltham, USA) as per manufacturer’s instructions with negative control of only vector (without insert) and positive control of ‘control fragment’ supplied with the kit. Following ligation, E. coli DH5α cells were transformed using the ligated vector (InsTAclone kit, Thermo Scientific). Transformed cells, with recombinant plasmid were selected through Blue-White colony screening on X-Gal (20 µg/ml)—IPTG (24 µg/ml)—ampicillin (50 µg/ml) containing agar plates. For transformation control, plasmids (with and without insert) provided in the kit were used as positive and negative control, respectively. Presence of blaNDM gene in cloned sample was confirmed by colony PCR as described above. Orientation of the insert was checked by colony PCR employing forward primer of NDM and T7 promoter universal primer. In order to eliminate the background resistance effect of the vector pTZ57R/T on the final result, E. coli DH5α cells were transformed only with the vector without any insert.
Transformed E. coli cells and the KOEC3 isolate were subjected to antibiotic susceptibility test by Pheonix™ 100 (Becton–Dickinson, Singapore) or by broth dilution method as per EUCAST guideline [24]. Results were interpreted according to manufacturer’s instructions/EUCAST recommendation.
Sequencing
Purified recombinant plasmids from transformed E. coli cells were subjected to bi-directional sequencing by using the BigDye Terminator cycle sequencing kit (Applied Biosystems, USA) in ABI 3500xL Genetic analyzer automated sequencer (Applied Biosystems, USA) as per the manufacturer’s instructions.
Sequence Analysis
The sequences obtained through bidirectional sequencing were then assembled and homology was searched against the blaNDM sequences available at Lahey Clinic database [7] using the NCBI blastn tool (available at http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Nucleotide sequence was further translated into corresponding amino acid sequence using the EMBOSS Transeq tool and was analysed for primary information using the ExPASyProtParam tool.
3D Model Building
3D model of the enzyme was built through homology modelling using Swiss-Model Workspace available at http://swissmodel.expasy.org/. Due to unavailability of crystal structure of NDM-5 in the protein data bank an initial template search was made and three structures of NDM-1 (PDB IDs 4eyb.1.A, 4h0d.1.A, 3q6x.1.A) were chosen. NDM-5 models were built using these templates and the final model was selected based on their QMEAN score.
Quality of the computed model was analysed at QMEAN server available at http://swissmodel.expasy.org/qmean/cgi/index.cgi. Ramachandran plot analysis, all atom clash score, and protein geometry of the structure were estimated using the MolProbity tool available at http://molprobity.biochem.duke.edu/.
Computed model was also put to molecular dynamic simulation for 2.5 ns using NAMD software [25]. For simulation, the model was minimized, solvated, added with sodium and chloride ions at 0.15 mol/l concentration.
Structure Analysis
Model structure was analysed at the PDBsum server available at http://www.ebi.ac.uk/pdbsum/. Secondary structure was analysed using ProMotif, and cleft analysis was conducted using SurfNet1.4 and metal binding was investigated using the Ligplot available at the same server. During analysis computed model, two other previously reported structures (PDB ID: 4EXY and 4EYL) were also included. Since our computed model was a monomer, only monomeric structures were chosen up to a resolution of 2 Å. In case(s) where ligand was already complexed, the same was removed and further processed.
Binding Study
Binding of five carbapenem antibiotics (doripenem, ertapenem, faropenem, imipenem, meropenem) were studied using the SwissDock server [26] under accurate mode. Ligand molecules (PubChem IDs: doripenem-73303; ertapenem-150610; faropenem-65894; imipenem-104838; meropenem-441130) were prepared using UCSF Chimera tool 1.10 [27]. Since energy minimization was known to optimize molecules towards stable state, the receptor molecule (NDM-5) was prepared by addition of H-ions and minimization of energy by AMBER force field 99SB using Chimera 1.10. Resulting molecule was submitted to SwissDock server for blind docking and results were visualized with Chimera 1.10. Similarly two other structures (PDB ID: 4EXY, 4EYL) of NDM-1 was also included in the analysis for further comparison.
Results and Discussion
Molecular Characterization
Molecular characterization of the E. coli isolate (KOEC3) targeting blaNDM gene resulted in amplification of an expected product with a molecular weight of 813 bp. Colony PCR of transformed cell resulted in amplification of an approximately 850 bp indicating correct orientation (Supplementary Figure 1). Alignment of obtained sequence with blaNDM sequences listed at Lahey database and NCBI nucleotide database [7] revealed 100 % sequence alignment with blaNDM-5 (JN104597, KF408074, KF408073, KF408072, KM598665). Therefore, the blaNDM gene of KOEC3 isolate was inferred as blaNDM-5. The sequence was deposited to NCBI (KC769583.2).
The isolate (KOEC3) was resistant to almost all antibiotics (19/22) tested (Table 1). The isolate was also resistant to cephalosporins (cefazolin, cefuroxime, ceftazidime, cefepime->16 µg/ml; ceftriaxone->32 µg/ml), carbapenems (imipenem, meropenem, doripenem, ertapenem->8 µg/ml; faropenem->16 µg/ml), other β-lactams (ampicillin, amoxicillin-clavulanate, aztreonam) but was susceptible to tetracycline, colistin and chloramphenicol. In contrast, transformed E. coli DH5α cells were resistant only to cefazolin (>16 µg/ml), ampicillin (>16 µg/ml) and amoxicillin-clavulanate (>16/8 µg/ml; Table 1). Similar results were also obtained for transformed E. coli DH5α cells without the insert. The loss of resistance to a majority of β-lactams in the transformants was also reported previously [10] suggesting the role of native promoter in expression of NDM-5. In the present study, the absence of native promoter in the transformed DH5α cells might have resulted in susceptibility to several antibiotics. Moreover, common cloning vectors are known to contain resistance marker genes under separate promoters, but to our knowledge none is known to contain carbapenem resistance as ‘resistance marker’ for selection of transformed cells. Therefore, the loss of resistance to carbapenems drugs was most likely due to absence of native promoter.
Table 1.
Minimum inhibitory concentrations (MIC) of E. coli isolate KOEC3 and transformants for various antimicrobials (µg/ml)
| Antimicrobials | KOEC3 | Transformed E. coli DH5α (with blaNDM-5 insert) |
|---|---|---|
| Amikacin | >32 | ≤8 |
| Gentamicin | >8 | ≤1 |
| Tobramycin | >8 | ≤1 |
| Imipenem | >8 | ≤1 |
| Meropenem | >8 | ≤1 |
| Doripenem | >8 | ≤1 |
| Faropenem | >16 | ≤1 |
| Ertapenem | >8 | ≤0.5 |
| Cefazolin | >16 | >16 |
| Cefuroxime | >16 | 8 |
| Ceftazidime | >16 | ≤0.5 |
| Ceftriaxone | >32 | ≤2 |
| Cefepime | >16 | ≤1 |
| Aztreonam | >16 | ≤2 |
| Ampicillin | >16 | >16 |
| Amoxicillin-Clavulanatea | >16/8 | >16/8 |
| Piperacillin-Tazobactamb | >64/4 | >64/4 |
| Trimethoprim-sulfamethoxazole | >2/38c | ≤0.5/9.5d |
| Nitrofurantoin | 32 | ≤16 |
| Ciprofloxacin | >2 | ≤0.5 |
| Levofloxacin | >4 | ≤1 |
| Moxifloxacin | >4 | ≤1 |
| Tetracycline | ≤2 | ≤2 |
aAmoxicillin (16 µg/ml) and clavulanate (8 µg/ml) in combination
bPiperacillin (64 µg/ml) and tazobactam (4 µg/ml) in combination
cTrimethoprim (2 µg/ml) and sulfamethoxazole (38 µg/ml) in combination
dTrimethoprim (0.5 µg/ml) and sulfamethoxazole (9.5 µg/ml) in combination
Amino Acid Sequence
Primary sequence of NDM-5 gene consisted of 270 amino acids, with molecular weight of 28495.4 and theoretically determined isoelectric point of 5.88 which was similar to NDM-4 [28]. Calculated instability index (36.99) indicated the protein to be stable as proteins with an instability index below 40 were considered as stable [29]. Comparison of amino acid sequence of NDM-5 (KOEC3) with other NDM sequences listed at Lahey database revealed varying degree of amino acid substitutions ranging from 1 to 7 (Supplementary Table 1). Two substitutions (Val88Leu, Met154Leu) observed in NDM-5 (KOEC3) was also reported previously [10]. Interestingly, the presence of leucine at position 88 was unique to NDM-5 and might serve as a signature for NDM-5. Though this substitution is believed to confer increased resistance to carbapenems [10], in our subsequent docking studies we did not observe any direct interaction between the drug molecule and the leucine residue at position 88.
Modelling of NDM-5
3D modelling of NDM-5 yielded three models and the final model (Fig. 1a, b) was chosen based on the highest QMEAN4 score of 0.79. Model quality checking (Supplementary Table 2) revealed that all atom clash score was 3.38 (97th percentile; 100th-best; 0th-worst) and overall MolProbity score was 1.18 (99th percentile) indicating reliable model quality.
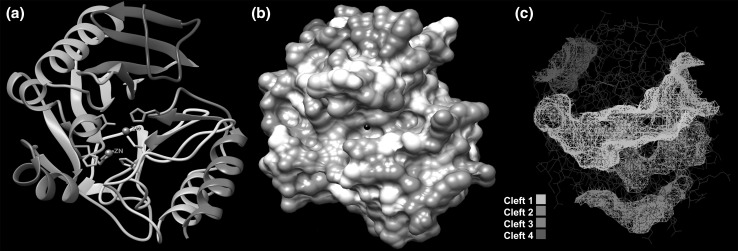
Fig. 1.
Computed three dimensional structure of NDM-5 (created with UCSF Chimera 1.10). a Ribbon structure of NDM-5 coloured in rainbow style. Metal ions coloured grey. b Surface topology of NDM-5 coloured by Coulombic surface colouring. Surface calculated using AMBER ff 14SB charge model on three colour scale of red, −10 kcal/(mol e), white, 0 kcal/(mol e), and blue, 10 kcal/(mol e), where e is unit electron charge. c Predicted binding clefts of NDM-5
Further quality check against non-redundant set of protein data bank structures at QMEAN server, indicated acceptable model quality (Supplementary Figure 2a, 2b, 2c) with QMEAN score of 0.796 and overall Z-score of 0.29. Per residue error plot of the model was visualized on a colour scale of blue (errors <1 Å) to red (errors above 3.5 Å). While our computed model was quite reliable, two error prone regions were also discovered.
Ramachandran plot analysis of NDM-5 structure revealed that 97.9 % of all residues were in favoured regions and 99.6 % of residues were in Ramachandran allowed regions with one outlier (phi, psi) at 90 Asp (Supplementary Figure 3).
Molecular dynamic simulation of computed NDM-5 model revealed consistent RMSD values (Supplementary Figure 4) which indicated reliable structure.
The NDM-5 chain was a monomer with two zinc atoms attached (Fig. 1a). Surface topology of the NDM-5 chain was computed using Chimera 1.10. The total solvent accessible surface area (SASA) was 10105 Å2 and solvent excluded surface area was 9057.37 Å2. SASA was also calculated from the molecular weight of the protein by the equation:
where, M is the molecular weight of protein [30]. Calculated SASA was 11761.8. From these two estimates we calculated relative SASA value for NDM-5 which was 0.86. Since relative SASA was known to be a dependable predictor of flexibility of monomeric proteins, our results indicated that our computed model of NDM-5 was reasonably flexible allowing conformational changes during catalytic activity of the enzyme [30]. Conformational changes in metallo-β-lactamases leads to a closure of loop 3 that aids in substrate interaction and can increase the substrate range [3].
Secondary structure analysis revealed that the NDM-5 chain (residue 30–270) was composed of 2 sheets (A and B), 4 beta–alpha–beta motifs, 12 strands, 8 helices. There were 24 instances of β-turns, 6 β-hairpins, 4 β-bulges, 1 γ-turn and 5 helix–helix interactions (Supplementary Figure 5).
Metal ions (zinc) are important for the activity of metallo-β-lactamases [3]. Results (Supplementary Figure 6) of metal binding analysis revealed that both the zinc atoms were coordinated in trigonal bipyramid geometry at His and Cys site. While the zinc atom at Cys site was held by aspartic acid (124), cystine (208) and histidine (250, also the closest; 2.03 Å), the other zinc atom at His site was coordinated by three histidine residues located at 120, 122 (closest at 1.96 Å), 189 position In our model for NDM-5, the distance between two zinc atoms was 4.54 Å which was less than a previous report on NDM-1 [31].
To identify, the most probable binding sites, we undertook cleft analysis of our computer generated model. Our analysis showed existence of four major clefts (Fig. 1c, Supplementary Table 3). The volume of the largest cleft was 2958.19 Å3 (bright green) followed by the second largest cleft with a volume of 934.45 Å3 (dull green). In case of single chain proteins, possession of large cleft and the ratio (ideally >2.0) of two largest clefts (R1) are important functional requirements as ligands tend to preferentially bind to the largest cleft [32]. Therefore, the largest cleft (cleft 1) is presumably the most probable binding site for our computed model of NDM-5. In case of our model of NDM-5, the ratio was 3.17, indicating that cleft 1 was the most probable and also the preferential binding site for ligands. Cleft analysis was also performed for two previously published structures of NDM-1 (PDB ID: 4EXY, 4EYL) employing same web server (PDBsum). Results revealed that compared to NDM-5 (KOEC3), the largest clefts of 4EXY and 4EYL were smaller by 4.35 and 13 %, respectively. Moreover, largest clefts of 4EXY and 4EYL were also less deep. Therefore, the results of our study indicating existence of a comparatively larger and deeper cleft in NDM-5 (cleft 1), along with higher R1 ratio, flexibility and stability of the molecule (see previous sections), possibly explained the greater catalytic activity by NDM-5 against wide range of antibiotics. Inclusion of bulky side chain to antibiotic molecule is a common strategy for circumventing antibiotic hydrolysis by bacterial enzymes. To investigate the possibility of such approach, we compared the molecular sizes of the carbapenem molecules (Doripenem −341.0 Å3, Ertapenem −390.2 Å3, Faropenem −223.6 Å3, Imipenem −261.2 Å3, Meropenem −326.5 Å3) with the predicted largest binding cleft (cleft 1) volume. Our analysis revealed very low drug volume-to-cleft volume ratio ranging from 0.076 for faropenem to 0.132 for ertapenem. These results indicated that due to existence of a quite large binding cleft the tactic to increase the bulk of the drug molecule may not be suitable for development of novel antibiotics against NDM-5 producing bacteria and other alternate strategies need to be explored.
Ligand Binding
Prediction of ligand binding (docking) by in silico molecular docking is a useful approach to study molecular interactions [22]. This has been successfully applied for identification of novel antimicrobial targets [33], putative antifungal analogues [34]. However, limitations of computational docking should not be ignored while interpreting results [35].In the present study we employed computational docking for analysis of interactions between NDM-5 and five carbapenems antibiotics.
Docking studies were performed for five carbapenem antibiotics at the SwissDock server using CHARMM force field parameters [26]. We assumed that the most likely ligand BMs should be the ones that bind within the largest cleft on the protein surface with best available full fitness value returned by the docking experiments. Based on this we attempted to identify most probable BMs for the antibiotics (Fig. 2a–e). Overall, our results corroborated with previous results on NDM-1 [36]. Free energy values (kcal/mol) and full fitness scores of ligands (Supplementary Table 4) revealed lowest ΔG for meropenem (−8.65) followed by imipenem (−8.24) and doripenem (−8.18). But highest full fitness score was obtained for doripenem (−1059.9) followed by imipenem (−1052.4) and meropenem (−917.5). The results indicated that among all BMs, BM for imipenem was most consistent.
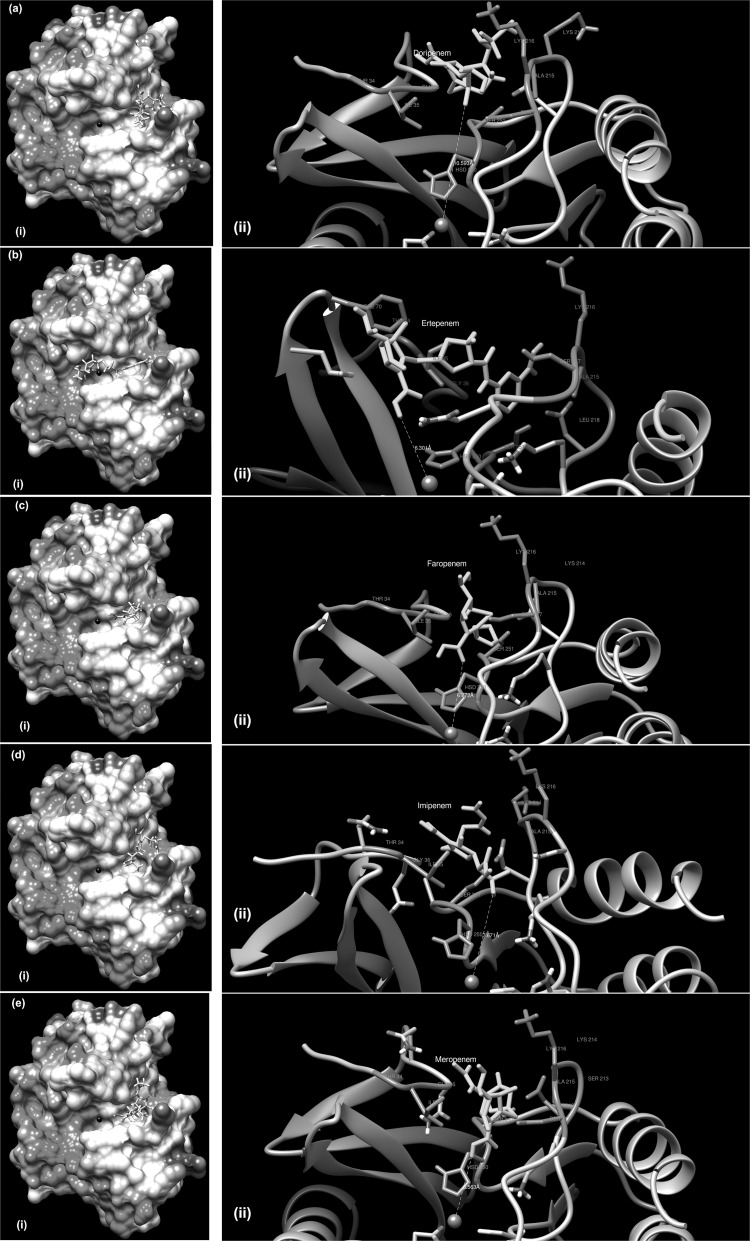
Fig. 2.
Binding of various antibiotic molecules with NDM-5. a Doripenem, b Ertapenem, c Faropenem, d Imipenem, and e Meropenem. (i) and (ii) in each panel indicate overview and involvements of residues (orange) and metal ions (grey), respectively
Further inspection of the BMs, revealed that the rim of the binding pocket was formed by Ile35, Gly36, Gln37, Lys214, Ala215 and Lys216. Residues at the rim of the binding pocket are known to influence the binding between the enzyme and drug molecules by van der Waals interactions and hydrogen bonding thus securing the drug molecules in binding cleft [31]. Due to unavailability of similar studies with NDM-5, we compared our results with previous report available for crystal structure of NDM-1 complexed with hydrolyzed meropenem [37]. In both cases, bindings were mediated by Ala215, Gly219, His250, and Lys216. However differences were observed from a previous study on NDM-1 [31], where binding of imipenem and meropenem were predicted closer (within 5 Å) to the His site of zinc coordination. However, comparison of docking results of NDM-5 (KOEC-3) with the docking results of two other previously reported structures (4EXY, 4EYL) revealed that the minimum distances over which the ligand binding took place were variable (Supplementary Table 5). While doripenem interacted with NDM-5 at farthest (10.593 Å), faropenem and imipenem interacted closer to NDM-5 compared to NDM-1 molecules (4EXY, 4EYL). Distances for ertapenem and meropenem were in between respective values for 4EXY and 4EYL.
Binding of multiple diverse ligands at a particular protein site is an emerging conceptual framework in protein science [38]. Proteins, if allowed to interact with an ensemble of ligands, might bind to multiple ligands with altered shapes and sizes [38]. In the present study, we observed a large preferentially binding cleft (cleft 1) on the NDM-5 molecule and the cleft volume was much greater than the molecular volumes of the ligands (carbapenem drugs) studied. Therefore, it is tempting to explore the possibility of binding of multiple ligands. However, it should be kept in mind that ligand binding in metallo-β-lactamases are mediated through zinc atoms which play critical role in initiation of catalysis and stabilization of hydroxide ion for nucleophilic attack on the carbonyl group of the drug molecule [3]. Unless, the zinc atoms are freed, binding of multiple ligands might not have biological implication.
Conclusions
In conclusion, our study reported molecular and structural characterization of the blaNDM-5 previously isolated from an E. coli isolate (KOEC3) of bovine origin. While molecular characterization indicated the likely importance of native promoter for expression of NDM-5, computer aided structural analysis generated a stable three dimensional structure of the protein with four major predicted binding sites, coordination of the metal atoms (zinc) at His and Cys sites. Existence of large binding cleft on the molecule posed hindrance for development of novel antimicrobial by incorporating bulky side chain into the existing carbapenems drugs. Computer aided ligand binding studies, on the other hand, identified possible binding poses and interactions of five carbapenem antibiotic molecules (ligand) with the receptor (NDM-5).
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
PCR detection of blaNDM-5 gene in Escherichia coli isolates (KOEC3 and transformed DH5α cells). Lane M: Molecular weight marker; Lane A: KOEC3 (positive control); Lane B: Transformed DH5α (blaNDM-5 forward and reverse primers); Lane C: Transformed DH5α (blaNDM-5 forward and T7 promoter primers); Lane D: Negative control. (PNG 342 kb)
Quality assessment of computed three dimensional model of NDM-5. (a) Comparison of NDM-5 computed model with non-redundant set of PDB structures. (b) Density plot of the QMEAN score of reference PDB set with NDM-5 (in red). (c) Per residue error plot NDM-5. Blue: errors < 1 Å, red: errors above 3.5 Å. (PNG 360 kb)
Ramachandran plot analysis of NDM-5 (PNG 336 kb)
RMSD plot of residues of NDM-5 following molecular dynamics simulation of 2.5 ns (PNG 98 kb)
Secondary structure of NDM-5 (PNG 8622 kb)
Zinc coordination of computed model of NDM-5. Hydrogen bondings are depicted as dashed lines. (PNG 1793 kb)
Acknowledgments
The authors are thankful to Director, ICAR RC for NEH Region, Meghalaya for providing necessary facilities and to Department of Biotechnology, New Delhi for partly funding the study under Twinning programme for NE (Sanction Order No. BT/363/NE/TBFJIL/12 dated March 21, 2013). Authors are thankful to Dr Manish Kakkar for his critical inputs into the manuscript.
Footnotes
D. Purkait, A. Ahuja and U. Bhattacharjee have contributed equally for this study.
References
- 1.World Health Organization (WHO) (2012) The evolving threat of antimicrobial resistance: options for action. WHO Press, World Health Organization, 20 Avenue Appia, 1211 Geneva 27, Switzerland
- 2.Makena A, Brem J, Pfeffer I, Geffen RE, Wilkins SE, Tarhonskaya H, Flashman E, Phee LM, Wareham DW, Schofield CJ. Biochemical characterization of New Delhi metallo-β-lactamase variants reveals differences in protein stability. J Antimicrob Chemother. 2015;70:463–469. doi: 10.1093/jac/dku403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palzkill T. Metallo-β-lactamase structure and function. Ann N Y Acad Sci. 2013;1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berrazeg M, Diene S, Medjahed L, Parola P, Drissi M, Raoult D &Rolain J (2014) New Delhi Metallo-beta-lactamase around the world: an eReview using Google Maps. Euro Surveill 19: 20809. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20809 [DOI] [PubMed]
- 6.Jones LS, Carvalho MJ, Toleman MA, White PL, Connor TR, Mushtaq A, Weeks JL, Kumarasamy KK, Raven KE, Török ME, Peacock SJ, Howe RA, Walsh TR. Characterization of plasmids in extensively drug-resistant Acinetobacter strains isolated in India and Pakistan. Antimicrob Agents Chemother. 2014;59:923–929. doi: 10.1128/AAC.03242-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lahey Clinic (2015) NDM-type ß-Lactamases. http://www.lahey.org/Studies/other.asp#table1. Accessed on 28 Oct 2015
- 8.Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. BioMed Res Int. 2014 doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang P, Xie Y, Feng P, Zong Z. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother. 2014;58:7548–7552. doi: 10.1128/AAC.03911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornsey M, Phee L, Wareham DW. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 2011;55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahman M, Shukla SK, Prasad KN, Ovejero CM, Pati BK, Tripathi A, Singh A, Srivastava AK, Gonzalez-Zorn B. Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents. 2014;44:30–37. doi: 10.1016/j.ijantimicag.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Toleman MA, Bugert JJ, Nizam SA. Extensively drug-resistant New Delhi metallo-β-lactamase–encoding bacteria in the environment, Dhaka, Bangladesh, 2012. Emerg Infect Dis. 2015;21:1027–1030. doi: 10.3201/eid2106.141578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, Zhu Y, Li Y, Mu X, You L, Xu C, Qin P, Ma J. Coexistence of SFO-1 and NDM-1 β-lactamase genes and fosfomycin resistance gene fosA3 in an Escherichia coli clinical isolate. FEMS Microbiol Lett. 2014;362:1–7. doi: 10.1093/femsle/fnu018. [DOI] [PubMed] [Google Scholar]
- 14.Seija V, Presentado JCM, Bado I, Ezdra RP, Batista N, Gutierrez C, Guirado M, Vidal M, Nin M, Vignoli R. Sepsis caused by New Delhi metallo-β-lactamase (blaNDM-1) and qnrD-producing Morganellamorganii, treated successfully with fosfomycin and meropenem: case report and literature review. Int J Infect Dis. 2015;30:20–26. doi: 10.1016/j.ijid.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Tran HH, Ehsani S, Shibayama K, Matsui M, Suzuki S, Nguyen MB, Tran DN, Tran VP, Tran DL, Nguyen HT, Dang DA, Trinh HS, Nguyen TH, Wertheim HFL. Common isolation of New Delhi metallo-beta-lactamase 1-producing Enterobacteriaceae in a large surgical hospital in Vietnam. Eur J Clin Microbiol Infect Dis. 2015;34:1247–1254. doi: 10.1007/s10096-015-2345-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharya D, Dey S, Kadam S, Kalal S, Jali S, Koley H, Sinha R, Nag D, Kholkute SD, Roy S. Isolation of NDM-1-producing multidrug-resistant Pseudomonas putida from a paediatric case of acute gastroenteritis, India. New Microbe New Infect. 2015;5:5–9. doi: 10.1016/j.nmni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitart C, Solé M, Roca I, Román A, Moreno A, Vila J, Marco F. Molecular characterization of blaNDM-5 carried on an IncFII plasmid in an Escherichia coli isolate from a non-traveler patient in Spain. Antimicrob Agents Chemother. 2015;59:659–662. doi: 10.1128/AAC.04040-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sassi A, Loucif L, Gupta SK, Dekhil M, Chettibi H, Rolain JM. NDM-5 carbapenemase-encoding gene in multidrug-resistant clinical isolates of Escherichia coli from Algeria. Antimicrob Agents Chemother. 2014;58:5606–5608. doi: 10.1128/AAC.02818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano R, Nakano A, Hikosaka K, Kawakami S, Matsunaga N, Asahara M, Ishigaki S, Furukawa T, Suzuki M, Shibayama K, Ono Y. First report of metallo-β-lactamase NDM-5-producing Escherichiacoli in Japan. Antimicrob Agents Chemother. 2014;58:7611–7612. doi: 10.1128/AAC.04265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho SY, Huh HJ, Baek JY, Chung NY, Ryu JG, Ki CS, Chung DR, Lee NY, Song JH. Klebsiellapneumoniae co-Producing NDM-5 and OXA-181 carbapenemases, South Korea. Emerg Infect Dis. 2015;21:1088–1089. doi: 10.3201/eid2106.150048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaheen BW, Nayak R, Boothe DM. Emergence of a New Delhi metallo-β-lactamase (NDM-1)-encoding gene in clinical Escherichiacoli isolates recovered from companion animals in the United States. Antimicrob Agents Chemother. 2013;57:2902–2903. doi: 10.1128/AAC.02028-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cain R, Narramore S, McPhillie M, Simmons K, Fishwick CWG. Applications of structure-based design to antibacterial drug discovery. Bioorgan Chem. 2014 doi: 10.1016/j.bioorg.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Ghatak S, Singha A, Sen A, Guha C, Ahuja A, Bhattacherjee U, Das S, Pradhan NR, Puro K, Jana C, Dey TK, Prashantkumar KL, Das A, Shakuntala I, Biswas U, Jana PS. Detection of New Delhi metallo-beta-Lactamase and extended-spectrum beta-lactamase genes in Escherichia coli isolated from mastitic milk samples. Transbound Emerg Dis. 2013;60:385–389. doi: 10.1111/tbed.12119. [DOI] [PubMed] [Google Scholar]
- 24.EUCAST (European Committee on Antimicrobial Susceptibility Testing) (2003) Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. EUCAST Discussion Document E Dis 5: 1–7. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/MIC_testing/Edis5.1_broth_dilution.pdf. Accessed on 20 Oct 2015
- 25.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Nordmann P, Boulanger AE, Poirel L. NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob Agents Chemother. 2012;56:2184–2186. doi: 10.1128/AAC.05961-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. New Jersey: Humana Press Inc; 2005. pp. 571–607. [Google Scholar]
- 30.Marsh JA, Teichmann SA. Relative solvent accessible surface area predicts protein conformational changes upon binding. Structure. 2011;19:859–867. doi: 10.1016/j.str.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JF, Chou KC. Insights from modelling the 3D structure of New Delhi metallo-β-lactamase and its binding interactions with antibiotic drugs. PLoS ONE. 2011;6:e18414. doi: 10.1371/journal.pone.0018414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laskowski RA, Luscombe NM, Swindells MB, Thornton JM. Protein clefts in molecular recognition and function. Protein Sci. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Ma S. Advances in the discovery of novel antimicrobials targeting the assembly of bacterial cell division protein FtsZ. Eur J Med Chem. 2015;95:1–15. doi: 10.1016/j.ejmech.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 34.Karumuri S, Singh PK, Shukla P. In silico analog design for Terbinafine against Trichophytonrubrum: a preliminary study. Indian J Microbiol. 2015;55:333–340. doi: 10.1007/s12088-015-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mihasan M. What in silico molecular docking can do for the ‘bench-working biologists’. J Biosci. 2012;37:1089–1095. doi: 10.1007/s12038-012-9273-8. [DOI] [PubMed] [Google Scholar]
- 36.Green VL, Verma A, Owens RJ, Phillips SE, Carr SB. Structure of New Delhi metallo-β-lactamase 1 (NDM-1) Acta Crystallogr, Sect F: Struct Biol Cryst Commun. 1996;67:1160–1164. doi: 10.1107/S1744309111029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim Y, Tesar C, Jedrzejczak R, Babnigg G, Sacchettini J, Joachimiak A. Crystal structure of New Delhi metallo-β-lactamase-1 in complex with hydrolyzed meropenem. PDB ID: 4RBS 2014; Midwest Center for Structural Genomics
- 38.Ma B, Shatsky M, Wolfson HJ, Nussinov R. Multiple diverse ligands binding at a single protein site: a matter of pre-existing populations. Protein Sci. 2002;11:184–197. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR detection of blaNDM-5 gene in Escherichia coli isolates (KOEC3 and transformed DH5α cells). Lane M: Molecular weight marker; Lane A: KOEC3 (positive control); Lane B: Transformed DH5α (blaNDM-5 forward and reverse primers); Lane C: Transformed DH5α (blaNDM-5 forward and T7 promoter primers); Lane D: Negative control. (PNG 342 kb)
Quality assessment of computed three dimensional model of NDM-5. (a) Comparison of NDM-5 computed model with non-redundant set of PDB structures. (b) Density plot of the QMEAN score of reference PDB set with NDM-5 (in red). (c) Per residue error plot NDM-5. Blue: errors < 1 Å, red: errors above 3.5 Å. (PNG 360 kb)
Ramachandran plot analysis of NDM-5 (PNG 336 kb)
RMSD plot of residues of NDM-5 following molecular dynamics simulation of 2.5 ns (PNG 98 kb)
Secondary structure of NDM-5 (PNG 8622 kb)
Zinc coordination of computed model of NDM-5. Hydrogen bondings are depicted as dashed lines. (PNG 1793 kb)