ABSTRACT
Human cytomegalovirus (HCMV) pUL93 and pUL77 are both essential for virus growth, but their functions in the virus life cycle remain mostly unresolved. Homologs of pUL93 and pUL77 in herpes simplex virus 1 (HSV-1) and pseudorabies virus (PRV) are known to interact to form a complex at capsid vertices known as the capsid vertex-specific component (CVSC), which likely stabilizes nucleocapsids during virus maturation and also aids in nuclear egress. In herpesviruses, nucleocapsids assemble and partially mature in nuclear replication compartments and then travel to the inner nuclear membrane (INM) for nuclear egress. The factors governing the recruitment of nucleocapsids to the INM are not known. Kinetic analysis of pUL93 demonstrates that this protein is expressed late during infection and localizes primarily to the nucleus of infected cells. pUL93 associates with both virions and capsids and interacts with the components of the nuclear egress complex (NEC), namely, pUL50, pUL53, and pUL97, during infection. Also, multiple regions in pUL93 can independently interact with pUL77, which has been shown to help retain viral DNA during capsid assembly. These studies, combined with our earlier report of an essential role of pUL93 in viral DNA packaging, indicate that pUL93 serves as an important link between nucleocapsid maturation and nuclear egress.
IMPORTANCE HCMV causes life-threatening disease and disability in immunocompromised patients and congenitally infected newborns. In this study, we investigated the functions of HCMV essential tegument protein pUL93 and determined that it interacts with the components of the nuclear egress complex, namely, pUL50, pUL53, and pUL97. We also found that pUL93 specifically interacts with pUL77, which helps retain viral DNA during capsid assembly. Together, our data point toward an important role of pUL93 in linking virus maturation to nuclear egress. In addition to expanding our knowledge of the process of HCMV maturation, information from these studies will also be utilized to develop new antiviral therapies.
INTRODUCTION
In herpes simplex virus 1 (HSV-1) and pseudorabies virus (PRV), pUL17 and pUL25, the homologs of human cytomegalovirus (HCMV) pUL93 and pUL77, respectively, form a complex called the capsid vertex-specific component (CVSC) (1–5). Five copies of this complex surround each of the 12 vertices of the capsids. This complex was originally termed the C-capsid-specific component (2) based on its presence on DNA containing C-capsids, but it was later found to be present on all capsid types (1). Nevertheless, some differences have been reported in the relative distribution of the CVSC on different capsid types in herpesviruses. In HSV-1, occupancy of the CVSC is approximately 50% on C-capsids and only 25% or less on A- (empty) and B-capsids (scaffold containing), while in PRV, occupancy of the CVSC is nearly 100% on C-capsids and 55% on B-capsids (2, 5). The CVSC is thought to aid in DNA packaging and retention by stabilizing capsids during capsid assembly and in nuclear egress, but these functions have yet to be studied (1, 6).
Nuclear egress, the process by which nucleocapsids escape from the nucleus into the cytoplasm of infected cells, is a crucial step in the life cycle of herpesviruses. During nuclear egress, nucleocapsids first undergo primary envelopment at the inner nuclear membrane (INM), enter the perinuclear space (PNS), where they are present as enveloped nucleocapsids, and finally lose this envelope as they bud through the outer nuclear membrane (ONM) into the cytoplasm (7, 8). Nuclear egress involves the disruption of the nuclear lamina via phosphorylation of lamins, which is mediated by viral protein kinase pUL97 in human cytomegalovirus (HCMV) (9). Viral proteins pUL50 and pUL53, which together make up the core of the nuclear egress complex (NEC), are essential for the recruitment of pUL97 to the nuclear envelope (9, 10). The recruitment of pUL97 by the NEC appears to be unique to HCMV, as herpes simplex virus 1 (HSV-1) and murine cytomegalovirus (MCMV) have been shown to recruit host protein kinase C (PKC) instead for the phosphorylation and subsequent dissolution of lamins (9). However, expression of pUL50 and pUL53 in the absence of any other viral proteins has been shown to be sufficient for the disruption of the nuclear lamina, suggesting that PKC is recruited for this purpose in the absence of pUL97 (11).
HSV-1 pUL31, the homolog of HCMV pUL53 and the functional arm of the NEC, has been shown to interact with pUL17 and pUL25 (12). It is hypothesized that this interaction between the NEC and the CVSC in HSV-1 selectively promotes the nuclear egress of C-capsids that have the potential to become infectious viral particles (5, 12). Moreover, HSV-1 and PRV pUL25, components of the CVSC and homologs of HCMV pUL77, are required for nuclear egress (13, 14). It is thought that PRV pUL25 specifically acts as a trigger that allows the process of primary envelopment to occur (14). In the absence of PRV pUL25, DNA is cleaved and packaged appropriately, as evidenced by the presence of C-capsids in the infected cell nucleus; however, these mutant capsids remain closely associated with the INM and do not exit the nucleus (14). Overall, these data suggest that one function of the CVSC is to mediate nuclear egress.
Although HCMV pUL93, a putative tegument protein, and pUL77, a putative capsid protein, are essential for virus growth (15, 16), there is little known about the function of these proteins and whether or not they form a structure similar to the CVSC found in other herpesviruses. We recently discovered that pUL93 is required for viral genome cleavage and packaging, similar to its homolog pUL17 in HSV-1 (17). These findings are supported by another recent study which found that both pUL93 and pUL77 are required for genome encapsidation (18). pUL77 has been found to bind double-stranded DNA and interact with DNA-packaging motor components as well as with major capsid protein, suggesting that pUL77 helps retain viral DNA during capsid assembly (19).
Here, we engineered a virus encoding fluorescent-tagged pUL93 (UL93-eYFP-Towne-BAC) to establish that pUL93 is a late protein that localizes to the infected cell nucleus and associates with nuclear capsids and virions. Also, pUL93 interacts either directly or indirectly with the core components of the NEC, namely, pUL50 and pUL53, as well as with pUL97, during infection. It is important to note that in HSV-1, pUL17 interacts with pUL31 but not with pUL34, the homolog of HCMV pUL50 and the component of the NEC that is responsible for membrane tethering (12). We also found that pUL93 interacts with pUL77, and multiple regions of pUL93 mediate this interaction. These data point toward a role of pUL93 in linking nucleocapsid maturation to nuclear egress.
MATERIALS AND METHODS
Cells.
Primary human foreskin-derived fibroblasts (HF) prepared from deidentified and discarded human foreskins (exempted by the Institutional Review Board) at the University of Mississippi Medical Center or 293T cells (CRL-3216; American Type Culture Collection, Manassas, VA) were cultured at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (MT10013CM; Fisher Scientific, Waltham, MA) containing 4.5 g/ml glucose, 1 mM sodium pyruvate, 2 mM l-glutamine and supplemented with 10% fetal bovine serum (NC0327704; Fisher Scientific, Waltham, MA) and 100 U/ml penicillin-streptomycin (MT30002CI; Fisher Scientific, Waltham, MA). HF between passages 5 and 15 were used for transfections and infections. The cell culture medium was changed (for transfections) or additional medium was added (for infections) every other day.
Plasmids.
For the construction of a Myc-UL77 expression plasmid, the full-length UL77 open reading frame (ORF) was amplified from the HCMV TB40/E genome (GenBank EF999921.1) by PCR and cloned in frame between EcoRI and KpnI sites of pCMV-Myc vector (Clontech, Mountain View, CA), resulting in the fusion of a c-Myc epitope tag at the N terminus of UL77 (Myc-UL77) (Table 1). For the construction of a FLAG-UL93 expression plasmid or several plasmids expressing truncated FLAG-UL93 fragments, the full-length UL93 ORF or truncated UL93 ORF, respectively, was amplified from the HCMV TB40/E genome by PCR and cloned in frame between NotI and XbaI sites of p3XFLAG-CMV-10 vector (Clontech, Mountain View, CA), resulting in a 3XFLAG epitope tag fusion at the N terminus of UL93 (Table 1). For the construction of a Myc-tagged pUL96, the full-length UL96 ORF was amplified from the HCMV Towne-BAC genome (GenBank accession number KF493877.1) and cloned in frame between the multiple cloning sites of pCDNA3.1/myc-His vector (Fisher Scientific, Waltham, MA).
TABLE 1.
Nucleotide sequences of primers used in this study
| Primer name | Sequence |
|---|---|
| UL77-EcoRI-F | GCGGAATTCGGATGAGTCTGTTGCAC |
| UL77-KpnI-R | CTGGTACCCAACACCGCCACGCTC |
| UL93FL-NotI-F | ATAAGAATGCGGCCGCTATGGAAACGCACCTG |
| UL93FL-XbaI-R | GCTCTAGACTAAAGATCGTCGAA |
| UL93T-100-1785-NotI-F | ATAAGAATGCGGCCGCTATGAACGAGGAGGCCGAG |
| UL93T-199-1785-Not-IF | ATAAGAATGCGGCCGCTATGCCGGCCTCCGACGAC |
| UL93T-298-1785-NotI-F | ATAAGAATGCGGCCGCTATGCCACTGGAGTTGCTC |
| UL93T-397-1785-NotI-F | ATAAGAATGCGGCCGCTATGGGTCTGCGCTGCCCC |
| UL93T-448-1785-NotI-F | ATAAGAATGCGGCCGCTATGAGACTGGTCTGGCCC |
| UL93T-496-1785-NotI-F | ATAAGAATGCGGCCGCTATGCGTGACCTGTTACGAG |
| UL93T-586-1785-NotI-F | ATAAGAATGCGGCCGCTATGCTGCGTAATAACTTAG |
| UL93T-1-580-XbaI-R | GCTCTAGAGCTCAATTACGCAGTCCGCC |
| UL93T-1-874-XbaI-R | GCTCTAGAGCTCAGTCGTGCGGATCGGT |
BAC mutagenesis and recombinant viruses.
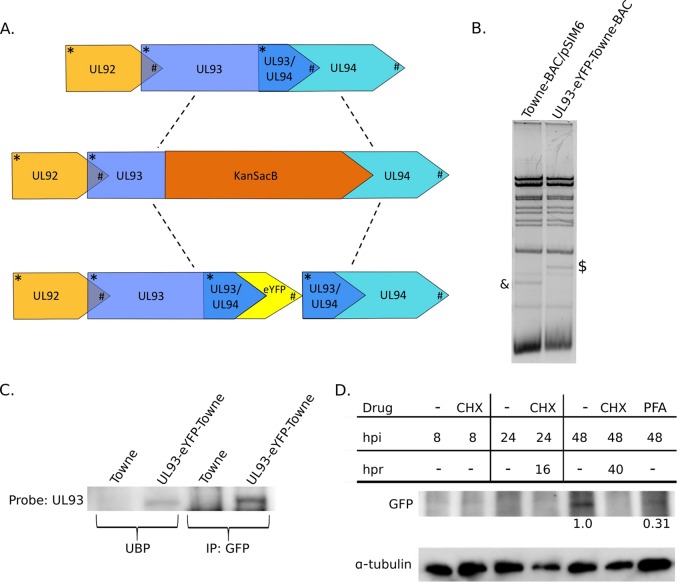
Bacterial artificial chromosome (BAC) recombineering protocols (20–22) were followed, with minor modifications, in order to engineer enhanced yellow fluorescent protein (eYFP) at the C terminus of UL93 in the Towne-BAC genome (GenBank accession number KF493877.1). Briefly, in the first step, a KanSacB cassette replacing the UL93 region was inserted into the BAC genome, and clones were selected on the basis of kanamycin resistance (Fig. 1A). Once verified by restriction fragment length polymorphism (RFLP), in the second step, a PCR fragment containing UL93-eYFP was introduced in the selected BAC clone, and subsequent screening for the loss of kanamycin resistance identified clones that contained this desired mutation (Fig. 1A). UL93 partially overlaps UL94, so to avoid interruption of UL94 by eYFP, the overlapping UL93-UL94 sequence was duplicated and added downstream of eYFP in the PCR fragment used in the second step of recombineering. Insertion of the mutation as well as the absence of any undesired changes in the proximity of the insertion site were confirmed by RFLP analysis and further by DNA sequencing of the UL93-UL94 region from these BAC. In a third round of BAC recombineering, a KanSacB cassette was inserted to knock out the GFP expression cassette that replaced US1 to US12 in the Towne-BAC genome (23) to enable the visualization of pUL93-eYFP fluorescence independent of the fluorescence from BAC-integrated GFP. The selected kanamycin-resistant clones were confirmed by RFLP (Fig. 1B) analysis and DNA sequencing. BAC were transfected in HF, and infected cells were harvested after cytopathogenic effect (CPE) reached 100%. Harvested cells were sonicated (Braun-Sonic 2000; B. Braun Biotech Inc., Allentown, PA) to release the virus, and the preparation was diluted 10- to 100,000-fold before plating on confluent HF for the determination of viral titers. At 10 days postinfection (dpi), cells were fixed in 100% methanol, stained with Giemsa stain (GS1L; Sigma-Aldrich, St. Louis, MO), and allowed to dry before enumerating PFU under an optical microscope. UL93-eYFP-Towne-BAC virus grew to high titers (1.38 × 108 PFU) in HF. After virus stocks were harvested and the titers were determined, the insertion of the desired mutations and absence of undesired changes in the UL93-UL94 region were verified by PCR amplification and DNA sequencing of this region of the virus genome. Upon sequencing, we found that UL93-eYFP-Towne-BAC virus stocks contained both eYFP-tagged UL93 virus and wild-type (untagged UL93) virus, indicating some mixing of the genomes (data not shown), most likely as a result of unintended recombining out of the eYFP cassette due to duplication of the overlapping UL93-UL94 sequence after transfection. After plaque purification of this virus, mixed populations became apparent again after two to three passages, further supporting that these mixed populations were due to recombination events. Virus was harvested and immunoblotting (IB) was performed, probing with the pUL93 antibody (described below) to demonstrate the presence of both the tagged and untagged versions of pUL93 (data not shown). We also used the same technique to tag UL93 with enhanced green fluorescent protein (eGFP) in the TB40/E strain, but mixed populations also arose after transfection with this BAC (data not shown).
FIG 1.
UL93 is a late viral protein. (A) Construction of a fluorescently tagged UL93 Towne virus (UL93-eYFP-Towne-BAC) using BAC recombineering (21). Schematic of the UL92-UL94 region in Towne-BAC (top line) with the intermediate and final genetic constructs shown underneath. UL93 overlaps both UL92 and UL94 open reading frames. A C-terminal region of UL93 overlapping UL94 first was replaced with a KanSacB cassette for selection with kanamycin (middle line). The KanSacB cassette was subsequently replaced with UL93 fused to eYFP flanked by duplicate sequences of the overlapping UL93/UL94 region in order to keep the UL94 open reading frame intact (bottom line). Start codons are marked by asterisks and stop codons by hashtags. (B) RFLP analysis of wild-type Towne-BAC and UL93-eYFP-Towne-BAC. The hash tag and asterisk mark expected changes in UL93-eYFP-Towne-BAC after recombineering. (C) HF were infected with UL93-eYFP-Towne-BAC virus at an MOI of 3.0 and harvested at 72 hpi, and immunoprecipitation (IP) with an anti-GFP antibody and immunoblot (IB) probing with an anti-UL93 antibody were performed to confirm the fusion of pUL93 to eYFP. UBP, unbound protein (supernatant of the immunoprecipitations). (D) Time course (hpi) of pUL93-eYFP expression following UL93-eYFP-Towne-BAC virus infection of HF using whole-cell lysates and detection with antibodies against GFP or α-tubulin (loading control) and depicting the impact of cycloheximide (CHX) or phosphonoformic acid (PFA) treatment on pUL93-eYFP expression. Shown are the accumulated levels of pUL93-eYFP or α-tubulin from UL93-eYFP-Towne-BAC virus-infected cells that were either cultured with the indicated drugs until harvest at the indicated times (hpi) or cultured with the indicated drugs for 8 h, washed, and then cultured in normal medium until harvest at the times, indicated as hours postrelease (hpr). The PFA-treated GFP band is 31% the density of the mock-treated GFP band at 48 hpi based on densitometry.
Co-IP analysis.
HF infected with UL93-eYFP-Towne-BAC virus at a multiplicity of infection (MOI) of 3.0 were harvested at 72 h postinfection (hpi) for coimmunoprecipitation (co-IP) with protein A/G plus-agarose (SC-2003; Santa Cruz Biotech, Dallas, TX) according to the manufacturer's protocol, followed by IB. 293T cells cotransfected with Myc-UL77 and FLAG-UL93 (full-length or truncated) plasmids were harvested at 48 hpi for co-IP with anti-c-Myc affinity gel (E6654; Sigma-Aldrich, St. Louis, MO) according to the manufacturer's protocol, followed by IB.
Drug inhibition and release experiments.
For determination of pUL93 kinetics, HF were pretreated with cycloheximide (CHX; 50 μg/ml; AC357420010; Fisher Scientific, Waltham, MA) for 1 h and then infected with UL93-eYFP-Towne-BAC virus at an MOI of 3.0 in medium containing CHX (50 μg/ml) for 1 h, washed, and then incubated for 8 h in the presence of CHX (50 μg/ml). After this, HF were washed four times with medium and incubated in drug-free medium. Phosphonoformic acid (PFA; 300 μg/ml; P6801; Sigma-Aldrich, St. Louis, MO) was added at 1 hpi and maintained until 48 hpi. Cells were harvested at designated time points and stored at −80°C before IB analyses.
Separation of cytoplasmic and nuclear fractions.
HF were infected with UL93-eYFP-Towne-BAC virus at an MOI of 3.0 or mock infected and were harvested at 0, 6, 24, 48, 72, 96, and 120 hpi. Nuclear and cytoplasmic extraction reagents (NE-PER; PI78833; Fisher Scientific, Waltham, MA) were used to separate nuclear and cytoplasmic fractions of infected cells according to the manufacturer's protocols before analysis by IB.
Virus and capsid purification.
Extracellular HCMV virions and nuclear B-capsids were purified using established protocols for herpesviruses (24–26), with some modifications. Briefly, HF were infected with UL93-eYFP-Towne-BAC virus or Towne virus at an MOI of 3.0 and harvested after 100% CPE. Medium was clarified of any cellular debris by low-speed centrifugation (2,000 × g, 10 min), and virus particles were pelleted (20,000 × g, 1 h). Extracellular virus (ECV) particles were purified on a 30% sucrose cushion (SW 32Ti rotor, Beckman L-90 ultracentrifuge, 24,000 rpm, 1 h), harvested by puncturing the side of the centrifuge tube with a 23-gauge needle, and diluted 1:3 in phosphate buffer before concentration by centrifugation (SW-41 rotor, Beckman L-80 ultracentrifuge, 24,000 rpm, 1 h). For purification of nuclear B-capsids, cell pellets were washed in 1× phosphate-buffered saline (PBS) and incubated in hypotonic buffer (20 mM Tris-HCl, pH 7.5) for 20 min to swell cells before the addition of Triton X-100 (1.5% final concentration) to lyse the cells for 30 min. Nuclei were spun at 2,000 × g for 10 min, resuspended in TNE (500 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, pH 8.0), sonicated (Braun-Sonic 2000; B. Braun Biotech Inc., Allentown, PA) for 10 s, and spun at 14,000 rpm for 30 s in a microcentrifuge. The above-described step was repeated once, and combined supernatants were loaded onto a 20 to 50% discontinuous sucrose-in-TNE gradient before centrifugation (SW-41 rotor, Beckman L-80 ultracentrifuge, 24,000 rpm, 1 h). Capsids were observed as visible-light-scattering bands with A-capsids forming a thin upper band at around 30% sucrose concentration and B-capsids forming a thick lower band at around 35%; we were unable to see a C-capsid band. The B-capsids were harvested by puncturing the side of the centrifuge tube with a 23-gauge needle and diluted in TNE before concentration by centrifugation (SW-41 rotor, Beckman L-80 ultracentrifuge, 24,000 rpm, 1 h). The capsids or ECV particles were resuspended in 2% SDS-containing lysis buffer and run on a 4 to 20% gradient polyacrylamide gel before transfer to a polyvinylidene difluoride (PVDF) membrane and analysis by IB.
Antibodies and immunoblotting.
Mouse monoclonal antibody to GFP (MAB1083; EMD-Millipore, Darmstadt, Germany) was used in co-IP and IB experiments. This anti-GFP antibody interacts with GFP as well as with its variants, including eYFP (27). Mouse monoclonal antibody to α-tubulin (MA1 19401; Fisher Scientific, Waltham, MA) was used to confirm equal loading of SDS gels in IB experiments. For co-IP and IB in the infection setting, mouse monoclonal anti-UL93 antibody, kindly provided by Eva Maria Borst (18), mouse monoclonal anti-lamin A/C antibody (MA3 1000; Fisher Scientific, Waltham, MA), and rabbit anti-UL50, anti-UL53, and anti-UL97 antibodies, kindly provided by Don Coen (9), were used as the primary antibodies. Peroxidase-labeled goat anti-mouse IgG (PI31444; Fisher Scientific, Waltham, MA) or peroxidase-labeled goat anti-rabbit IgG (PI31460; Fisher Scientific, Waltham, MA) was used as the secondary antibody in IB. For transient transfection in 293T cells and co-IP experiments, anti-c-Myc antibody (9E10.3; Fisher Scientific, Waltham, MA) was used for immunoprecipitation, anti-FLAG M2 antibody (200474; Agilent Technologies, Santa Clara, CA) was used as the primary antibody, and peroxidase-labeled goat anti-mouse IgG (PI31444; Fisher Scientific, Waltham, MA) was used as the secondary antibody for IB. All blots were detected using Clarity Western ECL substrate (1705061; Bio-Rad Laboratories, Hercules, CA).
Microscopy.
Samples were prepared using established protocols for fluorescence microscopy of HCMV-infected cells (22, 28). HF were grown on coverslip inserts in 24-well tissue culture dishes and subsequently infected with UL93-eYFP-Towne-BAC virus at an MOI of 3.0 or were mock infected. Cells were fixed for fluorescence microscopy at 0 or 96 hpi in 3.7% formaldehyde for 10 min and incubated in 50 mM NH4Cl in 1× PBS for 10 min to reduce autofluorescence. This was followed by washing in 1× PBS, incubation in 0.5% Triton X-100 for 20 min to permeabilize cells, and finally washing and staining with Hoechst 33258 (AnaSpec Corporation, San Jose, CA) to identify the nuclei. Coverslips were retrieved from wells, mounted on glass slides with a drop of mounting medium (Gel/Mount; Biomeda, Foster City, CA), and dried overnight before imaging. Images were acquired on an EVOS-FL microscope.
RESULTS
pUL93 is expressed late during infection and is localized primarily to the nucleus of infected cells.
To determine when pUL93 expression occurs during infection, we first engineered an eYFP tag at the C terminus of pUL93 in the Towne-BAC genome (UL93-eYFP-Towne-BAC) using BAC recombineering (Fig. 1A) (20–22). BAC constructs were validated by RFLP analysis as well as by PCR sequencing of the UL93-UL94 region of the BAC genome (Fig. 1B). To further confirm the fusion of eYFP to pUL93, HF were infected with UL93-eYFP-Towne-BAC virus and harvested at 72 hpi for immunoprecipitation (IP); IP with an anti-GFP antibody and IB probing with an anti-pUL93 antibody, which became available during the course of this study (18), were performed. The 95-kDa band seen in the IB matches the predicted size of pUL93-eYFP, confirming that pUL93 is fused to eYFP and attesting to the feasibility of co-IP with this anti-GFP antibody (Fig. 1C). UL93-eYFP-Towne-BAC was grown in HF, and final virus yields at 5 dpi were comparable to those of the parent Towne-BAC virus (data not shown), indicating no impact on pUL93 function due to eYFP fusion. HF were treated with either CHX or PFA and then infected with UL93-eYFP-Towne-BAC virus and probed with an anti-GFP antibody to monitor the time course of pUL93 expression. CHX blocks all protein synthesis, whereas PFA blocks viral DNA synthesis, thereby blocking only late viral protein expression. pUL93-eYFP was first detected at 48 hpi in the mock-treated sample but not in the CHX-treated samples and was greatly reduced (31% of the mock-treated sample) in the PFA-treated sample, indicating that pUL93 is a late protein (Fig. 1D). It is important to note that since PFA did not completely block protein expression, pUL93 is a leaky late protein, not a true late protein.
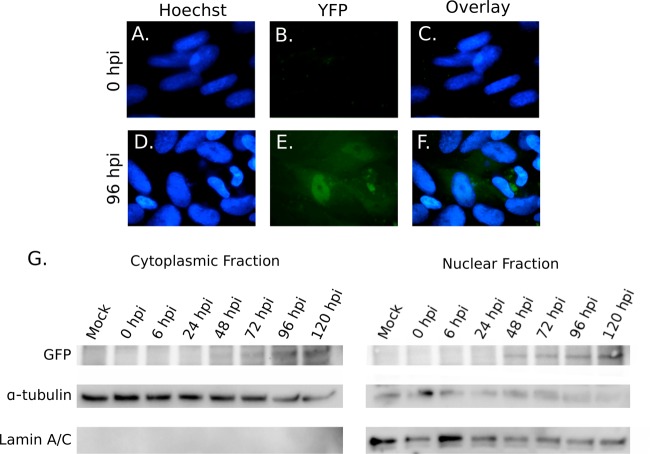
To determine the localization of pUL93 during infection, HF were infected with UL93-eYFP-Towne-BAC virus at an MOI of 3.0 and fixed and stained with Hoechst at 0 and 96 hpi. pUL93 localized primarily to the nucleus of infected cells (Fig. 2A to F). To confirm this localization, HF were infected with UL93-eYFP-Towne-BAC virus and harvested at 0, 6, 24, 48, 72, 96, and 120 hpi. Nuclear and cytoplasmic fractions were then separated and IB was performed using anti-GFP, anti-lamin A/C (nuclear control), and anti-α-tubulin (cytoplasmic control) antibodies. Based on IB probing for α-tubulin and lamin A/C, there was no nuclear contamination in the cytoplasmic fraction, but there was some cytoplasmic contamination in the nuclear fraction (Fig. 2G). The majority of pUL93 localized to the nuclear fraction, although some pUL93 was detected in the cytoplasmic fraction by 48 hpi. Overall, the levels of this protein increased through 120 hpi. These experiments confirm that pUL93 is located primarily in the nucleus during infection (Fig. 2G).
FIG 2.
pUL93 primarily localizes to the nucleus of infected cells. (A to F) HF were infected with UL93-eYFP-Towne-BAC virus at an MOI of 3.0, fixed at 0 or 96 hpi, and stained with Hoechst (blue). (G) HF were infected with UL93-eYFP-Towne-BAC virus at an MOI of 3.0 and harvested at 0, 6, 24, 48, 72, 96, and 120 hpi. Cytoplasmic and nuclear fractions were separated, and IB was performed using antibodies against GFP, α-tubulin, and lamin A/C.
pUL93 is associated with virions and B-capsids.
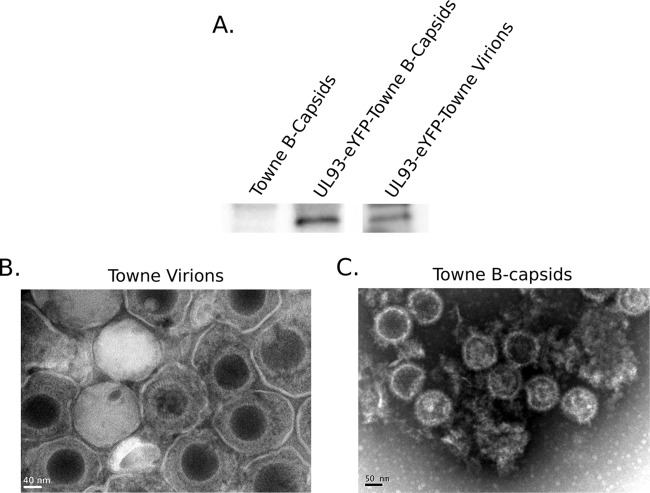
To determine if pUL93 is present in virions and capsids, UL93-eYFP-Towne-BAC virions and nuclear B-capsids as well as unlabeled Towne nuclear B-capsids were purified on sucrose gradients, run on SDS-PAGE gels, and transferred to PVDF membranes before probing for eYFP. The IB data show that pUL93-eYFP is associated with purified virions and nuclear B-capsids (Fig. 3A). Unlabeled Towne nuclear B-capsids did not have the pUL93-eYFP specific protein signal, confirming the specificity of eYFP-labeled pUL93 detection.
FIG 3.
pUL93 associates with virions and capsids. (A) UL93-eYFP-Towne-BAC virions and nuclear B-capsids or unlabeled Towne nuclear B-capsids were harvested from infected HF and purified on sucrose gradients and then run on an SDS-PAGE gel. IB was performed, probing for pUL93-eYFP. Also shown are typical transmission electron micrographs of purified Towne virions (B) and Towne B-capsids (C). The negative stain (uranyl acetate) penetrated some virions, visible as a dark-stained DNA core, but not all of the virions. Some debris in B-capsid preparation is visible as a result of disintegration of capsids, most likely as a result of sonication that was performed to reduce capsid aggregation.
pUL93 interacts with pUL77 and multiple regions of pUL93 mediate this interaction.
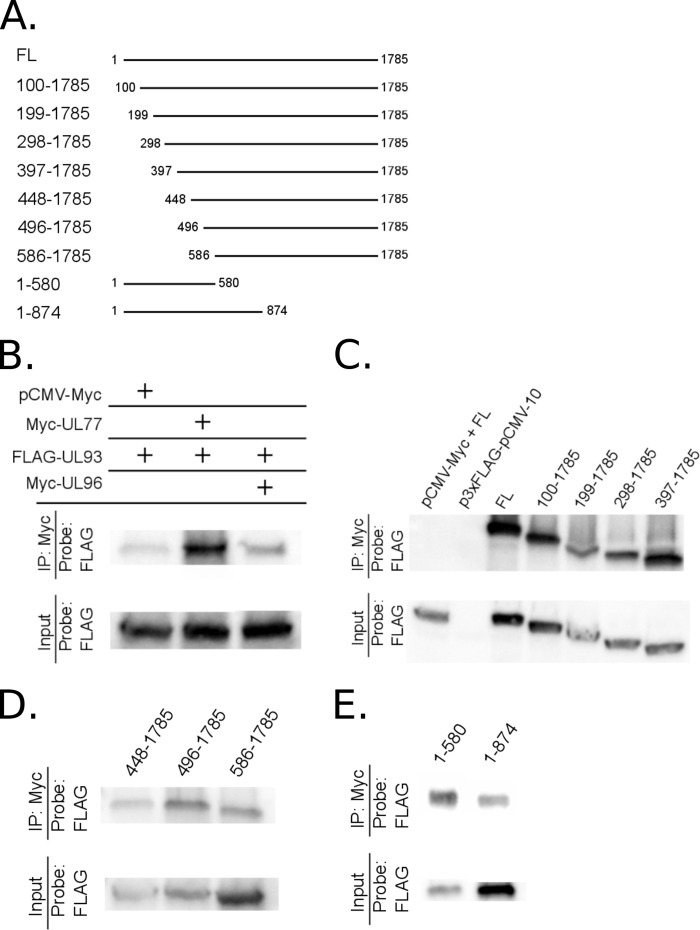
To determine if HCMV pUL77 and pUL93 interact with each other like their homologs in other herpesviruses, we constructed Myc-UL77 and FLAG-pUL93 expression plasmids. Interactions between Myc-UL77 and FLAG-UL93 were investigated in transfected 293T cells where anti-Myc antibody was used for co-IP and anti-FLAG antibody was used for IB. An empty pCMV-Myc plasmid and a Myc-UL96 (an unrelated protein) expression plasmid were used as noninteracting controls. Immunoblotting demonstrated that pUL93 and pUL77 interacted with each other and that this interaction was specific (Fig. 4B).
FIG 4.
Multiple regions of pUL93 independently interact with pUL77. 293T cells were cotransfected with plasmids expressing Myc-tagged pUL77 and FLAG-tagged pUL93 or FLAG-tagged pUL93 truncated from either the N terminus or the C terminus (A) and harvested at 48 hpi, and co-IP and IB were performed as indicated (B to E). Empty vector pCMV-Myc or a Myc-UL96 (an unrelated protein) expression plasmid was cotransfected with full-length FLAG-UL93 as a negative control in panel B. The empty vector pCMV-Myc was cotransfected with full-length FLAG-UL93 (FL) and the empty vector p3xFLAG-pCMV-10 was cotransfected with Myc-UL77 as negative controls in panel C.
To determine which regions of pUL93 are necessary for interaction with pUL77, we generated UL93 expression clones that were truncated to various lengths from the N terminus or the C terminus (Fig. 4A). Given that pUL93 is a relatively small protein (595 amino acids) and we could not identify any conserved domains, we progressively truncated UL93 from either the N or the C terminus. Interactions between Myc epitope-tagged UL77 and FLAG epitope-tagged UL93 clones were investigated using co-IP and IB. All of the N-terminal and C-terminal truncations of pUL93 interacted with pUL77, indicating that multiple regions of pUL93 can independently interact with pUL77 (Fig. 4C to E) in this overexpression setting.
pUL93 interacts with components of the nuclear egress complex.
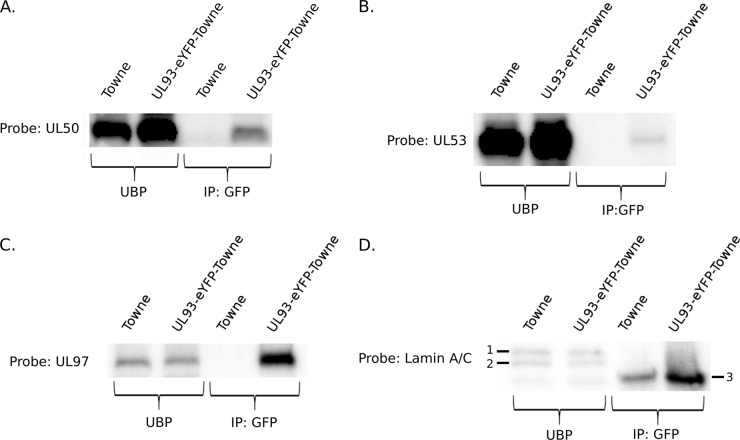
HSV-1 pUL17, the homolog of HCMV pUL93, interacts with pUL31, the homolog of HCMV pUL53 and a component of the NEC, possibly to aid in nuclear egress (12). To detect interaction of pUL93 with components of the NEC in HCMV, we infected HF with UL93-eYFP-Towne-BAC virus, harvested cells at 72 hpi, and then performed co-IP with anti-GFP antibody and IB with antibodies targeting either pUL50, pUL53, pUL97, or lamin A/C. Cells infected with unlabeled Towne virus were used as a control. The data demonstrated that pUL93 interacted with pUL50 and pUL53, the core components of the NEC (Fig. 5A and B). We also found that pUL93 interacted with pUL97 (Fig. 5C). pUL93 did not interact with lamin A/C in this assay (Fig. 5D).
FIG 5.
pUL93 interacts with components of the NEC. HF were infected with UL93-eYFP-Towne-BAC virus at an MOI of 3.0 and harvested at 72 hpi, and co-IP with an anti-GFP antibody was performed. IB was performed, probing for pUL50 (A), pUL53 (B), pUL97 (C), or lamin A/C (D). Bands labeled 1 and 2 are lamins A and C, and the band labeled 3 is IgG heavy chain. UBP, unbound protein (supernatant of the immunoprecipitation); IP, immunoprecipitation.
DISCUSSION
In the current study, we show that pUL93 is a leaky late protein that is localized primarily to the nucleus of infected cells (Fig. 1D and 2), which is consistent with our recent study implicating pUL93 in viral DNA packaging (17). An early-late promoter most likely is involved in the transcription of UL93 (29). Bioinformatics analysis shows that pUL93 contains at least two strong putative nuclear localization signals (NLS) at amino acids 177 to 193 (KRDRQHQLATATNHRRR) and 442 to 447 (RARRQR) (30) that could mediate its localization to the nucleus independent of other proteins. Based on HSV-1 data showing that homologs of pUL93 and pUL77 interact with pUL31 (a homolog of HCMV pUL53 and a component of the NEC) (12), it was hypothesized that pUL93 and pUL77 together (as the CVSC) are involved in nuclear egress, and localization of pUL93 primarily to the nucleus is in line with this hypothesis. We also found that pUL93 is associated with purified virions and nuclear capsids (Fig. 3), which is in agreement with a recent study (18). These data support an important structural role for pUL93.
To determine if pUL93 interacts with pUL77, we performed co-IP and IB in a transient-transfection setting. We found that pUL93 interacted with pUL77 (Fig. 4B). To further investigate this interaction, we tested the ability of pUL93 truncation mutations to interact with pUL77 and found that multiple regions in pUL93 independently interacted with pUL77 (Fig. 4A and C to E). This aroused our suspicion that pUL93 could be nonspecifically interacting with pUL77; however, a very weak interaction with pUL96, an unrelated protein, and no interaction with the Myc-expressing empty vector verified the specificity of this pUL93-pUL77 interaction (Fig. 4B). It is also important to note that these interactions were tested only in overexpression settings, not during infection. Our findings are in agreement with two recent studies that also found that pUL93 and pUL77 interact with each other (18, 31). These data support the hypothesis that pUL93 interacts with pUL77 to form a structure similar to the CVSC in other herpesviruses. The identity and structure of this complex are the subject of further investigation in our laboratory.
During nuclear egress, the NEC in HCMV recruits viral protein kinase pUL97 for the dissolution of the nuclear lamina, and it is the main entity involved in directing the budding of nucleocapsids through the nuclear membrane for primary envelopment and eventual egress of nucleocapsids from the nucleus (9, 10). The factors involved in the movement of nucleocapsids from the site of viral DNA packaging to the INM are not well understood, but in a recent study it was proposed that this movement occurs by diffusion of nucleocapsids rather than by directed movement (32). Given that the homologs of pUL93 and pUL77 in HSV-1 and PRV are known to interact to form the CVSC and that they individually interact with a component of the NEC in HSV-1, it has been hypothesized that the CVSC is responsible for the selection of capsids at the INM for nuclear egress (5, 12). To test the possibility of an interaction between pUL93 and the NEC during infection, we used co-IP and IB experiments. We demonstrate that pUL93 interacted with both components of the NEC in HCMV (pUL50 and pUL53) (Fig. 5A and B). It is worth noting that in HSV-1, both pUL17 and pUL25 interact with pUL31, but neither pUL17 nor pUL25 interacts with pUL34 (homolog of HCMV pUL50), which is responsible for nuclear membrane tethering (12). Interestingly, we also found that pUL93 interacted with pUL97 (Fig. 5C). We were unable to detect an interaction between pUL93 and lamin A/C (Fig. 5D), suggesting that pUL93 is not directly responsible for tethering of nucleocapsids to the nuclear lamina but would probably use an NEC interaction mechanism for this purpose, as reported for other herpesviruses (33–35). Given that these studies were done in an infection setting, it is possible that these are not direct interactions and might require additional factors present only during infection.
Given our findings that pUL93 interacted with pUL77 and also with several components of the NEC, we hypothesize that pUL93 and pUL77 interact to form a complex at the capsid surface that then interacts either directly or indirectly with pUL97, pUL50, and pUL53 to aid in nuclear egress. pUL93 is an essential structural protein, and targeting structural proteins has been shown to avoid the development of resistance (36). Once we know more about the interaction surfaces in play, novel tools such as nanobiotechnology (37) can be used to target those activities. Further studies are needed to establish the presence of a CVSC-like structure on HCMV nucleocapsid vertices and to delineate its roles in nucleocapsid stability and nuclear egress.
ACKNOWLEDGMENTS
We are thankful to Eva Maria Borst for providing antibody against pUL93 and Don Coen for providing antibodies against pUL50, pUL53, and pUL97. Madeline Archer and Leslie Davis helped with cell culture in the Tandon laboratory.
REFERENCES
- 1.Toropova K, Huffman JB, Homa FL, Conway JF. 2011. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J Virol 85:7513–7522. doi: 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell 26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL. 2011. Residues of the UL25 protein of herpes simplex virus that are required for its stable interaction with capsids. J Virol 85:4875–4887. doi: 10.1128/JVI.00242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conway JF, Cockrell SK, Copeland AM, Newcomb WW, Brown JC, Homa FL. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J Mol Biol 397:575–586. doi: 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Homa FL, Huffman JB, Toropova K, Lopez HR, Makhov AM, Conway JF. 2013. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J Mol Biol 425:3415–3428. doi: 10.1016/j.jmb.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thurlow JK, Murphy M, Stow ND, Preston VG. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J Virol 80:2118–2126. doi: 10.1128/JVI.80.5.2118-2126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt B. 2007. Maturation and egress. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, England. [PubMed] [Google Scholar]
- 8.Mettenleiter TC. 2002. Herpesvirus assembly and egress. J Virol 76:1537–1547. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma M, Kamil JP, Coughlin M, Reim NI, Coen DM. 2014. Human cytomegalovirus UL50 and UL53 recruit viral protein kinase UL97, not protein kinase C, for disruption of nuclear lamina and nuclear egress in infected cells. J Virol 88:249–262. doi: 10.1128/JVI.02358-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leigh KE, Sharma M, Mansueto MS, Boeszoermenyi A, Filman DJ, Hogle JM, Wagner G, Coen DM, Arthanari H. 2015. Structure of a herpesvirus nuclear egress complex subunit reveals an interaction groove that is essential for viral replication. Proc Natl Acad Sci U S A 112:9010–9015. doi: 10.1073/pnas.1511140112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camozzi D, Pignatelli S, Valvo C, Lattanzi G, Capanni C, Dal Monte P, Landini MP. 2008. Remodelling of the nuclear lamina during human cytomegalovirus infection: role of the viral proteins pUL50 and pUL53. J Gen Virol 89:731–740. doi: 10.1099/vir.0.83377-0. [DOI] [PubMed] [Google Scholar]
- 12.Yang K, Baines JD. 2011. Selection of HSV capsids for envelopment involves interaction between capsid surface components pUL31, pUL17, and pUL25. Proc Natl Acad Sci U S A 108:14276–14281. doi: 10.1073/pnas.1108564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn J, Leege T, Klupp BG, Granzow H, Fuchs W, Mettenleiter TC. 2008. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J Virol 82:5725–5734. doi: 10.1128/JVI.02441-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klupp BG, Granzow H, Keil GM, Mettenleiter TC. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J Virol 80:6235–6246. doi: 10.1128/JVI.02662-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeRussy BM, Tandon R. 2015. Human cytomegalovirus pUL93 is required for viral genome cleavage and packaging. J Virol 89:12221–12225. doi: 10.1128/JVI.02382-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borst EM, Bauerfeind R, Binz A, Stephan TM, Neuber S, Wagner K, Steinbruck L, Sodeik B, Lenac Rovis T, Jonjic S, Messerle M. 23 March 2016. The essential human cytomegalovirus proteins pUL77 and pUL93 are structural components necessary for viral genome encapsidation. J Virol doi: 10.1128/JVI.00384-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meissner CS, Koppen-Rung P, Dittmer A, Lapp S, Bogner E. 2011. A “coiled-coil” motif is important for oligomerization and DNA binding properties of human cytomegalovirus protein UL77. PLoS One 6:e25115. doi: 10.1371/journal.pone.0025115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 21.Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, Oppenheim A. 2007. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol Chapter 1:Unit 1.16. [DOI] [PubMed] [Google Scholar]
- 22.Brechtel TM, Mocarski ES, Tandon R. 2014. Highly acidic C-terminal region of cytomegalovirus pUL96 determines its functions during virus maturation independently of a direct pp150 interaction. J Virol 88:4493–4503. doi: 10.1128/JVI.03784-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchini A, Liu H, Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J Virol 75:1870–1878. doi: 10.1128/JVI.75.4.1870-1878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson W. 1981. Structural and nonstructural proteins of strain Colburn cytomegalovirus. Virology 111:516–537. doi: 10.1016/0042-6822(81)90354-8. [DOI] [PubMed] [Google Scholar]
- 25.Irmiere A, Gibson W. 1985. Isolation of human cytomegalovirus intranuclear capsids, characterization of their protein constituents, and demonstration that the B-capsid assembly protein is also abundant in noninfectious enveloped particles. J Virol 56:277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomsen DR, Newcomb WW, Brown JC, Homa FL. 1995. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J Virol 69:3690–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bossuyt J, Despa S, Martin JL, Bers DM. 2006. Phospholemman phosphorylation alters its fluorescence resonance energy transfer with the Na/K-ATPase pump. J Biol Chem 281:32765–32773. doi: 10.1074/jbc.M606254200. [DOI] [PubMed] [Google Scholar]
- 28.Tandon R, Mocarski ES. 2011. Cytomegalovirus pUL96 is critical for the stability of pp150-associated nucleocapsids. J Virol 85:7129–7141. doi: 10.1128/JVI.02549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wing BA, Huang ES. 1995. Analysis and mapping of a family of 3′-coterminal transcripts containing coding sequences for human cytomegalovirus open reading frames UL93 through UL99. J Virol 69:1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin JR, Hu J. 2013. SeqNLS: nuclear localization signal prediction based on frequent pattern mining and linear motif scoring. PLoS One 8:e76864. doi: 10.1371/journal.pone.0076864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koppen-Rung P, Dittmer A, Bogner E. 6 April 2016. Intracellular distributions of capsid-associated pUL77 of HCMV and interactions with packaging proteins and pUL93. J Virol doi: 10.1128/JVI.00351-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosse JB, Hogue IB, Feric M, Thiberge SY, Sodeik B, Brangwynne CP, Enquist LW. 2015. Remodeling nuclear architecture allows efficient transport of herpesvirus capsids by diffusion. Proc Natl Acad Sci U S A 112:E5725–E5733. doi: 10.1073/pnas.1513876112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J Virol 75:8803–8817. doi: 10.1128/JVI.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang L, Baines JD. 2005. Identification of an essential domain in the herpes simplex virus 1 UL34 protein that is necessary and sufficient to interact with UL31 protein. J Virol 79:3797–3806. doi: 10.1128/JVI.79.6.3797-3806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamauchi Y, Shiba C, Goshima F, Nawa A, Murata T, Nishiyama Y. 2001. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J Gen Virol 82:1423–1428. doi: 10.1099/0022-1317-82-6-1423. [DOI] [PubMed] [Google Scholar]
- 36.Tanner EJ, Liu HM, Oberste MS, Pallansch M, Collett MS, Kirkegaard K. 2014. Dominant drug targets suppress the emergence of antiviral resistance. eLife 3:e03830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeRussy BM, Aylward MA, Fan Z, Ray PC, Tandon R. 2014. Inhibition of cytomegalovirus infection and photothermolysis of infected cells using bioconjugated gold nanoparticles. Sci Rep 4:5550. [DOI] [PMC free article] [PubMed] [Google Scholar]