ABSTRACT
HIV-1 requires the CD4 receptor and a coreceptor (CCR5 [R5 phenotype] or CXCR4 [X4 phenotype]) to enter cells. Coreceptor tropism can be assessed by either phenotypic or genotypic analysis, the latter using bioinformatics algorithms to predict tropism based on the env V3 sequence. We used the Primer ID sequencing strategy with the MiSeq sequencing platform to reveal the structure of viral populations in the V1/V2 and C2/V3 regions of the HIV-1 env gene in 30 late-stage and 6 early-stage subjects. We also used endpoint dilution PCR followed by cloning of env genes to create pseudotyped virus to explore the link between genotypic predictions and phenotypic assessment of coreceptor usage. We found out that the most stringently sequence-based calls of X4 variants (Geno2Pheno false-positive rate [FPR] of ≤2%) formed distinct lineages within the viral population, and these were detected in 24 of 30 late-stage samples (80%), which was significantly higher than what has been seen previously by using other approaches. Non-X4 lineages were not skewed toward lower FPR scores in X4-containing populations. Phenotypic assays showed that variants with an intermediate FPR (2 to 20%) could be either X4/dual-tropic or R5 variants, although the X4 variants made up only about 25% of the lineages with an FPR of <10%, and these variants carried a distinctive sequence change. Phylogenetic analysis of both the V1/V2 and C2/V3 regions showed evidence of recombination within but very little recombination between the X4 and R5 lineages, suggesting that these populations are genetically isolated.
IMPORTANCE Primer ID sequencing provides a novel approach to study genetic structures of viral populations. X4 variants may be more prevalent than previously reported when assessed by using next-generation sequencing (NGS) and with a greater depth of sampling than single-genome amplification (SGA). Phylogenetic analysis to identify lineages of sequences with intermediate FPR values may provide additional information for accurately predicting X4 variants by using V3 sequences. Limited recombination occurs between X4 and R5 lineages, suggesting that X4 and R5 variants are genetically isolated and may be replicating in different cell types or that X4/R5 recombinants have reduced fitness.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) requires the CD4 receptor and a coreceptor to infect host cells. Entry is mediated by the viral envelope protein (Env), which is processed to give two associated subunits, gp120 and gp41, that assemble into a trimeric structure embedded in the viral envelope/membrane. The binding of gp120 to CD4 triggers a structural change that exposes its variable region 3 (V3) loop, which is part of the coreceptor domain (1, 2). The majority of transmitted/founder (T/F) viruses and viruses isolated from the clinically latent stage in HIV-infected subjects use CCR5 as the coreceptor, making the virus CCR5 tropic (or an R5 virus). The virus can evolve to use CXCR4 (CXCR4 tropic [or an X4 virus]), and viruses that have switched coreceptor usage are found mostly in late-stage subjects with low CD4+ T cell counts (3, 4). Viruses evolving an X4 phenotype often retain some capacity to use CCR5 for entry and are often referred to as X4/dual-tropic viruses.
Coreceptor tropism can be assessed by either phenotypic or genotypic analysis. Cell-based assays for entry phenotype test either virus isolates or viruses pseudotyped by using cloned viral env genes. Phenotyping consists of infecting cells expressing CD4 and either CCR5 or CXCR4 (5, 6) or inhibiting infection with antagonists of CCR5 and/or CXCR4. Alternatively, assignment of coreceptor tropism by sequence analysis has been based on sequencing of the V3 loop coding domain of the viral env gene. As the V3 loop of gp120 directly interacts with the coreceptor, the sequence of the V3 loop is the major determinant of the coreceptor tropism phenotype (7, 8). In contrast to V3, which appears to be variable due to the coreceptor switch, variable regions V1, V2, V4, and V5 are surface loops of the gp120 subunit involved in autologous antibody escape, and these regions are separated by relatively constant regions (i.e., C1, V1/V2, C2, V3, C4, V4, C5, V5, and C6).
Several bioinformatics interpretation systems have been developed to assess V3 sequence data to infer coreceptor tropism. The 11/25 charge rule is the simplest algorithm based on the presence of basic amino acids at positions 11 and/or 25 of the V3 loop (positions 1 and 35 are invariant cysteines that form a disulfide bond with each other, creating the intervening loop). However, the 11/25 rule lacks sensitivity in detecting X4 variants compared with phenotypic assays (9), with an Arg residue at position 25 being the least predictive. The position-specific scoring matrix (PSSM) analyzes the entire V3 sequence and predicts X4 variants based on a scoring of the probability of the amino acid at each position being overrepresented among X4 sequences versus R5 sequences (10). The Geno2Pheno (G2P) algorithm (11) also analyzes the entire V3 sequence and provides a quantitative score (false-positive rate [FPR]) representing the probability of falsely predicting an R5 variant as an X4 variant; the validity of this approach has been assessed in several clinical trials (12–16). G2P has been widely used in research and clinical settings, but it requires a preset FPR cutoff to call X4 variants. There is significant uncertainty about where to place the FPR cutoff and about the reliability of the FPR values in the intermediate range as representing the frequency of X4 and R5 variants with those intermediate FPR values, creating uncertainty about overcalls and undercalls, especially in the context of X4 evolution and coexisting viral lineages.
HIV-1 displays extensive genetic diversity and undergoes rapid evolution within individual hosts (17), and the env gene has the most sequence diversity of all of the HIV-1 genes, which can often include coexisting distinct lineages within a person. However, certain approaches to the analysis of env genes can confound the interpretation due to the introduction of artifacts into the data set. Bulk PCR amplification of a population of sequences can lead to PCR-mediated recombination, which significantly affects the quality of the phylogenetic analysis (18). Endpoint dilution PCR (EPD PCR) (also called single-genome amplification [SGA] or single-genome sequencing [SGS]) combined with Sanger sequencing avoids the confounding effect of PCR-mediated recombination and allows a true assessment of viral sequences as they appear in the viral population (18, 19). EPD PCR is time-consuming and labor-intensive, making it challenging to analyze more than a few dozen viral genomes per sample (20–22), which limits the depth of sampling of viral genomes when assessing the complexity of the viral population. Recently, viral population studies have started to incorporate deep-sequencing or next-generation sequencing (NGS) technologies that have the capacity to greatly extend the depth of sampling (12, 23–28). However, conventional approaches using NGS in HIV-1 population studies have serious limitations in representing an accurate sampling of the original viral population. Artifactual heterogeneity can be introduced due to the preceding PCR step, which introduces misincorporation and recombination (29, 30) as well as high rates of sequencing errors associated with NGS, while artifactual homogeneity can be introduced by PCR resampling of the same starting templates (31) and using reference sequences to construct consensus sequences for variable regions. We have overcome these limitations by using the Primer ID sequencing approach to define template sampling depth and reduce the error rate to around 1 in 10,000 nucleotides (32, 33).
Here we present a study in which we used the Primer ID approach coupled with Illumina MiSeq paired-end sequencing to reveal the structure of viral populations in the V1/V2 and C2/V3 regions of the HIV-1 env gene and to predict coreceptor tropism in sequence lineages found in chronically infected and post-acute-phase HIV-infected subjects. We also used EPD PCR followed by cloning of env genes for use in pseudotyping to explore the link between genotypic predictions and phenotypic assessment, focusing on distinguishing intermediate prediction values. Neither of these approaches (Primer ID NGS or EPD PCR) has been used to examine the presence of X4 variants in samples taken from late-stage subjects. We found that X4/dual-tropic variants form discrete lineages within the viral population, typically having G2P FPR scores of <2%. The coincident R5 populations in subjects with X4/dual-tropic viruses present were not skewed toward X4-like sequences compared to all-R5 populations or transmitted/founder viruses. The X4 and R5 populations were genetically isolated based on the inability to detect significant recombination between the R5 and X4 populations when V1/V2 and V3 markers were compared. Finally, we suggest sequence determinants of the V3-associated entry phenotype that could explain some of the uncertainty in making an R5 or X4 call associated with viruses with intermediate FPR genotypic scores.
MATERIALS AND METHODS
Study participants and clinical samples.
We examined 30 plasma samples from late-stage subjects (with CD4+ T cell counts of <100 cells/mm3) and 6 plasma samples from early-stage subjects (within the first 2 years of infection). All subjects were infected with subtype B HIV-1, and all subjects provided informed consent to participate in protocols that were approved by the Institutional Review Boards (IRBs) at Yale University, UCSF, and UNC. Late-stage samples were obtained at the entry time point from subjects who were enrolled in an early trial of ritonavir (34); some of the subjects had had prior exposure to zidovudine (AZT) and/or lamivudine (3TC) as monotherapy, and early-stage subjects were naive to antiretroviral therapy.
cDNA synthesis and MiSeq library construction.
We used the QIAamp viral RNA minikit (Qiagen, Valencia, CA) to extract viral RNA from the plasma samples and a previously described approach to construct the Primer ID MiSeq library after cDNA synthesis using the viral RNA as the template and PCR (33). The Primer ID cDNA primer was composed of the complement of the target region in the RNA template for cDNA synthesis (HXB2 [GenBank accession number K03455] numbering for the gene-specific region spanning positions 7209 to 7238, at the 3′ boundary of the V3 coding region of the env gene), an 8-base random sequence that served as the Primer ID and one random nucleotide as a sequencing buffer, and a 5′ tail of the primer sequence that served as the PCR primer. The sequence of the cDNA primer was 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNNNNCAGTCCATTTTGCTCTACTAATGTTACAATGTGC-3′. We ordered hand-mixing for the synthesis of the Primer ID cDNA primer (IDT, Coralville, IA) to maximize the randomness of the nine degenerate base positions. SuperScript III reverse transcriptase (Life Technologies, Grand Island, NY) was used for cDNA synthesis, with the chosen number of RNA templates (typically between 5,000 and 10,000 copies of viral RNA per reaction) being estimated based on viral load testing. After cDNA synthesis, all samples were purified by using Agencourt RNAClean XP magnetic beads (Beckman Coulter, Brea, CA) with a stringent wash protocol (the ratio of the volume of beads to cDNA was 0.6 to 1.0, with the beads being washed 4 times) to remove unused cDNA primers. All of the purified cDNA was used for the first of two rounds of PCR amplification. The first-round forward and reverse PCR primers were 5′-GCCTCCCTCGCGCCATCAGAGATGTGTATAAGAGACAGNNNNTTATGGGATCAAAGCCTAAAGCCATGTGTA-3′ (HXB2 numbering for the gene-specific region spanning positions 6555 to 6584, just upstream of the V1/V2 region of the env gene) and 5′-GTGACTGGAGTTCAGACGTGTGCTC-3′, respectively. PCR products from the first-round PCR were purified by using Agencourt AMPure XP PCR purification beads (Beckman Coulter) with a stringent protocol (the ratio of the volume of beads to DNA was 0.6 to 0.8, with the beads being washed 2 times). This step also allowed us to create heterogeneity near the left end of the amplicon (the N4 stretch in the primer) so that there was efficient cluster assignment during the sequencing step. We used the Illumina indexed primers as the second-round reverse PCR primers to tag each sample with a unique bar code, allowing us to multiplex up to 24 libraries in the same sequencing run. The forward and reverse primers for second-round PCR were 5′-AATGATACGGCGACCACCGAGATCTACACGCCTCCCTCGCGCCATCAGAGATGTG-3′ and 5′-CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCAGACGTGTGCTC-3′, respectively. The 6-nucleotide stretch of N′s represents the position of the 24 Illumina bar codes. Kapa Robust and Kapa High-Fidelity PCR kits (Kapa Biosystems, Woburn, MA) were used for first- and second-round PCR amplification, respectively. The second-round PCR products were gel purified by using a QIAquick MinElute gel extraction kit (Qiagen). The recovered DNA was then quantified by using a Qubit 2.0 fluorometer (Life Technologies), and the products were pooled in equal amounts. Library quality was assessed by using the Experion electrophoresis system (Bio-Rad, Hercules, CA).
Illumina sequencing and bioinformatics pipelines.
The UNC High Throughput Sequencing Facility performed Illumina MiSeq analysis by using either 250-nucleotide or 300-nucleotide paired-end sequencing. A random PhiX genomic DNA library was mixed with the HIV-1 library pool to increase the complexity between clusters on the flow cell. The Illumina pipeline (v.1.8.2) was used to generate sequence reads with assigned quality scores. In-house bioinformatics pipelines written in Ruby were used to filter out sequence reads of low quality, link sequence end pairs, capture Primer IDs, and create consensus sequences without a reference sequence. To avoid the inclusion of sequencing errors in the Primer ID sequence tag (offspring Primer IDs) that would create artifactual viral genomes, we calculated a cutoff for the number of raw reads needed for each Primer ID to form a template consensus sequence (TCS) based on a simulation to estimate the frequency of offspring Primer IDs with errors. We then used Primer IDs with a number of raw reads above this cutoff to create a TCS for each starting RNA template (33). Additionally, TCSs were filtered for large deletions and frameshifts.
EDP PCR and phenotyping of coreceptor usage.
env gene clones were generated as previously described (22). In brief, cDNA was generated by using an oligo(dT) primer from viral RNA isolated from blood plasma, followed by EDP PCR (3, 18, 35) to amplify full-length env genes for sequencing. DNA sequence alignments of the env genes were performed by using ClustalW (36). Sequences for each subject were codon aligned, and phylogenetic trees were generated by using a neighbor-joining method (MEGA5) (37). An additional round of amplification was performed to reamplify env genes for the purpose of cloning into the pcDNA 3.1D/V5-His_TOPO expression vector (Life Technologies), and these clones were used for pseudotype virus production.
293T cells were cultured in Dulbecco's modified Eagle medium (DMEM) with 4.5 g/liter glucose (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin, and 50 μg/ml streptomycin. Affinofile cells (38) were also cultured in DMEM but supplemented with 10% dialyzed FBS (12- to 14-kDa cutoff; Atlanta Biologicals, Flowery Branch, GA) and 50 mg/ml blasticidin (Life Technologies).
Env-pseudotyped luciferase reporter viruses were generated by cotransfection of 0.81 μg of the pNL4-3.LucR-E plasmid (obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH), 0.81 μg of an HIV-1 env clone, and 4.9 μl of FuGENE 6 transfection reagent (Promega, Madison, WI) in a total volume of 75 μl of medium added to 1 well of a 6-well dish seeded with 2.4 × 105 293T cells. At 5 h posttransfection, the medium was replaced, and the cells were incubated at 37°C for 36 to 42 h. Pseudotyped-virus stocks were isolated from the supernatants by filtration though a 0.45-μm filter (Millipore, Billerica, MA) prior to storage at −80°C. Virus stocks were not subjected to multiple freeze-thaw cycles.
Affinofile cell assays were performed as previously reported (38). In short, Affinofile cells were plated into black poly-l-lysine (Sigma)-treated 96-well plates at a concentration of 1.8 × 104 cells/well. Cells were induced for CCR5 and CD4 expression by using ponasterone A (PonA) (Life Technologies) and doxycycline (Doxy) (Sigma, St. Louis, MO) for 24 h before medium exchange and virus addition (Affinofile cells constitutively express CXCR4). Cells were centrifuged at 2,000 rpm for 2 h at 37°C in the presence of virus to enhance infection (i.e., spinoculated), followed by incubation for 48 h at 37°C, and then washed two times with phosphate-buffered saline (PBS) prior to being lysed with 50 μl of 1× reporter lysis buffer (Promega) and stored at −80°C. Viral entry was assessed using the luciferase assay system (Promega) by thawing the lysates and quantifying luciferase expression. Virus stocks were titrated by using Affinofile cells expressing maximum levels of CD4 and CCR5. The amount of virus used for an infection was determined by calculating the volume of virus needed to produce ∼800,000 relative light units (RLU) of luciferase readout, a value near the high end of the linear portion of the dose-response curve for luciferase activity in our assay.
Coreceptor usage was determined by infecting cells in the presence of maraviroc, AMD3100, or maraviroc plus AMD3100 (both obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH). The coreceptor phenotype was determined by using Affinofile cells expressing a single high level of CD4 (Doxy) and a high level of CCR5 (PonA). One hour prior to spinoculation, the coreceptor antagonist was added at a single high dose (50 μM maraviroc, 2 μM AMD3100, or 50 μM maraviroc plus 2 μM AMD3100). After spinoculation, the medium was removed and replaced with fresh medium, followed by a 48-h incubation at 37°C. The cells were then washed with PBS, and lysis buffer was added, followed by freezing, thawing, and then assaying for luciferase activity.
Phylogenetic analysis and coreceptor tropism prediction.
Full-length env sequences derived from the EPD PCR amplicons from each sample were trimmed to include the same region as that in the NGS consensus sequences. We used MUSCLE (v3.8.1) (39, 40) to align sequencing reads and construct neighbor-joining trees for the V1/V2 region and the C2/V3 region separately. Coreceptor genotyping of V3 sequences from both NGS and EPD PCR was performed by using the G2P algorithm (11) to generate an FPR value.
Nucleotide sequence accession numbers.
Raw MiSeq sequencing reads are available at the NCBI Sequence Read Archive (SRA) (SRX1628641, SRX1628643, SRX1628644, SRX1628655, SRX1628657–SRX1628660, SRX1628663, SRX1628670, SRX1628675, SRX1628682, SRX1628688–SRX1628690, SRX1628697, SRX1628704–SRX1628706, SRX1628709, SRX1628719, SRX1628720, SRX1628722, SRX1628729, SRX1628731–SRX1628736, SRX1628740, and SRX1628742–SRX1628746).
RESULTS
Development of an HIV-1 env gene amplicon for paired-end sequencing.
Next-generation sequencing offers new opportunities to examine the structure of genetically diverse populations, such as HIV-1. However, different platforms offer different advantages in read length, number of reads, and accuracy of reads. We have developed an amplicon to examine HIV-1 env populations using the MiSeq platform based on 250-nucleotide (now 300-nucleotide) paired-end reads. We chose to generate an 835-bp amplicon starting upstream of the V1 region (HBX2 position 6555) and extending through the V3 region (HBX2 position 7238). In the paired-end-read strategy, ∼200 to 250 nucleotides of HIV-specific sequence are generated from each end of the amplicon. This allows 216 nucleotides of the V1/V2 region and all of the V3 region and into C2 to be analyzed, and with the current 300-nucleotide paired-end sequencing, the entire V1/V2 region can be captured in most cases. In the current format, these paired-end reads do not overlap, although the MiSeq platform allows them to be linked as coming from the same template molecule. We targeted this region for deep-sequencing analysis since the V1/V2 region is the most diverse sequence within the HIV-1 genome, and the V3 region can be used to infer the X4 or R5 entry phenotype for the virus.
cDNA synthesis was done by using a primer that included a 9-nucleotide stretch of randomized bases, an indexing strategy that we have termed Primer ID (32, 33); the Primer ID cDNA primer was used just for cDNA synthesis and then removed prior to the PCR step. This allowed each RNA template to be linked to a unique sequence tag that could be identified in a subsequent sequence analysis. The cDNA was then amplified by PCR using a primer that was complementary to a region in the cDNA primer that was behind the Primer ID sequence tag, with the other PCR primer being a gene-specific upstream/forward primer. This approach largely resolves two significant limitations of the use of NGS for a genetically diverse population that is first amplified by PCR: it allows the recognition of PCR resampling, i.e., the repetitive sequencing of the original template sequences as represented many times in the PCR product, which is identified by tagging each template with a unique sequence tag, and it allows PCR resampling (i.e., sequences with the same Primer ID sequence) to be used to create a TCS for each original template to remove most of the PCR and sequencing errors.
Characteristics of HIV-positive subjects providing plasma samples for sequence analysis.
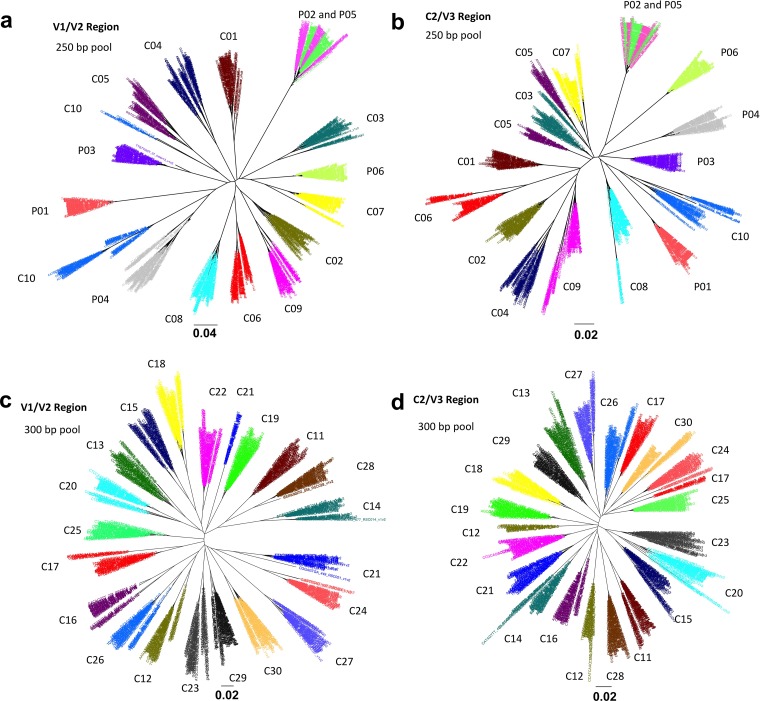
Table 1 shows the characteristics of the study subjects and the blood plasma samples provided as well as the number of TCSs that we obtained for each plasma sample. In this initial characterization of the V1-V3 env sequence amplicon, we examined samples from 30 subjects late in the course of their HIV-1 infection, as indicated by their low average CD4+ T cell counts (23 cells/μl) and relatively high average viral loads (5.2 log10 copies/ml) (Table 1). We also included samples from six subjects early in infection with a median estimated number of days of infection of 328, a median CD4+ T cell count of 600 cells/μl, and a median viral RNA load of 4.6 log10 copies/ml (Table 1). Ten of the late-stage and the six early-stage samples were sequenced by using the 250-nucleotide paired-end sequencing protocol in one library pool, and the other 20 late-stage samples were sequenced by using the 300-nucleotide paired-end sequencing protocol in a second pool. The median numbers of TCSs of late- and early-stage subjects were 1,357 and 1,532, respectively, derived from medians of 557,450 and 170,310 raw reads, respectively (Table 1); given the average number of reads per template in these runs, the low-end read cutoff averaged at least 20 reads per template to be used for TCS formation (calculated according to methods reported previously [33]). Also, this number of consensus sequences represented a rate of conversion of starting RNA templates to TCSs of between 10% and 30%. We estimate that for most samples, there is a 95% chance of seeing minor variants with a 0.1 to 0.5% abundance, depending on the exact number of TCSs obtained. Figure 1 shows the neighbor-joining trees of a subset of TCSs from each clinical sample of both V1/V2 (Fig. 1a) and C2/V3 (Fig. 1b) regions of the 250-nucleotide paired-end pool (100 consensus sequences per sample) and the 300-nucleotide paired-end pool (50 TCSs per library) (Fig. 1c and d). No cross-sample contamination was observed, although a linked pair of primary-infection cases was inferred (subjects P02 and P05). In addition, EPD PCR was used to generate env gene amplicons for 16 out of 30 late-stage samples and all 6 of the early-stage samples, with an average of 19 EPD amplicons being analyzed per sample and an average of 3 amplicons being cloned per subject for entry analysis of representatives of the major env sequence lineages.
TABLE 1.
Demographic and clinical characteristics of study subjects and numbers of sequencing readsa
| Characteristic | Value for group |
|
|---|---|---|
| Early-stage subjects (n = 6) | Late-stage subjects (n = 30) | |
| Median age (yr) (IQR) | 40.5 (27.5–49) | |
| No. (%) of male patients | 6 (100) | |
| Median most recent CD4 cell count (cells/μl) (IQR) | 600 (556–617) | 23 (13–41) |
| Median most recent HIV RNA level (log10 copies/ml) (IQR) | 4.64 (4.30–4.83) | 5.2 (5.0–5.4) |
| Median no. of days after infection (IQR) | 328 (135–448) | NA |
| Median no. of TCSs (IQR) | 1,532 (1,332–1,883) | 1,357 (992–1,982) |
| Median sequencing depth (%) (IQR)b | 0.2 (0.16–0.22) | 0.22 (0.15–0.33) |
IQR, interquartile range; TCS, template consensus sequence; NA, not applicable.
Sequencing depth is defined as the lowest abundance of a variant that can be detected with a 95% chance of a certain number of TCSs.
FIG 1.
Neighbor-joining trees for a pool of 100 (250-bp pool) or 50 (300-bp pool) randomly picked consensus sequences from each sample at both the V1/V2 and C2/V3 regions. P01 to P06 are six samples obtained from subjects early during infection. C01 to C30 are 30 samples from late-stage HIV-1-infected subjects. The bar for the phylogenetic trees indicates the number of nucleotide substitutions per site. (a) V1/V2 tree of the 250-bp sequencing pool. (b) C2/V3 trees of the 250-bp sequencing pool. (c) V1/V2 tree of the 300-bp sequencing pool. (d) C2/V3 trees of the 300-bp sequencing pool.
Characterization of major and minor lineages by coreceptor tropism prediction.
We used the G2P algorithm (11) to infer the presence of X4 or dual-tropic viruses (which we refer to collectively as X4 viruses) in these sequence populations under two levels of stringency, making X4 calls for sequences with an FPR of <5% and then at the more stringent FPR cutoff of <2%. Table 2 shows the numbers and percentages of X4 viruses predicted by G2P (FPR cutoffs of 2.0% and 5.0%) in both NGS and EPD PCR sequences. X4 viruses were detected in 24 out of 30 late-stage subjects by using the 2% FPR cutoff and in 26 samples by using the 5% cutoff. However, the two additional samples (samples C12 and C19) detected by using the 5% cutoff had 1% or less of the total TCSs with FPR scores below 5%, while the majority of the variants in these samples had FPR scores above 5%. Among the 24 samples predicted to have X4 variants at both cutoff stringencies, the 5% FPR cutoff predicted more X4 variants in 17 (71%) samples than did the 2% FPR cutoff. This analysis highlights the uncertainty of the choice of the FPR cutoff value in using this algorithm.
TABLE 2.
Numbers and percentages of X4 variants predicted by using the Geno2pheno[CORECEPTOR] algorithm on sequences assessed by both NGS and SGA
| Sample | No. of C2/V3 sequences |
No. (%) of X4 variants predicted by Geno2pheno[CORECEPTOR] |
||||
|---|---|---|---|---|---|---|
| NGSa | SGA | FPR of ≤2.0% |
FPR of ≤5.0% |
|||
| NGS | SGA | NGS | SGA | |||
| C01 | 1,116 | 20 | 0 | 0 | 0 | 0 |
| C02 | 381 | 17 | 0 | 0 | 0 | 0 |
| C03 | 1,031 | 23 | 154 (15) | 1 (4) | 155 (15) | 1 (4) |
| C04 | 502 | 15 | 102 (20) | 2 (13) | 424 (85) | 12 (87) |
| C05 | 1,438 | 17 | 670 (47) | 8 (47) | 672 (47) | 8 (47) |
| C06 | 616 | 29 | 170 (28) | 12 (41) | 170 (28) | 12 (41) |
| C07 | 1,248 | 19 | 152 (12) | 4 (21) | 962 (77) | 17 (89) |
| C08 | 1,610 | 38 | 217 (13) | 3 (8) | 217 (13) | 3 (8) |
| C09 | 2,108 | 38 | 401 (19) | 3(8) | 579 (28) | 6 (16) |
| C10 | 808 | 24 | 66 (8) | 10 (42) | 66 (8) | 10 (42) |
| C11 | 900 | 266 (30) | 267(30) | |||
| C12 | 1,353 | 14 | 0 | 0 | 13 (1) | 0 |
| C13 | 2,230 | 200 (9) | 201 (9) | |||
| C14 | 2,334 | 523 (22) | 524 (22) | |||
| C15 | 988 | 178 (18) | 178 (18) | |||
| C16 | 855 | 36 | 657 (77) | 28 (78) | 670 (78) | 28 (78) |
| C17 | 635 | 111 (17) | 111 (17) | |||
| C18 | 1,361 | 74 (5) | 164 (12) | |||
| C19 | 787 | 0 | 2 (0.3) | |||
| C20 | 2,626 | 154 (6) | 161 (6) | |||
| C21 | 1,141 | 12 | 30 (3) | 0 | 1,102 (97) | 8 (67) |
| C22 | 1,418 | 203 (14) | 207 (15) | |||
| C23 | 1,247 | 0 | 0 | |||
| C24 | 2,019 | 92 (5) | 92 (5) | |||
| C25 | 1,431 | 18 | 228 (16) | 2 (11) | 1,381 (17) | 2 (11) |
| C26 | 2,166 | 333 (15) | 336 (16) | |||
| C27 | 1,791 | 19 | 304 (5) | 1 (5) | 1,790 (100) | 19 (100) |
| C28 | 1,868 | 19 | 1,585 (85) | 17 (89) | 1,869 (100) | 19 (100) |
| C29 | 2,856 | 0 | 0 | |||
| C30 | 3,103 | 841 (27) | 841 (27) | |||
| P01 | 2,732 | 19 | 0 | 0 | 8 (0.1) | 0 |
| P02 | 1,464 | 17 | 0 | 0 | 4 (0.3) | 0 |
| P03 | 1,963 | 20 | 0 | 0 | 0 | 0 |
| P04 | 669 | 11 | 0 | 0 | 1 (0.1) | 0 |
| P05 | 1,552 | 9 | 0 | 0 | 4 (0.3) | 0 |
| P06 | 1,277 | 10 | 0 | 0 | 2 (0.2) | 0 |
Number of template consensus sequences.
The same algorithm was also used on the samples taken early during infection. G2P with a 2% FPR score did not call any X4 variants in these six samples, while G2P with a 5% FPR score made X4 calls in five samples but with only a small number of TCSs (<1% of the population) (Table 2). Thus, we conclude that small changes in the FPR can give discrepant results when estimating the relative abundance of the X4 variant population and can give discrepant results when examining samples that are likely to have a low abundance of or no X4 variants, such as early viral populations. Overall, a 5% FPR cutoff called additional X4 variants, largely in low abundance, in approximately two-thirds of the samples taken both early and late during infection.
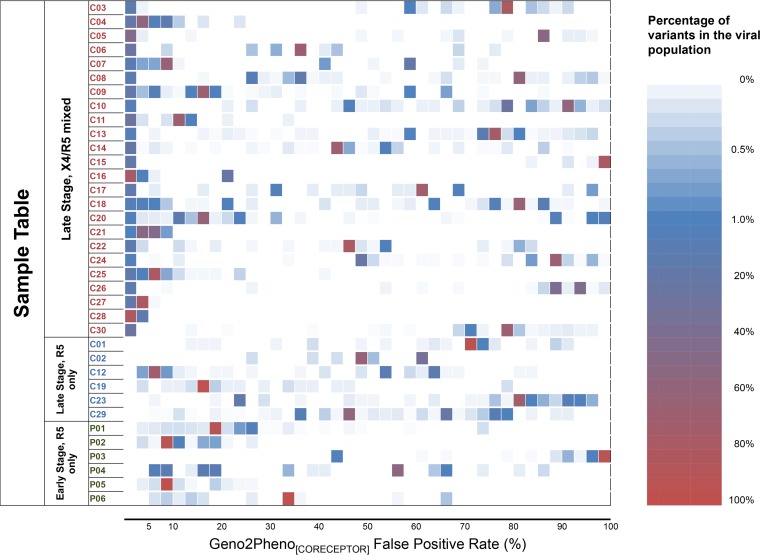
In the G2P algorithm, each sequence is assigned a specific FPR value to estimate the likelihood of a sequence being from an X4 variant. We plotted all of the assigned FPR values for these sequences from all of these samples. This analysis showed that groups of sequences within each subject clustered with their FPR values, with there being a number of groups with intermediate values and what appeared to be a tight clustering of low FPR values for those predicted to be X4 variants (Fig. 2). These results suggest that these sequence variants represent distinct lineages with similar FPR values within the host rather than a continuum of variability.
FIG 2.
Distribution of Geno2pheno[CORECEPTOR] false-positive rates among viral populations. P01 to P06 are six samples from subjects early during infection. C01 to C30 are 30 samples from late-stage HIV-1-infected subjects.
Linkage of FPR scores and phylogenetic lineages of viral populations based on C2/V3 sequences.
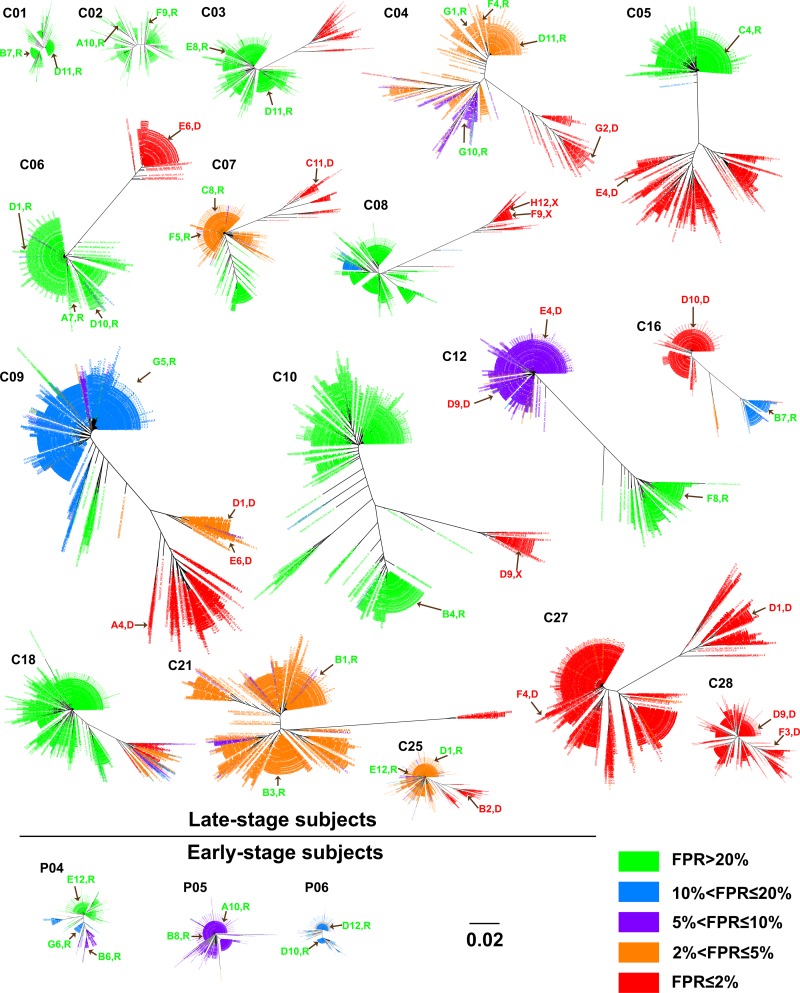
In order to examine the population structure of these sequences, we created neighbor-joining trees for the sequences from each subject. In this analysis, we color-coded the phylogenetic lineages with the FPR score for each sequence, binned as ≤2%, >2% and ≤5%, >5% and ≤10%, >10% and ≤20%, and >20%. We consider values of ≤2% to represent true X4 viruses, values of >20% to be true R5 viruses, and values between these two to be intermediate. As shown in Fig. 3 and Fig. S1 in the supplemental material, for the C2/V3 trees for the late-stage subjects, the most stringently called X4 sequences (≤2% FPR) are associated with one or more discrete lineages within the population. In some of these samples, there are distinct viral lineages with intermediate FPR values, with the clearest examples being samples C04, C07, C09, C20, and C25. In two samples (samples C27 and C28), almost all of the sequences have low FPR values (below 2%). In one sample (sample C18), there are FPR scores of between 1.7% and 16% for sequences within one distinct viral lineage, while the remaining portion of the population has FPR scores of >20%. After examining the sequences of the FPR score mixtures in this particular lineage, we found that all sequences in this lineage contain a V3 11K or -R mutation, a disruption in the GPGR crown (GPKR), and other changes not associated with R5 viruses, consistent with this lineage likely being an X4 lineage but poorly recognized by the G2P algorithm. In all other cases, the X4 lineages with a ≤2% FPR were relatively homogeneous in their FPR scores, and these lineages coexisted with other, most often relatively homogeneous, lineages with a range of FPR values usually strongly predicted to be R5, with FPR values of >20%. Thus, another feature of viral populations that carry strongly called X4 viruses is that these sequences represent a distinct lineage within the sequence population.
FIG 3.
Phylogenetic trees of the C2/V3 region for study samples showing different Geno2pheno[CORECEPTOR] FPRs among viral populations. Brown arrows indicate the identical or closest sequences in the tree of clones that we obtained by single-genome amplification and that were examined for entry phenotypes in vitro. Clone names and their phenotypic tropism (R, R5; X, X4; D, dual tropism) are indicted along with the arrows. The bar in the phylogenetic trees indicates the number of nucleotide substitutions per site.
Overall, we detected clear X4 lineages in 80% of these late-stage subjects (24 of 30). This frequency of X4 variants within people infected with subtype B HIV-1 is higher than previously reported estimates of around 50% (4). The fraction of the viral population in the X4 lineage within each person varied widely, ranging from 3% to 85%. Our ability to detect X4 variants at a higher level may be due to the use of NGS, as about half of the subjects with X4 viruses had a viral population where the X4 lineage comprised <20% of the total viral population. It is important to note that the use of Primer ID is essential in validating the number of genomes queried in this analysis to ensure that our assignment of lineage abundance is well above our subject-specific measured depth of sampling (i.e., the number of TCSs).
The remaining samples had no strong X4 lineages, including samples C01, C02, C12, C19, C23, and C29 from the late-stage subjects and samples P01 to P06, representing all of the early-stage subjects. However, the lineages within these viral populations in these subjects varied in their FPR values: samples C01, C02, C23, C29, and P03 had strongly R5 lineages; samples C19, P01, P02, P05, and P06 had relatively homogeneous populations with intermediate FPR scores; sample P04 had a mixture of lineages with intermediate FPR scores and R5 FPR scores, while the tree topology is relatively condensed; and sample C12 had two distant lineages, one with an FPR score above 20% and the other with an FPR score of around 6%. Based on data from the genotypic analysis alone, it is difficult to assess the significance of the lineages with intermediate FPR scores with respect to their contribution to an X4 phenotype, especially given that strong X4 calls were easily seen as discrete lineages.
Another layer of complexity in interpreting the genotypic calls for the entire population was the presence of one or two sequence variants with more X4-like FPR scores embedded within a lineage with a higher FPR score. For example, a majority of the sequences in samples P01, P02, and P05 had intermediate FPR scores of between 5% and 20%. However, a total of 7 distinct sequences had stronger X4 calls (FPR of 2 to 5%). None of these sequences formed a distinct lineage. Only one nucleotide substitution distinguished these sequences within each lineage, and none of them had an amino acid substitution typically associated with X4 viruses. Sample C07 had 3 major lineages with distinct FPR values that are labeled in red (≤2%), orange (2 to 5%), and green (>20%) in Fig. 3. Several sequences with FPR scores of ≤2% were found in the lineage with the intermediate FPR value. However, these sequences had just one nucleotide difference from the lineage consensus sequence and did not themselves form a discrete lineage. Thus, we conclude that the strongest X4 calls (≤2% FPR) are associated with discrete lineages of sequences within the population and that weaker X4 calls either identify lineages with intermediate values or represent occasional sequence variants intermingled with lineages with stronger R5 calls. The appearance of a single X4-like sequence variant differing by a single nucleotide from the consensus sequence of the lineage could be the result of a residual sequencing error, which should represent a total of about 25 single nucleotide polymorphisms introduced into a data set of 1,000 C2/V3 sequences based on an error rate of 1 in 10,000 nucleotides sequenced (33).
Entry phenotype associated with major lineages.
In an effort to link the X4, intermediate, and R5 FPR scores with the phenotype of specific lineages, we created full-length env gene expression vectors for the major lineages within these subjects. We first used EPD PCR to create a series of amplicons for sequencing and comparison to the lineages identified by using the C2/V3 MiSeq-generated sequences. The number of amplicons sequenced for each sample and the genotypic predictions are shown in Table 2. Given the comparatively low number of genomes analyzed by this method (∼20 per sample), we would not expect the percentage of X4 variants to recapitulate the percentage of X4 variants identified by the much larger sample size of sequences obtained by deep sequencing. However, we were interested in identifying variants from each of the major C2/V3 lineages. Amplicons were selected based on sequence similarity to the major C2/V3 sequence lineages, and env expression clones were generated.
The env clones were used to generate pseudotyped viruses with the expressed Env protein on the virion generated by coexpression with an env deletion genome that contained a reporter gene (luciferase) that was used to identify subsequent infectious events. Using Affinofile cells, we were able to test the entry phenotype predicted by the genotype by testing individually isolated env genes from the same samples that represented the major lineages identified by NGS. Affinofile cells have inducible CCR5 and CD4 but constitutively express CXCR4. Maraviroc and AMD3100, coreceptor inhibitors for R5 and X4 viruses, respectively, were used under the following three conditions to discern between CCR5-tropic, CXCR4-tropic, and dual-tropic viruses: (i) maraviroc only, (ii) AMD3100 only, or (iii) maraviroc plus AMD3100. Table 3 summarizes the phenotypic testing results as well as FPR scores for each clone. In Fig. 3, the brown arrows indicate the C2/V3 lineage with the sequence most similar to that of the clone. Clone names and their phenotypic tropisms are also labeled along with the brown arrows. In total, we tested 4 control env genes and 49 new env genes in these experiments, where 15 env clones encoded Env proteins that showed dual tropism, 3 encoded Env proteins that showed CXCR4 tropism, and 31 encoded Env proteins that showed CCR5 tropism. All of the 14 env clones with FPR scores of <2% encoded Env proteins that were X4 or dual-tropic viruses, consistent with the reliability of an inference of CXCR4 tropism associated with a low FPR score. All of the 13 env clones with FPR scores of >20% encoded Env proteins that were CCR5 tropic, consistent with the 20% cutoff being a reliable call for R5 viruses. In addition, no env clones from the early-stage subjects encoded Env proteins that were X4 variants.
TABLE 3.
Phenotyping of 4 env controls and 49 env isolates from 19 samples
| Sample | Clone | FPR (%) | Infectivity vs that for no-drug control (%) |
Phenotype | Description | ||
|---|---|---|---|---|---|---|---|
| Maraviroc | AMD3100 | AMD3100 + maraviroc | |||||
| BAL | NA | 24.7 | 0.18 | 66.18 | 0.11 | R5 | Control |
| JR-CSF | NA | 31.7 | 1.73 | 92.57 | 0.28 | R5 | Control |
| BV-5002 | NA | 2.0 | 145.64 | 0.80 | 0.03 | X4 | Control |
| NL43 | NA | 2.0 | 145.36 | 0.10 | 0.13 | X4 | Control |
| C01 | B7 | 56.5 | 0.81 | 159.62 | 1.26 | R5 | |
| D11 | 42.7 | 0.35 | 177.87 | 0.47 | R5 | ||
| C02 | A10 | 38.0 | 0.80 | 84.05 | 0.55 | R5 | |
| F9 | 38.0 | 0.18 | 62.28 | 0.17 | R5 | ||
| C03 | D11 | 68.6 | 0.20 | 75.55 | 0.22 | R5 | |
| E8 | 68.6 | 0.05 | 61.10 | 0.05 | R5 | ||
| C04 | D11 | 2.6 | 0.31 | 141.78 | 0.68 | R5 | |
| F4 | 2.9 | 0.08 | 63.45 | 0.12 | R5 | ||
| G1 | 2.7 | 0.08 | 103.96 | 0.24 | R5 | ||
| G10 | 6.9 | 0.77 | 91.48 | 0.30 | R5 | ||
| G2 | 0.7 | 84.34 | 39.04 | 0.40 | Dual | ||
| C05 | C4 | 64.0 | 0.22 | 103.64 | 0.17 | R5 | |
| E4 | 1.7 | 94.55 | 67.94 | 27.44 | Dual | ||
| C06 | A7 | 21.8 | 18.62 | 122.43 | 27.50 | R5 | |
| D1 | 24.7 | 16.40 | 76.27 | 6.81 | R5 | ||
| D10 | 24.7 | 15.43 | 96.58 | 9.94 | R5 | ||
| E6 | 0.5 | 88.37 | 64.79 | 39.04 | Dual | ||
| C07 | C11 | 1.7 | 102.89 | 42.17 | 3.19 | Dual | |
| C8 | 5.0 | 1.58 | 123.26 | 0.56 | R5 | ||
| F5 | 5.0 | 0.77 | 124.26 | 0.17 | R5 | ||
| C08 | F9 | 0.5 | 88.57 | 0.06 | 0.10 | X4 | |
| H12 | 0.5 | 158.23 | 3.70 | 0.17 | X4 | ||
| C09 | A4 | 1.1 | 123.05 | 50.92 | 0.42 | Dual | |
| D1 | 4.8 | 4.66 | 57.93 | 0.28 | Dual | ||
| E6 | 4.8 | 23.60 | 38.31 | 0.12 | Dual | ||
| G5 | 11.4 | 1.34 | 75.12 | 0.87 | R5 | ||
| C10 | D9 | 0.0 | 133.23 | 1.39 | 1.58 | X4 | |
| G4 | 67.5 | 3.51 | 79.59 | 2.15 | R5 | ||
| C12 | D9 | 5.3 | 99.92 | 39.14 | 6.00 | Dual | |
| E4 | 5.3 | 99.09 | 67.85 | 1.48 | Dual | ||
| F8 | 43.2 | 0.41 | 97.86 | 0.06 | R5 | ||
| C16 | D10 | 1.7 | 52.21 | 37.72 | 0.17 | Dual | |
| B7 | 16.6 | 0.08 | 104.19 | 0.05 | R5 | ||
| C21 | B1 | 5.0 | 0.08 | 104.19 | 0.05 | R5 | |
| B3 | 4.0 | 0.18 | 62.81 | 0.14 | R5 | ||
| C25 | B2 | 0.2 | 98.79 | 56.84 | 0.23 | Dual | |
| D1 | 5.0 | 0.05 | 96.29 | 0.08 | R5 | ||
| E12 | 5.0 | 0.07 | 95.85 | 0.44 | R5 | ||
| C27 | D1 | 0.2 | 80.77 | 57.49 | 0.30 | Dual | |
| F4 | 1.8 | 22.37 | 92.32 | 0.50 | Dual | ||
| C28 | D9 | 1.7 | 62.16 | 33.30 | 0.37 | Dual | |
| F3 | 1.9 | 76.52 | 71.27 | 0.70 | Dual | ||
| P04 | B6 | 6.0 | 0.12 | 89.75 | 0.08 | R5 | |
| E12 | 41.2 | 3.03 | 85.81 | 2.64 | R5 | ||
| G6 | 10.2 | 2.03 | 83.84 | 1.94 | R5 | ||
| P05 | A10 | 7.0 | 6.52 | 88.10 | 7.46 | R5 | |
| B8 | 7.0 | 4.48 | 86.71 | 4.70 | R5 | ||
| P06 | D10 | 16.9 | 0.26 | 62.59 | 0.28 | R5 | |
| D12 | 16.9 | 0.35 | 92.78 | 0.25 | R5 | ||
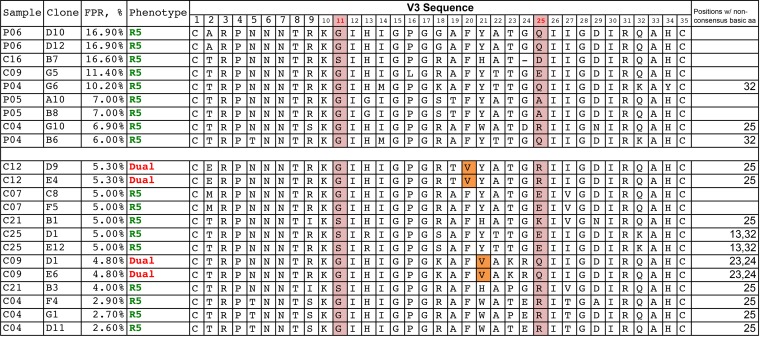
We next explored the env clones with intermediate FPR scores in an attempt to clarify the determinants of CXCR4 tropism in the X4 viruses with intermediate FPR scores. There were 22 viral env gene clones with FPR values that were spread between 2.1 and 20%; of these, 4 encoded Env proteins that were X4/dual tropic (the rest were CCR5 tropic), and all 4 of these clones had FPR values of <6% (as did 5 other R5 env clones). All of the 11 clones with FPR scores of between 6% and 20% encoded Env proteins that were CCR5-only viruses. We then inspected the V3 sequences associated with intermediate FPR scores (n = 22 clones). Figure 4 shows the V3 loop sequences of these intermediate clones as well as their FPR scores and phenotypes. While the data set is too small to allow statistical analysis, inspection of the sequences suggests several paths to consider concerning how to predict a phenotype from these intermediate genotypic scores/sequences. In general, sequences with intermediate values frequently have added basic amino acid substitutions, but these substitutions alone do not explain the pattern of measured phenotypes. Thus, it is useful to think of other key sites as potentially enabling the phenotypic expression of the basic substitutions, with these enabling mutations either being linked to the X4 phenotype or at least being a polymorphism that is compatible with the X4 phenotype. At one extreme, subject C04 had several clones that contained a 25R substitution and a low FPR score (<3.0%) but were R5 viruses based on their phenotype. Since 25R can appear in both R5 and X4 viruses, there must be additional determinants needed to realize the X4 phenotype. In this subject, both positions 20 and 21 were mutated to aliphatic amino acids in the cocirculating X4 viruses, in addition to other X4-related substitutions, suggesting that these positions may regulate the expression of the X4 phenotype for basic substitutions in some cases. While this analysis is very exploratory, among all 13 examples with intermediate FPR values of between 2 and 6%, X4 variants met the following criteria: the presence of one or more basic amino substitutions at positions 13, 23, 24, 25, and 32 (in addition to position 11) and the substitution of an aliphatic amino acid at either position 20 or 21 (and, based on skewing in X4 populations, perhaps including position 19), whereas the R5 variants with intermediate FPR scores did not meet these criteria.
FIG 4.
V3 amino acid (aa) sequences of cloned env sequences with intermediate FPRs and their coreceptor phenotypes.
Comparison of G2P FPR scores of transmitted/founder viruses and late-stage viral lineages.
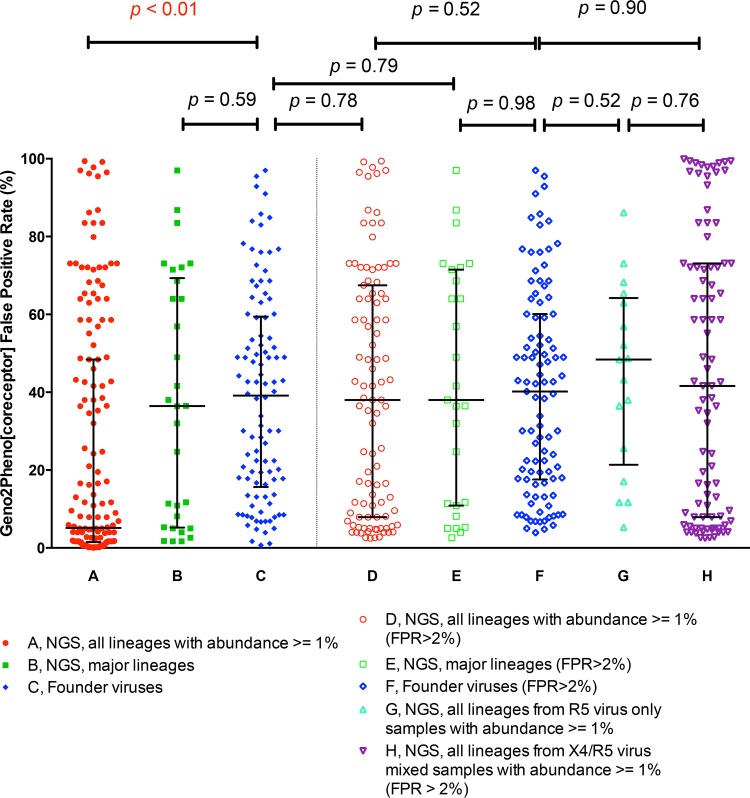
Most of our late-stage samples contained one or more X4 viral lineages. In contrast, the transmitted/founder (T/F) viruses are typically R5 viruses (3). We were interested in the possibility that the lineages with intermediate FPR scores in late-stage subjects are skewed toward the X4 population or in determining whether they represent a normal distribution of R5 lineages. In order to compare the predicted coreceptor usage/FPR scores of founder viruses and late-stage viral lineages, we downloaded aligned sequences from 104 acutely infected subjects, as described previously by Keele et al. (3). The consensus sequence was used to represent each T/F virus, and an FPR score was determined for each V3 sequence. For comparison, all viral lineages with abundances of >1% in the deep-sequencing analysis were used from the viral population from the late-stage subjects. A total of 153 lineages were identified from the 30 late-stage samples (Fig. 5A). The FPR scores for all of these lineages from the late-stage subjects were significantly lower (Fig. 5A) than those for T/F viruses (Fig. 5C). However, the range of FPR scores of the single major lineage in each subject sample (Fig. 5B) was comparable with that of the T/F viruses (Fig. 5C). When we excluded the X4 lineages from the late-stage subjects with FPR scores of ≤2%, the FPR scores of the remaining late-stage viral lineages (Fig. 5D) or the single major lineage from each subject (Fig. 5E) were comparable to the FPR scores of the T/F viruses, regardless of whether we included all of the T/F viral sequences (Fig. 5C) or excluded the few T/F viral sequences with FPR values of <2% (Fig. 5F). We further separated the late-stage viral lineages with FPR scores of >2% into those from R5-only samples (Fig. 5G) and those from mixed R5/X4 samples (Fig. 5H). The distributions of FPR scores of R5 lineages (FPR score of ≥2%) in the samples that also had an X4 lineage (Fig. 5H) were similar to those of the founder viruses (Fig. 5F). The distribution of FPR scores in the R5-only samples (Fig. 5G) also did not differ significantly from the distribution of FPR scores in the samples with X4 lineages when the lineages with FPR scores of ≤2% were excluded (Fig. 5H). Overall, we conclude that the distribution of FPR scores for the lineages with scores of >2% are similar between samples that have an X4 viral lineage (FPR score of ≤2%) and those that do not, regardless of whether the virus is isolated early or late during infection. This observation reinforces the ideas that the X4 lineages represent a specific clade within the viral population and that there is not a general shift toward X4-like sequences, as assessed by the FPR score, in subjects who evolve an X4 lineage.
FIG 5.
Comparison of Gene2pheno[CORECEPTOR] FPRs of the founder virus and lineages from late-stage clinical samples.
Restricted recombination between specific lineages within the viral population.
There are conflicting data as to whether X4 and R5 viruses replicate in the same cell type or in distinct cell types. One way in which to identify replication in the same cell type would be through the detection of recombinants, indicating infection by X4 and R5 viruses in the same cell, a prerequisite for the formation of heterodimeric genomic RNAs that are the basis for recombination during a subsequent round of infection. To address this question, we examined the linkage between the V1/V2 lineages from the deep-sequencing analysis and the C2/V3 lineages. Specifically, we determined if there were V1/V2 lineages that were shared among different C2/V3 lineages, indicative of recombination, or if there were discrete V1/V2 lineages that were linked to specific C2/V3 lineages, indicative of genetic isolation. In a control experiment, we constructed Primer ID sequencing libraries from mixed viral RNAs from two endpoint dilutions of a viral outgrowth assay mixture using same template copy number for each virus. These two endpoint dilutions had been sequenced separately and had 9 nucleotide differences in the V1-through-V3 region. After mixing, downstream steps of PCR amplification and sequence analysis were carried out. We found that recombinants between the two different samples had formed but only at a low level (∼0.1%) (S. K. Lee, S. Zhou, P. L. Baldoni, E. Spielvogel, N. M. Archin, M. G. Hudgens, D. M. Margolis, and R. Swanstrom, submitted for publication). While more recombinants were likely present after the PCR step, the construction of a TCS for each template obscures their presence, since they would rarely become the majority sequence for that template.
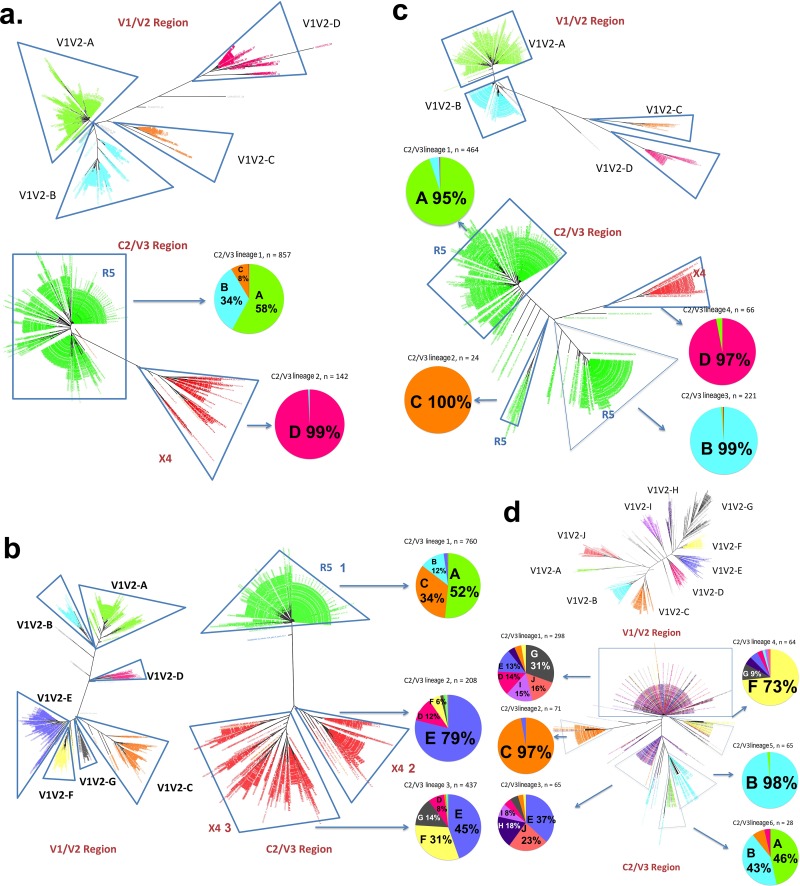
Figure 6 shows the neighbor-joining trees of the V1/V2 region and the C2/V3 region for four samples. X4 and R5 viral lineages are indicated, and the different V1/V2 lineages are color-coded. In sample C03 (Fig. 6A), there is a distinct lineage in the V1/V2 tree (V1/V2 lineage D [V1V2-D]) linked to the X4 C2/V3 lineage, and the three other V1/V2 lineages form the major R5 lineage in the C2/V3 tree. Thus, recombination between the X4 and R5 viruses between V1 and V3 was extremely rare, while different R5 V3 lineages were linked with several different V1/V2 lineages, consistent with recombination. Sample C05 (Fig. 6B) had seven distinct V1/V2 lineages (lineages A to G), three of which were associated with the major R5 V3 lineage (lineages V1V2-A, -B, and -C with C2/V3 R5 lineage 1) and four of which were associated with the two X4 virus lineages (lineages V1V2-D, -E, -F, and -G with C2/V3 X4 lineages 2 and 3); there appeared to be extensive mixing/recombination between the two C2/V3 X4 lineages and their V1/V2 lineages but very little mixing/recombination between the X4 lineages and the R5 lineage. In sample C10 (Fig. 6C), there were four major V1/V2 lineages, and each one was strongly associated with a separate C2/V3 lineage, three of which were R5 and one of which was X4. In this case, all four lineages appeared to be replicating independently without mixing through recombination. Linked G2P V3 scores were used to color-code the V1/V2 trees for all remaining samples with mixed R5 and X4 populations, and these trees are shown in Fig. S2 in the supplemental material. In summary, all X4 lineages with FPRs of ≤2% have little to no detectable recombination with R5 lineages within X4-containing samples.
FIG 6.
Lineages of the V1/V2 region and the C2/V3 region and their recombination in 4 subjects (subjects C03, C05, C10, and P04). For samples from subjects C03, C05, and C10, lineages have the same color-codes in the V1/V2 trees and the pie charts of the major lineages of the corresponding C2/V3 trees, while the C2/V3 trees are color-coded for coreceptor tropism (red for X4 and green for R5 variants). The percentages on the pie charts show the abundances of the V1/V2 lineages in the C2/V3 lineages. In subject P04, as all variants are R5 variants, lineages have the same color-codes in the corresponding V1/V2 and C2/V3 trees. (a) Subject C03; (b) subject C05; (c) subject C10; (d) subject P04.
In one early-stage sample, sample P04 (Fig. 6D), all of the sequences suggested R5 viruses, but there were a number of C2/V3 lineages based on distinct polymorphisms. Conversely, there were multiple distinct V1/V2 lineages (labeled in different colors in Fig. 6D). When these patterns were combined, it was clear that there were some C2/V3 lineages (lineages 1 and 3) that had sampled most (but not all) of the V1/V2 lineages, some C2/V3 lineages that had relatively few recombinants (lineages 2, 4, and 5), and one C2/V3 lineage largely composed of a linkage to just two V1/V2 lineages (C2/V3 lineage 6). These patterns suggest several stages of population mixing and isolation coexisting within this infected person.
DISCUSSION
We have used two approaches to analyze viral sequence populations that have not previously been applied to the examination of HIV-1 populations late in the disease course. The traditional use of PCR results in the introduction of PCR-mediated recombination (18), which can disrupt linkages within the population, confounding the analysis of phylogenetic relationships. PCR itself includes misincorporation, and NGS platforms suffer from both the problem of requiring a prior PCR step and having a relatively high rate of errors in the sequence data set. By using endpoint dilution PCR to generate full-length env genes and Primer ID NGS to sample the viral population in depth, we have been able to characterize these late-stage viral populations in the absence of these confounding technical artifacts. Using these approaches, we have found that most stringently called X4 variants are usually clustered in distinct lineages, that conventional prediction algorithms have difficulties in predicting X4 variants with intermediate scores, and that there is limited recombination between X4 and R5 variants.
Most current approaches using NGS to study HIV-1 populations have serious limitations in accurately sampling the population. First, most approaches usually require a preceding PCR amplification step. Thus, PCR errors, including nucleotide misincorporation by the polymerase, PCR recombination, and PCR resampling, may alter diversity as well as skew the allelic frequencies (29, 30). Second, in some approaches, the amplicons are first sheared before sequencing, and the sequencing reads are aligned with a reference sequence (41). With this approach, it is difficult to perform linkage analysis and estimate allelic frequencies. Third, sequencing errors can also introduce artificial diversity into the population. For instance, 454 pyrosequencing was broadly used in coreceptor tropism deep-sequencing studies, but it suffers from severe homopolymer errors, while the homopolymer issue is not a feature of the MiSeq platform used in this study (42, 43); furthermore, the homopolymer problem is a feature of all platforms that allow multiple or continuous additions of nucleotides where the number of additions in a homopolymer region must be interpreted by downstream software analysis. In addition, some studies use a reference sequence in their bioinformatics pipelines to complement the false insertions and deletions due to the homopolymer errors. This approach can be problematic when processing sequencing data from highly variable regions such as the V3 loop of the HIV-1 env gene, as it may create artificial alleles.
The Primer ID sequencing approach coupled with the MiSeq platform overcomes these serious limitations of conventional deep sequencing of genetically diverse viral populations by addressing the sampling depth, greatly reducing the sequencing error rate, and largely removing PCR-mediated recombination artifacts. This allowed us to perform a phylogenetic analysis where one TCS corresponds to one original viral template/genome, thus validating the level of viral population sampling for each patient sample analyzed. EDP PCR accomplishes these same features of characterizing viral populations and has the advantage of permitting the generation of longer amplicons (such as intact env genes) but has the limitation of a modest sampling depth compared to what is easily achieved by using next-generation sequencing. We took advantage of the strengths of these two approaches to examine X4 populations that evolve late in the HIV-1 disease course.
The most widely used entry-phenotype-predicting algorithm is Geno2Pheno[CORECEPTOR], which ascribes a false-positive rate value to a given V3 sequence to determine the probability of that sequence being from an X4 or dual-tropic virus based on an analysis of a large data set of viruses that were both sequenced and phenotyped (6). The MOTIVATE clinical trial showed that an FPR below 2% was strongly associated with treatment failure when the CCR5 inhibitor maraviroc was included in a multidrug regimen, and FPR values of between 2 and 5.75% were associated with a loss of antiviral activity of this inhibitor. Based on data from these trials, some guidelines suggest the use of an FPR value of 5.75% to predict X4/dual-tropic viruses or, even more conservatively, a 20% FPR cutoff. It seems clear that assignment of an X4 phenotype to a sequence with an FPR value of <2% has a very high probability of being correct (all 14 clones tested from 12 subjects in our data set were X4/dual tropic when the FPR value was <2%), and a similar argument can be made for an R5 phenotype when the FPR value is >20% (all 13 clones tested from 8 subjects in our data set were R5 when the FPR value was >20%). However, we were also interested in determining if detailed information about the population structure could add information about the sequences with FPR values of between 2% and 20%, where 4 of the 22 clones tested from 10 subjects were X4/dual tropic. Focusing just on lineages with FPR values of <2%, we found such lineages in 80% of this study population of subjects with late-stage disease, a value higher than what was previously seen, suggesting that next-generation sequencing can increase the sensitivity of detection of these variants.
We used several approaches to gain a better understanding of the meaning of variants with intermediate FPR values (between 2% and 20%). Overall, we observed that the distribution of FPR values of HIV-1 variants in individual subjects was not continuous; among late-stage subjects, the FPR values were similar within specific sequence lineages and could differ significantly between lineages within the same subject. Sequences with an FPR of ≤2% were usually, but not always, clustered in a single distinct lineage. We attempted to determine if there was additional information about the presence of X4 viruses with FPR values above 2% in these subjects. However, the distribution of variants with FPR values above 2% in subjects with X4 viruses (i.e., with FPR lineages of ≤2%) was not distinguishable from either a panel of transmitted/founder viruses or the lineages present in late-stage subjects who did not have an X4 lineage (defined as an FPR of ≤2%) (Fig. 2 and 5). We did not see evidence of a genotypic history of a gradual shift in FPR values resulting in the strong X4 lineage, suggesting that these viruses do not grow out to be an appreciable fraction of the population until they have a strong X4 signal. This conclusion is limited due to the fact this is a cross-sectional analysis of late-stage subjects, although in half of the subjects with X4 variants, this lineage represented <20% of the population, ranging down to 3%.
We also examined the V3 sequences themselves to see if there were clues that might distinguish between X4 and R5 variants within the FPR range of 2% to 20% in our data set. The premise was that the G2P database may be incomplete in its ability to assess rare V3 sequences, while other algorithms could be designed to compensate for this limitation. We also note that there were three env gene clones (from subjects C05 and C06) whose encoded proteins retained some infectivity in the presence of both the CCR5 and CXCR4 inhibitors. It was reported previously that CCR5-tropic but maraviroc-resistant variants can use inhibitor-bound CCR5 (44). The three env clones from this study may encode Env proteins that are examples of a naturally occurring CCR5-tropic maraviroc-resistant virus, or they may use coreceptors other than CCR5 or CXCR4.
Given a deep-sequencing view of the V3 sequence population, we propose the following steps for interpretation of the data. First, a clear lineage of sequences with FPR values of ≤2% are scored as X4/dual-tropic variants. We also propose that spurious or isolated sequences within an otherwise homogeneous FPR lineage should be interpreted as the bulk of the lineage rather than as isolated FPR values of single sequences, which could still be impacted by rare sequencing errors; thus, FPR assignments should be made for distinct lineages, which brings attention to the necessity of documenting the depth of sampling by using an approach like Primer ID. Conversely, lineages with an FPR of >20% should be uniformly treated as R5 lineages. This leaves the question of how to interpret lineages with FPR values of between 2% and 20%. We found one case where the entire lineage was of mixed FPR values, where inspection of the V3 sequences clearly suggested that this was an X4 lineage (with 11K and -R variants). Examination of the more homogeneous lineages of V3 sequences with FPR values of between 2% and 20% provided hints about how to improve the R5/X4 calls in this ambiguous region. Specifically, observing the presence or absence of basic amino acid substitutions at positions 11,13, 23, 24, 25, and 32 with an aliphatic amino acid (V or I) at position 20 or 21 correctly called 18 phenotyped clones with these in-between FPR values (>2% and <20%). The use of EDP PCR for generating our amplicons for cloning ensured no fortuitous recombination in the construction of the clones, but this requirement limits the number of phenotyped clones available to extend this initial correlation. The use of site-directed mutagenesis could provide some support for this suggestion, although the role of background mutations in defining the evolutionary pathways for V3 is poorly understood.
Several previously reported studies used deep sequencing to predict HIV-1 coreceptor usage by sequencing of the HIV-1 env V3 region (26, 45–48). Most of those studies used 454 sequencing, which was limited by homopolymer errors and relative low throughput compared to the Illumina or IonTorrent platform (25). In these studies, the presence of X4 variants was defined by the detection of >2% of sequences with FPR values of <3.5% in the viral population, as seen in the MERIT and MOTIVATE cohort studies (12, 13, 49). However, the predetermined FPR is arbitrary, and our data show that an FPR of between 2% to 6% can indicate either X4 or R5 variants. Meanwhile, without defining the sampling depth and correcting PCR resampling, the percentage of X4 variants detected is the percentage of X4 sequences in the PCR amplicon instead of the abundance of X4 variants among the viral genomes. Thus, the 2% cutoff in the viral population is also arbitrary and inaccurate, since there is no knowledge of how many templates/genomes were sampled. Several studies claimed that 10 to 20% of viruses from primary infection or transmitted viruses could be X4 variants by deep sequencing of the V3 loop (45, 48). We believe that these conclusions are compromised by the flawed sequencing methods and bioinformatics determination of X4 variants, and a more accurate approach, such as Primer ID sequencing coupled with phylogenetic analysis, should be used for such types of studies.
Each viral particle contains two copies of the viral genome. Typically, the two copies of the genome are identical, being transcribed from the same provirus. When multiple viruses infect the same host cell, virions containing two different genomes can be produced. When these heterodimeric viruses infect target cells, reverse transcriptase can switch back and forth between the two templates during DNA synthesis; thus, recombinant viruses can be produced (50). Recombination plays an important role in viral evolution (51). In this study, we found that the X4 and R5 variants in the same host displayed limited evidence of recombination between these variants. All X4 and R5 variants had distinct V1/V2 and C2/V3 regions. In contrast, in individuals with multiple X4 lineages or R5 lineages, we observed extensive recombination within X4 or R5 variants. This observation suggests that R5 and X4 variants are compartmentalized in different host cell types, or alternatively, their recombinants have low fitness and do not grow out in the viral population. Several previous studies have shown that X4 variants preferentially infect naive T cells and that R5 variants preferentially infect memory T cells (52–54). However, one study observed that the level of integration is lower in resting naive T cells than in memory T cells regardless of the coreceptor tropisms within the viral population (55).
In conclusion, Primer ID deep sequencing provides a novel approach to study genetic structures of viral populations. X4 variants may be more prevalent than previously reported when assessed by using NGS and with a sufficient depth of sampling. Phylogenetic analysis to identify lineages of sequences with low FPR values may provide additional information for accurately predicting X4 variants by using V3 sequences. Limited recombination occurs between X4 and R5 lineages, suggesting that X4 and R5 variants are genetically isolated and may be replicating in different cell types.
Supplementary Material
ACKNOWLEDGMENTS
UNC is pursuing IP protection for Primer ID, and R.S. is listed as a coinventor and has received nominal royalties.
We thank the UNC High Throughput Sequencing Facility for performing MiSeq sequencing.
R.S. supervised the project. S.Z. designed and performed deep sequencing. M.M.B. and C.B.S. performed endpoint dilution PCR and bulk sequencing. M.M.B. and B.M.H. performed the phenotypic entry assays. S.Z. performed deep-sequencing analysis and phylogenetic analysis. S.Z. and R.S. wrote the manuscript. M.M.B. provided critical revision to the manuscript.
This work was supported by NIH grants R21 AI108539 and R37 AI44667 to R.S. This work was also supported by the UNC Center for AIDS Research (NIH award P30 AI50410) and the UNC Lineberger Comprehensive Cancer Center (NIH award P30 CA16068).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00441-16.
REFERENCES
- 1.Chan DC, Kim PS. 1998. HIV entry and its inhibition. Cell 93:681–684. doi: 10.1016/S0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 3.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brumme ZL, Goodrich J, Mayer HB, Brumme CJ, Henrick BM, Wynhoven B, Asselin JJ, Cheung PK, Hogg RS, Montaner JS, Harrigan PR. 2005. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis 192:466–474. doi: 10.1086/431519. [DOI] [PubMed] [Google Scholar]
- 5.Su Z, Gulick RM, Krambrink A, Coakley E, Hughes MD, Han D, Flexner C, Wilkin TJ, Skolnik PR, Greaves WL, Kuritzkes DR, Reeves JD, AIDS Clinical Trials Group A5211 Team. 2009. Response to vicriviroc in treatment-experienced subjects, as determined by an enhanced-sensitivity coreceptor tropism assay: reanalysis of AIDS clinical trials group A5211. J Infect Dis 200:1724–1728. doi: 10.1086/648090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandekerckhove LP, Wensing AM, Kaiser R, Brun-Vezinet F, Clotet B, De Luca A, Dressler S, Garcia F, Geretti AM, Klimkait T, Korn K, Masquelier B, Perno CF, Schapiro JM, Soriano V, Sonnerborg A, Vandamme AM, Verhofstede C, Walter H, Zazzi M, Boucher CA, European Consensus Group on Clinical Management of Tropism Testing. 2011. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect Dis 11:394–407. doi: 10.1016/S1473-3099(10)70319-4. [DOI] [PubMed] [Google Scholar]
- 7.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol 66:6777–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Low AJ, Dong W, Chan D, Sing T, Swanstrom R, Jensen M, Pillai S, Good B, Harrigan PR. 2007. Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates. AIDS 21:F17–F24. doi: 10.1097/QAD.0b013e3282ef81ea. [DOI] [PubMed] [Google Scholar]
- 10.Garrido C, Roulet V, Chueca N, Poveda E, Aguilera A, Skrabal K, Zahonero N, Carlos S, Garcia F, Faudon JL, Soriano V, de Mendoza C. 2008. Evaluation of eight different bioinformatics tools to predict viral tropism in different human immunodeficiency virus type 1 subtypes. J Clin Microbiol 46:887–891. doi: 10.1128/JCM.01611-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. 2007. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol 25:1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 12.Swenson LC, Mo T, Dong WW, Zhong X, Woods CK, Jensen MA, Thielen A, Chapman D, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2011. Deep sequencing to infer HIV-1 co-receptor usage: application to three clinical trials of maraviroc in treatment-experienced patients. J Infect Dis 203:237–245. doi: 10.1093/infdis/jiq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swenson LC, Mo T, Dong WW, Zhong X, Woods CK, Thielen A, Jensen MA, Knapp DJ, Chapman D, Portsmouth S, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2011. Deep V3 sequencing for HIV type 1 tropism in treatment-naive patients: a reanalysis of the MERIT trial of maraviroc. Clin Infect Dis 53:732–742. doi: 10.1093/cid/cir493. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez V, Masia M, Robledano C, Padilla S, Lumbreras B, Poveda E, De Mendoza C, Soriano V, Gutierrez F. 2011. A highly sensitive and specific model for predicting HIV-1 tropism in treatment-experienced patients combining interpretation of V3 loop sequences and clinical parameters. J Acquir Immune Defic Syndr 56:51–58. doi: 10.1097/QAI.0b013e3181fc012b. [DOI] [PubMed] [Google Scholar]
- 15.McGovern RA, Thielen A, Portsmouth S, Mo T, Dong W, Woods CK, Zhong X, Brumme CJ, Chapman D, Lewis M, James I, Heera J, Valdez H, Harrigan PR. 2012. Population-based sequencing of the V3-loop can predict the virological response to maraviroc in treatment-naive patients of the MERIT trial. J Acquir Immune Defic Syndr 61:279–286. doi: 10.1097/QAI.0b013e31826249cf. [DOI] [PubMed] [Google Scholar]
- 16.Kagan RM, Johnson EP, Siaw MF, Van Baelen B, Ogden R, Platt JL, Pesano RL, Lefebvre E. 2014. Comparison of genotypic and phenotypic HIV type 1 tropism assay: results from the screening samples of Cenicriviroc Study 202, a randomized phase II trial in treatment-naive subjects. AIDS Res Hum Retroviruses 30:151–159. doi: 10.1089/aid.2013.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes EC, Zhang LQ, Simmonds P, Ludlam CA, Brown AJ. 1992. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc Natl Acad Sci U S A 89:4835–4839. doi: 10.1073/pnas.89.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol 82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol 73:10489–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ping LH, Joseph SB, Anderson JA, Abrahams MR, Salazar-Gonzalez JF, Kincer LP, Treurnicht FK, Arney L, Ojeda S, Zhang M, Keys J, Potter EL, Chu H, Moore P, Salazar MG, Iyer S, Jabara C, Kirchherr J, Mapanje C, Ngandu N, Seoighe C, Hoffman I, Gao F, Tang Y, Labranche C, Lee B, Saville A, Vermeulen M, Fiscus S, Morris L, Karim SA, Haynes BF, Shaw GM, Korber BT, Hahn BH, Cohen MS, Montefiori D, Williamson C, Swanstrom R, CAPRISA Acute Infection Study, Center for HIV-AIDS Vaccine Immunology Consortium. 2013. Comparison of viral Env proteins from acute and chronic infections with subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J Virol 87:7218–7233. doi: 10.1128/JVI.03577-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sturdevant CB, Dow A, Jabara CB, Joseph SB, Schnell G, Takamune N, Mallewa M, Heyderman RS, Van Rie A, Swanstrom R. 2012. Central nervous system compartmentalization of HIV-1 subtype C variants early and late in infection in young children. PLoS Pathog 8:e1003094. doi: 10.1371/journal.ppat.1003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. 2011. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog 7:e1002286. doi: 10.1371/journal.ppat.1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsibris AM, Korber B, Arnaout R, Russ C, Lo CC, Leitner T, Gaschen B, Theiler J, Paredes R, Su Z, Hughes MD, Gulick RM, Greaves W, Coakley E, Flexner C, Nusbaum C, Kuritzkes DR. 2009. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One 4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dybowski JN, Heider D, Hoffmann D. 2010. Structure of HIV-1 quasi-species as early indicator for switches of co-receptor tropism. AIDS Res Ther 7:41. doi: 10.1186/1742-6405-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Archer J, Weber J, Henry K, Winner D, Gibson R, Lee L, Paxinos E, Arts EJ, Robertson DL, Mimms L, Quinones-Mateu ME. 2012. Use of four next-generation sequencing platforms to determine HIV-1 coreceptor tropism. PLoS One 7:e49602. doi: 10.1371/journal.pone.0049602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunnik EM, Swenson LC, Edo-Matas D, Huang W, Dong W, Frantzell A, Petropoulos CJ, Coakley E, Schuitemaker H, Harrigan PR, van't Wout AB. 2011. Detection of inferred CCR5- and CXCR4-using HIV-1 variants and evolutionary intermediates using ultra-deep pyrosequencing. PLoS Pathog 7:e1002106. doi: 10.1371/journal.ppat.1002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Archer J, Braverman MS, Taillon BE, Desany B, James I, Harrigan PR, Lewis M, Robertson DL. 2009. Detection of low-frequency pretherapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS 23:1209–1218. doi: 10.1097/QAD.0b013e32832b4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raymond S, Saliou A, Nicot F, Delobel P, Dubois M, Cazabat M, Sandres-Saune K, Marchou B, Massip P, Izopet J. 2011. Frequency of CXCR4-using viruses in primary HIV-1 infections using ultra-deep pyrosequencing. AIDS 25:1668–1670. doi: 10.1097/QAD.0b013e3283498305. [DOI] [PubMed] [Google Scholar]
- 29.Meyerhans A, Vartanian JP, Wain-Hobson S. 1990. DNA recombination during PCR. Nucleic Acids Res 18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorzer I, Guelly C, Trajanoski S, Puchhammer-Stockl E. 2010. The impact of PCR-generated recombination on diversity estimation of mixed viral populations by deep sequencing. J Virol Methods 169:248–252. doi: 10.1016/j.jviromet.2010.07.040. [DOI] [PubMed] [Google Scholar]
- 31.Liu SL, Rodrigo AG, Shankarappa R, Learn GH, Hsu L, Davidov O, Zhao LP, Mullins JI. 1996. HIV quasispecies and resampling. Science 273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- 32.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. 2011. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci U S A 108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou S, Jones C, Mieczkowski P, Swanstrom R. 2015. Primer ID validates template sampling depth and greatly reduces the error rate of next-generation sequencing of HIV-1 genomic RNA populations. J Virol 89:8540–8555. doi: 10.1128/JVI.00522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron DW, Heath-Chiozzi M, Danner S, Cohen C, Kravcik S, Maurath C, Sun E, Henry D, Rode R, Potthoff A, Leonard J. 1998. Randomised placebo-controlled trial of ritonavir in advanced HIV-1 disease. The Advanced HIV Disease Ritonavir Study Group. Lancet 351:543–549. [DOI] [PubMed] [Google Scholar]
- 35.Abrahams MR, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping LH, Athreya GS, Treurnicht FK, Keele BF, Wood N, Salazar-Gonzalez JF, Bhattacharya T, Chu H, Hoffman I, Galvin S, Mapanje C, Kazembe P, Thebus R, Fiscus S, Hide W, Cohen MS, Karim SA, Haynes BF, Shaw GM, Hahn BH, Korber BT, Swanstrom R, Williamson C, CAPRISA Acute Infection Study Team, Center for HIV-AIDS Vaccine Immunology Consortium. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol 83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnston SH, Lobritz MA, Nguyen S, Lassen K, Delair S, Posta F, Bryson YJ, Arts EJ, Chou T, Lee B. 2009. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J Virol 83:11016–11026. doi: 10.1128/JVI.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Archer J, Rambaut A, Taillon BE, Harrigan PR, Lewis M, Robertson DL. 2010. The evolutionary analysis of emerging low frequency HIV-1 CXCR4 using variants through time—an ultra-deep approach. PLoS Comput Biol 6:e1001022. doi: 10.1371/journal.pcbi.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brockman W, Alvarez P, Young S, Garber M, Giannoukos G, Lee WL, Russ C, Lander ES, Nusbaum C, Jaffe DB. 2008. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res 18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang X, Feyertag F, Meehan CJ, McCormack GP, Travers SA, Craig C, Westby M, Lewis M, Robertson DL. 2015. Characterizing the diverse mutational pathways associated with R5-tropic maraviroc resistance: HIV-1 that uses the drug-bound CCR5 coreceptor. J Virol 89:11457–11472. doi: 10.1128/JVI.01384-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chalmet K, Dauwe K, Foquet L, Baatz F, Seguin-Devaux C, Van Der Gucht B, Vogelaers D, Vandekerckhove L, Plum J, Verhofstede C. 2012. Presence of CXCR4-using HIV-1 in patients with recently diagnosed infection: correlates and evidence for transmission. J Infect Dis 205:174–184. doi: 10.1093/infdis/jir714. [DOI] [PubMed] [Google Scholar]
- 46.Sede MM, Moretti FA, Laufer NL, Jones LR, Quarleri JF. 2014. HIV-1 tropism dynamics and phylogenetic analysis from longitudinal ultra-deep sequencing data of CCR5- and CXCR4-using variants. PLoS One 9:e102857. doi: 10.1371/journal.pone.0102857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raymond S, Saliou A, Delobel P, Cazabat M, Pasquier C, Jeanne N, Saune K, Massip P, Marchou B, Izopet J. 2014. Evolution of HIV-1 quasispecies and coreceptor use in cell reservoirs of patients on suppressive antiretroviral therapy. J Antimicrob Chemother 69:2527–2530. doi: 10.1093/jac/dku147. [DOI] [PubMed] [Google Scholar]
- 48.Raymond S, Saliou A, Nicot F, Delobel P, Dubois M, Carcenac R, Saune K, Marchou B, Massip P, Izopet J. 2013. Characterization of CXCR4-using HIV-1 during primary infection by ultra-deep pyrosequencing. J Antimicrob Chemother 68:2875–2881. doi: 10.1093/jac/dkt290. [DOI] [PubMed] [Google Scholar]
- 49.Swenson LC, Chui CK, Brumme CJ, Chan D, Woods CK, Mo T, Dong W, Chapman D, Lewis M, Demarest JF, James I, Portsmouth S, Goodrich J, Heera J, Valdez H, Harrigan PR. 2013. Genotypic analysis of the V3 region of HIV from virologic nonresponders to maraviroc-containing regimens reveals distinct patterns of failure. Antimicrob Agents Chemother 57:6122–6130. doi: 10.1128/AAC.01534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu WS, Temin HM. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci U S A 87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burke DS. 1997. Recombination in HIV: an important viral evolutionary strategy. Emerg Infect Dis 3:253–259. doi: 10.3201/eid0303.970301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cashin K, Paukovics G, Jakobsen MR, Ostergaard L, Churchill MJ, Gorry PR, Flynn JK. 2014. Differences in coreceptor specificity contribute to alternative tropism of HIV-1 subtype C for CD4(+) T-cell subsets, including stem cell memory T-cells. Retrovirology 11:97. doi: 10.1186/s12977-014-0097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishimura Y, Brown CR, Mattapallil JJ, Igarashi T, Buckler-White A, Lafont BA, Hirsch VM, Roederer M, Martin MA. 2005. Resting naive CD4+ T cells are massively infected and eliminated by X4-tropic simian-human immunodeficiency viruses in macaques. Proc Natl Acad Sci U S A 102:8000–8005. doi: 10.1073/pnas.0503233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Groot F, van Capel TM, Schuitemaker J, Berkhout B, de Jong EC. 2006. Differential susceptibility of naive, central memory and effector memory T cells to dendritic cell-mediated HIV-1 transmission. Retrovirology 3:52. doi: 10.1186/1742-4690-3-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai J, Agosto LM, Baytop C, Yu JJ, Pace MJ, Liszewski MK, O'Doherty U. 2009. Human immunodeficiency virus integrates directly into naive resting CD4+ T cells but enters naive cells less efficiently than memory cells. J Virol 83:4528–4537. doi: 10.1128/JVI.01910-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.