ABSTRACT
PA-X is a recently identified influenza virus protein that is composed of the PA N-terminal 191 amino acids and unique C-terminal 41 or 61 residues. We and others showed that PA-X has a strong ability to suppress host protein synthesis via host mRNA decay, which is mediated by endonuclease activity in its N-terminal domain (B. W. Jagger, H. M. Wise, J. C. Kash, K. A. Walters, N. M. Wills, Y. L. Xiao, R. L. Dunfee, L. M. Schwartzman, A. Ozinsky, G. L. Bell, R. M. Dalton, A. Lo, S. Efstathiou, J. F. Atkins, A. E. Firth, J. K. Taubenberger, and P. Digard, 2012, Science 337:199–204, http://dx.doi.org/10.1126/science.1222213, and E. A. Desmet, K. A. Bussey, R. Stone, and T. Takimoto, 2013, J Virol 87:3108–3118, http://dx.doi.org/10.1128/JVI.02826-12). However, the mechanism of host mRNA degradation, especially where and how PA-X targets mRNAs, has not been analyzed. In this study, we determined the localization of PA-X and the role of the C-terminal unique region in shutoff activity. Quantitative subcellular localization analysis revealed that PA-X was located equally in both cytoplasm and nucleus. By characterizing a series of PA-X C-terminal deletion mutants, we found that the first 9 amino acids were sufficient for nuclear localization, but an additional 6 residues were required to induce the maximum shutoff activity observed with intact PA-X. Importantly, forced nuclear localization of the PA-X C-terminal deletion mutant enhanced shutoff activity, highlighting the ability of nuclear PA-X to degrade host mRNAs more efficiently. However, PA-X also inhibited luciferase expression from transfected mRNAs synthesized in vitro, suggesting that PA-X also degrades mRNAs in the cytoplasm. Among the basic amino acids in the PA-X C-terminal region, 3 residues, 195K, 198K, and 199R, were identified as key residues for inducing host shutoff and nuclear localization. Overall, our data indicate a critical role for the 15 residues in the PA-X C-terminal domain in degrading mRNAs in both the cytoplasm and nucleus.
IMPORTANCE Influenza A viruses express PA-X proteins to suppress global host gene expression, including host antiviral genes, to allow efficient viral replication in infected cells. However, little is known about how PA-X induces host shutoff. In this study, we showed that PA-X localized equally in both the cytoplasm and nucleus of the cells, but the nuclear localization of PA-X mediated by its C-terminal region has a significant impact on shutoff activity. Three basic residues at the C-terminal region play a critical role in nuclear localization, but additional basic residues were required for maximum shutoff activity. Our findings indicate that PA-X targets and degrades mRNAs in both the nucleus and cytoplasm, and that the first 15 residues of the PA-X unique C-terminal region play a critical role in shutoff activity.
INTRODUCTION
Influenza virus infection induces suppression of host protein synthesis, called “host shutoff,” during viral replication in infected cells. This host shutoff activity has been thought to contribute to efficient virus replication in the cells by dampening host antiviral response. Earlier studies have suggested that the cap-snatching activity and degradation of RNA polymerase II mediated by viral RNA polymerase (PA, PB1, and PB2) play a role in host shutoff (1, 2). In addition, the NS1 protein, a well-known antagonist against host antiviral interferon responses, has been shown to induce host shutoff in infected cells (3). Mechanistically, NS1 binds a cellular 30-kDa subunit of the cleavage and polyadenylation specificity factor (CPSF30), which is an essential component for 3′-end cleavage and polyadenylation of host pre-mRNAs (4, 5). The binding of NS1 to CPSF30 blocks general posttranscriptional processing of cellular pre-mRNA, which results in degradation of mRNA and suppression of general cellular protein synthesis. However, binding of NS1 to CPSF30 is not conserved among all influenza virus strains (3). NS1 proteins from laboratory-adapted human H1N1 virus A/Puerto Rico/8/34 (PR8), 2009 pandemic H1N1 (pH1N1) virus, and novel H7N9 human influenza virus are unable to bind CPSF30 and inhibit general host gene expression due to mutations in the CPSF30 binding domain (6–8). Importantly, a PR8 mutant strain lacking the NS1 gene was able to suppress host protein synthesis at a level similar to that of the PR8 wild-type (WT) strain in infected cells (9). We also showed that pH1N1 virus effectively induced host shutoff in infected cells, even though its NS1 protein is unable to bind CPSF30 (8, 10). Taken together, these findings suggested the presence of other viral factors that are crucial for host shutoff.
Recently, a novel influenza virus protein, termed PA-X, was identified and found to possess an ability to suppress host protein synthesis (11). PA-X is composed of the PA N-terminal domain (amino acids 1 to 191) fused to a unique C-terminal region, which is produced by the +1 reading frame of PA mRNA via ribosomal frameshift (11, 12). Most influenza A virus strains possess PA-X proteins with 252 amino acids and a unique C-terminal 61 amino acids. However, some strains of influenza A viruses, such as pH1N1 viruses, harbor shorter 232-amino-acid PA-X proteins with a unique C-terminal 41 amino acids due to the presence of an early stop codon (12). PA-X and PA possess a common endonuclease active site (P107D108X10E119K134) in their N-terminal domains (11, 13). Previous studies by us and others revealed that transfection of a PA-X mutant containing a replacement of one of the residues of the endonuclease active sites with alanine (K134A or D108A) failed to induce host shutoff, suggesting that PA-X suppresses host protein synthesis via host mRNA decay (11, 14). In line with this, we previously reported that a recombinant pH1N1 A/California/04/2009 (Cal) PA-XFS mutant virus expressing a reduced amount of PA-X due to mutations in the frameshift motif of the PA gene induced less mRNA degradation and suppression of host protein synthesis than the WT strain (10). Our previous study also showed that PA-X contributes to viral growth through inhibition of the host antiviral innate response both in vitro and in vivo and suppression of the host adaptive immune response (10, 15), although the effect of PA-X expression on viral growth and host responses in vivo may vary between nonpathogenic and highly pathogenic viruses (11, 15–18).
We and others have shown that PA-X suppressed coexpressed protein production much more effectively than full-length PA despite both proteins possessing a common endonuclease active domain, suggesting the importance of the unique C-terminal region of PA-X for its activity (11, 14). In this study, we further characterized PA-X-mediated mRNA degradation using Cal PA-X and addressed the role of the PA-X C-terminal region in its function. We found that PA-X equally localizes in both cytoplasm and nucleus, and the first 9 amino acids in the C-terminal unique domain (residues 192 to 200) are responsible for nuclear localization of PA-X. However, we also found that an additional 6 amino acids (residues 201 to 206) are required for maximum shutoff activity compared to WT PA-X. Strikingly, forced nuclear transport of Cal PA-X lacking the entire C-terminal region, which is otherwise localized in the cytoplasm, significantly increased shutoff activity, suggesting a major impact for mRNA degradation by Cal PA-X in the nucleus. However, we also found that PA-X targeted and degraded mRNAs in the cytoplasm as well. Further analysis using site-directed mutagenesis indicates that the 6 basic amino acids within the first 15 residues of the PA-X C-terminal region were essential for shutoff activity. In particular, the first 3 residues (195K/198K/199R) were found to be responsible for nuclear localization. Our studies indicate that PA-X is capable of inducing mRNA degradation and host shutoff at different sites in the cells and that the unique C-terminal domain plays a major role in efficient shutoff activity.
MATERIALS AND METHODS
Virus and cells.
Recombinant Cal WT and PA-XFS viruses were rescued previously (10). 293T cells and A549 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Corning) supplemented with 8% fetal bovine serum (FBS; Life Technologies or Seradigm) and 1× GlutaMAX (Life Technologies).
Plasmids.
pCAGGS-Luc, pCAGGS-CalNS1flag, pCAGGS-WSNPA-Xflag, pCAGGS-CalPA-XWTflag, pCAGGS-CalPA-XK134Aflag, and pCAGGS-CalPA-XK134A/c0flag were constructed previously (10, 14). For the construction of pCAGGS-WSNNS1flag or pCAGGS-AichiNS1flag, the NS1 gene of A/WSN/33 (WSN) or A/Aichi/2/68 (Aichi) was amplified by PCR from pPolI-WSNNS (19) or pPolI-AichiNS, respectively, and subcloned into pCAGGS. For the construction of pCAGGS-CalPA-X252aaflag and pCAGGS-CalPA-XK134A/252aaflag, a point mutation (A698G) in the PA genes was introduced by site-directed mutagenesis as described previously (20). For the construction of pCAGGS expressing CalPA-Xc0flag, CalPA-Xc9flag, CalPA-Xc15flag, CalPA-Xc22flag, and CalPA-Xc34flag and the related K134A mutants, the corresponding regions were amplified by PCR from pCAGGS-CalPA-XWTflag or pCAGGS-CalPA-XK134Aflag as the template, and the resultant PCR products were subcloned into pCAGGS. For the construction of pCAGGS expressing CalPA-XNLS/WTflag, CalPA-XNLS/c0flag, and CalPA-XNLS/c9flag and the related K134A mutants, the corresponding regions were amplified by PCR using pCAGGS-CalPA-XWTflag or pCAGGS-CalPA-XK134Aflag as the template. The forward primer encodes simian virus 40 (SV40) tag nuclear localization signal (NLS) sequences (CCAAAAAAGAAAAGGAAGGTT) followed by the first 20 nucleotides of PA-X coding sequences, as described previously (21), and the PCR products were subcloned into pCAGGS. For the construction of pcDNA-Luc, pcDNA-eGFPflag, and pcDNA-CalPA-XK134Aflag, the corresponding regions were amplified by PCR using pCAGGS-Luc, pCAGGS-eGFPflag (14), or pCAGGS-CalPA-XK134Aflag as the template, and the PCR products were subcloned into pcDNA 3.1(−). For the construction of pCAGGS-T7-CalPA-Xc0flag, pCAGGS-T7-CalPA-Xc9flag, pCAGGS-T7-CalPA-Xc15flag, pCAGGS-T7-CalPA-Xc22flag, pCAGGS-T7-CalPA-Xc34flag, and pCAGGS-T7-CalPA-XWTflag, the corresponding regions were amplified by PCR using pCAGGS-CalPA-XWTflag as a template. The forward primer encodes the promoter sequence of T7 polymerase (TAATACGACTCACTATAGGG) followed by the first 20 nucleotides of PA-X coding sequences, and the PCR products were subcloned into pCAGGS. Mutations in Cal PA-X or Cal NS1 genes were created by site-directed mutagenesis. The coding regions of all constructed plasmids were sequenced, and we confirmed that there were no additional mutations. All primer sequences for plasmid constructions are available upon request.
In vitro transcription.
5′ Capped and polyadenylated RNA transcripts encoding enhanced green fluorescent protein (eGFP), Cal PA-X, or its variants were synthesized with the linearized plasmid carrying the corresponding gene using restriction enzymes. We used HindIII for pcDNA-Luc and pcDNA-eGFPflag, AflII for pcDNA-CalPA-XK134Aflag, and XhoI for pCAGGS-T7-CalPA-Xc0flag, pCAGGS-T7-CalPA-Xc9flag, pCAGGS-T7-CalPA-Xc15flag, pCAGGS-T7-CalPA-Xc22flag, pCAGGS-T7-CalPA-Xc34flag, pCAGGS-T7-CalPA-XWTflag, pCAGGS-T7-CalPA-X3E-1flag, pCAGGS-T7-CalPA-X3E-2flag, and pCAGGS-T7-CalPA-X6Eflag. We synthesized 5′ capped and polyadenylated RNAs from these linearized templates using an mMessage mMachine T7 ultra kit (Ambion), as reported previously (22, 23).
Virus rescue.
Recombinant Cal viruses (CalPA-X-c9 and CalPA-X-c15) were rescued using the 12-plasmid rescue system (24) as reported previously (10). Briefly, 293T and MDCK cocultures in a 6-well plate were transfected with 0.1 μg each pPol-I plasmid carrying the 8 segments from Cal together with 0.4 μg of each pCAGGS plasmid carrying the Cal PA, PB1, PB2, or NP gene using Lipofectamine 2000 (Life Technologies). To rescue CalPA-X-c9 or CalPA-X-c15, we used pPol-I-CalPA-T603A or pPol-I-CalPA-T621A, respectively, to generate a stop codon (TAG) in the PA-X open reading frame (ORF) without changing the PA ORF. Rescued viruses were plaque purified, and stock virus was propagated in 10-day-old embryonated chicken eggs. Mutations in the PA gene were confirmed by sequence analysis. Virus titers were determined by immunofluorescence assay detecting viral nucleoprotein in infected A549 cells.
Luciferase assay.
For plasmid transfection, 293T cells in 12-well plates were transfected with 0.4 μg of pCAGGS containing the indicated genes together with 0.4 μg of pCAGGS-Luc for 24 h using Lipofectamine 2000 or polyethylenimine (PEI) reagent (Polysciences). For mRNA transfection, 293T cells in 12-well plates were transfected with 2 μg of the indicated mRNAs together with 2 or 4 μg of luciferase mRNA for 24 h using PEI reagent. The cells were lysed with the passive lysis buffer included in the kit (Dual-Luciferase reporter assay system; Promega). Luciferase production was measured using the Dual-Luciferase reporter assay system as described previously (14).
Fractionation.
293T cells in 12-well plates were transfected with pCAGGS containing the indicated genes for 24 h. For detection of luciferase mRNA, the cells were collected and fractionated into nuclear and cytoplasmic materials using a nuclear/cytosol fractionation kit according to the manufacturer's instructions (BioVision). RNAsecure reagent (Life Technologies) was added to cytoplasmic and nuclear extraction buffer to protect RNAs from RNase. RNAs in both fractions were extracted using the illustra RNAspin minikit (GE Healthcare). For detection of PA-X protein, the cells were collected and lysed with cytoplasmic extract buffer (10 mM HEPES [pH 7.9], 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], and 0.5% NP-40) supplemented with protein inhibitor cocktail (Calbiochem) on ice for 10 min, followed by centrifugation for 5 min at 4°C at 12,000 rpm. The supernatant was saved as the cytoplasmic fraction. The pellet (nuclear fraction) was washed once with phosphate-buffered saline (PBS) and then lysed with 1× Laemmli sample buffer (Bio-Rad).
Northern blotting.
For plasmid transfection, 293T cells in 12-well plates were transfected with pCAGGS containing the indicated genes together with pCAGGS-Luc for 24 h. For viral infection, A549 cells in 6-well plates were infected with Cal viruses at a multiplicity of infection (MOI) of 1.5 for 1 h and incubated in DMEM containing 0.15% bovine serum albumin for an additional 11 h. The extracted total, cytoplasmic, or nuclear RNA was subjected to Northern blot analysis using a DIG (digoxigenin) Northern starter kit (Roche) as described previously (10). To detect luciferase mRNA, the DIG-labeled antisense luciferase RNA probe, corresponding to nucleotides 736 to 1370, was synthesized by in vitro transcription using the DIG Northern starter kit according to the manufacturer's protocol (10). Detection of actin mRNA was performed as described previously (10).
RT-PCR.
Cytoplasmic and nuclear RNAs (50 ng) were used in reverse transcription-PCR (RT-PCR) with the RevertAid first-strand cDNA synthesis kit (Thermo Scientific) and Taq DNA polymerase (BioLabs) with primers specific for human mitochondrial 12S rRNA (5′-GCTAAGACCCAAACTGGGATTAG-3′ and 5′-ACAGGCTCCTCTAGAGGGATAT-3′) or human U1 small nuclear RNA (snRNA; 5′-ATACTTACCTGGCAGGGGAGATA-3′ and 5′-ACTACCACAAATTATGCAGTCG-3′). Samples were collected after 13 or 15 cycles for detection of 12S rRNA or U1 snRNA, respectively. The PCR products were electrophoresed on a 2% agarose gel and stained with ethidium bromide.
Western blotting.
Western blot analysis was performed as described previously (10, 25). For total cell lysate, 293T or A549 cells were lysed with passive lysis buffer (Dual-Luciferase reporter assay system) or Triton-X lysis buffer (20 mM HEPES [pH 7.5], 1.5 mM MgCl2, 500 mM NaCl, 0.2 mM EDTA, 1% Triton X-100, and 20% glycerol), respectively. The total, cytoplasmic, or nuclear cell lysate was separated by SDS-PAGE (4 to 12% gel [Life Technologies] or 12% gel [Bio-Rad]). The proteins were then transferred onto a polyvinylidene difluoride membrane (Millipore). The membrane was blocked in 2% dry milk and incubated with the following primary antibodies: mouse anti-CalPA-X monoclonal antibody (MAb), which reacts with the common N-terminal region between PA and PA-X (clone F17-112; 1:1,000) (10), mouse anti-Flag MAb (1:1,500; Cell Signaling), mouse anti-influenza A virus hemagglutinin (HA) MAb (1:1,000; NR42019; BEI Resources), mouse anti-influenza A virus M1 MAb (1:1,000; GA2B; Thermo Scientific), rabbit anti-influenza A virus NS1 serum (1:3,000), kindly provided by L. Martínez-Sobrido (University of Rochester), rabbit anti-α/β-tubulin antibody (1:3,000; Cell Signaling), mouse anti-lamin A/C MAb (1:1,000; Cell Signaling), and mouse anti-β-actin MAb (1:10,000; clone 8H10D10; Cell Signaling). The membrane was then incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:10,000; Bio-Rad) or HRP-conjugated goat anti-rabbit IgG (1:5,000; Bio-Rad). Target proteins were visualized using SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific). Images were taken using the ChemDoc XRS system (Bio-Rad), and band intensities were quantified using Quantity One 1-D analysis software (Bio-Rad).
Metabolic labeling.
293T cells in 12-well plates were transfected with 2 μg of the indicated mRNAs for 24 h. After 1 h posttransfection, the cells were left untreated (dimethyl sulfoxide) or were treated with 4 μg/ml actinomycin D (ActD; Sigma) for 8.5 h. The cells were then labeled with [35S]Met/Cys (PerkinElmer) at 50 μCi/ml for 30 min in medium lacking methionine and cysteine (Corning). The cells were lysed with Triton-X lysis buffer, and the radiolabeled lysates were separated by SDS-PAGE. Dried gels were exposed onto a phosphor screen and visualized using the Bio-Rad personal molecular imager (Bio-Rad) as described previously (10, 23).
Statistical analysis.
Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Tukey's multiple-comparison test (GraphPad Prism 5.0 software). A P value of <0.05 was considered statistically significant.
RESULTS
PA-X proteins possess stronger shutoff activity than NS1 proteins.
Both PA-X and NS1 proteins have been shown to have an ability to suppress host protein synthesis (8, 11, 14). However, their activities have not been directly compared. To perform this comparison, the shutoff activities of PA-X and NS1 proteins from various influenza A virus strains were analyzed. We constructed pCAGGS encoding Cal PA-X, WSN PA-X, Cal NS1, WSN NS1, and Aichi NS1. Aichi NS1 possesses the five amino acids (103F, 106M, 108K, 125D, and 189D) known to be involved in CPSF30 binding (Fig. 1A), while Cal and WSN NS1 proteins contain 3 and 1 mutated amino acids, respectively (Fig. 1A). We also constructed a Cal NS1 mutant plasmid (Cal NS1 3mut) containing 3 substitutions (R108K, E125D and G189D) so that it expresses the NS1 capable of binding CPSF30 (8). 293T cells were cotransfected with a pCAGGS plasmid encoding luciferase (pCAGGS-Luc) and the individual plasmids for 24 h, and the effect of the PA-X and NS1 on luciferase production was determined (Fig. 1B). Consistent with previous reports (8, 14), the expression of Cal NS1 did not effectively reduce luciferase expression (2.2-fold reduction), whereas Cal NS1 3mut suppressed luciferase expression by 20-fold, similar to Aichi NS1 (Fig. 1B). WSN NS1 reduced the expression at an intermediate level between Cal NS1 and Aichi NS1 (5.7-fold reduction). These results confirmed a previous report that NS1 containing CPSF30 binding sites inhibits protein expression (8). In sharp contrast to NS1, Cal PA-X showed much stronger shutoff activity (233-fold reduction) (Fig. 1B). Although WSN PA-X was 5.3-fold less active than Cal PA-X (14), it was more active than all NS1 proteins examined (Fig. 1B). Western blot analysis showed that Cal PA-X, WSN PA-X, Cal NS1 3mut, and Aichi NS1, all of which suppressed luciferase expression efficiently, were barely detectable due to strong suppression of its own expression (Fig. 1B) (8, 26). These data clearly indicate that PA-X proteins have an enhanced capacity for host shutoff activity compared to NS1 proteins.
FIG 1.
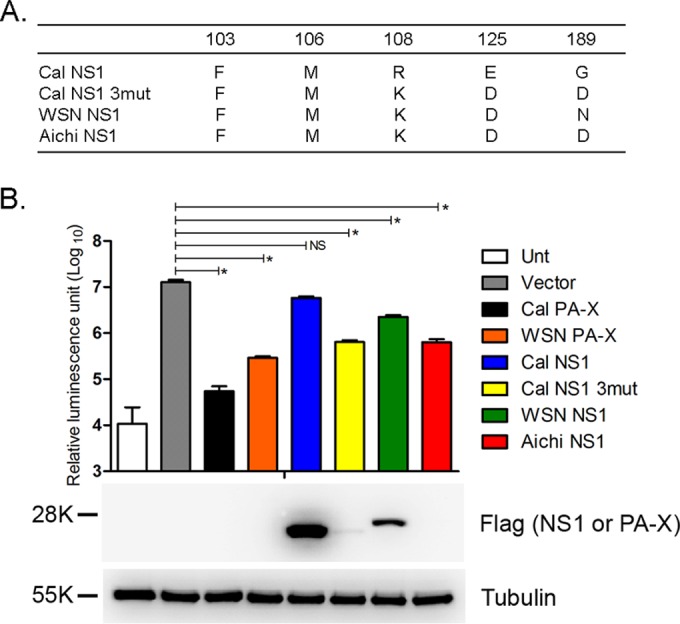
Comparison of shutoff activities of PA-X and NS1. (A) Differences of amino acids involved in NS1 binding to CPSF30. (B) Luciferase expression from pCAGGS-Luc in 293T cells cotransfected with either empty pCAGGS plasmid (Vector) or pCAGGS containing the indicated genes for 24 h. The results are shown as means plus the standard deviations (n = 3). Asterisks indicate statistically significant differences (*, P < 0.05 by one-way ANOVA followed by Tukey's multiple-comparison test). NS, not significant (P > 0.05). Expression of PA-X or NS1 protein in the transfected cells was detected using an anti-Flag antibody. Expression of tubulin was detected using an anti-tubulin antibody. Unt, untransfected.
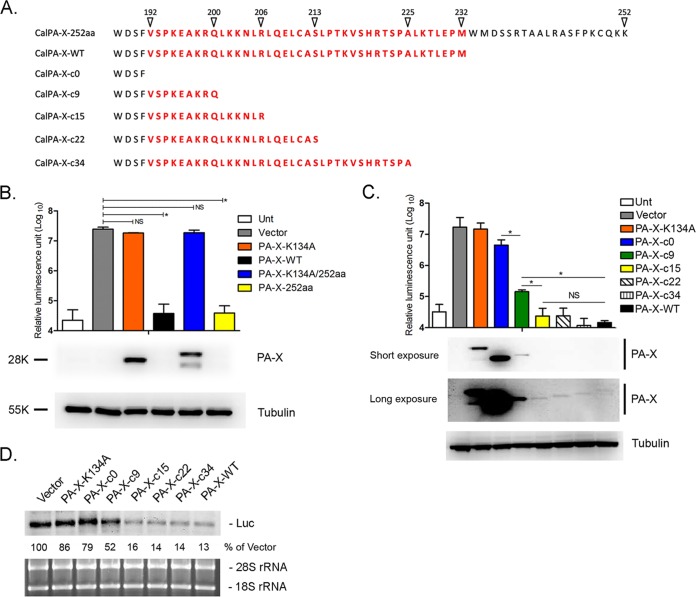
The first 15 amino acids in the PA-X C-terminal region contribute to suppression of luciferase expression and degradation of mRNA.
Most influenza A viruses, such as human and avian strains, possess a unique C-terminal 61 amino acids in their PA-X proteins, whereas pH1N1 viruses, including Cal, express PA-X proteins with a unique C-terminal 41 amino acids. To determine the effect of the additional 20 residues at the C terminus on activity, we created a plasmid expressing Cal PA-X containing 252 amino acids (CalPA-X-252aa) (Fig. 2A) and compared the shutoff activity to that of the native 232-amino-acid Cal PA-X (CalPA-X-WT). CalPA-X-252aa shut off luciferase expression as efficiently as CalPA-X-WT (P > 0.05) (Fig. 2B), suggesting that the additional C-terminal 20 amino acids do not have a significant impact on the host shutoff activity of Cal PA-X. As we previously showed, a mutation at the endonuclease active site (K134A) eliminated shutoff activity, suggesting that PA-X degrades mRNAs (14). To map the C-terminal regions responsible for suppression of luciferase expression and degradation of mRNA, we next created a series of C-terminal deletion mutants of Cal PA-X (Fig. 2A). Expression of CalPA-X-c15, CalPA-X-c22, or CalPA-Xc-34 strongly suppressed coexpressed luciferase at a level similar to that of CalPA-X-WT, suggesting that the C-terminal 26 residues have little impact on shutoff activity (Fig. 2C). CalPA-X-c0, lacking the entire C-terminal region, showed only a slight reduction in luciferase expression (3.8-fold reduction), whereas CalPA-X-c9 showed intermediate shutoff activity between CalPA-X-c0 and CalPA-X-WT (P < 0.05) (Fig. 2C). As expected, highly active PA-X proteins (CalPA-X-c15, CalPA-X-c22, CalPA-X-c34, and CalPA-X-WT) were barely detected in the cell lysates, while PA-X proteins with no or little activity (CalPA-X-K134A and CalPA-X-c0) were expressed well (Fig. 2C). We next measured luciferase mRNA levels in the cells cotransfected with pCAGGS-Luc and the individual PA-X plasmids by Northern blotting. Consistent with the luciferase protein expression result (Fig. 2C), CalPA-X-c15, CalPA-X-c22, CalPA-X-c34, or CalPA-XWT strongly reduced luciferase mRNA levels by over 80% compared to that of the vector control, while CalPA-X-c9 partially reduced expression (52%) (Fig. 2D). These data demonstrated that the first 15 amino acids in the PA-X C-terminal region are sufficient to induce host mRNA degradation and shutoff.
FIG 2.
Shutoff activity and mRNA degradation by PA-X mutants. (A) C-terminal amino acid sequences of Cal PA-X variants constructed in this study. Cal PA-X-252aa contains an additional 20 amino acids at its C terminus. Cal PA-X-c0 lacks the entire C-terminal region. Cal PA-X-c9, Cal PA-X-c15, Cal PA-X-c22, and Cal PA-X-c34 contain the C-terminal 9, 15, 22, and 34 amino acids, respectively. (B and C) Luciferase expression from pCAGGS-Luc in 293T cells cotransfected with either empty pCAGGS plasmid (Vector) or pCAGGS containing the indicated genes for 24 h. The results are shown as means plus standard deviations from three independent experiments. Asterisks indicate statistically significant differences (*, P < 0.05 by one-way ANOVA followed by Tukey's multiple-comparison test). NS, not significant (P > 0.05). Expression of PA-X in the transfected cells was detected using an anti-PA-X antibody. Expression of tubulin was detected as a loading control. (D) 293T cells were transfected with either empty vector (Vector) or pCAGGS containing the indicated genes together with pCAGGS-Luc for 24 h. The total RNA was extracted from the cell lysate and subjected to Northern blot analysis using a DIG-labeled antisense luciferase RNA probe. 18S rRNAs and 28S rRNAs stained with ethidium bromide are shown as a loading control. The intensities of luciferase mRNA in each sample were normalized by the intensities of 28S rRNA. The relative level of vector control was set to 100%. The data shown are representative of two independent experiments.
The first 9 amino acids in the PA-X C-terminal region contain an NLS.
So far, the localization of PA-X in the cytoplasm and nucleus has not been determined quantitatively. It remains unclear if the PA-X C-terminal region affects localization in the cells. Therefore, we determined the subcellular localization of a series of PA-X deletion mutants (Fig. 3). Since active PA-X proteins are barely detectable due to self-suppression (Fig. 2B and C) (26), we mutated a residue in the endonuclease active site (K134A) to allow efficient expression. Nuclear and cytoplasmic fractions of the transfected cells were prepared, and the quantities of the PA-X proteins in each fraction were determined by Western blotting (Fig. 3). CalPA-X-WT was detected equally in both cytoplasm and nucleus. Similarly, CalPA-X-c9, CalPA-X-c15, CalPA-X-c22, and CalPA-X-c34 localized in both fractions at almost the same ratio as CalPA-X-WT. In sharp contrast, CalPA-X-c0 predominantly localized in the cytoplasm. These data indicate that amino acids within residues 192 to 200 in the PA-X C-terminal region function as an NLS.
FIG 3.
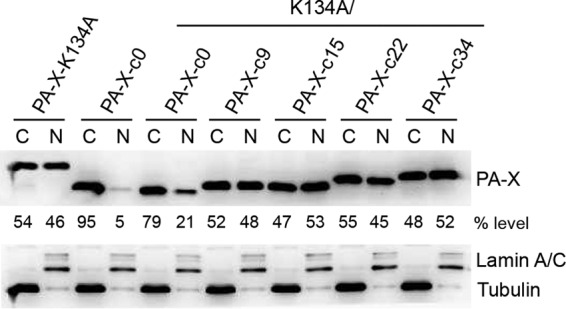
Subcellular localization of Cal PA-X protein. 293T cells were transfected with the indicated genes in pCAGGS. After 24 h, the cells were fractionated into the nuclear and cytoplasmic materials followed by Western blotting. Expression of PA-X protein was detected using anti-PA-X antibody. Expression of lamin A/C and tubulin was detected as a nuclear (N) and cytoplasmic (C) marker, respectively. The ratios of PA-X in the cytoplasm and nucleus are also shown. The data shown are representative of two independent experiments.
Nuclear localization of PA-X protein contributes to shutoff activity.
Cal PA-X lacking the entire unique C-terminal region showed low shutoff activity and was predominantly localized in the cytoplasm (Fig. 2C and 3). These results led us to hypothesize that the nuclear localization of PA-X is required for efficient shutoff activity. To address this question, we introduced the SV40 tag NLS to the N terminus of CalPA-X-c0 (CalPA-X-NLS/c0) and compared shutoff activity with that of parental CalPA-X-c0. To ascertain the effect of NLS on PA-X localization, we quantified the expression of the PA-X constructs in the cytoplasm and nucleus (Fig. 4A). As expected, the introduction of NLS to CalPA-X-K134A, CalPA-X-K134A/c0, or CalPA-X-K134A/c9 significantly shifted their localizations into the nucleus (Fig. 4A). We next compared the shutoff activity of these PA-X constructs. CalPA-X-NLS/c0 showed enhanced shutoff activity compared to the same construct without an NLS (7.6-fold difference) (Fig. 4B). Consistent with the luciferase level, expression of CalPA-X-NLS/c0 was lower than that of CalPA-X-c0 due to higher shutoff activity (Fig. 4B). In contrast, the addition of NLS to CalPA-X-WT or CalPA-X-c9 did not further enhance shutoff activities (Fig. 4B). Taken together, these results indicate that nuclear localization of PA-X contributes to efficient shutoff activity of the protein.
FIG 4.
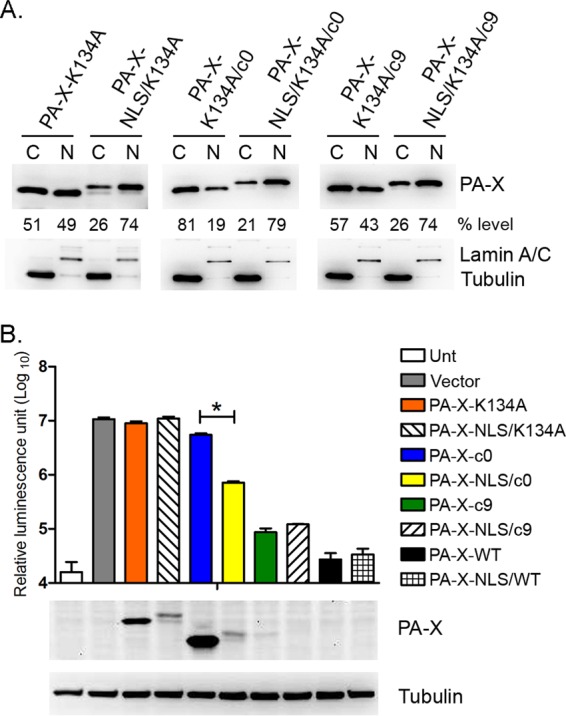
Effect of PA-X nuclear localization on shutoff activity. (A) Localization of PA-X in the nuclear and cytoplasmic fractions was determined by Western blotting as described for Fig. 3. The data shown are representative of two independent experiments. (B) Luciferase expression from pCAGGS-Luc in 293T cells cotransfected with either empty plasmid (Vector) or pCAGGS containing the indicated genes for 24 h. The results are shown as means plus standard deviations (n = 3). Asterisks indicate statistically significant differences (*, P < 0.05 by one-way ANOVA followed by Tukey's multiple-comparison test). Expression of PA-X and tubulin was determined by Western blotting as described for Fig. 2B.
PA-X induces mRNA degradation in the nucleus.
To directly assess if PA-X degrades mRNAs in the nucleus, we measured luciferase mRNA levels in the cytoplasmic and nuclear fractions by Northern blotting. First, we detected cytoplasmic mitochondrial 12S rRNA and nuclear U1 snRNA in fractionated samples by RT-PCR, which confirmed the appropriate fractionation of the samples (Fig. 5). Expression of CalPA-X-WT almost completely degraded luciferase mRNAs in the nucleus and cytoplasm compared to that of CalPA-X-K134A (Fig. 5). Expression of Cal PA-X without the entire C-terminal region (CalPA-X-c0) partially reduced the mRNA level in the cytoplasm but not that in the nucleus (Fig. 5). This is consistent with the result of localization in the cytoplasm (Fig. 3). As expected, introduction of an NLS to CalPA-X-c0 (PA-X-NLS-c0) enhanced mRNA degradation in the nucleus (Fig. 5). These data indicate that the PA-X C-terminal region is required for localization and mRNA degradation in the nucleus.
FIG 5.
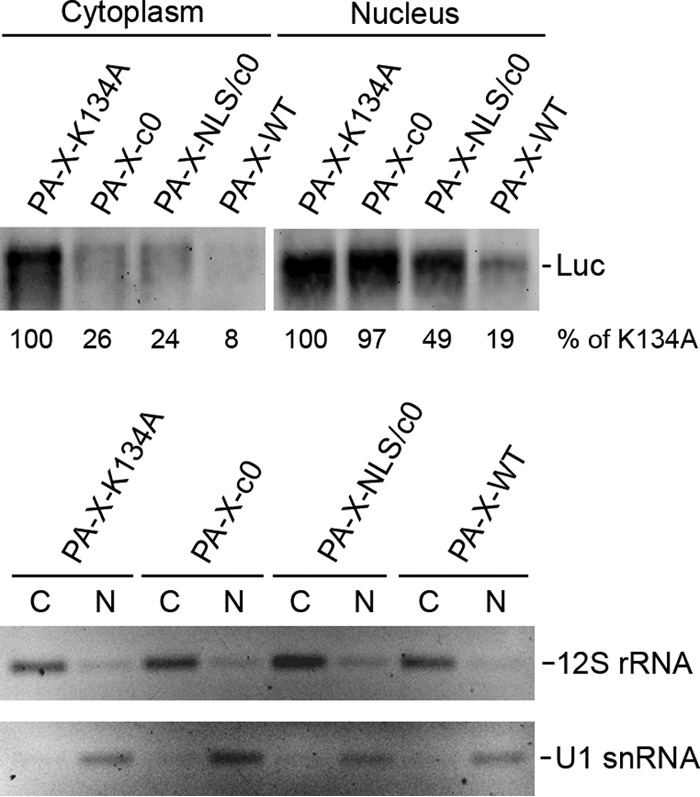
Luciferase mRNA levels in the nuclear and cytoplasmic fractions in PA-X-transfected cells. 293T cells were transfected with pCAGGS containing the indicated genes together with pCAGGS-Luc for 24 h. The cells were then fractionated into the nuclear and cytoplasmic materials. The total RNA from each fraction was extracted and subjected to Northern blot analysis as described for Fig. 2D. The luciferase mRNA level of the PA-X K134A transfection sample of each fraction was set to 100%. The level of U1 snRNA or 12S rRNA was detected as a nuclear (N) or cytoplasmic (C) marker, respectively, by RT-PCR.
The first 15 amino acids in PA-X C-terminal regions contribute to targeting mRNA in the cytoplasm.
Our results strongly suggested that Cal PA-X degrades mRNAs in the nucleus to induce host shutoff (Fig. 4 and 5). However, since Cal PA-X localizes to both the nucleus and cytoplasm (Fig. 3), there remains the possibility that PA-X also targets mRNAs in the cytoplasm as well. In fact, we detected partial degradation of luciferase mRNAs in the cytoplasmic fraction expressing CalPA-X-c0, which localizes mainly in the cytoplasm (Fig. 3 and 5). To address this question, we synthesized 5′ capped and polyadenylated RNA transcripts in vitro encoding luciferase, eGFP, Cal PA-X, and its variants. We transfected cells with mRNAs encoding Cal PA-X, Cal PA-X deletion mutants, or eGFP (control) together with either pCAGGS-Luc or in vitro-synthesized luciferase mRNAs. Compared to eGFP, expression of Cal PA-X significantly reduced luciferase expression from cDNA (197-fold reduction) (Fig. 6A) as well as that from in vitro-synthesized mRNAs (13.3-fold reduction) (Fig. 6B), demonstrating that PA-X is capable of targeting mRNAs directly introduced into the cytoplasm. The suppressive effect of PA-X was significantly reduced by the loss of the entire C-terminal region (PA-X-c0), while deletion of 26 amino acid residues (PA-X-c15) did not affect shutoff activity in both the luciferase plasmid- and mRNA-transfected cells (Fig. 6). These data suggest that PA-X degrades mRNAs in both the nucleus and cytoplasm, and that the first 15 amino acids in the PA-X C-terminal region are sufficient for targeting and degrading mRNAs in the cytoplasm.
FIG 6.
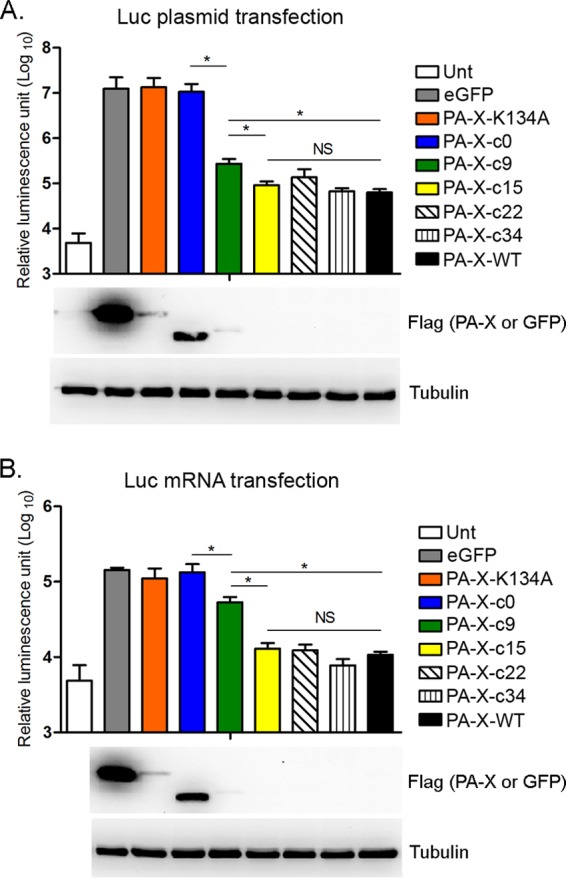
Shutoff of luciferase expression from plasmid cDNA or mRNAs by PA-X mutants. Luciferase expression in 293T cells transfected with pCAGGS-Luc (A) or luciferase mRNA (B) together with the indicated PA-X mRNAs for 24 h is shown. The results are shown as means plus standard deviations from three independent experiments. Asterisks indicate statistically significant differences (*, P < 0.05 by one-way ANOVA followed by Tukey's multiple-comparison test). NS, not significant (P > 0.05). Expression of PA-X, eGFP, or tubulin was determined by Western blotting as described for Fig. 1B.
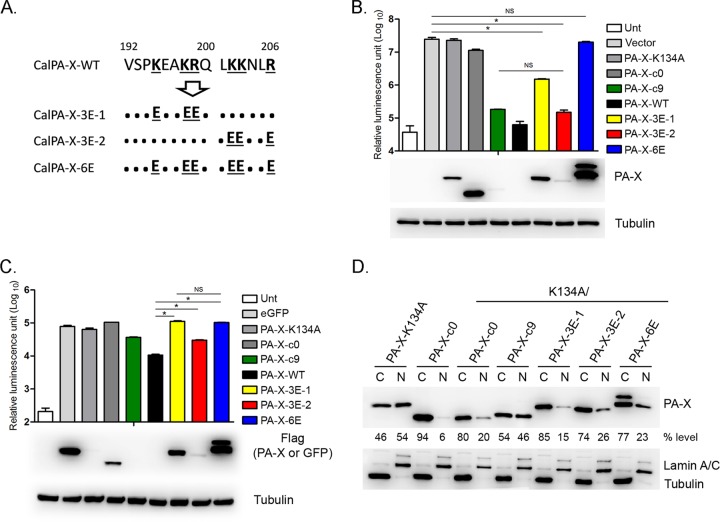
Six basic amino acids in the PA-X C-terminal region play a key role in nuclear localization and host shutoff.
Our analysis using a series of PA-X C-terminal deletion mutants revealed that residues 192 to 200 are sufficient for nuclear localization of PA-X, but an additional 6 residues are necessary for maximum shutoff activity (Fig. 2, 3, and 6). Recently, Oishi et al. reported that replacement of the 6 basic amino acids (195R/198K/199R/202K/203K/206K) with glutamic acid (E), aspartic acid (D), or alanine (A) reduced shutoff activity of WSN PA-X (26). Since these 6 basic amino acids are conserved in Cal PA-X, we tested if they affect the localization of Cal PA-X. We replaced the basic amino acids with E but not A, because WSN PA-X containing the replacement of 6 basic amino acids with A (WSNPA-X6A) was barely detectable (26), which makes it difficult to perform a quantitative localization analysis. We made plasmids or in vitro-synthesized mRNAs encoding CalPA-X-3E-1 with the first 3 basic amino acids replaced with E (K195E/K198E/R199E), CalPA-X-3E-2 with the second 3 basic amino acids replaced with E (K202E/K203E/R206E), and CalPA-X-6E with all 6 residues replaced with E (Fig. 7A). In cells transfected with luciferase cDNAs, CalPA-X-3E-1 showed only a 16.8-fold reduction in luciferase expression, in contrast to CalPA-X-WT, causing a 357-fold reduction (Fig. 7B). CalPA-X-3E-2 showed a 175-fold reduction, which was a level of reduction similar to that of CalPA-X-c9, consistent with the data showing the contribution of the additional 6 residues for maximum shutoff activity (Fig. 2C). Importantly, replacement of all 6 basic amino acids with E (CalPA-X-6E) almost completely abolished shutoff activity (Fig. 7B). Similarly, in luciferase mRNA-transfected cells, shutoff activity of Cal PA-X was almost completely abolished by substitution of the first 3 or all 6 basic amino acids (Fig. 7C). Substitutions of the second 3 basic amino acids (CalPA-X-3E-2) partially reduced the activity of PA-X (Fig. 7C). These results strongly suggest that residues 195K/198K/199R play a critical role in targeting and degrading mRNA in both the cytoplasm and nucleus, but the second 3 basic residues also contribute to host shutoff. We next determined the localization of these PA-X mutants in the cytoplasm and nucleus (Fig. 7D). We mutated position 134 (K134A) in these PA-X mutants, and the localizations of the proteins were quantified. As expected from the results with the truncated mutants (Fig. 3), CalPA-X-3E-1 and CalPA-X-6E were localized predominantly in the cytoplasm (Fig. 7D). The same mutants without the K134A mutation mainly localized in the cytoplasm as well (data not shown). These results suggest that residues 195K/198K/199R are key residues for nuclear localization of PA-X. Unexpectedly, CalPA-X-3E-2 mainly localized in the cytoplasm (Fig. 7D); the replacement of basic amino acids with acidic acids may have interfered with the NLS in residues 192 to 200 of the PA-X C-terminal region. We consistently detected double bands in the cells transfected with CalPA-X-6E with or without the K134A mutation for an unknown reason (Fig. 7), as reported with WSNPA-X-6E (26). Taken together, our data demonstrated that 6 basic amino acids, especially the 3 basic residues 195K/198K/199R, play an important role in nuclear localization of PA-X as well as mRNA degradation in both the nucleus and cytoplasm.
FIG 7.
Role of the 6 basic residues in the PA-X C-terminal region in shutoff activity and localization. (A) Mutations in the PA-X C-terminal region created in this study. The 6 basic residues are shown in boldface and are underlined. Cal PA-X-3E-1 and Cal PA-X-3E-2 contain K195E/K198E/R199E and K202E/K203E/R206E, respectively. Cal PA-X-6E contains all 6 mutations. (B) Luciferase expression from pCAGGS-Luc in 293T cells cotransfected with either empty pCAGGS plasmid (Vector) or the indicated genes in pCAGGS for 24 h. The results are shown as means plus standard deviations (n = 3). Asterisks indicate statistically significant differences (*, P < 0.05 by one-way ANOVA followed by Tukey's multiple-comparison test). NS, not significant (P > 0.05). Expression of PA-X and tubulin was determined by Western blotting as described for Fig. 2B. (C) Luciferase expression from mRNAs in 293T cells cotransfected with the indicated mRNAs for 24 h. The results are shown as means plus standard deviations (n = 3). Asterisks indicate statistically significant differences (*, P < 0.05 by one-way ANOVA followed by Tukey's multiple-comparison test). NS, not significant (P > 0.05). Expression of PA-X, eGFP, or tubulin was determined by Western blotting as described for Fig. 1B. (D) Localization of the indicated PA-X proteins in nuclear and cytoplasmic fractions determined by Western blotting as described for Fig. 3. The data shown are representative of two independent experiments.
PA-X C-terminal region plays a critical role in endogenous host mRNA degradation and shutoff.
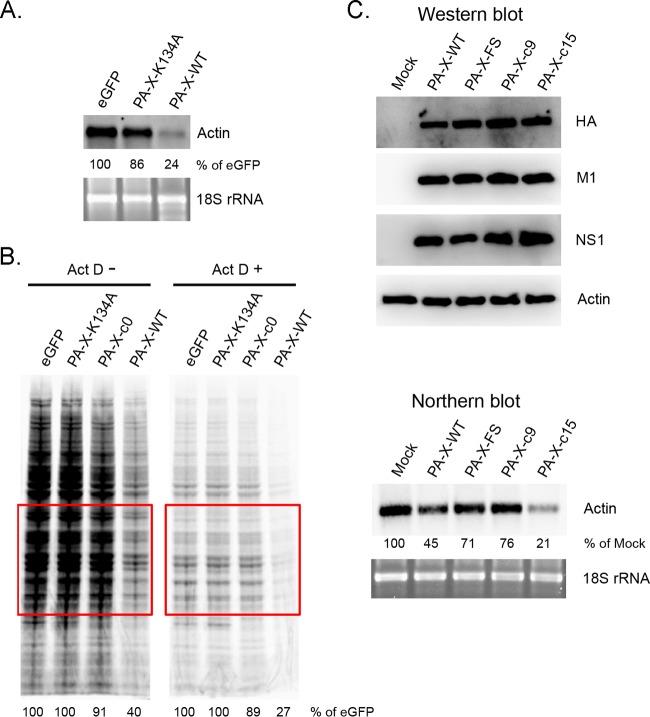
To assess if PA-X induces endogenous host mRNA degradation, we transfected cells with mRNAs encoding Cal PA-X, Cal PA-X K134A mutant, or eGFP (control) and measured host actin mRNA levels by Northern blotting. Consistent with the results of the luciferase mRNAs (Fig. 2D), PA-X-WT, but not the PA-X-K134A mutant, strongly reduced actin mRNA expression compared to that of the eGFP control (Fig. 8A), indicating that PA-X is capable of degrading endogenous host mRNA. We next determined the role of the PA-X C-terminal domain in host protein synthesis. Expression of PA-X WT, but not the K134A mutant or PA-X-c0, markedly suppressed host protein synthesis in the absence or presence of ActD, which blocks host mRNA synthesis (Fig. 8B). This result is consistent with reduced expression of the luciferase gene from transfected cDNA, indicating that PA-X targets and degrades host mRNAs and induces host shutoff, and the C-terminal region is essential for this function.
FIG 8.
Effect of PA-X expression on actin mRNA levels and host protein synthesis. (A) 293T cells were transfected with the indicated mRNAs for 24 h. The extracted total RNA was subjected to Northern blot analysis using a DIG-labeled antisense human β-actin probe. The intensities of actin mRNA in each sample were normalized by the intensities of 18S rRNA. The relative level of eGFP was set to 100%. The data shown are representative of two independent experiments. (B) 293T cells were transfected with the indicated mRNAs for 1 h, followed by incubation in the absence or presence of ActD. After 8.5 h of incubation, the cells were labeled with [35S]Met/Cys for 30 min and the lysates were resolved by SDS-PAGE. The intensities of proteins in the indicated area were quantified and normalized to that of control cells expressing eGFP. (C) A549 cells were either left uninfected or were infected with Cal viruses at an MOI of 1.5. At 12 hpi, HA, M1, and NS1 proteins in total cell lysate were detected by Western blotting using specific antibodies. The total RNA was extracted and subjected to Northern blot analysis as described for panel A.
Finally, we determined the effect of the PA-X C-terminal region on host mRNA degradation in the context of viral infection. For this purpose, we rescued mutant Cal viruses whose PA-X proteins possess C-terminal domains composed of either 9 (CalPA-X-c9) or 15 amino acids (CalPA-X-c15). We analyzed the actin mRNA levels in infected cells and compared these data with the results for Cal WT or Cal PA-XFS, which expresses a reduced amount of PA-X (10). Western blot analysis of infected cells indicates that all of the viruses expressed HA, M1, and NS1 proteins at similar levels (Fig. 8C). Consistent with our previous results (10), CalPA-X WT, but not CalPA-XFS, strongly reduced actin mRNA levels (Fig. 8C). CalPA-X-c9 did not efficiently induce actin mRNA degradation while CalPA-X-c15 retained its ability (Fig. 8C), demonstrating that the first 15 amino acids in the PA-X C-terminal region play a key role in host mRNA degradation in the context of viral infection, which is in accordance with the plasmid transfection data (Fig. 2D). Taken together, our data clearly indicate that the first 15 amino acids in the PA-X C-terminal region contribute to host mRNA degradation and suppression of host protein synthesis.
DISCUSSION
Despite both PA and PA-X having common N-terminal domains, including the endonuclease active site, PA-X induces much stronger shutoff activity than PA (11, 14), suggesting the role of the PA-X unique C-terminal region in the activity. In this study, we assessed the functional role of the PA-X C-terminal region to gain insights into the mechanism of PA-X-induced host shutoff. Our data indicate that PA-X is distributed equally in both the cytoplasm and nucleus and that PA-X targets and degrades mRNAs in both locations (Fig. 3 to 6). The first 9 amino acids in the PA-X C-terminal region contained residues involved in nuclear localization, while an additional 6 residues were required for maximum shutoff activity (Fig. 2C and 3). Similar to influenza A virus, several other viruses express host shutoff proteins to suppress host protein synthesis via mRNA decay (27, 28). However, the location where the viral protein targets and degrades mRNAs varies among the viruses. Kaposi's sarcoma-associated herpesvirus SOX localizes in both the cytoplasm and nucleus, while it targets and degrades mRNAs only in the cytoplasm (27, 29, 30). Severe acute respiratory syndrome coronavirus (SARS-CoV) nsp1 localizes exclusively in the cytoplasm, where it binds the 40S ribosome and thereby induces mRNA cleavage and inhibition of translation (27, 31). In contrast to SARS-CoV nsp1, Middle East respiratory syndrome coronavirus nsp1 is distributed both in the cytoplasm and the nucleus but selectively targets mRNAs transcribed in the nucleus while sparing those of cytoplasmic origin by an unknown mechanism (23). Our findings strongly suggest that, unlike any other viral host shutoff endonucleases, PA-X utilizes a unique host shutoff mechanism by targeting mRNAs in both the cytoplasm and nucleus.
Although the size of the C-terminal region varies between viruses, the addition of 20 residues common to most conventional human viruses did not enhance or reduce shutoff activity of pH1N1 PA-X (Fig. 2B), suggesting that the 20 residues of the C-terminal end are not important for shutoff activity. In fact, our analysis of various truncated PA-X mutants indicates that the first 15 amino acids in the unique C-terminal region are sufficient for maximum shutoff activity and degradation of mRNAs (Fig. 2C and D), which is consistent with the data reported by Oishi et al. (26). However, a previous report suggested a role of the C-terminal 20 residues in the nuclease activity as determined biochemically in vitro using purified proteins (32). Another report indicated a difference in the shutoff activity between PA-X containing C-terminal domains composed of 61 and 41 residues (20), but the difference was minimal (∼50%) compared to our results showing an over 100-fold difference in shutoff activity between the constructs lacking the entire C-terminal domain and that containing the first (N-terminal) 15 residues (Fig. 2C). Our data strongly indicate that the first 15 residues play a critical role in shutoff activity of PA-X.
Quantitative subcellular localization analysis revealed that the 3 basic amino acids (195K/198K/199R) in the PA-X C-terminal region play a major role in PA-X nuclear localization (Fig. 3 and 7D). We also found that forced nuclear accumulation of PA-X-c0 tagged with an NLS, which normally localizes in the cytoplasm, significantly enhanced the ability to suppress coexpressed luciferase production (Fig. 4B). This result indicates that PA-X without the unique C-terminal domain functions better in the nucleus than the cytoplasm and suggests that nuclear PA-X plays a major role in host shutoff. However, our data also indicate that PA-X targets luciferase mRNA directly introduced into the cytoplasm, suggesting that cytoplasmic PA-X also contributes to mRNA degradation and host shutoff (Fig. 6). Although both PA-X 3E-1 and 3E-2 predominantly localize in the cytoplasm, 3E-2 retains high shutoff activity compared to 3E-1 in cells transfected with luciferase mRNAs (Fig. 7C). This result suggests that the first 3 basic residues are also involved in targeting mRNAs for digestion in the cytoplasm in addition to its role in PA-X nuclear localization. At this stage, it is unclear how these basic residues contribute to mRNA decay. It is possible that the essential 6 basic amino acids in the PA-X C-terminal region play a key role in targeting mRNAs through either direct binding to the mRNAs or interaction with host proteins associated with the mRNAs. The fact that these 6 basic amino acids (K or R) in the PA-X C-terminal region are highly conserved among all influenza A strains suggests their essential function in host mRNA decay and shutoff (26).
Recently, Khaperskyy et al. demonstrated a selective degradation of host RNA polymerase II-transcribed RNAs by PA-X in the nucleus (33). They reported that PA-X degrades mature mRNAs possessing a 5′ cap and poly(A) tail, while it spares pre-mRNAs and mRNAs that lack the poly(A) tail, suggesting that PA-X targets the 3′-end maturation step of host mRNAs in the nucleus or interact with cellular factors associated with the mature mRNAs (33). Interestingly, they also showed that PA-X did not inhibit protein expression from luciferase transcripts driven by T7 RNA polymerase in the cytoplasm. These transcripts include an internal ribosomal entry site from encephalomyocarditis virus at the 5′ end and 3′-untranslated regions of hepatitis C virus (33–35). Because we detected that PA-X shutoff protein expression from the 5′ capped and polyadenylated luciferase mRNAs directly transfected into cells (Fig. 6), it is possible that PA-X interacts directly with RNA components unique to the host mature mRNAs or host factors associated with the mRNAs. Further analysis of PA-X interactions with mRNAs will be necessary to elucidate the molecular mechanism by which PA-X induces host shutoff.
Among the viral proteins expressed in infected cells, PA-X and NS1 have been identified to contribute to the host shutoff activity of influenza A viruses. Comparison of shutoff activity between PA-X and NS1 from several influenza A viruses suggests that PA-X possesses a much stronger ability to shut off protein expression than NS1, which binds CPSF30 for mRNA decay (Fig. 1). However, it should be noted that the amount of PA-X expressed in infected cells is much less than that of NS1 due to poor ribosomal frameshift efficiency (11). Unlike other human viruses, pH1N1 NS1 possesses 3 mutations (108R, 125E, and 189G) in CPSF30 binding sites and lacks shutoff activity (Fig. 1A) (8). However, pH1N1 PA-X has much stronger shutoff activity than other human virus PA-X proteins (14). Therefore, PA-X is likely the major factor which induces host shutoff in pH1N1-infected cells. Since virus-induced host shutoff is considered to play a role in the inhibition of the host antiviral response, the ability of NS1 and PA-X to induce host shutoff seems to be well balanced among influenza A virus strains.
ACKNOWLEDGMENTS
This work was supported by the New York Influenza Center of Excellence, a member of the NIAID CEIRS network, under NIH contract HHSN266200700008C.
We thank Brian M. Ward, Luis Martinez-Sobrido, and Stephen Dewhurst for reagents. We also thank Leslie A. MacDonald for technical assistance and Raychel Stone for editing the manuscript.
REFERENCES
- 1.Vreede FT, Fodor E. 2010. The role of the influenza virus RNA polymerase in host shut-off. Virulence 1:436–439. doi: 10.4161/viru.1.5.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boivin S, Cusack S, Ruigrok RW, Hart DJ. 2010. Influenza A virus polymerase: structural insights into replication and host adaptation mechanisms. J Biol Chem 285:28411–28417. doi: 10.1074/jbc.R110.117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Sastre A. 2011. Induction and evasion of type I interferon responses by influenza viruses. Virus Res 162:12–18. doi: 10.1016/j.virusres.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nemeroff ME, Barabino SML, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol Cell 1:991–1000. doi: 10.1016/S1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 5.Das K, Ma L-C, Xiao R, Radvansky B, Aramini J, Zhao L, Marklund J, Kuo R-L, Twu KY, Arnold E, Krug RM, Montelione GT. 2008. Structural basis for suppression of a host antiviral response by influenza A virus. Proc Natl Acad Sci U S A 105:13093–13098. doi: 10.1073/pnas.0805213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kochs G, García-Sastre A, Martínez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol 81:7011–7021. doi: 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayllon J, Domingues P, Rajsbaum R, Miorin L, Schmolke M, Hale BG, Garcia-Sastre A. 2014. A single amino acid substitution in the novel H7N9 influenza A virus NS1 protein increases CPSF30 binding and virulence. J Virol 88:12146–12151. doi: 10.1128/jvi.01567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hale BG, Steel J, Medina RA, Manicassamy B, Ye J, Hickman D, Hai R, Schmolke M, Lowen AC, Perez DR, Garcia-Sastre A. 2010. Inefficient control of host gene expression by the 2009 pandemic H1N1 influenza A virus NS1 protein. J Virol 84:6909–6922. doi: 10.1128/JVI.00081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salvatore M, Basler CF, Parisien JP, Horvath CM, Bourmakina S, Zheng H, Muster T, Palese P, Garcia-Sastre A. 2002. Effects of influenza A virus NS1 protein on protein expression: the NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J Virol 76:1206–1212. doi: 10.1128/JVI.76.3.1206-1212.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi T, MacDonald LA, Takimoto T. 2015. Influenza A virus protein PA-X contributes to viral growth and suppression of the host antiviral and immune responses. J Virol 89:6442–6452. doi: 10.1128/JVI.00319-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. 2012. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J Virol 86:12411–12413. doi: 10.1128/JVI.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 14.Desmet EA, Bussey KA, Stone R, Takimoto T. 2013. Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J Virol 87:3108–3118. doi: 10.1128/JVI.02826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi T, Chaimayo C, Takimoto T. 2015. Impact of influenza PA-X on host response. Oncotarget 6:19364–19365. doi: 10.18632/oncotarget.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao H, Xu G, Sun Y, Qi L, Wang J, Kong W, Sun H, Pu J, Chang KC, Liu J. 2015. PA-X is a virulence factor in avian H9N2 influenza virus. J Gen Virol 96:2587–2594. doi: 10.1099/jgv.0.000232. [DOI] [PubMed] [Google Scholar]
- 17.Hu J, Mo Y, Wang X, Gu M, Hu Z, Zhong L, Wu Q, Hao X, Hu S, Liu W, Liu H, Liu X, Liu X. 2015. PA-X decreases the pathogenicity of highly pathogenic H5N1 influenza A virus in avian species by inhibiting virus replication and host response. J Virol 89:4126–4142. doi: 10.1128/JVI.02132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao H, Sun Y, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng LG, Sun H, Pu J, Chang KC, Liu X, Liu J. 2015. The contribution of PA-X to the virulence of pandemic 2009 H1N1 and highly pathogenic H5N1 avian influenza viruses. Sci Rep 5:8262. doi: 10.1038/srep08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bialas KM, Bussey KA, Stone RL, Takimoto T. 2014. Specific nucleoprotein residues affect influenza virus morphology. J Virol 88:2227–2234. doi: 10.1128/JVI.03354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao H, Sun H, Hu J, Qi L, Wang J, Xiong X, Wang Y, He Q, Lin Y, Kong W, Seng LG, Pu J, Chang KC, Liu X, Liu J, Sun Y. 2015. Twenty amino acids at the C-terminus of PA-X are associated with increased influenza A virus replication and pathogenicity. J Gen Virol 96:2036–2049. doi: 10.1099/vir.0.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cros JF, Garcia-Sastre A, Palese P. 2005. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 6:205–213. doi: 10.1111/j.1600-0854.2005.00263.x. [DOI] [PubMed] [Google Scholar]
- 22.Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, Makino S. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lokugamage KG, Narayanan K, Nakagawa K, Terasaki K, Ramirez SI, Tseng CT, Makino S. 2015. Middle east respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J Virol 89:10970–10981. doi: 10.1128/JVI.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A 96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi T, Wills S, Bussey KA, Takimoto T. 2015. Identification of influenza A virus PB2 residues involved in enhanced polymerase activity and virus growth in mammalian cells at low temperatures. J Virol 89:8042–8049. doi: 10.1128/JVI.00901-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oishi K, Yamayoshi S, Kawaoka Y. 2015. Mapping of a region of the PA-X protein of influenza A virus that is important for its shutoff activity. J Virol 89:8661–8665. doi: 10.1128/JVI.01132-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abernathy E, Glaunsinger B. 2015. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology 479–480:600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayanan K, Makino S. 2013. Interplay between viruses and host mRNA degradation. Biochim Biophys Acta 1829:732–741. doi: 10.1016/j.bbagrm.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glaunsinger B, Chavez L, Ganem D. 2005. The exonuclease and host shutoff functions of the SOX protein of Kaposi's sarcoma-associated herpesvirus are genetically separable. J Virol 79:7396–7401. doi: 10.1128/JVI.79.12.7396-7401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Covarrubias S, Richner JM, Clyde K, Lee YJ, Glaunsinger BA. 2009. Host shutoff is a conserved phenotype of gammaherpesvirus infection and is orchestrated exclusively from the cytoplasm. J Virol 83:9554–9566. doi: 10.1128/JVI.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narayanan K, Ramirez SI, Lokugamage KG, Makino S. 2015. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res 202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bavagnoli L, Cucuzza S, Campanini G, Rovida F, Paolucci S, Baldanti F, Maga G. 2015. The novel influenza A virus protein PA-X and its naturally deleted variant show different enzymatic properties in comparison to the viral endonuclease PA. Nucleic Acids Res 43:9405–9417. doi: 10.1093/nar/gkv926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khaperskyy DA, Schmaling S, Larkins-Ford J, McCormick C, Gaglia MM. 2016. Selective degradation of host RNA polymerase II transcripts by influenza A virus PA-X host shutoff protein. PLoS Pathog 12:e1005427. doi: 10.1371/journal.ppat.1005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoki Y, Aizaki H, Shimoike T, Tani H, Ishii K, Saito I, Matsuura Y, Miyamura T. 1998. A human liver cell line exhibits efficient translation of HCV RNAs produced by a recombinant adenovirus expressing T7 RNA polymerase. Virology 250:140–150. doi: 10.1006/viro.1998.9361. [DOI] [PubMed] [Google Scholar]
- 35.Shi ST, Lai MMC. 2006. HCV 5′ and 3′UTR: when translation meets replication. In Tan SL. (ed), Hepatitis C viruses: genomes and molecular biology. Horizon Bioscience, Norfolk, United Kingdom. [Google Scholar]