ABSTRACT
Although respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infections in infants, a safe and effective vaccine is not yet available. Live-attenuated vaccines (LAVs) are the most advanced vaccine candidates in RSV-naive infants. However, designing an LAV with appropriate attenuation yet sufficient immunogenicity has proven challenging. In this study, we implemented reverse genetics to address these obstacles with a multifaceted LAV design that combined the codon deoptimization of genes for nonstructural proteins NS1 and NS2 (dNS), deletion of the small hydrophobic protein (ΔSH) gene, and replacement of the wild-type fusion (F) protein gene with a low-fusion RSV subgroup B F consensus sequence of the Buenos Aires clade (BAF). This vaccine candidate, RSV-A2-dNS-ΔSH-BAF (DB1), was attenuated in two models of primary human airway epithelial cells and in the upper and lower airways of cotton rats. DB1 was also highly immunogenic in cotton rats and elicited broadly neutralizing antibodies against a diverse panel of recombinant RSV strains. When vaccinated cotton rats were challenged with wild-type RSV A, DB1 reduced viral titers in the upper and lower airways by 3.8 log10 total PFU and 2.7 log10 PFU/g of tissue, respectively, compared to those in unvaccinated animals (P < 0.0001). DB1 was thus attenuated, highly immunogenic, and protective against RSV challenge in cotton rats. DB1 is the first RSV LAV to incorporate a low-fusion F protein as a strategy to attenuate viral replication and preserve immunogenicity.
IMPORTANCE RSV is a leading cause of infant hospitalizations and deaths. The development of an effective vaccine for this high-risk population is therefore a public health priority. Although live-attenuated vaccines have been safely administered to RSV-naive infants, strategies to balance vaccine attenuation with immunogenicity have been elusive. In this study, we introduced a novel strategy to attenuate a recombinant RSV vaccine by incorporating a low-fusion, subgroup B F protein in the genetic background of codon-deoptimized nonstructural protein genes and a deleted small hydrophobic protein gene. The resultant vaccine candidate, DB1, was attenuated, highly immunogenic, and protective against RSV challenge in cotton rats.
INTRODUCTION
Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infections in infants (1). Globally, RSV causes an estimated 3.4 million (1) hospitalizations and 234,000 deaths per year in children under the age of 5 years (2). Almost all children have been infected with RSV by the age of 2 years, with clinical manifestations ranging from upper respiratory tract infections to pneumonia with respiratory failure. Despite the striking burden of RSV disease in children worldwide, no RSV-specific therapies or vaccines are commercially available. The development of a safe and effective RSV vaccine is therefore a public health priority.
The initial attempt to develop an RSV vaccine by formalin inactivation (FI-RSV) not only failed to protect against infection but also primed RSV-naive infants for enhanced respiratory disease upon natural infection (3). Subsequent animal studies also demonstrated enhanced disease following vaccination with some RSV protein-based vaccines (4, 5). Although many protein-based vaccines have not caused enhanced disease in animals, the risk of this outcome has hindered their administration to seronegative infants to date. In contrast, RSV live-attenuated vaccines (LAVs) have never been associated with enhanced disease in animal models or in humans (6). Thus, LAVs are the only RSV vaccines which have been safely administered to the target population of RSV-naive infants. LAVs offer multiple advantages over nonreplicating vaccines, including intranasal administration and the ability to broadly stimulate cellular and humoral immune responses. However, one major limitation of LAVs is the relatively poor immunogenicity and incomplete protection conferred by natural RSV infection. A successful LAV must therefore maintain its immunogenicity yet be sufficiently attenuated so as not to cause symptoms in recipients.
RSV reverse genetics has enabled the rational design of LAVs which incorporate genetic modifications designed to balance attenuation and immunogenicity. One such genetic modification we recently described is the codon deoptimization of RSV nonstructural proteins NS1 and NS2 (dNS), which are virulence proteins that antagonize the host interferon responses (7). Codon deoptimization (8, 9) and codon pair deoptimization (10, 11) are strategies to decrease viral protein expression by incorporating the least used codons or least used codon pairs in the human genome, respectively. In previous studies, deletion of NS1 was overattenuating in nonhuman primates (12), whereas deletion of NS2 was underattenuating (13). However, we demonstrated that codon deoptimization reduced expression of NS1 and NS2 by 70 to 90%, which resulted in an LAV that was moderately attenuated in vivo, with enhanced immunogenicity in mice compared to that of wild-type virus (7). Similarly, the deletion of the small hydrophobic (SH) protein has been introduced into multiple RSV LAV candidates because of its potential to attenuate site-specific viral replication in vivo (13, 14) without compromising immunogenicity (14). Importantly, the codon deoptimization of nonstructural proteins and the deletion of SH do not attenuate viral replication in Vero cells, which could allow for LAV production in this cell line (7, 14, 15).
In this study, our objective was to implement reverse genetics to design an RSV LAV which was both attenuated and immunogenic. To accomplish this, we first identified an RSV subgroup B F protein consensus sequence of the Buenos Aires clade (BAF) with poor fusogenicity compared to that of wild-type F protein. We then incorporated BAF into the genetic background of RSV-A2 with codon-deoptimized nonstructural protein genes and a deletion of the small hydrophobic protein gene. The resultant vaccine candidate, RSV-A2-dNS-ΔSH-BAF (DB1), was attenuated, highly immunogenic, and protective against RSV A challenge in cotton rats. Additionally, DB1 elicited broadly neutralizing antibodies against a novel panel of recombinant reporter viruses representing diverse RSV A and B subgroup isolates.
MATERIALS AND METHODS
Cell lines and primary cell cultures.
HEp-2 (ATCC CCL-23), Vero (ATCC CCL-81), and 293T (ATCC, CRL-3216) cells were maintained in minimal essential medium (MEM) with Earle's salts and l-glutamine (Gibco) supplemented with 10% fetal bovine serum (FBS; HyClone) and 1 μg/ml of penicillin, streptomycin sulfate, and amphotericin B solution (PSA; Life Technologies, Grand Island, NY). BEAS-2B cells were maintained in RPMI 1640 (Cellgro) with 10% FBS and 1 μg/ml of PSA as described previously (16). BSR T7/5 cells (a gift from Ursula Buchholz, National Institutes of Health, Bethesda, MD) were cultured in Glasgow's minimal essential medium (GMEM) containing 10% FBS, 1 μg/ml of PSA, and, every other passage, Geneticin at 1 mg/ml as described previously (17).
Normal human bronchial epithelial (NHBE) cells (Lonza, Allendale, NJ) were cultured at the air-liquid interface (ALI) according to the recommended protocols as described previously (7). Human airway epithelial (HAE) cells were isolated from airway specimens of patients without underlying lung disease by the University of North Carolina (UNC) Marsico Lung Institute Tissue Culture Core, which obtained airway specimens with informed consent under UNC at Chapel Hill Institutional Review Board-approved protocols from the National Disease Research Interchange (NDRI; Philadelphia, PA). Primary cells were expanded on plastic for one passage and plated at a density of 3 × 105 per well on permeable Transwell-Col supports. HAE cell cultures were grown at an ALI for 4 to 6 weeks to form differentiated, polarized cultures, as described previously (18).
Cloning and rescue of recombinant viruses.
To generate the BAF consensus sequence, the full-length F gene nucleotide sequences from 5 BA isolates deposited under GenBank accession numbers JF714712, JN032115, JN032117, JN032119, and JN032120 were obtained (http://www.ncbi.nlm.nih.gov/GenBank/index.html). These sequences were aligned using MegAlign within the DNASTAR, Inc., Lasergene Suite (Madison, WI), and the consensus BAF gene was synthesized by GeneArt (Thermo Fisher Scientific, Inc., Waltham, MA).
We next generated recombinant viruses kRSV-A2, kRSV-A2-line19F, kRSV-A2-BAF, kRSV-A2-dNS-ΔSH-BAF (kDB1), and RSV-A2-dNS-ΔSH-BAF (DB1) and a panel of viruses expressing mKate2 and the G and F surface glycoproteins of diverse RSV clinical isolates within an A2 backbone using reverse genetics as previously described (17). To do this, we made modifications to the pSynkRSV-line19F bacterial artificial chromosome (BAC), which contains the antigenome of kRSV-A2-line19F under the control of a T7 promoter with far-red fluorescent protein monomeric Katushka-2 (mKate2) in the first gene position to facilitate virus detection and quantification (17). We previously described modifications of pSynkRSV-A2-line19F to yield pSynkRSV-A2 and pSynkRSV-A2-dNS, which produce kRSV-A2 and kRSV-A2-dNS, respectively, where dNS represents complete codon deoptimization of NS1 and NS2 incorporating the least used codons in humans (7). To generate kRSV-A2-BAF, we cloned the BAF gene into pSynkRSV-A2 using SacII and SalI restriction sites.
To generate kDB1, we first removed the SH gene from pSynkRSV-A2-dNS using recombination-mediated mutagenesis (recombineering) with positive and negative selection of the galactokinase expression cassette (galK) as previously described (17, 19). To generate the galK casette, the following oligonucleotides were synthesized (Integrated DNA Technologies, Coralville, IA): dSH50f (5′-AGATCTAGTACTCAAATAAGTTAATAAAAAATATACACATGGACGTCCATCCTGTTGACAATTAATCATCGGCA-3′) and dSH50r (5′-GTCTTAGCGGTGCGTTGGTCCTTGTTTTTGGACATGTTTGCATTTGCCCCTCAGCACTGTCCTGCTCCTT-3′). We then amplified the sequence using PCR to generate the galK cassette flanked by RSV homology arms. Upon recombination, the galK cassette replaced the SH sequence to be deleted. For the second recombination step, to replace the galK cassette with the desired SH deletion, we used annealed oligonucleotide adapters with the following sequences: dSH100f (5′-AGATCTAGTACTCAAATAAGTTAATAAAAAATATACACATGGACGTCCATGGGGCAAATGCAAACATGTCCAAAAACAAGGACCAACGCACCGCTAAGAC-3′) and dSH100r (5′-GTCTTAGCGGTGCGTTGGTCCTTGTTTTTGGACATGTTTGCATTTGCCCCATGGACGTCCATGTGTATATTTTTTATTAACTTATTTGAGTACTAGATCT-3′). Following recombineering, the deletion of the SH gene from pSynkRSV-A2-dNS was confirmed by sequencing.
We then used SacII-SalI restriction sites to clone the synthetic BAF sequence in place of the A2F sequence to generate pSynkRSV-A2-dNS-ΔSH-BAF, which produced kDB1. A version of DB1 without the mKate2 gene was also generated from pSynkRSV-dNS-ΔSH-BAF by excising the coding region containing the mKate2 gene with KpnI and AvrII. The resultant fragment containing mKate2 flanked by identical BlpI sites was then excised using BlpI, and the flanking fragments were ligated to generate pSynkRSV-dNS-ΔSH-BAF without mKate2.
To construct the panel of recombinant viruses expressing mKate2 and clinical G and F surface glycoproteins within an A2 backbone, we selected 6 distinct RSV clinical strains with published G and F nucleotide sequences representing both A and B subgroups isolated from diverse temporal and geographic settings. The strains included (GenBank numbers are in parentheses) RSV A strains RSVA/human/USA/A1998-12-21 (JX069802), A/2001/2-20 (JF279545 and JF279544), Riyadh 91/2009 (JF714706 and JF714710), and RSV B strains NH1276 (JQ680988 and JQ736678), 9320 (AY353550), and TX11-56 (JQ680989 and JQ736679). We obtained the synthetic G and F nucleotide sequences of these clinical isolates, flanked by SacI-SacII and SacII-SalI restriction sites, respectively. We used these restriction sites to clone the corresponding G and F genes into pSynkRSV-A2-line19F. We also generated a recombinant RSV strain without G protein expression (kRSV-A2-220F-G-stop) as previously described (20, 21).
We recovered all recombinant viruses by cotransfecting the RSV antigenomic BACs with four human codon-optimized helper plasmids that expressed RSV N, P, M2-1, or L protein into BSR T7/5 cells (17). Master and working virus stocks of vaccine strains were subsequently propagated and harvested in Vero cells (7). The panel of viruses expressing clinical G and F surface glycoproteins was propagated and harvested in HEp2 cells. For cotton rat studies, RSVA/Tracy and RSVB/18537 stocks were generated in HEp-2 cells as previously described (22).
DSP fusion assay.
A dual-split-protein (DSP) cell-to-cell fusion assay was used to quantify fusion activity of expressed RSV F proteins as described previously (23–25). The consensus BAF gene was obtained in mammalian codon optimized form from GeneArt and cloned into pcDNA3.1. Subconfluent 293T cells in 12-well plates were transfected using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA). One population of cells was transfected with DSP1-7 and a plasmid encoding the RSV F protein of interest (BAF, A2F, or line19F, all codon optimized), while another population of cells was transfected with DSP8-11. All plasmids were transfected at a concentration of 1 μg per well in the presence of the fusion inhibitor BMS-433771 (provided by Jin Hong, Alios Biopharma, San Francisco, CA) (26). Twenty-four hours posttransfection, cells were resuspended, mixed at a 1:1 ratio, and supplemented with EnduRen live-cell luciferase substrate (Promega, Madison, WI) at 34 μg/ml in a 96-well opaque plate. Mixed cells were maintained at 37°C, and luciferase activity was quantified using a TopCount NXT microplate scintillation and luminescence counter (PerkinElmer, Waltham, MA) at the indicated time points.
In vitro growth analyses.
Subconfluent BEAS-2B cells in 6-well plates were infected in triplicate with either kRSV-A2 or kRSV-BAF at a multiplicity of infection (MOI) of 0.01 fluorescent focus unit (FFU)/cell in a volume of 500 μl. After rocking at room temperature for 1 h, the inoculum was removed, the cells were washed with phosphate-buffered saline (PBS), 2 ml of complete growth medium was added to each well, and plates were incubated at 37°C in 5% CO2. Then, at specified time points postinfection (p.i.), cells were scraped into medium and aliquots were stored at −80°C until analysis.
Titrations were performed by FFU assay as previously described (7), with slight modifications. Briefly, HEp-2 cells at 70% confluence in 96-well plates were inoculated in duplicate with 50 μl of 10-fold serial dilutions of samples. Plates were spinoculated at 2,095 × g for 30 min at 4°C, and 0.75% methylcellulose (EMD, Gibbstown, NJ) dissolved in MEM supplemented with 10% FBS and 1% PSA was added. Cells were incubated for 48 h at 37°C in 5% CO2 before counting of FFU per well. The limit of detection was defined as 2 FFU per well, corresponding to 40 FFU/ml.
Similarly, subconfluent Vero cells in 6-well plates were infected in triplicate with kRSV-A2-line19F or kDB1 at an MOI of 0.01 FFU/cell. Following the above-described protocol, cells were scraped into medium at specified time points p.i. and aliquots were stored at −80°C until titration by FFU assay.
Differentiated NHBE cells from two donors were infected at an MOI of 2.6 FFU/cell. Virus inoculum (100 μl) was applied apically after PBS wash, and samples were incubated for 2 h at 37°C. The inoculum was then removed by three apical PBS washes. To collect virus for specified time points p.i., differentiated medium without an inducer (150 μl) was added to the apical chamber and incubated for 10 min at 37°C. This step was repeated twice for each well (300 μl total), and the apical supernatant was snap-frozen in liquid nitrogen and stored at −80°C until analysis (7).
HAE cells from two donors were infected with an MOI of 6.0 50% tissue culture infective doses (TCID50)/cell after washing the apical surfaces with PBS and supplying fresh medium to the basolateral compartments. Cells were incubated for 2 h at 37°C. The inoculum was then removed, and cells were washed three times for 5 min each with PBS and incubated at 37°C. Virus released into the apical compartment was harvested by performing apical washes with 425 μl of medium for 30 min at 37°C. Samples were collected on days 1 to 7 p.i. and stored at −80°C until analysis (18).
Western blotting.
BEAS-2B cells at 70% confluence were infected with kRSV-A2line19F or kDB1 at an MOI of 1 FFU/cell or were mock infected. Cell lysates were harvested at 24 h p.i. in radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich, St. Louis, MO) and were cleared by centrifugation at 12,000 × g for 5 min. Proteins were separated by SDS-PAGE and were transferred to polyvinylidene fluoride (PVDF) membranes. Western blot analyses were performed by sequential probing with polyclonal rabbit antisera against NS1 and NS2 (gifts from Michael Teng, USF Health), motavizumab (anti-RSV F; provided by Nancy Ulbrandt, MedImmune), or D14 (anti-RSV N; a gift from Edward Walsh, University of Rochester) as a loading control, followed by the appropriate peroxidase-conjugated anti-rabbit, anti-human, or anti-mouse secondary antibodies (Jackson ImmunoResearch, West Grove, PA). The chemiluminescent signal was developed using Western Bright Quantum substrate (Advansta, Menlo Park, CA) and detected on a ChemiDoc XRS analyzer (Bio-Rad, Hercules, CA). Antibodies were stripped from membranes using Restore Western protein stripping buffer (Thermo Scientific Inc., Waltham, MA) and reprobed.
Animals.
Inbred Sigmodon hispidus cotton rats were obtained from colonies maintained at Sigmovir Biosystems, Inc. (Rockville, MD) for kA2-BAF and histopathology experiments or Baylor College of Medicine (Houston, TX) for DB1 experiments. Four- to 8-week-old animals were used for all experiments. Animals were housed in large polycarbonate cages and were fed a standard diet of rodent chow and water ad libitum. All studies were conducted under applicable laws and guidelines and after approval from the Sigmovir Biosystems, Inc., or Baylor College of Medicine Institutional Animal Care and Use Committee, respectively.
Viral attenuation in cotton rats.
Cotton rats (5 per group) were lightly anesthetized with isoflurane and inoculated intranasally (i.n.) with 105 FFU of either kRSV-A2-line19F or kRSV-A2-BAF. Following CO2 euthanasia on day 4 p.i., the left lobe of the lung was homogenized in 1 ml of Hanks balanced salt solution (HBSS) plus 10% sucrose-phosphate-glutamate plus 1% amphotericin B (Fungizone) plus 0.1% gentamicin for determination of lung viral titers as previously described (27).
To measure lung lavage and nasal wash titers of viruses kRSV-A2 and kDB1, groups of 3 cotton rats were similarly inoculated i.n. with 105 PFU of virus. On day 4 p.i., animals were sacrificed and nasal washes and lung lavages were performed. For nasal washes, the jaws were disarticulated, the head was removed, and 1 ml of Iscove's medium with 15% glycerin mixed with 2% FBS-MEM (1:1, vol/vol) was pushed through each naris (total of 2 ml). For lung lavages, the left lung and one of the large lobes of the right lung were removed and rinsed in sterile water to remove external blood contamination. The lung lobes were transpleurally lavaged using 3 ml of Iscove's medium with 15% glycerin mixed with 2% FBS-MEM. Fluid was collected and stored on ice until analysis. Viral titration of these specimens was performed by standard plaque assays using 24-well tissue culture plates as described previously (22). The plaques in wells containing between 20 and 100 plaques were enumerated, the average was obtained, and virus titers were calculated as the total log10 number of PFU for nasal wash fluid or log10 number of PFU/g of tissue for lungs. The lower limit of detection by this method is 0.7 log10 total PFU or approximately 1.4 log10 PFU/g of lung tissue, respectively.
Evaluation for enhanced histopathology in cotton rats.
To evaluate for enhanced histopathology attributable to vaccination following challenge, we vaccinated groups of 5 cotton rats intranasally with 105 FFU of either kDB1 or MEM (mock treatment), or we intramuscularly injected them with 100 μl of FI-RSV (lot 100; 1:125). On day 21 p.i., we boosted FI-RSV-vaccinated rats with a second identical vaccination. Then, on day 42 p.i., we challenged all rats intranasally with 106 FFU (100 μl) of kRSV-A2-line19F. Six days later, we harvested the lungs and scored them for histopathology as previously described (28). Briefly, we inflated the lungs intratracheally with 10% neutral buffered formalin, embedded specimens in paraffin, cut 4-μm sections, and stained them with hematoxylin and eosin. We performed blinded histopathologic scoring for four parameters: peribronchiolitis (inflammatory cells, primarily lymphocytes, surrounding bronchioles), perivasculitis (inflammatory cells surrounding blood vessels), interstitial pneumonitis (increased thickness of alveolar walls associated with inflammatory cells), and alveolitis (inflammatory cells within alveolar spaces). Each parameter was scored separately for each histopathologic section, with a maximum value of 4 and a minimum of 0 (28).
Viral immunogenicity and efficacy in cotton rats.
For immunogenicity and efficacy studies, groups of 6 cotton rats each were inoculated i.n. with 105 PFU of RSVA/Tracy or DB1. Prior to infection on day 0, blood was obtained from the orbital plexus of one cotton rat in each group. On days 28, 42, and 46 p.i., blood was obtained from all of the cotton rats. On day 42 p.i., cotton rats were challenged i.n. with 1.35 × 105 PFU (100 μl) of RSVA/Tracy. Lung lavage and nasal wash specimens were obtained and titrated 4 days postchallenge as described above.
Tests for serum neutralizing antibody (nAb) responses to RSVA/Tracy and RSVB/18537 were performed in 96-well plates with HEp-2 cells. Serum samples were heat inactivated at 56°C for 30 min, and serial 2-fold dilutions in duplicates starting at 3 log2 were performed. The nAb titer was defined as the serum dilution at which ≥50% reduction in viral cytopathic effect (CPE) was observed. CPE was defined as tissue destruction and was determined visually after the wells were fixed with 10% neutral buffered formalin and stained with crystal violet. Neutralizing antibody titers were categorical log numbers, and the lowest detectable nAb titer was 2.5 log2. Samples with nondetectable nAb titers were assigned a value of 2 log2.
Tests for serum nAb to the panel of RSV strains expressing mKate2 and G and F surface glycoproteins from clinical strains on days 0, 28, and 42 p.i. were performed by an FFU assay as previously described (7). Briefly, serum samples were heat inactivated at 56°C for 30 min. Serial 2-fold dilutions were performed in duplicate starting at 2 log2, and equal volumes were added to 50 to 100 FFU of each of the RSV strains on the panel. The virus and serum mixtures were incubated at 37°C for 1 h. Then, 50 μl of each mixture was transferred onto subconfluent HEp-2 cell monolayers in 96-well plates in duplicate. Plates were spinoculated at 2,095 × g for 30 min at 4°C, and FFU were counted after 48 h of incubation at 37°C and 5% CO2. The 50% effective concentration (EC50) was calculated using nonlinear regression analysis with four-parameter fitting in GraphPad Prism version 6.0. Nondetectable nAb titers were assigned a value of 2 log2.
Serum anti-RSV IgG antibodies on day 46 p.i. were determined by enzyme immunoassay (EIA) against RSV A2 lysate or mock control lysate. First, 96-well plates were coated with lysate and incubated at room temperature overnight. After blocking in 5% bovine serum albumin (BSA) in PBS containing 0.05% Tween 20 (PBST) for 1 h at room temperature, 100-μl volumes of 2-fold serial dilutions of serum samples (ranging from 1:512 to 1:65,536) in PBST were added to appropriate wells and incubated for 2 h at room temperature. Next, plates were washed 3 times with PBST and incubated for 1 h at 37°C with horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin antibody (diluted 1:1,000; Thermo Fisher Scientific, Waltham, MA). After washing 3 times with PBST, color was developed with an R&D Systems (Minneapolis, MN) substrate reagent pack (stabilized hydrogen peroxide and stabilized tetramethylbenzidine) as indicated by the manufacturer. The enzyme-substrate reaction was terminated by the addition of a sulfuric acid solution, and absorbance was measured at a wavelength of 450 nm. The cutoff value to determine antibody titer was chosen as the mean optical density (OD) + 1 standard deviation (SD) for the lowest dilution (1:512) of serum from mock-infected cotton rats. The highest serum dilution with absorbance above the cutoff was defined as the antibody titer.
Anti-RSV IgA antibodies from cotton rat nasal washes obtained on day 46 p.i. were similarly determined by EIA against RSV A2 or mock stock lysates. After coating plates overnight and blocking in 5% BSA, 100-μl volumes of 2-fold serial dilutions of nasal wash samples (ranging from 1:1 to 1:128) in PBST were added to appropriate wells and incubated for 2 h at room temperature. Wells were then washed with PBST and incubated for 1 h at 37°C with HRP-conjugated goat anti-mouse IgA (Southern Biotech, Birmingham, AL). Plates were then washed and incubated with R&D Systems substrate reagents as described above. The enzyme-substrate reaction was terminated with a sulfuric acid solution, and absorbance was measured at 450 nm. The cutoff to determine antibody titer was again chosen as the mean OD + 1 SD of the lowest dilution (1:1) of nasal wash from mock-infected cotton rats. The highest nasal wash dilution with absorbance above the cutoff was defined as the antibody titer.
Statistical analyses.
All statistical analyses were performed using GraphPad Prism software version 6.0 (San Diego, CA). Data are presented as means with SDs or standard errors of the means (SEMs) as indicated below. Student's t test and one-way or two-way analyses of variance (ANOVA) were used as indicated below, with Tukey's post hoc multiple-comparison test. For all analyses, a P value of 0.05 was considered significant.
RESULTS
Identification and characterization of a low-fusion F protein.
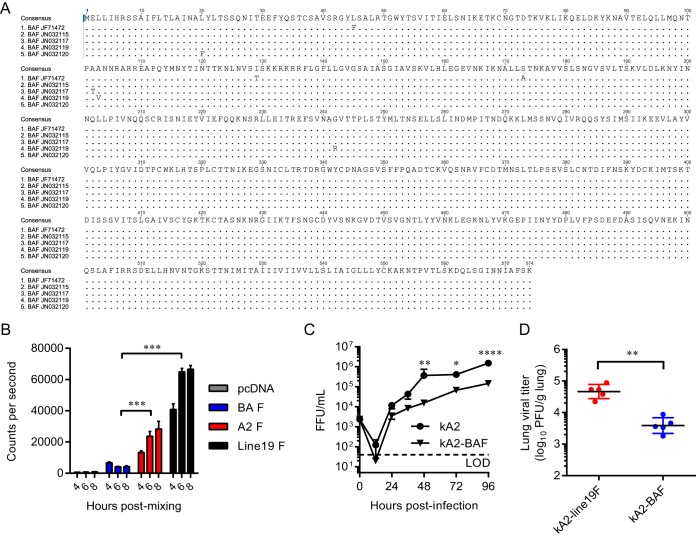
We generated an RSV F protein consensus sequence of the Buenos Aires (BA) clade by aligning the full-length F nucleotide sequences from 5 BA clinical isolates in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) (Fig. 1A). Using a dual-split protein (DSP) cell-to-cell fusion assay with 293T cells as previously described (23–25), we found that BAF, when expressed alone, was poorly fusogenic compared to A2F and line19F proteins (Fig. 1B). Based on these results, we hypothesized that BAF may confer attenuation to a vaccine candidate based on its low fusogenicity. To test this hypothesis, we cloned BAF in place of A2F in a bacterial artificial chromosome (BAC) construct containing the antigenome of strain kRSV-A2, where k represents the red fluorescent protein mKate2 gene (17). We rescued this recombinant virus and then performed multistep growth curve analyses with BEAS-2B cells to compare growth of kRSV-A2-BAF to that of kRSV-A2 in vitro. Although the two viruses replicated to similar levels at earlier time points p.i., growth of kRSV-A2-BAF was attenuated at 48, 72, and 96 h p.i. (Fig. 1C). We then measured the effects of BAF on viral attenuation in vivo by infecting groups of 5 cotton rats intranasally with 105 FFU of either kRSV-A2-line19F or kRSV-A2-BAF and measuring the lung viral titer 4 days later. kRSV-A2-BAF was attenuated by 1.1 log10 PFU/g of lung (P = 0.0054) compared to kRSV-A2-line19F in the lower respiratory tracts of the cotton rats (Fig. 1D). Taken together, these results demonstrated that BAF was poorly fusogenic and attenuating in vitro and in vivo.
FIG 1.
Fusion activity of the Buenos Aires F protein (BAF) and its effect on attenuation in vitro and in cotton rats. (A) The consensus sequence of the BAF protein was obtained by alignment of 5 clinical isolates from the GenBank database. Disagreements from the consensus sequence are indicated. (B) BAF fusion activity was measured using a dual-split protein (DSP) cell-to-cell fusion assay. A representative graph from three experimental replicates shows means + SEMs of 4 replicate wells. ***, P < 0.0005 using two-way ANOVA and Tukey multiple-comparison test. (C) BEAS-2B cells were infected at an MOI of 0.01 FFU/cell with either kRSV-A2 or kRSV-A2-BAF. Growth curve data are presented as means ± SEMs from 3 experimental replicates titrated in triplicate. *, P < 0.05; **, P < 0.005; ****, P < 0.00005 using two-way ANOVA and Tukey multiple-comparison test. LOD, limit of detection. (D) Groups of 5 cotton rats were infected with either kRSV-A2-line19F or kRSV-A2-BAF. On day 4 postinfection (p.i.), lungs were harvested and viral load was measured by standard plaque assay. Each dot represents one animal, and data represent means ± SDs. **, P < 0.005 by two-tailed Student's t test.
Design of RSV live-attenuated vaccine DB1 and expression of viral proteins.
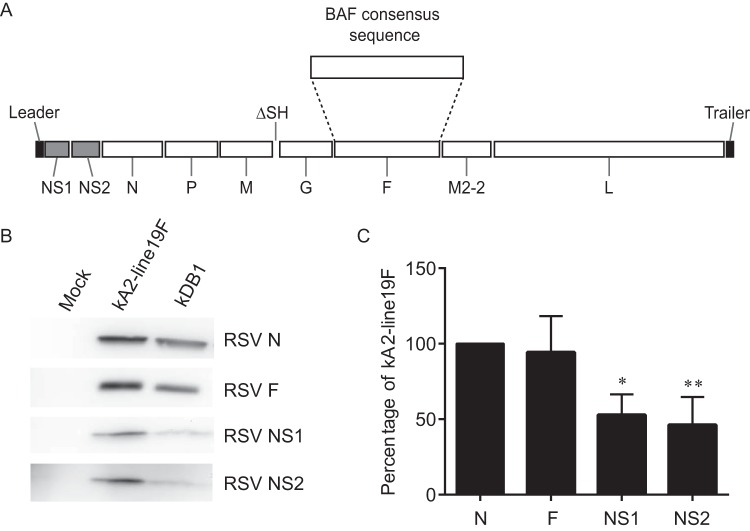
We next generated an RSV LAV candidate, DB1, by replacing the A2F gene with BAF, codon deoptimizing the genes for the nonstructural proteins NS1 and NS2 (dNS) (7), and deleting the small hydrophobic protein gene (ΔSH) in an RSV-A2 backbone (Fig. 2A). Our rationale for this vaccine design was to combine the novel low-fusion BAF protein with two attenuating genetic modifications known to preserve immunogenicity, namely, the codon deoptimization of NS1 and NS2 and the deletion of SH protein. We generated DB1 by cloning BAF in place of A2F in the bacterial artificial chromosome (BAC) construct containing the antigenome of RSV-A2-dNS-ΔSH. We similarly generated an antigenomic version of DB1 containing mKate2 in the first gene position (kDB1) and rescued these recombinant viruses (17). To analyze viral protein levels, we performed Western blotting on lysates from infected HEp-2 cells. The results demonstrated that expression levels of NS1 and NS2 proteins in kDB1-infected cells were less than in kRSV-A2-line19F-infected cells, as expected, whereas F protein expression levels were equivalent (Fig. 2B and C).
FIG 2.
Schematic of RSV live-attenuated vaccine candidate DB1 and expression of modified genes. (A) DB1 has codon-deoptimized nonstructural protein NS1 and NS2 genes (shaded), a deletion of small hydrophobic protein (SH) gene, and a subgroup B fusion glycoprotein consensus sequence gene from the Buenos Aires clade (BAF). (B) To analyze viral protein levels, HEp-2 cells were infected with either kRSV-A2-line19F or kDB1 or were mock infected at an MOI of 1 FFU/cell. Western blotting was performed for the F, NS1, or NS2 protein using the RSV nucleoprotein (N) as a loading control. A representative blot from three experimental replicates is shown. (C) Western densitometry of F, NS1, and NS2 expression was normalized to N for all data sets. Data represent three experimental replicates, expressed as means + SDs, *, P < 0.05 by one-way ANOVA and Tukey multiple-comparison test.
DB1 viral growth kinetics in vitro.
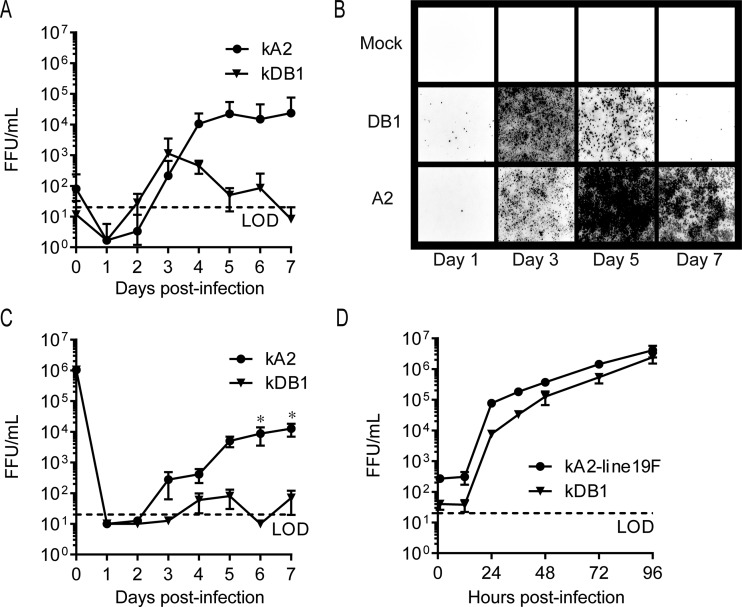
We next characterized kDB1 growth kinetics in vitro by performing multistep growth curve analyses with well-differentiated primary HAE (18) and NHBE (7) cells grown at the air-liquid interface. We employed these two models because well-differentiated human airway epithelial cells more accurately predict RSV attenuation in seronegative children than immortalized cell lines or nonhuman primates (29). In both models of well-differentiated airway epithelial cells, kDB1 was attenuated compared to kRSV-A2, with significant differences observed in NHBE cells on days 6 and 7 p.i. (Fig. 3A to C). In contrast, kDB1 had growth kinetics similar to those of kRSV-A2-line19F in Vero cells, an interferon-deficient cell line approved for vaccine production (Fig. 3D). The lack of kDB1 attenuation in Vero cells may be in part explained by the diminished impact of codon-deoptimized nonstructural proteins on this interferon-deficient cell line. In summary, these data demonstrated that kDB1 was attenuated in primary cells in vitro but grew to sufficient levels for rescue and production in Vero cells.
FIG 3.
kDB1 viral growth kinetics in vitro. Human airway epithelial (HAE) cells (A and B), normal human bronchial epithelial (NHBE) cells (C), or Vero cells (D) were infected at an MOI of 6 TCID50/cell, 2.6 FFU/cell, or 0.01 FFU/cell, respectively, with either kDB1, kRSV-A2, or kRSV-A2-line19F. Virus titers were measured at each time point by FFU assay. Growth curve data are presented as means ± SEMs of 2 or 3 experimental replicates. Representative photomicrographs of infected HAE cells show mKate2 positivity (with red color converted to black). *, P < 0.05 using two-way ANOVA and Tukey multiple-comparison test.
DB1 attenuation and histopathology in cotton rats.
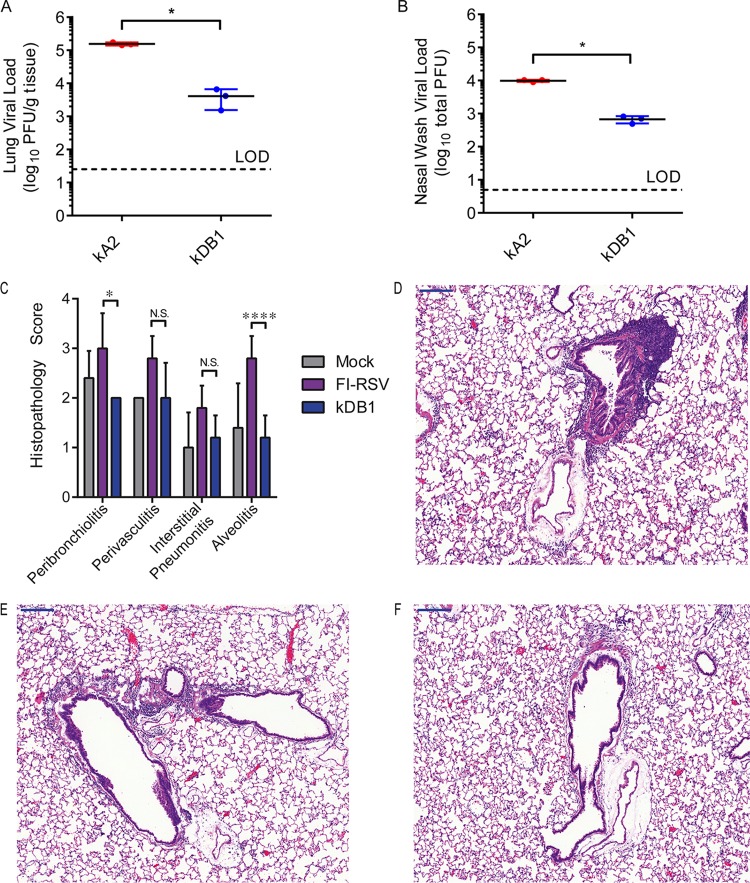
To determine the level of DB1 attenuation in vivo in cotton rats, we infected groups of 3 animals with either DB1 or RSV A2 (Fig. 4A and B). On day 4 p.i., we measured viral titers in lung lavage and nasal wash specimens. The DB1 titers were 1.7 log10 total PFU (P = 0.0208) and 1.2 log10 PFU/g lung (P = 0.0022) lower than those of RSV A2 in the lung lavage and nasal wash, respectively. These results demonstrated that DB1 was attenuated in cotton rats compared to RSV A2.
FIG 4.
kDB1 attenuation and histopathology following challenge in cotton rats. Groups of 3 cotton rats were inoculated intranasally (i.n.) with either kRSV-A2 or kDB1. Lung lavage (A) and nasal wash (B) were performed on day 4 p.i., and viral titers were measured by plaque assay. To evaluate for vaccine-attributable enhanced disease postchallenge, groups of 5 cotton rats were inoculated i.n. with either a control or kDB1 or intramuscularly (i.m.) with FI-RSV. Animals vaccinated with FI-RSV also received a boost on day 21 p.i. All animals were challenged with kRSV-A2-line19F on day 46 p.i., lungs were harvested 6 days later, and histopathology scores were determined (C). Representative hemotoxylin and eosin stains for FI-RSV-vaccinated (D), mock-vaccinated (E), and kDB1-vaccinated (F) rats are shown. Data are presented as means + SDs. *, P < 0.05.
To evaluate for enhanced disease attributable to vaccination following challenge, we next vaccinated groups of 5 cotton rats intranasally with kDB1 or a control, or intramuscularly with FI-RSV (lot 100; 1:125). On day 21 p.i., we boosted FI-RSV-vaccinated rats with a second identical vaccination. On day 42 p.i., we challenged all rats intranasally with 106 FFU of kRSV-A2-line19F. Six days later, we harvested the lungs for histopathology and performed scoring for peribronchiolitis, perivasculitis, interstitial pneumonitis, and alveolitis (Fig. 4C to F). The average histopathology scores for kDB1-vaccinated rats were statistically equivalent to those obtained with mock treatment for all four parameters. kDB1-vaccinated rats also had significantly lower histopathology scores than FI-RSV-vaccinated rats for peribronchiolitis (P = 0.0155) and alveolitis (P < 0.0001). These results demonstrated that kDB1 does not cause enhanced disease following challenge in the standard cotton rat model.
DB1 immunogenicity in cotton rats.
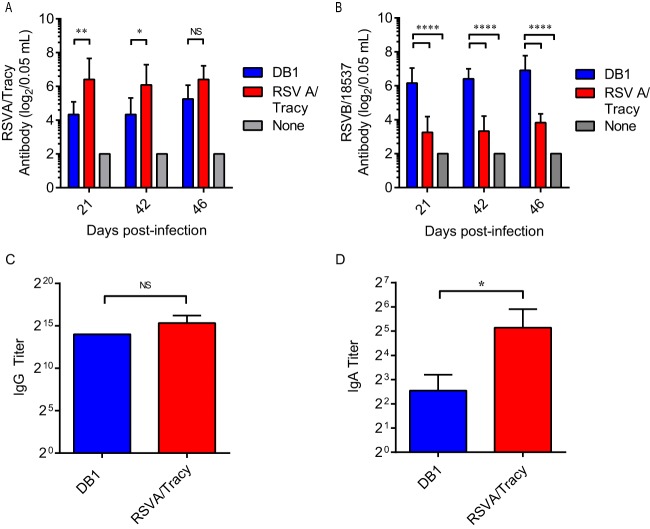
To analyze DB1 immunogenicity in cotton rats, we mock infected rats or infected groups of 6 cotton rats with 105 PFU of DB1 or RSVA/Tracy (GA1 genotype) and measured serum titers of nAb against RSVA/Tracy and RSVB/18537 by microneutralization assays on days 21, 42, and 46 p.i. (Fig. 5A and B). DB1 generated nAb to both RSVA/Tracy and RSVB/18537, and DB1 generated significantly higher serum titers of nAb to RSVB/18537 than did RSVA/Tracy.
FIG 5.
DB1 immunogenicity in cotton rats. Groups of 6 cotton rats were infected i.n. with either RSVA/Tracy, DB1, or a control. Serum titers of neutralizing antibody against RSVA/Tracy (A) and RSVB/18537 (B) were measured at the indicated days postinfection by a microneutralization assay. Serum obtained on day 46 p.i. was then pooled and analyzed by IgG ELISA to RSV-A2 (C). Total IgG titers are represented as means + SDs of three pooled replicates. Total IgA titers of individual nasal wash specimens collected on day 46 p.i. were also analyzed by IgA ELISA to RSV-A2 (D). Data represent means + SDs. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 by two-way ANOVA with Tukey multiple-comparison test (A and B) or two-tailed Student's t test (C and D). NS, not significant.
We then pooled cotton rat serum samples from day 46 p.i. and measured anti-RSV IgG binding antibodies via enzyme immunoassay (EIA) against kRSV-A2 (Fig. 5C). DB1 generated anti-RSV IgG titers of >213, which were statistically equivalent to titers generated by RSVA/Tracy (P = 0.4169). DB1 also generated detectable virus-specific IgA antibodies in cotton rat nasal washes obtained on day 46 p.i. (Fig. 5D).
Breadth of neutralizing antibody responses elicited by DB1.
To further characterize the spectrum of nAb generated by DB1, we designed a panel of chimeric RSV reporter strains expressing the attachment (G) and F glycoproteins of a broad range of clinical isolates in an RSV-A2 background with mKate2 in the first gene position. The panel of viruses included glycoproteins from RSV B strains TX11-56, NH1276, and 9320 and RSV A strains 1998/12-21, Riyadh 91/2009, and 2-20. These strains were chosen to represent phylogenetically distant RSV strains from both subgroups and multiple clades. We additionally generated a recombinant virus, kRSV-A2-2-20F/G-stop, which contained a premature stop codon in the G glycoprotein as previously described, in order to measure the relative effects of anti-G antibodies on neutralization (21). We also included kRSV-A2 in the panel and rescued the recombinant viruses.
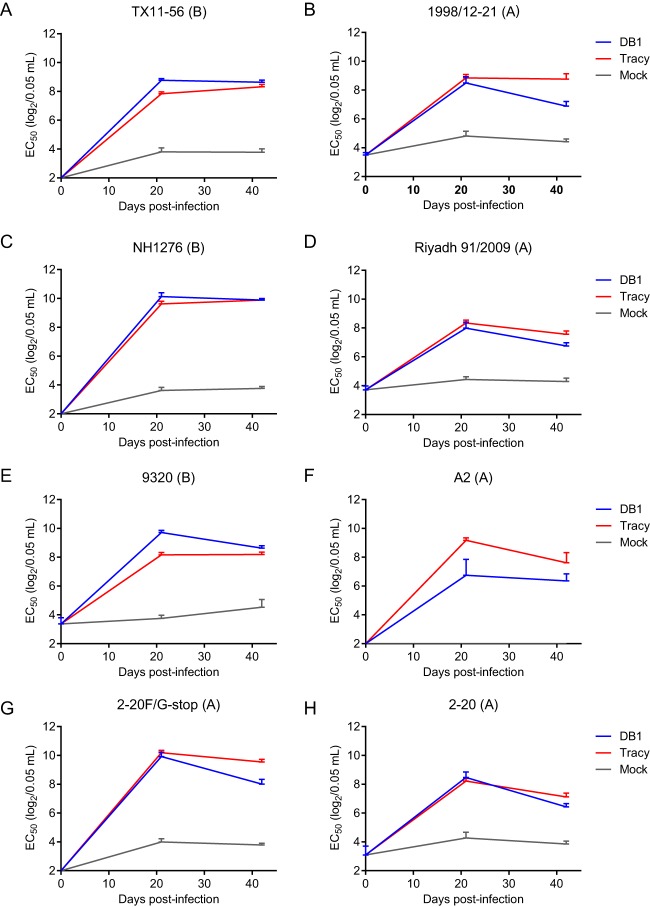
We subsequently pooled the cotton rat serum samples collected on days 0, 21, and 42 p.i. and measured nAb titers at each time point against the panel of viruses by an FFU reduction assay (Fig. 6) (7). DB1 generated broadly neutralizing Ab responses against both RSV A and B representative viruses that were similar in titer to those induced by RSVA/Tracy. Antibody titers against the chimeric clinical strains ranged from EC50s of 28 to 210 per 0.05 ml and were highest against RSV B strain NH1276, followed by B strains 9320 and TX11-56. Antibody titers were lowest against the laboratory-adapted wild-type strain RSV-A2, which may be less representative of currently circulating RSV clinical strains. Notably, nAb titers were higher against kRSV-A2-2-20F/G-stop than kRSV-A2-2-20, demonstrating that the neutralization generated by DB1 was not dependent on antibodies directed against G.
FIG 6.
Neutralizing antibody responses generated by DB1 against a panel of recombinant RSV viruses. Cotton rat sera were pooled and neutralizing antibody titers at each time point were measured against a panel of recombinant RSV viruses with the G and F surface glycoproteins from diverse clinical isolates. Data represent the EC50 + upper limit of the 95% confidence interval (CI) measured by FFU reduction assay from two experimental replicates of pooled sera. EC50, effective concentration at which 50% virus is neutralized.
Taken together, these data demonstrated that DB1 was highly immunogenic and generated broadly neutralizing Ab responses against both RSV A and B strains, with the highest titers observed against RSV B strains.
DB1 efficacy in cotton rats.
To analyze vaccine efficacy (Fig. 7), we next challenged the cotton rats described above with 1.35 × 105 PFU of RSVA/Tracy on day 42 postvaccination and measured RSVA/Tracy viral titers in lung lavage and nasal wash fluids by standard plaque assay 4 days later. In lung lavage samples (Fig. 7A), vaccination with DB1 reduced viral titers after challenge by 2.7 log10 PFU/g tissue compared to titers in untreated cotton rats (P < 0.0001). In the nasal washes, DB1 reduced titers by 3.8 log10 total PFU compared to those in untreated cotton rats (P < 0.0001), which were statistically equivalent to those of RSVA/Tracy (P = 0.3710) (Fig. 7B). DB1 was thus effective in reducing viral titers after RSV challenge by >99% in both the upper and lower airways of cotton rats.
FIG 7.
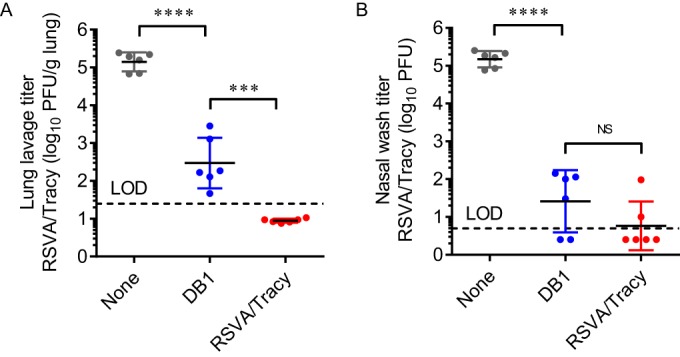
Vaccine efficacy of DB1 in cotton rats. Groups of 6 cotton rats were infected i.n. with DB1 or RSVA/Tracy or served as uninfected controls. Animals were challenged with RSVA/Tracy on day 42 p.i., and viral loads were measured 4 days later by standard plaque assays of lung lavage (A) and nasal wash (B) fluids. Each symbol represents a single animal. Data represent means ± SDs. Dotted lines indicate the limits of detection (LOD). ***, P < 0.0005; ****, P < 0.00005 by one-way ANOVA and Tukey multiple-comparison test.
DISCUSSION
We generated an RSV live-attenuated vaccine, DB1, which was both attenuated and highly immunogenic in cotton rats. This vaccine incorporated a low-fusion F protein as a novel strategy to attenuate viral replication without compromising immunogenicity. DB1 additionally incorporated two genetic modifications known to balance attenuation and immunogenicity, the codon deoptimization of nonstructural proteins (7) and the deletion of SH (13). The resultant vaccine was >10-fold attenuated in cotton rat upper and lower airways, yet it still elicited high titers of broadly neutralizing Ab to a diverse panel of RSV A and B recombinant strains. DB1 also generated mucosal immunity in the form of RSV-specific IgA antibodies in cotton rat nasal wash specimens. When vaccinated animals were challenged with RSV, DB1 reduced challenge strain titers by >99% in both the nasal wash and lung lavage specimens. Thus, DB1 was attenuated, highly immunogenic, and efficacious at protecting against RSV challenge in cotton rats.
To characterize the breadth of neutralization generated by DB1, we designed a novel panel of recombinant RSV viruses expressing mKate2 and the G and F surface glycoproteins of diverse RSV A and B clinical isolates. We found that intranasal inoculation with DB1 generated broadly neutralizing Ab against our panel of recombinant viruses, with the highest titers observed against the RSV B strains. We additionally observed subgroup-specific immune responses to the lab strain RSVB/18537 when analyzed by a standard plaque reduction neutralization test (PRNT). Notably, DB1 generated significantly higher neutralization of RSVB/18537 than did infection with wild-type RSVA/Tracy. Conversely, RSVA/Tracy also generated significantly higher neutralization of RSVA/Tracy (homologous) than did DB1 on days 28 and 42 postinfection. These subgroup-specific responses must have been attributable to differences within the fusion protein, as these were the principal antigenic differences between DB1 and RSVA/Tracy. Thus, our data implicate the fusion protein as an important mediator of subgroup-specific immunity, despite its highly conserved neutralizing epitopes (30). The data also validate that RSV elicits cross-neutralizing Ab responses to heterosubtypic viruses that are broad but variable in magnitude, which may have implications for vaccine design (31).
The titers of nAb elicited by DB1 against our panel of recombinant viruses fell in the range of what is known to be protective against lower respiratory tract infections in cotton rats. Previous studies with cotton rats demonstrated that serum nAb titers of 1:100 conferred partial pulmonary protection, whereas titers of 1:380 completely protected against lower respiratory tract disease (32). These results correlated with observations in human infants <2 months of age, which showed that maternally derived antibody titers of 1:400 offered protection against RSV bronchiolitis and pneumonia (33). The serum nAb titers elicited by DB1 in cotton rats to our panel of recombinant clinical viruses ranged from 1:256 to 1:1,024 by FFU reduction assay. In comparison, RSV LAV candidate RSV-ΔM2-2, which is under evaluation in clinical trials, generated nAb titers of 1:32 to 1:64 by the plaque reduction neutralization titer necessary to reduce the number of plaques by 60% (PRNT60) in cotton rats (34). With regard to efficacy, DB1 was >99% efficacious in reducing cotton rat nasal wash and lung lavage titers postchallenge. DB1 efficacy was thus comparable to that of palivizumab, which is dosed in humans based on the serum trough concentration necessary to reduce the cotton rat lung viral load by 99% (35).
With regard to attenuation, DB1 was highly attenuated in two models of primary human airway epithelial cells, which more accurately gauge levels of RSV LAV attenuation in humans than immortalized cell lines (29). However, the vaccine was only 15-fold attenuated in cotton rat upper airways and 45-fold attenuated in lung lavage specimens. We suspect that this discrepancy was observed because the dNS and ΔSH mutations exerted minimal effects in the cotton rat model. In fact, the BAF mutation alone conferred a reduction in viral titer in cotton rat lower airways (12-fold) nearly equivalent to those conferred by the combined mutations in DB1. This is consistent with previous studies which found that the ΔSH mutation conferred attenuation in chimpanzee lower airways (13) but not in cotton rats (15). Additionally, the dNS mutations have been characterized only in mice and primary cells (7), so their effects in cotton rats are unknown. Thus, the applicability of the cotton rat model to approximate human levels of attenuation with these specific mutations remains unclear.
In conclusion, we generated an RSV LAV DB1 that incorporated a low-fusion F protein as a novel strategy to attenuate viral replication without compromising immunogenicity. We combined this low-fusion F protein with codon-deoptimized nonstructural proteins and deleted the SH protein in an RSV-A2 backbone to generate a multifaceted vaccine that balanced attenuation and immunogenicity. DB1 also elicited broadly neutralizing antibodies against a panel of RSV A and B strains and protected against RSV challenge in cotton rats. DB1 is a promising RSV vaccine candidate that merits further investigation.
ACKNOWLEDGMENTS
We thank Ursula Buchholz and Karl-Klaus Conzelmann for BSR-T7/5 cells, Michael Teng for polyclonal rabbit antisera against NS1 and NS2, Nancy Ulbrandt for motavizumab, Edward Walsh for D-14, Jin Hong for the fusion inhibitor BMS-433771, and the Emory+Children's Pediatric Research Center Flow Cytometry Core.
We declare the following competing financial interests. J.C.G.B. is an employee of Sigmovir Biosystems, Inc., a contract research organization that uses the cotton rat model of infectious diseases. M.L.M. cofounded Meissa Vaccines, Inc., and serves as chief scientific officer for the company. C.A.R., C.C.S., A.L.H., J.M., and M.L.M. are coinventors of the RSV vaccine evaluated in this study.
C.A.R., C.C.S., B.E.G., R.J.P., A.L.H., J.M., J.C.G.B., and P.A.P. performed the experiments. S.M.M. and B.S.G. contributed reagents and to concepts of the experimental design. C.A.R. and M.L.M. designed the studies and analyzed data. C.A.R. wrote the paper.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lozano R, et al. . 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89:422–434. [DOI] [PubMed] [Google Scholar]
- 4.Murphy BR, Sotnikov AV, Lawrence LA, Banks SM, Prince GA. 1990. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3-6 months after immunization. Vaccine 8:497–502. doi: 10.1016/0264-410X(90)90253-I. [DOI] [PubMed] [Google Scholar]
- 5.Connors M, Collins PL, Firestone CY, Sotnikov AV, Waitze A, Davis AR, Hung PP, Chanock RM, Murphy BR. 1992. Cotton rats previously immunized with a chimeric RSV FG glycoprotein develop enhanced pulmonary pathology when infected with RSV, a phenomenon not encountered following immunization with vaccinia-RSV recombinants or RSV. Vaccine 10:475–484. doi: 10.1016/0264-410X(92)90397-3. [DOI] [PubMed] [Google Scholar]
- 6.Wright PF, Karron RA, Belshe RB, Shi JR, Randolph VB, Collins PL, O'Shea AF, Gruber WC, Murphy BR. 2007. The absence of enhanced disease with wild type respiratory syncytial virus infection occurring after receipt of live, attenuated, respiratory syncytial virus vaccines. Vaccine 25:7372–7378. doi: 10.1016/j.vaccine.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng J, Lee S, Hotard AL, Moore ML. 2014. Refining the balance of attenuation and immunogenicity of respiratory syncytial virus by targeted codon deoptimization of virulence genes. mBio 5:e01704-14. doi: 10.1128/mBio.01704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. 2006. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J Virol 80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns CC, Shaw J, Campagnoli R, Jorba J, Vincent A, Quay J, Kew O. 2006. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J Virol 80:3259–3272. doi: 10.1128/JVI.80.7.3259-3272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Nouën C, Brock LG, Luongo C, McCarty T, Yang L, Mehedi M, Wimmer E, Mueller S, Collins PL, Buchholz UJ, DiNapoli JM. 2014. Attenuation of human respiratory syncytial virus by genome-scale codon-pair deoptimization. Proc Natl Acad Sci U S A 111:13169–13174. doi: 10.1073/pnas.1411290111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teng MN, Whitehead SS, Bermingham A, St Claire M, Elkins WR, Murphy BR, Collins PL. 2000. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J Virol 74:9317–9321. doi: 10.1128/JVI.74.19.9317-9321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitehead SS, Bukreyev A, Teng MN, Firestone CY, St Claire M, Elkins WR, Collins PL, Murphy BR. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol 73:3438–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukreyev A, Whitehead SS, Murphy BR, Collins PL. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J Virol 71:8973–8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin H, Zhou H, Cheng X, Tang R, Munoz M, Nguyen N. 2000. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273:210–218. doi: 10.1006/viro.2000.0393. [DOI] [PubMed] [Google Scholar]
- 16.Stokes KL, Chi MH, Sakamoto K, Newcomb DC, Currier MG, Huckabee MM, Lee S, Goleniewska K, Pretto C, Williams JV, Hotard A, Sherrill TP, Peebles RS Jr, Moore ML. 2011. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J Virol 85:5782–5793. doi: 10.1128/JVI.01693-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hotard AL, Shaikh FY, Lee S, Yan D, Teng MN, Plemper RK, Crowe JE Jr, Moore ML. 2012. A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology 434:129–136. doi: 10.1016/j.virol.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickles RJ, McCarty D, Matsui H, Hart PJ, Randell SH, Boucher RC. 1998. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J Virol 72:6014–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teng MN, Whitehead SS, Collins PL. 2001. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289:283–296. doi: 10.1006/viro.2001.1138. [DOI] [PubMed] [Google Scholar]
- 21.Meng J, Hotard AL, Currier MG, Lee S, Stobart CC, Moore ML. 14 October 2015. The respiratory syncytial virus attachment glycoprotein contribution to infection depends on the specific fusion protein. J Virol doi: 10.1128/JVI.02140-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piedra PA, Wyde PR, Castleman WL, Ambrose MW, Jewell AM, Speelman DJ, Hildreth SW. 1993. Enhanced pulmonary pathology associated with the use of formalin-inactivated respiratory syncytial virus vaccine in cotton rats is not a unique viral phenomenon. Vaccine 11:1415–1423. doi: 10.1016/0264-410X(93)90170-3. [DOI] [PubMed] [Google Scholar]
- 23.Stokes KL, Currier MG, Sakamoto K, Lee S, Collins PL, Plemper RK, Moore ML. 2013. The respiratory syncytial virus fusion protein and neutrophils mediate the airway mucin response to pathogenic respiratory syncytial virus infection. J Virol 87:10070–10082. doi: 10.1128/JVI.01347-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hotard AL, Lee S, Currier MG, Crowe JE Jr, Sakamoto K, Newcomb DC, Peebles RS Jr, Plemper RK, Moore ML. 2015. Identification of residues in the human respiratory syncytial virus fusion protein that modulate fusion activity and pathogenesis. J Virol 89:512–522. doi: 10.1128/JVI.02472-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa H, Meng F, Kondo N, Iwamoto A, Matsuda Z. 2012. Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP. Protein Eng Des Sel 25:813–820. doi: 10.1093/protein/gzs051. [DOI] [PubMed] [Google Scholar]
- 26.Cianci C, Langley DR, Dischino DD, Sun Y, Yu KL, Stanley A, Roach J, Li Z, Dalterio R, Colonno R, Meanwell NA, Krystal M. 2004. Targeting a binding pocket within the trimer-of-hairpins: small-molecule inhibition of viral fusion. Proc Natl Acad Sci U S A 101:15046–15051. doi: 10.1073/pnas.0406696101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prince GA, Jenson AB, Horswood RL, Camargo E, Chanock RM. 1978. The pathogenesis of respiratory syncytial virus infection in cotton rats. Am J Pathol 93:771–791. [PMC free article] [PubMed] [Google Scholar]
- 28.Prince GA, Curtis SJ, Yim KC, Porter DD. 2001. Vaccine-enhanced respiratory syncytial virus disease in cotton rats following immunization with Lot 100 or a newly prepared reference vaccine. J Gen Virol 82:2881–2888. doi: 10.1099/0022-1317-82-12-2881. [DOI] [PubMed] [Google Scholar]
- 29.Wright PF, Ikizler MR, Gonzales RA, Carroll KN, Johnson JE, Werkhaven JA. 2005. Growth of respiratory syncytial virus in primary epithelial cells from the human respiratory tract. J Virol 79:8651–8654. doi: 10.1128/JVI.79.13.8651-8654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sande CJ, Mutunga MN, Medley GF, Cane PA, Nokes DJ. 2013. Group- and genotype-specific neutralizing antibody responses against respiratory syncytial virus in infants and young children with severe pneumonia. J Infect Dis 207:489–492. doi: 10.1093/infdis/jis700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prince GA, Horswood RL, Chanock RM. 1985. Quantitative aspects of passive immunity to respiratory syncytial virus infection in infant cotton rats. J Virol 55:517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parrott RH, Kim HW, Arrobio JO, Hodes DS, Murphy BR, Brandt CD, Camargo E, Chanock RM. 1973. Epidemiology of respiratory syncytial virus infection in Washington, DC. II. Infection and disease with respect to age, immunologic status, race and sex. Am J Epidemiol 98:289–300. [DOI] [PubMed] [Google Scholar]
- 34.Jin H, Cheng X, Zhou HZ, Li S, Seddiqui A. 2000. Respiratory syncytial virus that lacks open reading frame 2 of the M2 gene (M2-2) has altered growth characteristics and is attenuated in rodents. J Virol 74:74–82. doi: 10.1128/JVI.74.1.74-82.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saez-Llorens X, Castano E, Null D, Steichen J, Sanchez PJ, Ramilo O, Top FH Jr, Connor E. 1998. Safety and pharmacokinetics of an intramuscular humanized monoclonal antibody to respiratory syncytial virus in premature infants and infants with bronchopulmonary dysplasia. The MEDI-493 Study Group. Pediatr Infect Dis J 17:787–791. doi: 10.1097/00006454-199809000-00007. [DOI] [PubMed] [Google Scholar]