ABSTRACT
The western honeybee (Apis mellifera) is the most important commercial insect pollinator. However, bees are under pressure from habitat loss, environmental stress, and pathogens, including viruses that can cause lethal epidemics. Slow bee paralysis virus (SBPV) belongs to the Iflaviridae family of nonenveloped single-stranded RNA viruses. Here we present the structure of the SBPV virion determined from two crystal forms to resolutions of 3.4 Å and 2.6 Å. The overall structure of the virion resembles that of picornaviruses, with the three major capsid proteins VP1 to 3 organized into a pseudo-T3 icosahedral capsid. However, the SBPV capsid protein VP3 contains a C-terminal globular domain that has not been observed in other viruses from the order Picornavirales. The protruding (P) domains form “crowns” on the virion surface around each 5-fold axis in one of the crystal forms. However, the P domains are shifted 36 Å toward the 3-fold axis in the other crystal form. Furthermore, the P domain contains the Ser-His-Asp triad within a surface patch of eight conserved residues that constitutes a putative catalytic or receptor-binding site. The movements of the domain might be required for efficient substrate cleavage or receptor binding during virus cell entry. In addition, capsid protein VP2 contains an RGD sequence that is exposed on the virion surface, indicating that integrins might be cellular receptors of SBPV.
IMPORTANCE Pollination by honeybees is needed to sustain agricultural productivity as well as the biodiversity of wild flora. However, honeybee populations in Europe and North America have been declining since the 1950s. Honeybee viruses from the Iflaviridae family are among the major causes of honeybee colony mortality. We determined the virion structure of an Iflavirus, slow bee paralysis virus (SBPV). SBPV exhibits unique structural features not observed in other picorna-like viruses. The SBPV capsid protein VP3 has a large C-terminal domain, five of which form highly prominent protruding “crowns” on the virion surface. However, the domains can change their positions depending on the conditions of the environment. The domain includes a putative catalytic or receptor binding site that might be important for SBPV cell entry.
INTRODUCTION
The western honeybee Apis mellifera plays a vital role in agriculture by providing pollination services for numerous food crops, especially those with high nutritional and economic value (1). Honeybees are also critical for maintaining the ecological and genetic diversity of wild flowering plants (2). In addition, bumblebees and several other solitary bee species are becoming increasingly important commercial pollinators of specific crops (3). However, bees and the pollination services they provide are under increasing stress due to habitat loss, intensified agricultural management, pesticides, parasites, and pathogens, including numerous viruses (3). Annual honeybee colony mortality has been increasing in North America and Europe over the last couple of decades (4), which, coupled with a long-term decline in beekeeping, has become a serious threat to the adequate provision of pollination services and food security (4–6).
Honeybees are hosts to a large number of viruses, most of which persist covertly within the honeybee population interrupted by occasional outbreaks. Such outbreaks of some of the viruses can have fatal consequences for individual workers and whole colonies (7). Colony collapse disorder (CCD), a still largely unexplained rapid loss of adult bees from colonies, has been linked to virus infections (8, 9). Much of winter honeybee colony mortality is also associated with viruses (10, 11). The viruses that have the greatest impact on honeybee populations are small icosahedral picorna-like viruses from the families Dicistroviridae and Iflaviridae, including slow bee paralysis virus (SBPV), sacbrood virus (SBV), deformed wing virus (DWV), and varroa destructor virus 1 (VDV-1) (7). SBPV was discovered in 1974 (12) and was linked to honeybee colony mortality in the United Kingdom in the 1980s (13). Despite its efficient transmission by Varroa destructor (14), SBP is a rare disease of honeybees (15). However, it is common in bumblebees (16, 17), and therefore, honeybees may be an incidental, secondary host.
Viruses from the order Picornavirales have nonenveloped icosahedral virions containing a single-stranded positive-sense RNA genome about 10,000 nucleotides long (18). Picornavirus genomes are translated into polyproteins that are co- and posttranslationally cleaved by viral proteases to produce structural (capsid-forming) and nonstructural proteins. The capsid proteins originating from a single polyprotein form a protomer—the basic building block of the capsid. The entire capsid consists of 60 such protomers, arranged in 12 pentamer units of five protomers each. The major capsid proteins VP1 to VP3 are arranged in a pseudo-T3 icosahedral capsid.
The only structural information available on Iflaviridae family members is the 25-Å resolution cryo-electron microscopy structure of the Chinese sacbrood virus (19). The structure confirmed the pseudo-T3 icosahedral symmetry of the capsid and revealed a smooth outer surface of the virion. Iflaviruses were proposed to harbor short VP4 subunits consisting of only about 20 residues (15, 20). However, because of the low molecular weight of the peptides, the existence of VP4 subunits has not been unequivocally established (15, 20). Previous genetic and proteomic analyses of iflaviruses revealed a C-terminal extension of about 160 residues in length of one of the capsid proteins (15, 20, 21). Here we present the structure of SBPV determined from two crystal forms to resolutions of 3.4 Å and 2.6 Å. The structures offer the first high-resolution snapshots of a virus from the family Iflaviridae and of a viral pathogen of the honeybee.
MATERIALS AND METHODS
Virus propagation in honeybee pupae.
Propagations of SBPV were carried out as described in the COLOSS BEEBOOK (22). Brood areas with Apis mellifera white-eyed pupae were identified by color and structural features of the cell caps. White-eyed pupae were carefully extracted from the brood combs, so as not to injure the pupae. The pupae were placed on paper furrows with their ventral side up. In total, 544 pupae were used for the SBPV propagation. A virus inoculum (1 μl) was injected into pupae with a Hamilton micropipette with a 30-gauge 22-mm-long needle through the intersegmental cuticle between the 4th and 5th sternites. Pupae that leaked hemolymph after the injection were discarded. The optimal concentration of the virus in the inoculum for virus production was determined experimentally by comparing virus yields when using different virus concentrations in the injection inoculum. Inoculated pupae were placed into petri dishes with the paper furrows and incubated at 30°C and 75% humidity for 5 days. After incubation, the pupae were frozen at −20°C. For long-term storage, the pupae were kept at −80°C.
Virus purification.
Fifty to 70 experimentally infected honeybee pupae were homogenized with a Dounce homogenizer in 30 ml of phosphate-buffered saline (PBS), pH 7.5 (Sigma-Aldrich). The nonionic detergent NP-40 was added to a final concentration of 0.5%, and the homogenate was incubated for 1 h at room temperature. The extract was centrifuged at 8,000 × g for 30 min. The pellet was discarded and the supernatant was centrifuged at 150,000 × g for 3 h in a Ti50.2 fixed-angle rotor (Beckman-Coulter). The resulting pellet was resuspended in PBS to a final volume of 5 ml. MgCl2 was added to a final concentration of 5 mM, along with 20 μg/ml of DNase I and 20 μg/ml of RNase. The solution was incubated at room temperature for 30 min and centrifuged at 4,000 × g for 15 min. The resulting supernatant was loaded onto a CsCl (0.6-g/ml) solution prepared in PBS. The ultracentrifugation proceeded for 16 h to establish the CsCl gradient. Virus bands were collected by gentle piercing of the ultracentrifuge tubes with an 18-gauge needle. The viruses were transferred to PBS by several rounds of concentration and dilution using centrifuge filter units with a 100-kDa-molecular-mass cutoff. This procedure yielded about 300 μg of virus with a purity sufficient for sparse-matrix crystallization screening experiments. Sample purity with respect to contaminating honeybee viruses was checked by reverse transcription-quantitative PCR (RT-qPCR) using previously reported virus-specific assays (22). In both preparations, the total sum of contaminating viruses was less than 1% of the virus of interest. The nucleotide sequences of the virus preparations were determined by sequencing 300 ng of RNA, purified using a Qiagen RNA purification kit, by IonTorrent technology and standard protocols for library preparation and sequencing. The IonTorrent reads were mapped to the SBPV GenBank reference sequence EU035616 (SBPV) using Tmap v4.4.8, included in TorrentSuite 4.4.2, with Life Technologies-recommended parameters. Variability and consensus sequences were created using mpileup from samtools v.0.1.8 and an in-house script.
SBPV crystallization.
SBPV crystallization screening was conducted at 4°C and 20°C with virus concentrations of 5 mg/ml and 10 mg/ml. In total, 2,100 conditions were tested in a 96-well, sitting-drop vapor diffusion format. The initial crystals that formed in 0.1 M sodium citrate (pH 6.5)–5% (wt/vol) polyethylene glycol 4000 (PEG 4000) after 7 days of incubation at 20°C were spherical, with diameters of less than 0.03 μm. The crystallization conditions were optimized by using a 96-well additive screen (Hampton Research Inc.). Optimized crystals with cubic morphology grew under the starting conditions with extra 0.2 M NDSB-221 (nondetergent sulfobetaine) and could be reproduced in a hanging-drop format by mixing 1.5 μl of 10-mg/ml purified virus solution with 0.5 μl of the reservoir solution. The optimized crystals were cubic and required 3 weeks to reach their final size of about 0.1 μm. The best diffraction was obtained when crystals were transferred to a reservoir solution containing 10% ethylene glycol prior to flash-freezing in liquid nitrogen. Out of approximately 200 crystals tested, two crystals diffracted X rays to a resolution of 3.4 Å.
Another crystal form was discovered at 4°C in 0.1 M sodium acetate (pH 4.5)–5% PEG 10000 and contained rectangular crystals of about 0.1 mm. The crystals could be reproduced in a hanging-drop format, with some crystals reaching a length of 0.3 mm. The crystals were subjected to dehydration by gradually transferring the coverslip containing the hanging drop to the reservoir solution containing increasing concentrations of sodium acetate (pH 4.5) and of PEG 10000 as described previously (23). At 20% PEG 10000, crystals were harvested, cryoprotected in mother liquor solution containing 20% glycerol, and flash-frozen in liquid nitrogen. Out of 50 crystals screened, two crystals diffracted X rays to a resolution of 2.6 Å.
SBPV structure determination and refinement.
Diffraction data from SBPV crystal form 1 were collected at the Swiss Light Source X06SA beamline equipped with Pilatus-6M detector at the wavelength of 1.00003 Å at 100 K using a 0.1° rotation per image. The crystals were of space group I23. Unit cell size and packing considerations indicated that one pentamer of capsid protein protomers occupied a crystallographic asymmetric unit. There are two possibilities for superimposing icosahedral 532 symmetry with the 23 symmetry of the crystal, which are perpendicular to each other. The orientation of the virion was determined from a plot of the 5-fold rotation function, calculated with the program GLRF (24). Reflections between 5.0-Å and 4.5-Å resolutions were used for the calculations. Because of the superposition of the icosahedral and crystallographic symmetry, the center of the particle had to be positioned at the intersection of the 2-fold and 3-fold symmetry axes of the crystal. The triatoma virus (TrV) structure (PDB code 3NAP), converted to polyalanine, was used as a molecular replacement model. The model was placed into the orientation and position in the unit cell as described above and used to calculate phases for reflections at up to a 10-Å resolution, using the program CNS (25). The model-derived phases were refined by 25 cycles of 5-fold real-space electron density map averaging using the program ave (26). The mask for electron density averaging was generated by including all voxels within 5 Å of any atom of the TrV model, using the program mama from the package USF (27). Phase extension was applied in order to obtain phases for higher-resolution reflections. The addition of a small fraction of higher-resolution data (one index at a time) was followed by three cycles of averaging. This procedure was repeated until phases were obtained for all the reflections, up to a resolution of 3.4 Å. Inspection of the map showed that the mask used for electron density averaging cut the electron density of the capsid in an area around the icosahedral 5-fold axis. Thus, a new mask was prepared based on a correlation map calculated by comparing the electron density distributions among the five noncrystallographic symmetry (NCS)-related icosahedral asymmetric units. The correlation map was calculated using the program coma from Uppsala Software Factory (USF) (28). A cutoff value of 0.5 was used for the inclusion of voxels into the mask. The surface of the correlation mask was smoothened using the program mama (28). The phase extension procedure was repeated using the new mask. The resulting map was of sufficient quality to allow model building.
The program Buccaneer was used for automated model building, utilizing the 5-fold NCS present in the crystal (29, 30). The model from the automated building was about 50% complete, with assigned amino acid sequences. The initial model was subjected to iterative manual rebuilding using the programs Coot and O (31, 32) and coordinate and B-factor refinement using the programs CNS (25) and Phenix (33). No water molecules were added due to the limited resolution of the diffraction data.
Diffraction data from SBPV crystal form 2 crystals were collected at the synchrotron Soleil Proxima-1 beamline equipped with the Pilatus-6M detector at a wavelength of 0.97857 Å at 100 K using a 0.1° rotation per image. The crystals were of space group I222. The unit cell dimensions and the virus packaging considerations indicated that the crystallographic asymmetric unit consists of three pentamers of capsid protein protomers. Initially, a pentamer corresponding to the entire atomic model of crystal form I was used as a molecular replacement model to find the orientation and translation of the three pentamers in the crystallographic asymmetric unit using the program Phaser (34). The initial electron density map was subjected to 30 cycles of noncrystallographic symmetry averaging using the program AVE (26) and employing the mask based on the model from crystal form I. The averaged map lacked the electron density corresponding to the protruding domain altogether, which suggested that the molecular mask did not cover the correct part of the map. Therefore, a correlation map was calculated, as described for crystal form I, and the mask based on the correlation map was used for averaging. This map was used for the automated model building in the program Buccaneer (29) from the CCP4i software suite (35). The geometry of the model was adjusted manually using the program Coot (32). The coordinate and B-factor refinement were carried out using the program CNS (25) employing strict NCS constraints.
In order to improve the structure of the P domain in crystal form I, the P domain determined from crystal form II was positioned in crystal I using the program Phaser (34). The model of the icosahedral asymmetric unit with the properly positioned P domain was then used to generate a new mask for real-space electron density averaging in the program mama (27). Thirty cycles of real-space electron density averaging were carried out using the program AVE (26). P domain residues with no corresponding density in the averaged map were manually removed using the program Coot (32). The model was subjected to coordinate and B-factor DEN-assisted refinement using the atomic model of crystal form 2 as a reference structure using the software package CNS (25, 36).
Protein structure accession numbers.
The atomic coordinates of the SBPV virion in crystal forms 1 and 2, together with the structure factors and phases obtained by phase extension, were deposited into the Protein Data Bank under codes 5J96 and 5J98, respectively.
RESULTS AND DISCUSSION
Structure of SBPV virion and capsid proteins.
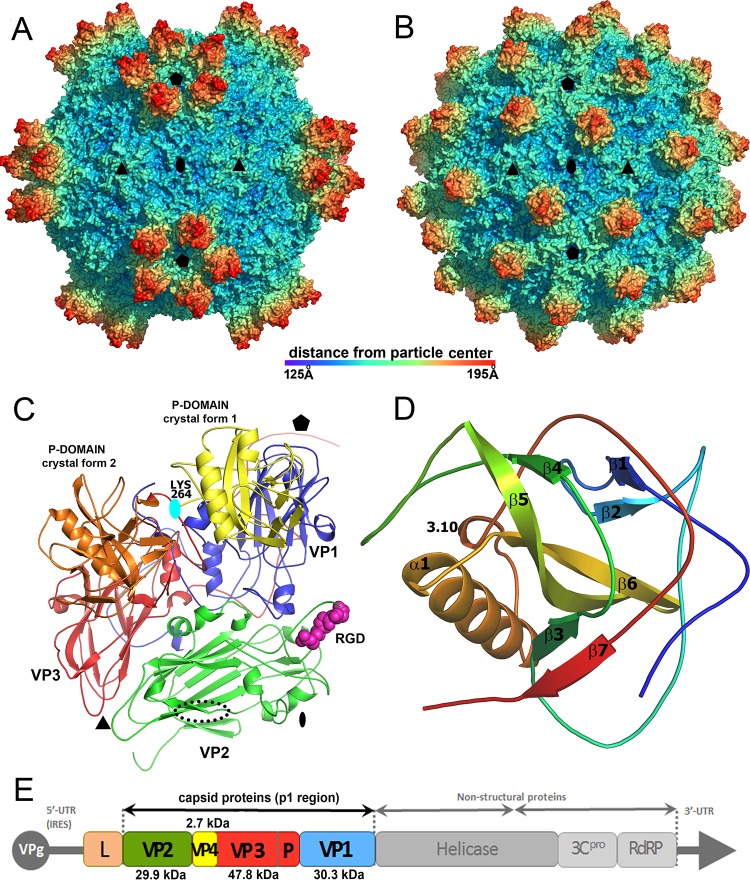
The structure of SBPV was determined from two crystal forms to resolutions of 3.4 Å and 2.6 Å (Table 1). The two structures are similar, with a Cα-atom root mean square deviation (RMSD) of 0.27 Å; however, they differ in the positions of protruding (P) domains of the VP3 subunits on the virion surface (Fig. 1A and B). The maximum outer diameter of the virion is 388 Å. The virion is bigger than those of other picornaviruses because of the P domains. The organization of capsid proteins within the SBPV virion is similar to that of other viruses from the order Picornavirales (Fig. 1C). The capsid is built from major capsid proteins VP1 to 3 arranged in pseudo-T3 icosahedral symmetry (Fig. 1). The major capsid proteins have jellyroll β-sandwich folds with β-strands named according to the picornavirus convention B to I (37). The two antiparallel β-sheets forming the β-sandwich fold contain the strands BIDG and CHEF, respectively. The structures of the major capsid proteins could be built except for residues 253 to 266 of VP1, 92 to 100 and 261 of VP2, and 418 to 430 of VP3. The electron density corresponding to VP4 could not be identified in either of the two structures.
TABLE 1.
Crystallographic data collection and refinement statistics
| Parameter | Crystal form 1 | Crystal form 2 |
|---|---|---|
| Crystallization conditions | Sodium citrate (pH 6.5), 5% (vol/vol) PEG 4000, 0.2 M NDSB-221 | Sodium acetate (pH 4.5), 5% (vol/vol) PEG 10000 |
| Space group | I23 | I222 |
| a, b, c (Å) | 360.7, 360.7, 360.7 | 340.0, 396.8, 431.7 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å)a | 70.7–3.41 (3.45–3.41) | 49.5–2.6 (2.64–2.60) |
| Rmergea | 0.31 (1.26) | 0.20 (0.98) |
| <I>/<σI>a | 5.6 (0.4) | 6.0 (0.9) |
| Completeness (%)a | 87.4 (43.7) | 88.3 (69.3) |
| Redundancy | 6.0 | 6.8 |
| No. of reflections | 92,015 | 780,730 |
| Rworkb | 0.339 | 0.274 |
| No. of atomsc | ||
| Protein | 7,029 | 7,369 |
| Water | 0 | 75 |
| Average B factors | ||
| Protein | 73 | 32 |
| Water | NAd | 30 |
| RMSD | ||
| Bond lengths (Å) | 1.04 | 1.10 |
| Bond angles (°) | 0.004 | 0.004 |
| Ramachandrane | ||
| Favored (%) | 94.37 | 94.47 |
| Allowed (%) | 5.40 | 5.19 |
| Outliers (%) | 0.23 | 0.11 |
| Poor rotamers (%)e | 1.59 | 0.74 |
| Cβ deviations (%)e | 0 | 0 |
| Clash scoree | 11.57 | 10.47 |
| Molprobity scoree | 2.11 (100th percentile) | 1.92 (98th percentile) |
The values in parentheses are for the highest-resolution shell.
If calculated, the Rfree value would have been very close to the Rwork value due to the 5- and 15-fold NCS (79). Thus, all measured reflections were used in the crystallographic refinement.
The values are for the icosahedral asymmetric unit.
NA, not applicable.
According to the criterion of Molprobity (80).
FIG 1.
Structure of SBPV virion and icosahedral asymmetric unit. Surface representations of SBPV virions determined in crystal form 1 (A) and crystal form 2 (B) show differences in the positioning of the P domains. The surfaces of the particles are rainbow-colored based on the distance from the particle center. Depressions are shown in blue and peaks in red. (C) Cartoon representation of SBPV icosahedral asymmetric unit. VP1 is shown in blue, VP2 in green, and VP3 in red. The P domain positioned as in crystal form 1 is shown in yellow and in crystal form 2 in orange. Locations of 5-fold, 3-fold, and 2-fold icosahedral symmetry axes are indicated by a pentagon, triangle, and oval, respectively. The RGD motif found in the GH loop of VP2 subunit is shown as space-filling model in magenta. The position of the RGD motif in FMDV is indicated with a dotted black oval. The cyan oval indicates the position of rotation axis relating the two P domain positions. (D) Cartoon representation of P domain rainbow colored from the N terminus in blue to the C terminus in red. Names of secondary structure elements are indicated. (E) Diagram of SBPV genome organization. Capsid proteins VP1, VP2, and VP3 were identified based on their location in the capsid according to the picornavirus convention. Predicted molecular masses of capsid proteins are specified. The location of the P domain of VP3 is indicated. VPg, viral protein, genome linked; L, leader peptide; IRES, internal ribosome entry site; UTR, untranslated region; 3CPRO, 3C protease; RdRP, RNA-dependent RNA polymerase.
Structure of the VP3 P domain.
The SBPV virion represents the first atomic structure of a virus from the family Iflaviridae. Unlike in the previously structurally characterized viruses from the order Picornavirales, the SBPV capsid protein VP3 contains a C-terminal extension of residues 267 to 430 (15) that fold into the globular P domain positioned on the capsid surface (Fig. 1C and D). The domain consists of a central twisted antiparallel β-sheet formed from strands β4, β5, and β6 surrounded by the 14-residue-long α-helix α1, 3-residue-long 3.10 helix, and two shorter β-sheets containing strands β1 and β2 and β3 to β7 (Fig. 1D). The β-strands are connected by loops that vary in length between 6 and 23 residues. In both of the crystal forms, the residues of the P domain have higher average B factors (crystal 1 B = 110 Å2; crystal 2 B = 57 Å2) than the average B factors of the rest of the capsid (crystal 1 B = 57 Å2; crystal 2 B = 16 Å2), indicating a higher mobility of the P domain. The P domains in the two crystal forms are similar, with an RMSD of 0.32 Å for 144 Cα atoms.
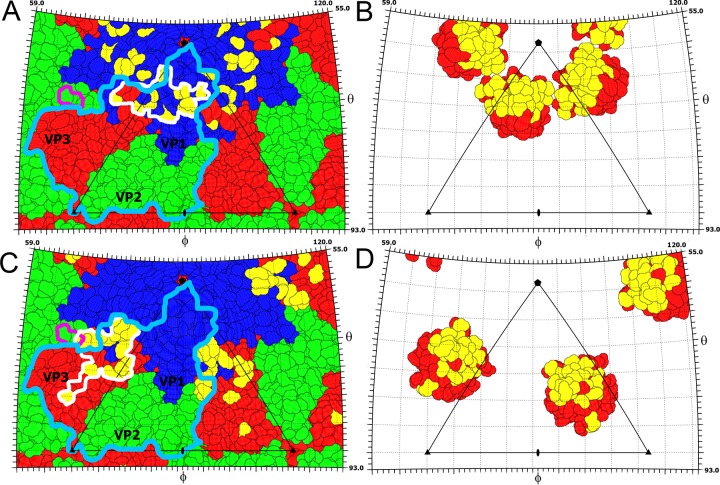
The P domains are positioned in different locations on the virion surface in the two crystal forms (Fig. 1 and 2). It is important to note that the domains are not held in position by crystal contact in either of the crystal forms. In crystal form 1, five P domains related by one icosahedral 5-fold axis form a “crown” on the virion surface (Fig. 1A and 3A). The crowns have a diameter of 90 Å and protrude 50 Å above the capsid surface, giving the SBPV virion its characteristic shape (Fig. 1A). Residues from loop β2-β3 as well as the N- and C-terminal loops and β2 of the P domain interact with the BC, CD, and EF loops of VP1, forming an interface with a buried surface area of 850 Å2 (Fig. 2A and B). P domains within the same crown do not interact with each other (Fig. 1A and 3A). In crystal form 1, the electron density map corresponding to the P domains is less well ordered than that of the rest of the SBPV virion, indicating an increased mobility of the crown.
FIG 2.
Interactions of P domain with the core of the SBPV capsid. (A and B) P domain footprints on the SBPV surface in crystal forms 1 (A) and 2 (B). The images show two-dimensional projections of the SBPV virion surface without the P domains. Residues of capsid proteins VP1, VP2, and VP3 are outlined in blue, green, and red, respectively. Residues involved in interaction with the P domain are shown in yellow. The P domain footprints are outlined by white lines. The border of one VP2-VP3-VP1 protomer is indicated by a light blue line. RGD residues of VP2 are indicated by a magenta line. (C and D) Inner surfaces of P domains in crystal forms 1 (C) and 2 (D), viewed from inside the particle. Residues interacting with the core of the capsid are shown in yellow and the remaining residues in red. Positions of 2-fold, 3-fold, and 5-fold icosahedral symmetry axes are shown as ovals, triangles, and pentagons, respectively. One icosahedral asymmetric unit is outlined by a triangle.
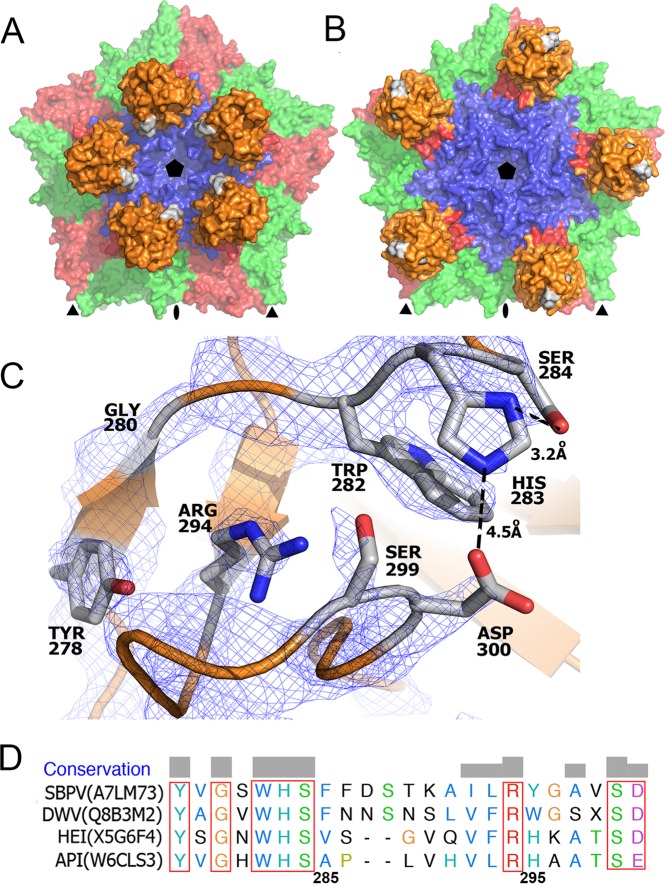
FIG 3.
The P domain contains a putative Ser-His-Asp active site that is part of a patch of residues that are conserved among iflaviruses. The conserved residues are highlighted in gray in pentamers of capsid protein protomers in the conformation from crystal form 1 (A) and crystal form 2 (B). Detail of the putative active site with electron density contoured at 2σ (C). A sequence alignment of residues forming the conserved patch in the P domain is also shown (D). HEI, heliconius erato iflavirus; API, antherae pernyi iflavirus. Uniprot accession numbers of the sequences used in the alignment are provided.
In crystal form 2, the P domain is positioned approximately equal distances from the icosahedral 5-fold, 3-fold, and 2-fold axes (Fig. 1B and 3B). Residues from α1, β3, β5, β7, and loops β2-β3, β3-β4, and β4-β5 of the P domain interact with the CD and GH loops of VP3, the C terminus of VP1, and the GH loop of VP2, forming an interface with a buried surface area of 1,150 Å2 (Fig. 2C and D). The density of the P domain is better resolved than in crystal form 1, indicating that the P domain forms more stable interactions with the capsid surface at the interface observed in crystal form 2. The transition between the two alternative positions of the P domain on the virion surface requires a 122° rotation of the domain around the axis, which passes through Lys266 (Fig. 1C). The center of mass of the P domain in crystal form 2 is shifted 36 Å toward the 3-fold axis relative to its position in crystal form 1 (Fig. 1). This movement of the domain is possible due to a 23-residue-long flexible linker that connects the P domain to the core of the VP3 subunit.
The crystallization conditions that produced the two crystal forms of SBPV differed in terms of solution components and pH, which was 6.5 for crystal form 1 and 4.5 for crystal form 2 (Table 1). We speculate that the differences in localization of the P domains might be induced by the differences in the crystallization conditions. Furthermore, it is possible that the two observed locations of the P domain on the virion surface reflect movements of the domain required for SBPV cell entry in vivo. Similar mobility of the protruding domain was previously reported for capsid proteins of mammalian caliciviruses, where it was speculated to facilitate virus-receptor interactions (38–40). The cell entry of iflaviruses has not been studied, but it is likely to involve receptor-mediated endocytosis as has been described for mammalian picornaviruses (41, 42). The endosomal entry involves exposure of the virions to low pH that could trigger movements of the P domain that might be required for cleavage of substrate by the putative catalytic triad within the P domain as described below.
The P domain contains a putative receptor-binding or catalytic site.
Residues Ser284, His283, and Asp300 from the P domain of VP3 are located close to each other, indicating the presence of a putative catalytic triad (43) that might be involved in the cleavage of an as-yet-unknown substrate. These residues face the interior of the crown in crystal form 1; however, they constitute the apex of the P domain in crystal form 2 (Fig. 3A and B). The distances between the side chains of the putative reactive site are larger than ideal for catalyzing the hydrolytic reaction (Fig. 3C) (43). Nevertheless, it is possible that the optimal configuration of the active site might be achieved upon binding the unknown substrate to the P domain. This type of catalytic triad has been previously identified in proteases, lipases, and esterases (43–45). The residues constituting the putative active site are conserved among other iflaviruses that have P domains, including DWV, VDV-1, and Kakugo virus (20, 46, 47). However, the iflaviruses Sacbrood and Perina nuda virus lack P domains altogether (48, 49). Catalytic activity of the putative active site might be required for the virions to escape from endosomes in a manner analogous to the lipase activity present in the N-terminal domain of capsid proteins of parvoviruses (50). There are five additional conserved residues located in the vicinity of the putative active site in strand β1 and loops connecting strands β1-β2 and β2-β3 (Fig. 3C). This is in contrast to the overall 12% sequence identity of the P domains. The conservation of the residues reinforces the possibility that they may constitute a receptor or substrate-binding site. Furthermore, a similar conserved patch of residues in P domains of noroviruses was shown to bind glycans (51, 52). Additional experiments are required to identify the putative receptor of SBPV and to determine whether the catalytic triad cleaves it.
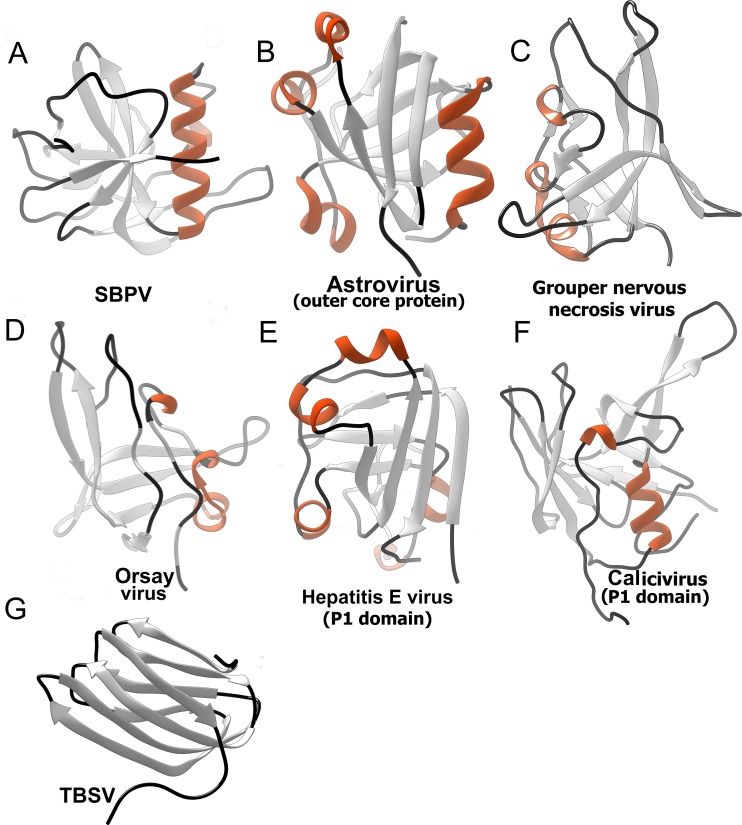
The DALI server was used to identify structures similar to the P domain (Table 2) (53). Most of the top hits were domains of virus capsid proteins that are exposed on the virion surface and therefore might be involved in receptor binding or cell entry. A common feature of these domains is a core formed of β-strands that is in some cases complemented by one or more short α-helices located at the periphery of the domain (Fig. 4). Furthermore, the P domains were also found in plant picorna-like viruses from the family Tombusviridae (54). In these species, however, the protrusions exhibit a β-jellyroll fold. Even though the surface domains could be identified in the DALI search, the structures of the domains are quite different and cannot be meaningfully superimposed. The surface domains were identified in viruses from the families Tombusviridae, Nodaviridae, Hepeviridae, and Astroviridae (54–57). All these viruses have positive-sense single-stranded RNA (ssRNA) genomes and similar overall virion architectures. It is therefore possible that a common ancestor of these viruses contained the P domain. However, the P domains were retained in the evolution of only some of the viruses.
TABLE 2.
DALI search identification of proteins similar to the SBPV P domain
| Structure | PDB code | DALI Z score | RMSD | Sequence identity (%) |
|---|---|---|---|---|
| Human astrovirus capsid protein | 5ewn | 4.5 | 3.6 | 9 |
| P domain of grouper nervous necrosis virus | 4rfu | 4.2 | 3.6 | 5 |
| Orsay virus | 4nww | 3.3 | 4.5 | 9 |
| Hepatitis E virus capsid protein | 2zzq | 3.0 | 3.3 | 13 |
FIG 4.
Protruding domains of viruses identified in a DALI search based on similarity to SBPV P domain. (A) SBPV; (B) human astrovirus outer coat protein (5EWN) (55); (C) grouper nervous necrosis virus (4RFU) (56); (D) orsay virus (4NWW) (57); (E) P1 domain of human hepatitis E virus (3HAG) (76); (F) P1 domain of human calcivirus (2GH8) (77). Protruding domain of tomato bushy stunt virus (2TBV) (78) is shown for comparison, however, it was not identified in the DALI server search. β-Strands are shown in light gray, helices in orange, and loops in black.
Putative SBPV integrin receptor binding site.
Currently there is no information about the cell entry of honeybee viruses, and the putative receptors remain to be identified. However, the VP2 subunit of SBPV contains the integrin recognition motif Arg-Gly-Asp (RGD) in the GH loop (Fig. 1C). The GH loop is exposed on the virion surface in crystal form 1 but is partly covered by the P domain in crystal form 2 (Fig. 2A and B). Integrins serve as cell entry receptors for numerous viruses, including human picornaviruses such as the foot-and-mouth disease virus (FMDV) and several parechoviruses (58–60). The RGD motif within FMDV is located in the VP2 subunit, similar to the case with SBPV, although closer to the icosahedral 2-fold axis (Fig. 1C). The RGD motif is not conserved across different iflaviruses and may confer specific tissue tropism to SBPV. Even though honeybees encode a number of integrins (61), their involvement in virus cell entry has not been demonstrated so far.
Decreased pH does not induce formation of SBPV A particles.
Picornaviruses enter cells through receptor-mediated endocytosis. The receptor binding and low pH of endosomes were shown to trigger the formation of expanded A particles and the subsequent genome release of many picornaviruses (62). The A particles are characterized by a 5 to 10% increase in virion radius and the formation of holes in the capsid (42, 63–66). However, the SBPV virion structures determined at pH 6.5 and 4.5 are nearly identical in size (Table 3). Therefore, it appears that the pH (4.5) of the crystallization condition was not sufficient to induce formation of the SBPV A particles. The induction of SBPV genome release might require binding to a receptor, or iflaviruses might use an entirely different mechanism for genome release.
TABLE 3.
Comparison of size and volume of SBPV particles determined in crystal forms 1 and 2
Determined as distance of the center of mass of the icosahedral asymmetric unit from the particle center.
Volume of virion cavity calculated based on virion structures. The space occupied by the unstructured parts of the capsid proteins located on the inside of the capsid was calculated based on average amino acid volumes and subtracted from the cavity volume.
Comparison to virion structures of dicistroviruses.
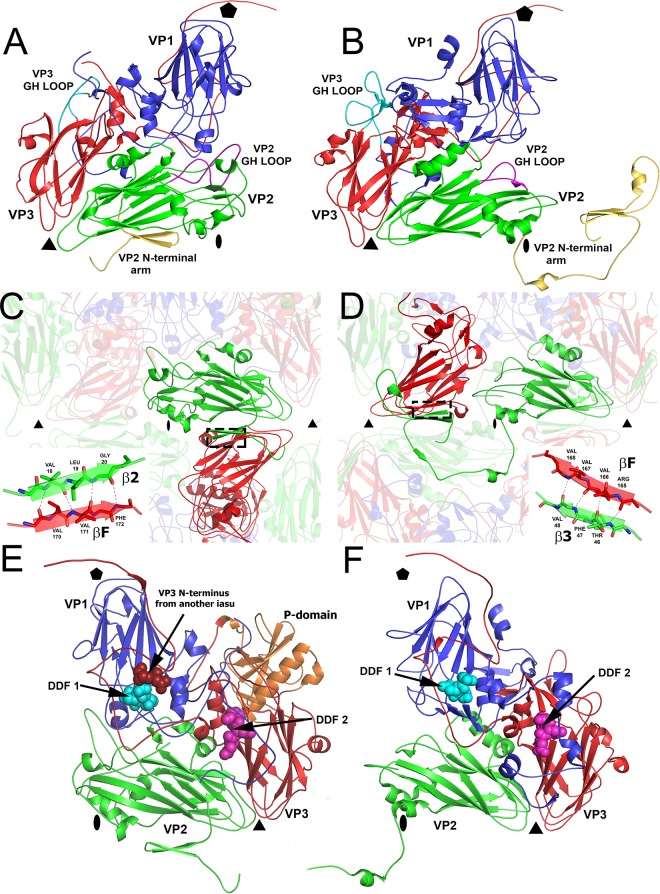
The most notable difference between SBPV and structurally characterized dicistroviruses, besides the P domain, is in the positioning of the N-terminal arm of the VP2 protein, which contributes to the interpentamer contacts within the capsid (Fig. 5A to D). In SBPV, two β-strands from the N-terminal arm of VP2 extend the β-sheet CHEF of a VP3 from the neighboring pentamer (Fig. 5C). In contrast, in dicistroviruses represented by TrV and cricket paralysis virus (CrPV), the N-terminal arm of the VP2 subunit reaches around an icosahedral 2-fold axis into the neighboring pentamer, approaches a 3-fold axis, and forms two β-strands that extend the β-sheet CHEF of a VP3 subunit from the same pentamer (Fig. 5D) (67, 68). Thus, the VP2 N-terminal arms of SBPV and dicistroviruses mediate interactions between VP2 and VP3 subunits in different relative positions within their virions. However, the type of interaction, i.e., extension of the β-sheet CHEF of VP3, is the same for both the viruses, representing domain swapping of the VP2 N-terminal arms. It was speculated previously that the observation of domain swapping among homologous complexes is indicative of hinge movements of structural units connected by the swapped domains. The alternative placements of the N-terminal arms of VP2 subunits therefore indicate that pentamers of capsid proteins could move relative to each other.
FIG 5.
Comparison of SBPV structure to that of dicistrovirus TrV. (A and B) Cartoon representations of icosahedral asymmetric units of SBPV (A) and TrV (B). VP1 subunits are shown in blue, VP2 in green, and VP3 in red. The GH loop of VP2 is highlighted in magenta, the GH loop of VP3 is in cyan, and the N-terminal arms of VP2 are in yellow. (C and D) Domain swapping between SBPV (C) and TrV (D) N-terminal arms of VP2 subunits that mediate interpentamer interactions. The insets show details of hydrogen bonds between β2 of VP2 and βF of VP3. (E and F) Location of DDF sequences, which might be involved in the cleavage of VP0 to VP4 and VP3, on the inside of the capsid of SBPV (E) and TrV (F).
Additional differences between SBPV and dicistroviruses can be found on the capsid surface. The RGD containing the GH loop of the SBPV VP2 subunit contains 30 residues, while in TrV and CrPV it is only 17 residues long (Fig. 5A and B) (67, 68). The SBPV loop therefore elevates higher above the surface of the virion, which might be required for binding to the putative integrin receptor (Fig. 1C). On the other hand, the GH loop of the VP3 subunit is longer in TrV, containing 36 residues in comparison to 24 in SBPV (Fig. 5A and B) (68).
The maturation of capsids of viruses from the order Picornavirales is connected to a cleavage of capsid protein VP4 from the N terminus of a precursor subunit, called VP0. In picornaviruses, VP0 cleavage generates the proteins VP4 and VP2, while it was suggested that in iflaviruses the precursor cleavage produces VP4 and VP3 (67, 68). It has been proposed that a conserved Asp-Asp-Phe (DDF) motif, present in parts of capsid proteins that are exposed to the virion cavity, is involved in the VP0 cleavage (67–69). The dicistroviruses CrPV and TrV contain the DDF sequence in a loop immediately following β-strand I of VP1, while TrV has an additional DDF sequence, in a loop following β-strand I of VP3 (Fig. 5F) (67, 68). SBPV also has two DDF sequences. One is in VP1, residues 226 to 228, and the second one is formed by residues 239 to 241 of VP3 (Fig. 5E). Therefore, the locations of the DDF sequences in SBPV are similar to those in TrV (Fig. 5E and F). The DDF site in VP1 subunit of SBPV is located within 4 Å of the N terminus of VP3 subunit from a neighboring protomer, suggesting that it might mediate the VP0 maturation cleavage (Fig. 5E).
Absence of a hydrophobic pocket in VP1.
The VP1 subunits of enteroviruses and several other vertebrate picornaviruses were indicated to contain a hydrophobic pocket that might bind a putative lipid-like molecule called the “pocket factor” (70, 71). Pocket factor mimetics that bind into the VP1 pocket with high affinity were shown to inhibit the infection of some picornaviruses (72–75). However, such a hydrophobic pocket is not formed within the VP1 subunits of SBPV. This suggests that capsid binding inhibitors may not be effective as antivirals against honeybee viruses. However, compounds targeting the putative His-Ser-Asp catalytic or receptor binding site in the P domain may prevent the infection of iflaviruses containing P domains.
ACKNOWLEDGMENTS
We thank Jiří Svoboda for technical assistance with pupa preparation. We thank the beamline scientists at SLS and Soleil synchrotrons for their outstanding technical assistance with crystal screening and data collection. We thank Christian Tellgren-Roth from LifeSciLabs at Uppsala University, Sweden, for assembling virus genome sequences and Emilia Semberg at SLU for technical assistance. We thank Jana Moravcová for her help with the preparation of the manuscript for submission. We thank Charles Sabin for his help with preparation of the figure depicting the genome of SBPV.
This research was carried out under the project CEITEC 2020 (LQ1601) with financial support from the Ministry of Education, Youth and Sports of the Czech Republic under National Sustainability Program II. We acknowledge the CF Cryo-Electron Microscopy and Tomography, CF X-ray Diffraction and Bio-SAXS, and CF Biomolecular Interactions and Crystallization supported by the CIISB research infrastructure (LM2015043 funded by MEYS CR) for their support with obtaining scientific data presented in this paper. The research leading to these results received funding from the European Research Council under the European Union's Seventh Framework Program (FP/2007-2013)/ERC Grant Agreement no. 355855′ and from EMBO installation grant no. 3041 to Pavel Plevka.
REFERENCES
- 1.Allsopp MH, de Lange WJ, Veldtman R. 2008. Valuing insect pollination services with cost of replacement. PLoS One 3:e3128. doi: 10.1371/journal.pone.0003128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biesmeijer JC, Roberts SP, Reemer M, Ohlemuller R, Edwards M, Peeters T, Schaffers AP, Potts SG, Kleukers R, Thomas CD, Settele J, Kunin WE. 2006. Parallel declines in pollinators and insect-pollinated plants in Britain and the Netherlands. Science 313:351–354. doi: 10.1126/science.1127863. [DOI] [PubMed] [Google Scholar]
- 3.Vanengelsdorp D, Meixner MD. 2010. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J Invertebr Pathol 103(Suppl 1):S80–S95. [DOI] [PubMed] [Google Scholar]
- 4.Dainat B, Vanengelsdorp D, Neumann P. 2012. Colony collapse disorder in Europe. Environ Microbiol Rep 4:123–125. doi: 10.1111/j.1758-2229.2011.00312.x. [DOI] [PubMed] [Google Scholar]
- 5.van Engelsdorp D, Hayes J Jr, Underwood RM, Pettis J. 2008. A survey of honey bee colony losses in the U.S., fall 2007 to spring 2008. PLoS One 3:e4071. doi: 10.1371/journal.pone.0004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith KM, Loh EH, Rostal MK, Zambrana-Torrelio CM, Mendiola L, Daszak P. 2013. Pathogens, pests, and economics: drivers of honey bee colony declines and losses. Ecohealth 10:434–445. doi: 10.1007/s10393-013-0870-2. [DOI] [PubMed] [Google Scholar]
- 7.Chen YP, Siede R. 2007. Honey bee viruses. Adv Virus Res 70:33–80. doi: 10.1016/S0065-3527(07)70002-7. [DOI] [PubMed] [Google Scholar]
- 8.Vanengelsdorp D, Evans JD, Saegerman C, Mullin C, Haubruge E, Nguyen BK, Frazier M, Frazier J, Cox-Foster D, Chen Y, Underwood R, Tarpy DR, Pettis JS. 2009. Colony collapse disorder: a descriptive study. PLoS One 4:e6481. doi: 10.1371/journal.pone.0006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams GR, Tarpy DR, vanEngelsdorp D, Chauzat MP, Cox-Foster DL, Delaplane KS, Neumann P, Pettis JS, Rogers RE, Shutler D. 2010. Colony collapse disorder in context. Bioessays 32:845–846. doi: 10.1002/bies.201000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berthound HIA, Haueter M, Radloff S, Neumann P. 2010. Virus infections and winter losses of honey bee colonies (Apis mellifera). J Apic Res 49:60–65. doi: 10.3896/IBRA.1.49.1.08. [DOI] [Google Scholar]
- 11.Genersh EvdO W, Kaatz H, Schroeder A, Otten C, Buchler R, Berg S, Ritter W, Muhlen W, Gisder S, Meixner M, Leibig G, Rosenkranz P. 2010. The German bee monitoring project: a long term study to understand periodically high winter losses of honey bee colonies. Adipologie 41:332–352. [Google Scholar]
- 12.Bailey L, Woods RD. 1974. Three previously undescribed viruses from the honey bee. J Gen Virol 25:175–186. doi: 10.1099/0022-1317-25-2-175. [DOI] [PubMed] [Google Scholar]
- 13.Carreck NL, B B, Wilson JK, Allen MF. 2005. The epodemiology of slow paralysis virus in honey bee colonies infested by Varroa destructor in the UK, p 32–33. Proceedings of XXXIXth International Apicultural Congress, Dublin, Ireland. [Google Scholar]
- 14.Santillan-Galicia MT, Ball BV, Clark SJ, Alderson PG. 2010. Transmission of deformed wing virus and slow paralysis virus to adult bees (Apis mellifera L.) by Varroa destructor. J Apic Res 49:141–148. doi: 10.3896/IBRA.1.49.2.01. [DOI] [Google Scholar]
- 15.de Miranda JR, Dainat B, Locke B, Cordoni G, Berthoud H, Gauthier L, Neumann P, Budge GE, Ball BV, Stoltz DB. 2010. Genetic characterization of slow bee paralysis virus of the honeybee (Apis mellifera L.). J Gen Virol 91:2524–2530. doi: 10.1099/vir.0.022434-0. [DOI] [PubMed] [Google Scholar]
- 16.Fürst MA, McMahon DP, Osborne JL, Paxton RJ, Brown MJ. 2014. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 506:364–366. doi: 10.1038/nature12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMahon DP, Furst MA, Caspar J, Theodorou P, Brown MJ, Paxton RJ. 3 March 2015. A sting in the spit: widespread cross-infection of multiple RNA viruses across wild and managed bees. J Anim Ecol doi: 10.1111/1365-2656.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Gall O, Christian P, Fauquet CM, King AM, Knowles NJ, Nakashima N, Stanway G, Gorbalenya AE. 2008. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T =3 virion architecture. Arch Virol 153:715–727. doi: 10.1007/s00705-008-0041-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Feng J, Liang Y, Chen D, Zhou ZH, Zhang Q, Lu X. 2001. Three-dimensional structure of the Chinese Sacbrood bee virus. Sci China C Life Sci 44:443–448. doi: 10.1007/BF02879612. [DOI] [PubMed] [Google Scholar]
- 20.Lanzi G, de Miranda JR, Boniotti MB, Cameron CE, Lavazza A, Capucci L, Camazine SM, Rossi C. 2006. Molecular and biological characterization of deformed wing virus of honeybees (Apis mellifera L.). J Virol 80:4998–5009. doi: 10.1128/JVI.80.10.4998-5009.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng P, Li W, Lin L, de Miranda JR, Emrich S, An L, Terenius O. 2014. Genetic characterization of a novel iflavirus associated with vomiting disease in the Chinese oak silkmoth Antheraea pernyi. PLoS One 9:e92107. doi: 10.1371/journal.pone.0092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Miranda JR, Bailey L, Ball BV, Blanchard P, Budge G, Chejanovsky N, Chen Y-P, Gauthier L, Genersch E, De Graaf D, Ribière M, Ryabov E, De Smet L, van der Steen JJM. 2013. Standard methods for Apis mellifera pest and pathogen research. In Dietemann V, Ellis JD, Neumann P (ed), The COLOSS BEEBOOK, vol II IBRA, Treforest, United Kingdom. [Google Scholar]
- 23.Newman J. 2006. A review of techniques for maximizing diffraction from a protein crystal in stilla. Acta Crystallogr D Biol Crystallogr 62:27–31. doi: 10.1107/S0907444905032130. [DOI] [PubMed] [Google Scholar]
- 24.Tong L, Rossmann MG. 1997. Rotation function calculations with GLRF program. Methods Enzymol 276:594–611. doi: 10.1016/S0076-6879(97)76080-4. [DOI] [PubMed] [Google Scholar]
- 25.Brunger AT. 2007. Version 1.2 of the Crystallography and NMR system. Nat Protoc 2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 26.Kleywegt GJ, Read RJ. 1997. Not your average density. Structure 5:1557–1569. doi: 10.1016/S0969-2126(97)00305-5. [DOI] [PubMed] [Google Scholar]
- 27.Kleywegt GJ, Jones TA. 1999. Software for handling macromolecular envelopes. Acta Crystallogr D Biol Crystallogr 55:941–944. doi: 10.1107/S0907444999001031. [DOI] [PubMed] [Google Scholar]
- 28.Kleywegt GJ. 1999. Experimental assessment of differences between related protein crystal structures. Acta Crystallogr D Biol Crystallogr 55:1878–1884. doi: 10.1107/S0907444999010495. [DOI] [PubMed] [Google Scholar]
- 29.Cowtan K. 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 30.Cowtan K. 2008. Fitting molecular fragments into electron density. Acta Crystallogr D Biol Crystallogr 64:83–89. doi: 10.1107/S0907444907033938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47(Part 2):110–119. [DOI] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54:905–921. [DOI] [PubMed] [Google Scholar]
- 37.Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, Johnson JE, Kamer G, Luo M, Mosser AG, Rueckert RR, Sherry B, Vriend G. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 38.Katpally U, Voss NR, Cavazza T, Taube S, Rubin JR, Young VL, Stuckey J, Ward VK, Virgin HW IV, Wobus CE, Smith TJ. 2010. High-resolution cryo-electron microscopy structures of murine norovirus 1 and rabbit hemorrhagic disease virus reveal marked flexibility in the receptor binding domains. J Virol 84:5836–5841. doi: 10.1128/JVI.00314-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taube S, Rubin JR, Katpally U, Smith TJ, Kendall A, Stuckey JA, Wobus CE. 2010. High-resolution x-ray structure and functional analysis of the murine norovirus 1 capsid protein protruding domain. J Virol 84:5695–5705. doi: 10.1128/JVI.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 41.Bergelson JM, Coyne CB. 2013. Picornavirus entry. Adv Exp Med Biol 790:24–41. doi: 10.1007/978-1-4614-7651-1_2. [DOI] [PubMed] [Google Scholar]
- 42.Fuchs R, Blaas D. 2012. Productive entry pathways of human rhinoviruses. Adv Virol 2012:826301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dodson G, Wlodawer A. 1998. Catalytic triads and their relatives. Trends Biochem Sci 23:347–352. doi: 10.1016/S0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 44.Canaan S, Roussel A, Verger R, Cambillau C. 1999. Gastric lipase: crystal structure and activity. Biochim Biophys Acta 1441:197–204. doi: 10.1016/S1388-1981(99)00160-2. [DOI] [PubMed] [Google Scholar]
- 45.Vaquero ME, Barriuso J, Martinez MJ, Prieto A. 2016. Properties, structure, and applications of microbial sterol esterases. Appl Microbiol Biotechnol 100:2047–2061. doi: 10.1007/s00253-015-7258-x. [DOI] [PubMed] [Google Scholar]
- 46.Ongus JR, Peters D, Bonmatin JM, Bengsch E, Vlak JM, van Oers MM. 2004. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J Gen Virol 85:3747–3755. doi: 10.1099/vir.0.80470-0. [DOI] [PubMed] [Google Scholar]
- 47.Fujiyuki T, Takeuchi H, Ono M, Ohka S, Sasaki T, Nomoto A, Kubo T. 2004. Novel insect picorna-like virus identified in the brains of aggressive worker honeybees. J Virol 78:1093–1100. doi: 10.1128/JVI.78.3.1093-1100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailey L, Gibbs AJ, Woods RD. 1964. Sacbrood virus of the larval honey bee (Apis Mellifera Linnaeus). Virology 23:425–429. doi: 10.1016/0042-6822(64)90266-1. [DOI] [PubMed] [Google Scholar]
- 49.Wu CY, Lo CF, Huang CJ, Yu HT, Wang CH. 2002. The complete genome sequence of Perina nuda picorna-like virus, an insect-infecting RNA virus with a genome organization similar to that of the mammalian picornaviruses. Virology 294:312–323. doi: 10.1006/viro.2001.1344. [DOI] [PubMed] [Google Scholar]
- 50.Farr GA, Zhang LG, Tattersall P. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc Natl Acad Sci U S A 102:17148–17153. doi: 10.1073/pnas.0508477102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol 81:5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansman GS, Shahzad-Ul-Hussan S, McLellan JS, Chuang GY, Georgiev I, Shimoike T, Katayama K, Bewley CA, Kwong PD. 2012. Structural basis for norovirus inhibition and fucose mimicry by citrate. J Virol 86:284–292. doi: 10.1128/JVI.05909-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morgunova E, Dauter Z, Fry E, Stuart DI, Stel'mashchuk V, Mikhailov AM, Wilson KS, Vainshtein BK. 1994. The atomic structure of carnation mottle virus capsid protein. FEBS Lett 338:267–271. doi: 10.1016/0014-5793(94)80281-5. [DOI] [PubMed] [Google Scholar]
- 55.York RL, Yousefi PA, Bogdanoff W, Haile S, Tripathi S, DuBois RM. 2015. Structural, mechanistic, and antigenic characterization of the human astrovirus capsid. J Virol 90:2254–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen NC, Yoshimura M, Guan HH, Wang TY, Misumi Y, Lin CC, Chuankhayan P, Nakagawa A, Chan SI, Tsukihara T, Chen TY, Chen CJ. 2015. Crystal structures of a piscine betanodavirus: mechanisms of capsid assembly and viral infection. PLoS Pathog 11:e1005203. doi: 10.1371/journal.ppat.1005203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo YR, Hryc CF, Jakana J, Jiang H, Wang D, Chiu W, Zhong W, Tao YJ. 2014. Crystal structure of a nematode-infecting virus. Proc Natl Acad Sci U S A 111:12781–12786. doi: 10.1073/pnas.1407122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson T, Mould AP, Sheppard D, King AM. 2002. Integrin alphavbeta1 is a receptor for foot-and-mouth disease virus. J Virol 76:935–941. doi: 10.1128/JVI.76.3.935-941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boonyakiat Y, Hughes PJ, Ghazi F, Stanway G. 2001. Arginine-glycine-aspartic acid motif is critical for human parechovirus 1 entry. J Virol 75:10000–10004. doi: 10.1128/JVI.75.20.10000-10004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalynych S, Palkova L, Plevka P. 2016. The structure of human parechovirus 1 reveals an association of the RNA genome with the capsid. J Virol 90:1377–1386. doi: 10.1128/JVI.02346-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinstock GM, et al. 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Groppelli E, Tuthill TJ, Rowlands DJ. 2010. Cell entry of the aphthovirus equine rhinitis A virus is dependent on endosome acidification. J Virol 84:6235–6240. doi: 10.1128/JVI.02375-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bakker SE, Groppelli E, Pearson AR, Stockley PG, Rowlands DJ, Ranson NA. 2014. Limits of structural plasticity in a picornavirus capsid revealed by a massively expanded equine rhinitis A virus particle. J Virol 88:6093–6099. doi: 10.1128/JVI.01979-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garriga D, Pickl-Herk A, Luque D, Wruss J, Caston JR, Blaas D, Verdaguer N. 2012. Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid. PLoS Pathog 8:e1002473. doi: 10.1371/journal.ppat.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levy HC, Bostina M, Filman DJ, Hogle JM. 2010. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J Virol 84:4426–4441. doi: 10.1128/JVI.02393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bostina M, Levy H, Filman DJ, Hogle JM. 2011. Poliovirus RNA is released from the capsid near a twofold symmetry axis. J Virol 85:776–783. doi: 10.1128/JVI.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tate J, Liljas L, Scotti P, Christian P, Lin T, Johnson JE. 1999. The crystal structure of cricket paralysis virus: the first view of a new virus family. Nat Struct Biol 6:765–774. doi: 10.1038/11543. [DOI] [PubMed] [Google Scholar]
- 68.Squires G, Pous J, Agirre J, Rozas-Dennis GS, Costabel MD, Marti GA, Navaza J, Bressanelli S, Guerin DM, Rey FA. 2013. Structure of the Triatoma virus capsid. Acta Crystallogr D Biol Crystallogr 69:1026–1037. doi: 10.1107/S0907444913004617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agirre J, Aloria K, Arizmendi JM, Iloro I, Elortza F, Sanchez-Eugenia R, Marti GA, Neumann E, Rey FA, Guerin DM. 2011. Capsid protein identification and analysis of mature Triatoma virus (TrV) virions and naturally occurring empty particles. Virology 409:91–101. doi: 10.1016/j.virol.2010.09.034. [DOI] [PubMed] [Google Scholar]
- 70.Smyth M, Pettitt T, Symonds A, Martin J. 2003. Identification of the pocket factors in a picornavirus. Arch Virol 148:1225–1233. doi: 10.1007/s00705-002-0974-4. [DOI] [PubMed] [Google Scholar]
- 71.Hadfield AT, Lee W, Zhao R, Oliveira MA, Minor I, Rueckert RR, Rossmann MG. 1997. The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle. Structure 5:427–441. doi: 10.1016/S0969-2126(97)00199-8. [DOI] [PubMed] [Google Scholar]
- 72.Smith TJ, Kremer MJ, Luo M, Vriend G, Arnold E, Kamer G, Rossmann MG, McKinlay MA, Diana GD, Otto MJ. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- 73.Hadfield AT, Diana GD, Rossmann MG. 1999. Analysis of three structurally related antiviral compounds in complex with human rhinovirus 16. Proc Natl Acad Sci U S A 96:14730–14735. doi: 10.1073/pnas.96.26.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grant RA, Hiremath CN, Filman DJ, Syed R, Andries K, Hogle JM. 1994. Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design. Curr Biol 4:784–797. [DOI] [PubMed] [Google Scholar]
- 75.Hiremath CN, Grant RA, Filman DJ, Hogle JM. 1995. Binding of the antiviral drug WIN51711 to the sabin strain of type 3 poliovirus: structural comparison with drug binding in rhinovirus 14. Acta Crystallogr D Biol Crystallogr 51:473–489. [DOI] [PubMed] [Google Scholar]
- 76.Guu TS, Liu Z, Ye Q, Mata DA, Li K, Yin C, Zhang J, Tao YJ. 2009. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc Natl Acad Sci U S A 106:12992–12997. doi: 10.1073/pnas.0904848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen R, Neill JD, Estes MK, Prasad BV. 2006. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc Natl Acad Sci U S A 103:8048–8053. doi: 10.1073/pnas.0600421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hopper P, Harrison SC, Sauer RT. 1984. Structure of tomato bushy stunt virus. V. Coat protein sequence determination and its structural implications. J Mol Biol 177:701–713. [DOI] [PubMed] [Google Scholar]
- 79.Kleywegt GJ, Brunger AT. 1996. Checking your imagination: applications of the free R value. Structure 4:897–904. doi: 10.1016/S0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- 80.O'Flaherty S, Coffey A, Edwards R, Meaney W, Fitzgerald GF, Ross RP. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J Bacteriol 186:2862–2871. doi: 10.1128/JB.186.9.2862-2871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]