ABSTRACT
Reconstitution of T cell immunity is absolutely critical for the effective control of virus-associated infectious complications in hematopoietic stem cell transplant (HSCT) recipients. Coinfection with genetic variants of human cytomegalovirus (CMV) in transplant recipients has been linked to clinical disease manifestation; however, how these genetic variants impact T cell immune reconstitution remains poorly understood. In this study, we have evaluated dynamic changes in the emergence of genetic variants of CMV in HSCT recipients and correlated these changes with reconstitution of antiviral T cell responses. In an analysis of single nucleotide polymorphisms within sequences encoding HLA class I-restricted CMV epitopes from the immediate early 1 gene of CMV, coinfection with genetically distinct variants of CMV was detected in 52% of patients. However, in spite of exposure to multiple viral variants, the T cell responses in these patients were preferentially directed to a limited repertoire of HLA class I-restricted CMV epitopes, either conserved, variant, or cross-reactive. More importantly, we also demonstrate that long-term control of CMV infection after HSCT is primarily mediated through the efficient induction of stable antiviral T cell immunity irrespective of the nature of the antigenic target. These observations provide important insights for the future design of antiviral T cell-based immunotherapeutic strategies for transplant recipients, emphasizing the critical impact of robust immune reconstitution on efficient control of viral infection.
IMPORTANCE Infection and disease caused by human cytomegalovirus (CMV) remain a significant burden in patients undergoing hematopoietic stem cell transplantation (HSCT). The establishment of efficient immunological control, primarily mediated by cytotoxic T cells, plays a critical role in preventing CMV-associated disease in transplant recipients. Recent studies have also begun to investigate the impact genetic variation in CMV has upon disease outcome in transplant recipients. In this study, we sought to investigate the role T cell immunity plays in recognizing and controlling genetic variants of CMV. We demonstrate that while a significant proportion of HSCT recipients may be exposed to multiple genetic variants of CMV, this does not necessarily lead to immune control mediated via recognition of this genetic variation. Rather, immune control is associated with the efficient establishment of a stable immune response predominantly directed against immunodominant conserved T cell epitopes.
INTRODUCTION
Allogeneic hematopoietic stem cell transplantation (HSCT) can be curative of life-threatening hematological malignancies. However, due to the underlying immunodeficiency associated with HSCT and as a consequence of the immunosuppressive regimes used to prevent graft-versus-host disease following HSCT, infectious complications remain a significant burden to the treatment modality. One significant infectious complication following HSCT is caused by the ubiquitous pathogen human cytomegalovirus (CMV) (1). A member of the human betaherpesvirus family, CMV is highly prevalent across populations and is typically established as a lifelong asymptomatic infection in immunocompetent individuals. However, CMV is a leading cause of viral complications in immunocompromised individuals (2). This is particularly evident in the absence of CMV-specific immunological memory, including in CMV-seropositive HSCT recipients (R+) who receive a transplant from a seronegative donor (D−) and are at a higher risk of CMV reactivations and associated complications, including enterocolitis and pneumonitis (3–5). Current therapeutic strategies to control CMV reactivation in HSCT recipients predominantly involve the preemptive administration of ganciclovir to control CMV following detection of viral reactivation (6). Through the use of immunological monitoring approaches, it is becoming apparent that the prevention of viral reactivation and the long-term control of CMV infection are dependent upon the induction of robust and stable CMV-specific immunological memory (7–10).
Recent studies have suggested that in addition to the efficiency of immunological control of CMV, exposure to genotypically distinct variants of CMV may also have an impact on clinical outcome following transplant. Genotypic analyses of surface CMV glycoproteins have shown that immunocompromised patients, both HSCT and solid-organ transplant (SOT) recipients, are commonly coinfected with multiple genotypically distinct CMV variants (11, 12). It has also been demonstrated that SOT recipients show an increased duration of viremia following reactivation of multiple genotypic isolates (12), suggesting potentially reduced immunological control following coinfection. Despite these observations, and considering the critical role T cell immunity plays in the control of CMV, very little research has been performed that specifically examines the impact of genetically distinct variants of CMV on CMV-specific T cell immunity (13–15). While this is particularly relevant for R+/D− HSCT patients, who are at increased risk of CMV-associated complications, immune control of CMV infection in R+/D+ recipients could also be impacted by exposure to distinct genetic viral variants of the recipient that are not efficiently controlled by preexisting donor immunity. To address the impact genetic variation has upon T cell immunity, we focused upon the immunodominant immediate early 1 gene (IE-1) of CMV, which has previously been shown to encode significant genetic variation, including within immunodominant CD8+ T cell epitopes (13–15). Using pyrosequencing analysis to identify genetic variation within IE-1, and IE-1-encoded epitope-specific T cell analysis, we sought to determine the impact of genetic variation and exposure to multiple viral variants on the induction of CMV-specific T cell immunity in a cohort of HSCT recipients.
MATERIALS AND METHODS
Study subjects.
The study subjects were from a cohort of 46 allogeneic HSCT recipients who were recruited on an immune monitoring study approved by the Royal Brisbane and Women's Hospital Human Research Ethics Committees (reference number 2006/192) (9, 16). All patients provided informed written consent. As described previously (9), all patients were monitored for CMV viral load using the COBAS Amplicator CMV Monitor test (Roche Diagnostics, Basel, Switzerland) and CMV-specific T cell immunity using the QuantiFERON-CMV assay (Cellestis, Carnegie, VIC, Australia). CMV reactivation was defined as the detection of >600 copies/ml of CMV DNA.
Detection of IE-1 variants using pyrosequencing.
DNA was extracted from plasma samples using the QIAamp DNA blood minikit (Qiagen, USA). DNA PCR amplifications were performed using the PyroMark PCR kit (Qiagen, USA) in a standard 25-μl reaction for 45 cycles. PCR amplification primers and the target sequences are as follows: IE1Start, forward primer GGAGATGTGGATGGCTTGTATT, reverse primer GCAGCCATTGGTGGTCTTA, and sequencing primer YATTCCTGTAGCACATATA (target sequence: MATCATCTTTCTCYTAAGTTCRTCCTT); IE1Middle, forward primer TAAGACCACCAATGGCTGC, reverse primer CATACAAGCGTCACTRGTGACCT, and sequencing primer AATCTTAAAKATYTTCTG (target sequence: GGMATAAGYCATAATCTCATCAGGG); and IE1end, forward primer TYTGTCGRGTGCTGTGCTGYT, reverse primer CACCAGCGGTGGCCAAAGTGTAG, and sequencing primers GRGTGCTGTGCTGYTA and AGGAGTCAGATGAGGAAR (target sequences: TRTCTTAGAGGAGACTAGTGTGWTGCTGG and AKGCTATTGYAGCCTACACTTTGGCC). The IUPAC nucleotide code is shown for ambiguous sites. PCR cycling conditions consisted of an initial 15 min of denaturation at 95°C and 45 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 40 s. Pyrosequencing reactions were performed according to the manufacturer's instructions using a Qiagen PyroMark Q24 system. Amplification products were washed in a series of buffers, and single-stranded, biotinylated DNA products were hybridized to sequencing primers in a 24-well plate and used at a final concentration of 0.375 μM in 20 μl of annealing buffer. PCR amplification bias in patient samples was corrected through pyrosequencing analysis of DNA from three well-characterized strains of human CMV (HCMV): AD169, Toledo, and TB40E. The limit of detection in this system is 5%; therefore, only values greater than this threshold were considered significant.
Establishment and maintenance of cell lines.
Polyclonal T cell lines specific for the IE-1-encoded variant epitopes listed in Table 2 and for CMV-encoded conserved T cells, epitopes (HLA-A1-restricted VTEHDTLLY and YSEHPTFTSQY, HLA-A2-restricted NLVPMVATV and FMDILTTCV, HLA-B7-restricted RPHERNGFTVL and TPRVTGGGAM, and HLA-B8-restricted QIKVRVDMV) were generated following stimulation of peripheral blood mononuclear cells (PBMC) with 1 μg/ml of cognate peptide. Polyclonal T cell cultures were maintained in growth medium containing recombinant interleukin-2 (IL-2) and assessed for T cell specificity after 2 weeks.
TABLE 2.
List of IE-1 epitope variants used in this study
| Epitope | HLA restriction | Sequence position | Major epitope varianta | Amino acid variant(s) (position) | Reference(s) |
|---|---|---|---|---|---|
| KARAKKDELR | A31 | 192–201 | KARAKKDELK | R/K (10) | This study |
| ARAKKDELR | B27 | 193–201 | ARAKKDELK | R/K (9) | This study |
| DELRRKMMY | B18, B44 | 198–206 | DELKRKMIY | R/K (4), M/I (8) | 17 |
| ELRRKMMYM | B8 | 199–207 | ELKRKMIYM | R/K (3), M/I (7) | 19 |
| RRKMMYMYCR | B27 | 201–210 | KRKMIYMYCR | R/K (1), M/I (5) | This study |
| AYAQKIFKIL | A23 | 248–257 | TYSQKIFKIL | A/T (1), A/S (3) | 20, 21 |
| VLEETSVML | A2 | 316–324 | YILEETSVML | V/I (1 or 2) | 18 |
| EEAIVAYTL | B18, B44 | 381–390 | EDAIAAYTL | E/D (2), V/A (5) | 17 |
Changes are indicated by bold type and underlining.
Intracellular cytokine staining.
Expanded polyclonal T cell lines were stimulated with 1 μg/ml of peptide and incubated for 4 h in the presence of brefeldin A (BD Biosciences, USA). For functional-avidity assays, T cells were stimulated in duplicate with 10-fold serial dilutions of peptide (ranging from 1 μg/ml to 0.1 ng/ml). Cells were then incubated with peridinin chlorophyll protein (PerCP)-Cy5.5-labeled anti-CD8 (eBioscience, USA) and fluorescein isothiocyanate (FITC)-labeled anti-CD4 (BD Biosciences, USA), fixed and permeabilized using a BD Cytofix/Cytoperm kit, and incubated with phycoerythrin (PE)-labeled anti-gamma interferon (anti-IFN-γ; BD Biosciences). Cell acquisition was performed using a BD LSRFortessa (BD Biosciences). Postacquisition analysis was performed using FlowJo software (TreeStar, USA).
Statistical analysis.
All statistical analysis was performed using Prism 6 software (GraphPad Software, USA). Statistical differences were assessed using the nonparametric Mann-Whitney test. Data were considered statistical significant when the P value was <0.05.
RESULTS
Dynamics of the emergence of genetic variants of CMV following viral reactivation in HSCT recipients.
Twenty-six patients undergoing allogeneic HSCT were enrolled on this study following informed consent (9, 16). The clinical characteristics of these patients are listed in Table 1. All patients received a T cell-replete bone marrow or granulocyte colony-stimulating factor (G-CSF)-mobilized peripheral blood stem cell graft, and none had in vivo T cell depletion. CMV-seropositive patients or patients who received a transplant from a seropositive donor were treated prophylactically with high-dose acyclovir from day −5 to day 28 or until discharge and then with valganciclovir until day 100. Patients with CMV DNAemia in plasma of >600 copies/ml were treated with ganciclovir at 5 mg/kg (of body weight) twice daily for 14 days, followed by once-daily maintenance until plasma DNAemia was <600 copies/ml, or were treated with valganciclovir at 900 mg twice daily followed by 900 mg once daily for maintenance. Foscarnet was used in patients who were nonresponsive or displayed significant toxicity from ganciclovir. Of the 26 HSCT recipients enrolled for this study, 17 had viral reactivation as defined by CMV DNAemia of >600 copies/ml. All of these patients were CMV seropositive prior to transplant: 12 had a CMV-seronegative donor (characterized as R+/D− recipients), while the remaining five had a CMV-seropositive donor (characterized as R+/D+ recipients). Early CMV reactivation developed in 16 of these patients, while 4 patients had late CMV reactivation, which occurred beyond the first 100 days posttransplant. Two of the late-CMV reactivation patients developed CMV-associated disease: one had colitis and one had enteritis. Fourteen of the 17 displayed an unstable CMV-specific immune response, as assessed by the CMV-QuantiFERON assay, and characterized by a failure to generate a stable CMV-specific IFN-γ response by 59 days posttransplant (9). All nine patients included in the current study who demonstrated CMV-immune reconstitution also were without evidence of viral reactivation.
TABLE 1.
Clinical characteristics of HSCT recipients included in this study
| Code | Recipient/donor serostatus | HLA type | No. of episodes of CMV reactivation | Maximal CMV titer | Days posttransplant that the CMV load was >600 copies/ml | CMV disease |
|---|---|---|---|---|---|---|
| Patients with CMV reactivationa | ||||||
| 04 | R+/D− | A2 A29 B44 B51 Cw1 | 4 | 10,000 | 60–70, 144–158, 189–195, 363–391 | Yes: CMV colitis |
| 06 | R+/D− | A23 A26 B39 B51 Cw2 | 1 | 900 | 64–71 | No |
| 13 | R+/D− | A2 A29 B44 B62 Cw3 Cw16 | 2 | 12,000 | 33–67, 77–84 | No |
| 14 | R+/D+ | A11 A31 B7 B60 | 6 | 120,000 | 46–55, 139–178, 192–196, 213–217, 249–269, 286–314 | Yes: CMV enteritis |
| 16 | R+/D− | A2 A24 B15 B27 Cw2 Cw3 | 1 | 870 | 69 | No |
| 17 | R+/D− | A1 A24 B08 B39 Cw7 | 2 | 40,000 | 37, 44–68 | No |
| 19 | R+/D+ | A2 A24 B44 Cw5 | 3 | 55,000 | 32–64, 73–80, 88–92 | No |
| 25 | R+/D− | A2 A3 B35 B62 Cw3 Cw10 | 2 | 2,400 | 59, 95–102 | No |
| 26 | R+/D− | A2 A33 B14 B15 Cw3 Cw8 | 3 | 4,100 | 35–60, 81–88, 273–277 | No |
| 28 | R+/D− | A2 A24 B44 Cw5 Cw6 | 1 | 6,800 | 46–67 | No |
| 30 | R+/D+ | A2 A24 B13 B60 Cw3 Cw4 | 1 | 64,000 | 314–332 | No |
| 32 | R+/D− | A2 B13 B40 Cw3 Cw6 | 5 | 22,000 | 39, 49–63, 151–157, 179, 192–237 | No |
| 34 | R+/D− | A1 A33 B8 B14 Cw7 Cw8 | 1 | 2,000 | 57–64 | No |
| 38 | R+/D+ | A1 A24 B41 B57 Cw6 Cw17 | 1 | 1,400 | 75–92 | No |
| 39 | R+/D− | A2 A29 B44 Cw5 | 1 | 6,900 | 45–62 | No |
| 44 | R+/D+ | A2 A32 B18 B44 Cw5 Cw7 | 1 | 1,000 | 43–48 | No |
| 46 | R+/D− | A2 B27 B44 Cw2 Cw5 | 2 | 2,800 | 32–35, 53 | No |
| Patients without CMV reactivation | ||||||
| 01 | R+/D− | A1 A3 B27 B60 Cw2 Cw3 | NAb | NA | NA | No |
| 07 | R−/D+ | A1 A2 B08 B15 Cw3 Cw7 | NA | NA | NA | No |
| 15 | R+/D− | A3 A31 B7 B60 Cw3 Cw7 | NA | NA | NA | No |
| 36 | R+/D− | A1 A2 B35 B62 Cw3 Cw4 | NA | NA | NA | No |
| 37 | R+/D− | A2 A23 B15 B44 Cw4 Cw7 | NA | NA | NA | No |
| 42 | R+/D+ | A2 A23 B15 B44 Cw4 Cw7 | NA | NA | NA | No |
| 43 | R+/D+ | A1 A26 B44 B13 Cw7 | NA | NA | NA | No |
| 45 | R+/D− | A1 A2 B37 B44 Cw5 Cw6 | NA | NA | NA | No |
| 47 | R+/D+ | A2 B7 B44 Cw5 Cw7 | NA | NA | NA | No |
CMV reactivation is defined as CMV DNAemia of >600 copies/ml.
NA, not applicable.
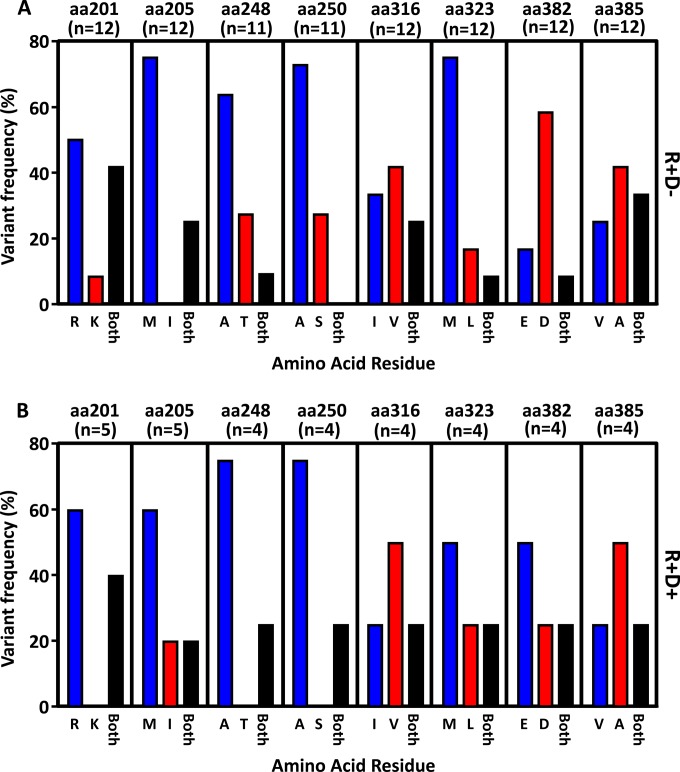
To delineate the impact of the emergence of genetic variants on T cell immune reconstitution in this cohort of HSCT recipients, we focused on eight HLA class I-restricted CD8+ T cell epitopes from the immediate early (IE-1) protein of CMV (Table 2). Three novel epitopes were mapped during this study (Table 2), and five epitopes have been previously described (17–21). Using the GenBank database, we were able to identify a series of variant sequences for each of these epitopes. We designed a pyrosequencing analysis to identify the single nucleotide polymorphisms (SNPs) within the CMV-encoded CD8+ T cell epitopes. Initially, these SNP analyses were carried out at the peak of viral load for all HSCT recipients who showed CMV reactivation. The amino acid residue at each variant position was extrapolated based upon the nucleotide sequence. Data in Fig. 1 represent the proportion of recipients showing either one or both amino acids at each position. Data were corrected for error rates at each position as outlined in Materials and Methods. Although we observed bias in amino acid usage at certain positions, we noted that the preferential usages of particular amino acid residues were similar in the R+/D− (Fig. 1A) and the R+/D+ (Fig. 1B) cohorts. This analysis also revealed that a high proportion of HSCT recipients had multiple IE-1 variants following reactivation, whereby 6 to 40% of the samples demonstrated both variant amino acids and 9 of 17 HSCT recipients (5 of 12 R+/D− and 2 of 5 R+/D+) showed definitive evidence of mixed infection characterized by the concurrent detection of both variant residues at least one position.
FIG 1.
Pyrosequencing analysis of the IE-1 sequence variants in HSCT recipients. DNA was extracted from plasma samples of 17 HSCT recipients during CMV reactivation. Following DNA PCR amplification, pyrosequencing analysis of the panel of SNPs was performed as outlined in Materials and Methods. The nucleotide data were extrapolated to determine the proportion of each amino acid residue for the 8 positions tested. (A) Data represent the proportion of R+/D− recipient samples encoding either a dominant single amino acid residue at each position or both amino acid residues at each position. (B) Data represent the proportion of R+/D+ recipient samples encoding either a dominant single amino acid residue at each position or both amino acid residues at each position.
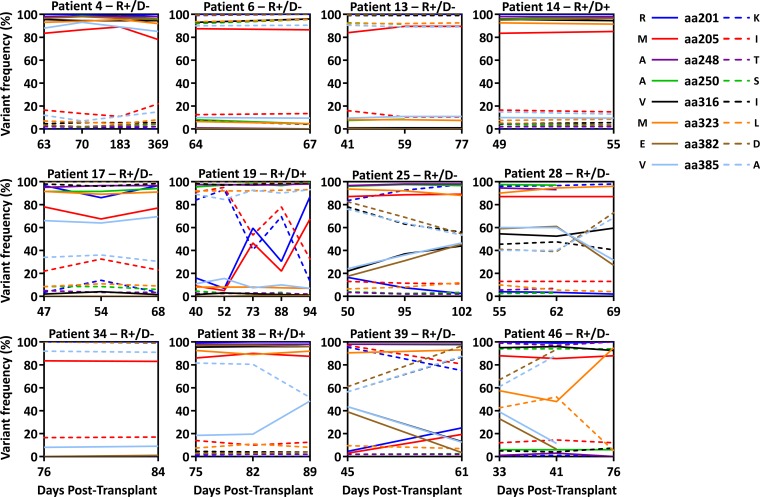
We subsequently assessed the stability of the viral variants over time, using longitudinal plasma samples during viral reactivation from 16 of the 17 HSCT recipients. Representative longitudinal analysis of all SNPs assessed in individual patients is shown in Fig. 2. While some HSCT recipients, including both R+/D− and R+/D+ patients, showed very little change in the pattern of SNP expression following detection of either predominantly single variants (recipient 4) or multiple variants (recipient 17), other HSCT recipients demonstrated changes in SNP frequency during periods of viral reactivation. This is particularly evident for the D+/R+ patient 19.
FIG 2.
Longitudinal pyrosequencing analysis in HSCT recipients. Longitudinal pyrosequencing analysis was performed with HSCT patients from whom more than a single time point of viral reaction was available. Each data line represents individual SNPs over time following a single or multiple rounds of viral reactivation.
Impact of coinfection on the T cell kinetics.
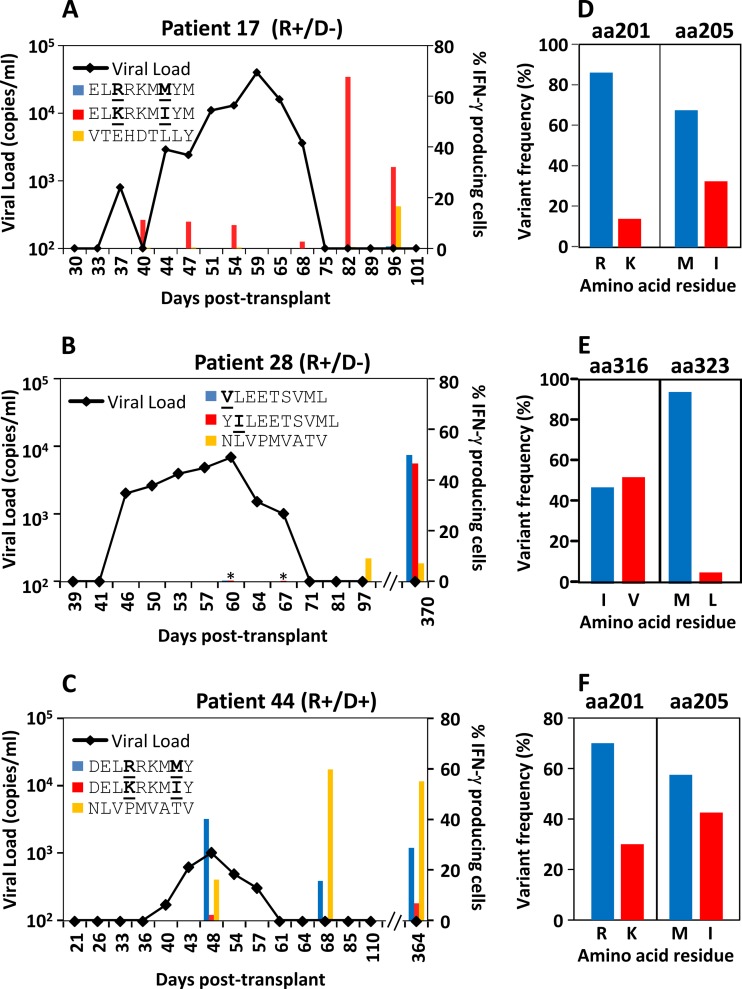
We next sought to assess the impact of epitope variation and coinfection on IE-1-specific T cell immunity. As the frequency of IE1-specific T cells was too low in the majority of patients for direct ex vivo analysis, PBMC from HSCT recipients showing evidence of viral reactivation were stimulated with all potentially HLA-matched variant peptide epitopes (Table 2) and then cultured in vitro for 2 weeks in the presence of IL-2. PBMC from nine HSCT recipients showing immune reconstitution with no evidence of CMV reactivation were also stimulated with HLA-matched variant peptide epitopes (Table 2). As a control, PBMC were stimulated with at least two conserved HLA-matched epitopes. Representative longitudinal analysis from three of these patients overlaid with viral reactivation kinetics is shown in Fig. 3A to C. An overall summary of the number of HSCT recipients tested for each epitope and the number of responding HSCT recipients is shown in Table 3. Interestingly, these observations suggested that while some patients could efficiently recognize multiple viral variants detected by pyrosequencing analysis (represented by patient 28 [Fig. 3B and E]), others showed preferential recognition, in some instances targeted against subdominant epitope variants. As evidenced in Fig. 3D, pyrosequencing analysis revealed that the IE-1 sequence in recipient 17 at amino acid positions 201 and 205 was dominated by the residues R and M, which would correspond to the ELRRKMMYM epitope in HLA-B8 individuals. Despite this, recipient 17 only generated a T cell response against the subdominant ELKRKMIYM variant (Fig. 3A). Interestingly, recipient 17 also showed the absence of a detectable response against the immunodominant conserved T cell epitope, VTEHDTTLY, during viral reactivation and failed to generate a T cell response against the dominant ELRRKMMYM variant even after resolution of viral infection. Similar observations were made for recipient 44 (Fig. 3F), for whom we could detect sequences encoding both of the HLA-B44 variants but were unable to detect a response against the DELKRKMIY variant during viral reactivation. Interestingly, these observations were also made for other HLA-B44-positive HSCT recipients for both of the HLA-B44-restricted epitopes (Table 3). This was particularly evident for the EDAIAAYTL variant, which could be detected in 6 of 7 HLA-B44-positive HSCT recipients but failed to induce a significant T cell response in any recipient. It is important to mention that we initially aimed to perform longitudinal analysis throughout the course of viral reactivation in all patients; however, in the majority of CMV reactivation patients tested, we were unable to see CMV-specific immune reconstitution until convalescence. The peak CD8+ T cell response of each patient to each epitope tested is presented in Table S1 in the supplemental material.
FIG 3.
Kinetics of variant-specific T cell activation following viral reactivation in HSCT transplant recipients. Longitudinal PBMC from HSCT recipients during and after CMV reactivation were stimulated with HLA-matched IE-1-encoded variant peptide epitopes and control nonvariant peptides and then cultured in vitro for 2 weeks in the presence of IL-2. Two weeks later, T cell cultures were recalled with cognate peptide and assessed for the intracellular expression of IFN-γ. Representative data from three HSCT recipients overlaid with the kinetics of viral reactivation are shown. (A) PBMC from recipient 14 were assessed for T cell responses on days 40, 47, 54, 68, 82, and 96 posttransplant. (B) PBMC from recipient 28 were assessed for T cell responses on days 41, 60, 67, 97, and 370 posttransplant. (C) PBMC from recipient 44 were assessed for T cell responses on days 48, 68, and 364 posttransplant. Representative data of the frequency of each variant amino acid residue relevant to the T cell responses shown in panels A to C at the peak of viral reactivation are shown for recipient 17 (D), recipient 28 (E), and recipient 44 (F). *, no response detected.
TABLE 3.
Summary of CMV-specific peptide epitope recognition by HSCT recipients
| Peptide sequencec | Reactivation |
No reactivation |
|||
|---|---|---|---|---|---|
| No. of HLA-matched recipients | No. of HLA-matched recipients with sequence detected | No. of respondersa | No. of HLA-matched recipients | No. of respondersa | |
| VLEETSVML | 12 | 7 | 3 | 5 | 1 |
| YILEETSVML | 12 | 5 | 4 | 5 | 2 |
| DELRRKMMY | 7 | 5 | 2 | 4 | 0 |
| DELKRKMIY | 7 | 3 | 1 | 4 | 0 |
| EEAIAVAYL | 7 | 4 | 2 | 4 | 0 |
| EDAIAAYTL | 7 | 6 | 0 | 4 | 0 |
| ELRRKMMYM | 2 | 2 | 1 | 2 | 2 |
| ELKRKMIYM | 2 | 1 | 2 | 2 | 2 |
| AYAQKIFKIL | 1 | 0 | 1 | 1 | 0 |
| TYSQKIFKIL | 1 | 1 | 1 | 1 | 1 |
| KARAKKDELR | 1 | 1 | 0 | 1 | 0 |
| KARAKKDELK | 1 | 0 | 0 | 1 | 0 |
| ARAKKDELK | 1 | 1 | 1 | 1 | 0 |
| ARAKKDELR | 1 | 1 | 1 | 1 | 0 |
| KRKMIYMCYR | 1 | 0 | 0 | 1 | 1 |
| RRKMMYMCYR | 1 | 1 | 1 | 1 | 1 |
| FMDILTTCV | 12 | NDb | 5 | 5 | 0 |
| NLVPMVATV | 12 | ND | 8 | 5 | 3 |
| RPHERNGFTVL | 1 | ND | 1 | 1 | 1 |
| TPRVTGGGAM | 1 | ND | 1 | 1 | 1 |
| VTEHDTLLY | 3 | ND | 3 | 4 | 3 |
| QIKVRVDMV | 2 | ND | 1 | 1 | 1 |
| YSEHPTFTSQY | 0 | ND | 0 | 2 | 2 |
Patients with >5% of CD8+ T cells producing IFN-γ following recall after 2 weeks of culture were considered responders.
ND, not done.
Underlining represents variant amino acids.
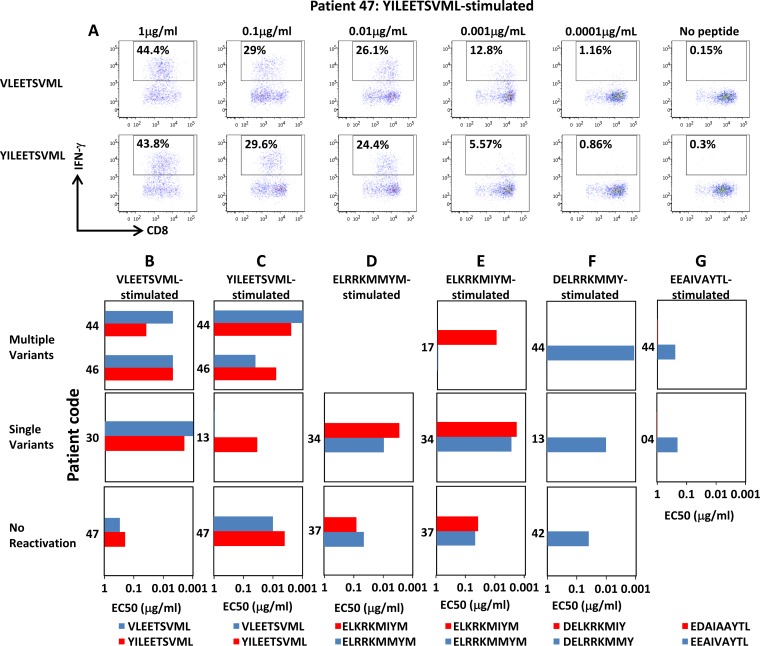
To further assess the recognition of epitope variants in our recipient cohort, cultured T cells from all HSCT recipients were stimulated with serial dilutions of both the cognate and variant peptide and assessed for the production of IFN-γ. The 50% effective concentration (EC50) was then calculated based upon the concentration of peptide required to induce 50% of maximal IFN-γ production. Representative analysis following recall of a YILEETSVML-stimulated T cell culture with 10-fold serial dilutions of the VLEETSVML and YILEETSVML epitope variants is shown in Fig. 4A. While T cells specific for HLA-A2-restricted epitopes (VLEETSVML and YILEETSVML) consistently recognized these variants with similar efficiencies (Fig. 3B and C), cross-reactivity toward the HLA-B8 epitopes, ELRRKMMYM and ELKRKMIYM, was patient dependent, characterized by preference for a single variant in some individuals (recipient 17) and cross-reactive in others (recipients 34 and 37) (Fig. 4D and E). We saw no evidence of cross-reactivity in T cells specific for the two B44-restricted epitopes, DELRRKMMY and EEAIVAYTL, which displayed preferential bias for a single variant, irrespective of evidence for exposure to multiple variants (Fig. 4F and G). These observations further demonstrate that exposure to multiple viral variants does not automatically lead to the efficient induction of cross-reactive T cell immunity and that repertoire “holes” may exist across genetically unrelated individuals.
FIG 4.
Functional-avidity analysis of IE-1 variant-specific T cell populations. Following in vitro expansion for 2 weeks in the presence of cognate peptide and IL-2, IE-1 epitope-specific T cells were incubated for 4 h with 10-fold serial dilutions of both the cognate peptide and the epitope variant. IFN-γ expression was then assessed using an intracellular cytokine assay. The EC50 was calculated based upon the peptide concentration required to induce activation in 50% of the maximal number of IFN-γ-producing cells. (A) Representative peptide titration from YILEETSVML-stimulated T cell cultures from patient 47 recalled with VLEETSVML and YILEETSVML is shown. Data in bottom rows correspond to T cells stimulated ex vivo with VLEETSVML (B), YILEETSVML (C), ELRRKMMYM (D), ELKRKMIYM (E), DELRRKMMY (F), and EEAIVAYTL (G). Color keys at the bottom of each row correspond to the cognate and variant peptides used to recall the T cells response after 2 weeks in culture.
Impact of exposure to multiple viral variants on viral control.
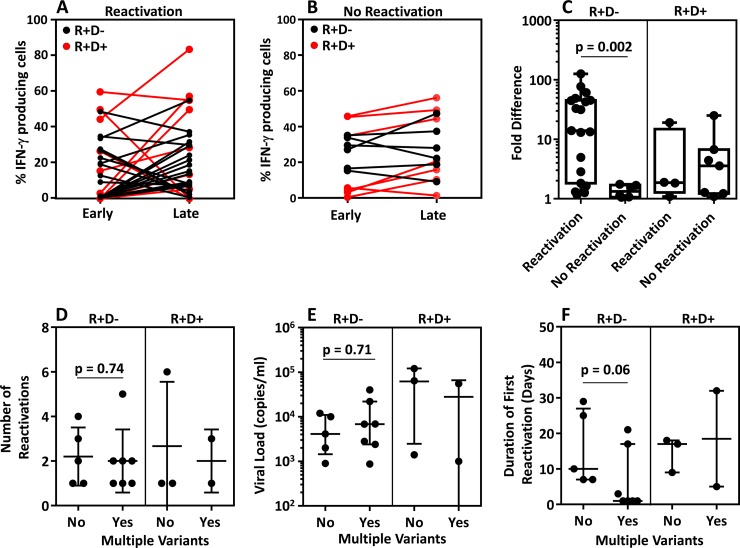
We next sought to determine if the reconstitution of the CMV-specific T cell response directed toward both variant IE-1 and/or conserved epitopes was associated with viral reactivation. We compared the frequencies of CD8+ T cells specific for both IE-1 variant epitopes and conserved epitopes early (90 to 106 days) and late (>180 days) posttransplant in HSCT recipients with and without evidence of reactivation. Pairwise analysis of the frequency of all detectable CMV-specific T cell responses early and late posttransplant demonstrated that HSCT recipients with evidence of viral reactivation (Fig. 5A) showed less stability in their T cell responses than HSCT recipients without reactivation (Fig. 5B). To contrast the responses in R+/D− and R+/D+ patients, we assessed the fold change in the responses early and late posttransplant in these two cohorts. While R+/D− recipients with reactivation showed significantly greater fold differences in the frequency of CMV-specific T cells between early and late responses than did R+/D− recipients with no reactivation, we did not see significant differences in the R+/D+ patients (Fig. 5C). To further assess the impact of reactivation with multiple viral variants on viral control, we compared (i) the number of viral reactivations, (ii) the peak viral load, and (iii) duration of the first viral reactivations in HSCT R+/D− and R+/D+ recipients with evidence of single or multiple variants in their peripheral blood. These analyses revealed no significant differences in the number of viral reactivations (Fig. 5D), in the peak viral load (Fig. 5E), or in the duration of reactivation (Fig. 5F) from patients with and without evidence of multiple viral variants. These observations suggest that while the induction of variant specific immunity may play a role in the control of viral reactivation following reactivation with multiple variants of CMV, the capacity to induce stable CMV-specific immune reconstitution to either conserved epitopes or via cross-reactive responses was more relevant for the efficient control of CMV reactivation following HSCT.
FIG 5.
Effect of coinfection on viral reactivations and association of viral reactivation with overall T cell immunity. (A to C) Pairwise analyses of the frequency of IFN-γ-producing T cells generated against individual epitopes from HSCT recipients showing evidence of reactivation (A) or with no evidence of reactivation (B) are shown. Responses are only shown when epitope-specific T cells were detected in at least one time point. Data in red represent R+/D+ patients, and data in black represent R+/D− patients. Fold difference (C) was calculated by dividing the higher frequency of IFN-γ-producing T cells, at either time point, by the lower frequency detected at either time point. (D to F) HSCT recipients were grouped based upon whether they showed evidence for exposure to multiple variants. (D) Data represent the number of reactivations in HSCT recipients with and without evidence of coinfection. (E) Data represent the viral load during primary reactivation. (F) Data represent the duration, in days, of the primary reactivation.
DISCUSSION
Observations over the last 2 decades, particularly with human immunodeficiency virus (HIV) and other retroviruses, have demonstrated that genetic variation in viral sequences can have a significant impact upon long-term viral control (22–24). Unlike the case with these rapidly mutating retroviruses, T cell immunity to CMV and other human herpesviruses has typically been shown to be stable, with little change in the T cell repertoire (25–27). However, there is emerging evidence that multiple CMV variants can be found in a single individual that encode a significant amount of genetic diversity (28, 29). In this study, we sought to assess the impact of genetic diversity and exposure to multiple CMV variants on immunomediated control of CMV in HSCT recipients. These analyses revealed that while a large proportion of HSCT recipients undergoing viral reactivation carry multiple viral variants, the long-term control of CMV infection is primarily mediated through the efficient induction of stable reconstitution of T cell immunity irrespective of the nature of the antigenic target. However, these observations also indicate that the impact of CMV genetic variation on immunity is complex, and larger sample sizes with greater sequencing depth will likely be require to thoroughly delineate the impact genetic variation has upon the immunological control of CMV.
As a major viral complication that has arisen since the advent of HSCT, CMV can lead to significant morbidity and mortality in immunocompromised patients (7). Complications associated with CMV infection are most evident in an immunologically naive setting; however, observations have shown that exposure to CMV can still cause disease irrespective of prior immunological exposure in immunocompromised individuals (3). It has been suggested that genotypic variation with CMV and exposure to multiple genetic variants may play a role in clinical outcome. Recent observations have demonstrated that the detection of multiple CMV genotypes in transplant recipients is common and can be associated with an increased duration of viral reactivation (12). Although we also detected evidence of multiple genetic variants of CMV in our cohort of HSCT recipients, we did not see any evidence of an impact on viral reactivation. However, it should be noted that previous studies were carried out with predominantly SOT recipients using genotypic analysis of surface glycoproteins, while our observations were generated with a cohort of HSCT recipients using genotypic analysis of IE-1. It could be speculated that differences in these observations could be attributable to (i) differences in immunogenicity/protection between glycoproteins and IE-1 targets, (ii) the different impact of coinfection in SOT versus HSCT recipients, or (iii) the limited size of our cohort.
We did observe an association between the stability of CMV-specific T cell immunity in our R+/D− cohort and viral reactivation. CMV-reactivation patients in this cohort were less likely to have stable epitope specific T cell responses irrespective of the conserved or variant nature of the target epitope than patients with no evidence of viral reactivation. These observations are consistent with previous studies, using different immunological approaches, demonstrating the association between CMV reactivation in transplant patients and poor or unstable CMV-specific T cell immunity (7, 9, 30, 31). Furthermore, the stability of this T cell response did not appear to be influenced by the nature of viral reactivation. CMV reactivation with both multiple or single viral strain was similarly associated with unstable T cell responses. We were unable to see a similar correlation between reactivation and T cell immunity in our R+D+ cohort. However, this cohort of R+D+ patients was small, impacting the ability to detect significant differences and the potential influence of other factors, such as genetic variation between the recipient and donor CMV isolates. It is also important to appreciate that CMV-specific CD8+ and CD4+ T cell immunity is directed against a diverse array of antigens, and genetic variation within a single CMV gene may have limited impact on overall immune control. While these observations suggest that the induction of a robust T cell response is more critical for immune control than the generation of multivariant specific immunity, in some individuals we could detect the induction of a non-cross-reactive T cell response only during viral reactivation, tentatively suggesting that in some instances an absence of cross-reactivity could be affecting viral control.
Although we did not see any definitive evidence that the reactivation of multiple variants impacted CMV disease, interestingly, we did observe that some variant epitopes failed to induce detectable T cell responses which were either cross-reactive or variant-specific, despite their detection in a large proportion of HSCT recipients (Table 3). This was particularly evident for the B44-restricted epitope variant EDAIAAYTL, for which we detected no T cells responses, despite the detection of T cells specific for EEAIVAYTL variants in 50% of HSCT recipients in which the variant sequences were detected. Previous studies in a number of settings have shown that amino acid sequence changes can restrict variant peptide recognition, often as a consequence of changes in major histocompatibility complex (MHC) anchor residues that result in poor MHC binding or due to restricted T cell repertoire diversity (32–34). Given that the EDAIAAYTL amino acid sequence changes do not occur in MHC anchor residues, our observations suggest that these variant epitopes may have reduced immunogenicity for other reasons, such as limitations in the T cell repertoire. While the implications for these observations in the control of viral reactivation are not clear, in settings of adoptive immunotherapy whereby donor-derived, autologous, or third-party T cells are used (35–37), limited cross-reactivity against variant epitopes could potentially limit the effectiveness of these T cells for pathogen surveillance.
In conclusion, the observations in this study provide evidence that exposure to multiple viral variants in an immunocompromised setting is common and does not necessarily lead to the automatic induction of cross-reactive immunity. This study also provides evidence that protection against genetically distinct variants of CMV in infected individuals is not necessarily dependent upon the induction of cross-reactive T cell populations against variant epitopes but can be efficiently mediated via the recognition of conserved T cell epitopes. These observations further demonstrate the importance of robust stable immune reconstitution in the long-term control of CMV following HSCT.
Supplementary Material
ACKNOWLEDGMENTS
We thank Linda Jones and Jacqueline Burrows for technical assistance.
C.S. and R.K. designed the study. C.S. and R.M.B. conducted various experimental studies. S.-K.T., M.J.S., S.R.B., J.J.M., and G.R.H. provided critical intellectual input into the design of the study. S.-K.T. and G.R.H. were responsible for recruitment and clinical management of the patients enrolled in the study. All authors contributed to the writing of the manuscript.
We declare no conflicts of interest.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00297-16.
REFERENCES
- 1.Boeckh M, Nichols WG, Papanicolaou G, Rubin R, Wingard JR, Zaia J. 2003. Cytomegalovirus in hematopoietic stem cell transplant recipients: current status, known challenges, and future strategies. Biol Blood Marrow Transplant 9:543–558. doi: 10.1016/S1083-8791(03)00287-8. [DOI] [PubMed] [Google Scholar]
- 2.Crough T, Khanna R. 2009. Immunobiology of human cytomegalovirus: from bench to bedside. Clin Microbiol Rev 22:76–98. doi: 10.1128/CMR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeckh M, Nichols WG. 2004. The impact of cytomegalovirus serostatus of donor and recipient before hematopoietic stem cell transplantation in the era of antiviral prophylaxis and preemptive therapy. Blood 103:2003–2008. doi: 10.1182/blood-2003-10-3616. [DOI] [PubMed] [Google Scholar]
- 4.Ramanathan M, Teira P, Battiwalla M, Barrett J, Ahn KW, Chen M, Green J, Laughlin M, Lazarus HM, Marks D, Saad A, Seftel M, Saber W, Savani B, Waller E, Wingard J, Auletta JJ, Lindemans CA, Boeckh M, Riches ML. 4 April 2016. Impact of early CMV reactivation in cord blood stem cell recipients in the current era. Bone Marrow Transplant doi: 10.1038/bmt.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teira P, Battiwalla M, Ramanathan M, Barrett AJ, Ahn KW, Chen M, Green JS, Saad A, Antin JH, Savani BN, Lazarus HM, Seftel M, Saber W, Marks D, Aljurf M, Norkin M, Wingard JR, Lindemans CA, Boeckh M, Riches ML, Auletta JJ. 2016. Early cytomegalovirus reactivation remains associated with increased transplant related mortality in the current era: a CIBMTR analysis. Blood 127:2427–2438. doi: 10.1182/blood-2015-11-679639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollack M, Heugel J, Xie H, Leisenring W, Storek J, Young JA, Kukreja M, Gress R, Tomblyn M, Boeckh M. 2011. An international comparison of current strategies to prevent herpesvirus and fungal infections in hematopoietic cell transplant recipients. Biol Blood Marrow Transplant 17:664–673. doi: 10.1016/j.bbmt.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeckh M, Leisenring W, Riddell SR, Bowden RA, Huang ML, Myerson D, Stevens-Ayers T, Flowers ME, Cunningham T, Corey L. 2003. Late cytomegalovirus disease and mortality in recipients of allogeneic hematopoietic stem cell transplants: importance of viral load and T-cell immunity. Blood 101:407–414. doi: 10.1182/blood-2002-03-0993. [DOI] [PubMed] [Google Scholar]
- 8.Sacre K, Nguyen S, Deback C, Carcelain G, Vernant JP, Leblond V, Autran B, Dhedin N. 2008. Expansion of human cytomegalovirus (HCMV) immediate-early 1-specific CD8+ T cells and control of HCMV replication after allogeneic stem cell transplantation. J Virol 82:10143–10152. doi: 10.1128/JVI.00688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tey SK, Kennedy GA, Cromer D, Davenport MP, Walker S, Jones LI, Crough T, Durrant ST, Morton JA, Butler JP, Misra AK, Hill GR, Khanna R. 2013. Clinical assessment of anti-viral CD8+ T cell immune monitoring using QuantiFERON-CMV(R) assay to identify high risk allogeneic hematopoietic stem cell transplant patients with CMV infection complications. PLoS One 8:e74744. doi: 10.1371/journal.pone.0074744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith C, Khanna R. 2013. Immune regulation of human herpesviruses and its implications for human transplantation. Am J Transplant 13(Suppl 3):9–23; quiz, 23. [DOI] [PubMed] [Google Scholar]
- 11.Dieamant DC, Bonon SH, Peres RM, Costa CR, Albuquerque DM, Miranda EC, Aranha FJ, Oliveira-Duarte G, Fernandes VC, De Souza CA, Costa SC, Vigorito AC. 2013. Cytomegalovirus (CMV) genotype in allogeneic hematopoietic stem cell transplantation. BMC Infect Dis 13:310. doi: 10.1186/1471-2334-13-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manuel O, Asberg A, Pang X, Rollag H, Emery VC, Preiksaitis JK, Kumar D, Pescovitz MD, Bignamini AA, Hartmann A, Jardine AG, Humar A. 2009. Impact of genetic polymorphisms in cytomegalovirus glycoprotein B on outcomes in solid-organ transplant recipients with cytomegalovirus disease. Clin Infect Dis 49:1160–1166. doi: 10.1086/605633. [DOI] [PubMed] [Google Scholar]
- 13.Smith C, Gras S, Brennan RM, Bird NL, Valkenburg SA, Twist KA, Burrows JM, Miles JJ, Chambers D, Bell S, Campbell S, Kedzierska K, Burrows SR, Rossjohn J, Khanna R. 2014. Molecular imprint of exposure to naturally occurring genetic variants of human cytomegalovirus on the T cell repertoire. Sci Rep 4:3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prod'homme V, Retiere C, Imbert-Marcille BM, Bonneville M, Hallet MM. 2003. Modulation of HLA-A*0201-restricted T cell responses by natural polymorphism in the IE1(315-324) epitope of human cytomegalovirus. J Immunol 170:2030–2036. doi: 10.4049/jimmunol.170.4.2030. [DOI] [PubMed] [Google Scholar]
- 15.Prod'homme V, Retiere C, Valtcheva R, Bonneville M, Hallet MM. 2003. Cross-reactivity of HLA-B*1801-restricted T-lymphocyte clones with target cells expressing variants of the human cytomegalovirus 72kDa-IE1 protein. J Virol 77:7139–7142. doi: 10.1128/JVI.77.12.7139-7142.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tey SK, Davenport MP, Hill GR, Kennedy GA, Durrant ST, Khanna R, Cromer D. 2015. Post transplant CMV-specific T-cell immune reconstitution in the absence of global T-cell immunity is associated with a high risk of subsequent virus reactivation. Bone Marrow Transplant 50:315–316. doi: 10.1038/bmt.2014.265. [DOI] [PubMed] [Google Scholar]
- 17.Khan N, Best D, Bruton R, Nayak L, Rickinson AB, Moss PA. 2007. T cell recognition patterns of immunodominant cytomegalovirus antigens in primary and persistent infection. J Immunol 178:4455–4465. doi: 10.4049/jimmunol.178.7.4455. [DOI] [PubMed] [Google Scholar]
- 18.Khan N, Cobbold M, Keenan R, Moss PA. 2002. Comparative analysis of CD8+ T cell responses against human cytomegalovirus proteins pp65 and immediate early 1 shows similarities in precursor frequency, oligoclonality, and phenotype. J Infect Dis 185:1025–1034. doi: 10.1086/339963. [DOI] [PubMed] [Google Scholar]
- 19.Elkington R, Walker S, Crough T, Menzies M, Tellam J, Bharadwaj M, Khanna R. 2003. Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J Virol 77:5226–5240. doi: 10.1128/JVI.77.9.5226-5240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burrows SR, Elkington RA, Miles JJ, Green KJ, Walker S, Haryana SM, Moss DJ, Dunckley H, Burrows JM, Khanna R. 2003. Promiscuous CTL recognition of viral epitopes on multiple human leukocyte antigens: biological validation of the proposed HLA A24 supertype. J Immunol 171:1407–1412. doi: 10.4049/jimmunol.171.3.1407. [DOI] [PubMed] [Google Scholar]
- 21.Kuzushima K, Hayashi N, Kimura H, Tsurumi T. 2001. Efficient identification of HLA-A*2402-restricted cytomegalovirus-specific CD8(+) T-cell epitopes by a computer algorithm and an enzyme-linked immunospot assay. Blood 98:1872–1881. doi: 10.1182/blood.V98.6.1872. [DOI] [PubMed] [Google Scholar]
- 22.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, Weinhold KJ, Moore S, B CCC, Letvin N, Haynes BF, Cohen MS, Hraber P, Bhattacharya T, Borrow P, Perelson AS, Hahn BH, Shaw GM, Korber BT, McMichael AJ. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med 206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goulder PJ, Watkins DI. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol 4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 24.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 25.Klarenbeek PL, Remmerswaal EB, ten Berge IJ, Doorenspleet ME, van Schaik BD, Esveldt RE, Koch SD, ten Brinke A, van Kampen AH, Bemelman FJ, Tak PP, Baas F, de Vries N, van Lier RA. 2012. Deep sequencing of antiviral T-cell responses to HCMV and EBV in humans reveals a stable repertoire that is maintained for many years. PLoS Pathog 8:e1002889. doi: 10.1371/journal.ppat.1002889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neller MA, Burrows JM, Rist MJ, Miles JJ, Burrows SR. 2013. High frequency of herpesvirus-specific clonotypes in the human T cell repertoire can remain stable over decades with minimal turnover. J Virol 87:697–700. doi: 10.1128/JVI.02180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miles JJ, Silins SL, Brooks AG, Davis JE, Misko I, Burrows SR. 2005. T-cell grit: large clonal expansions of virus-specific CD8+ T cells can dominate in the peripheral circulation for at least 18 years. Blood 106:4412–4413. doi: 10.1182/blood-2005-06-2261. [DOI] [PubMed] [Google Scholar]
- 28.Görzer I, Guelly C, Trajanoski S, Puchhammer-Stockl E. 2010. Deep sequencing reveals highly complex dynamics of human cytomegalovirus genotypes in transplant patients over time. J Virol 84:7195–7203. doi: 10.1128/JVI.00475-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renzette N, Bhattacharjee B, Jensen JD, Gibson L, Kowalik TF. 2011. Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog 7:e1001344. doi: 10.1371/journal.ppat.1001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar D, Chernenko S, Moussa G, Cobos I, Manuel O, Preiksaitis J, Venkataraman S, Humar A. 2009. Cell-mediated immunity to predict cytomegalovirus disease in high-risk solid organ transplant recipients. Am J Transplant 9:1214–1222. doi: 10.1111/j.1600-6143.2009.02618.x. [DOI] [PubMed] [Google Scholar]
- 31.Gratama JW, Boeckh M, Nakamura R, Cornelissen JJ, Brooimans RA, Zaia JA, Forman SJ, Gaal K, Bray KR, Gasior GH, Boyce CS, Sullivan LA, Southwick PC. 2010. Immune monitoring with iTAg MHC tetramers for prediction of recurrent or persistent cytomegalovirus infection or disease in allogeneic hematopoietic stem cell transplant recipients: a prospective multicenter study. Blood 116:1655–1662. doi: 10.1182/blood-2010-03-273508. [DOI] [PubMed] [Google Scholar]
- 32.Meyer-Olson D, Shoukry NH, Brady KW, Kim H, Olson DP, Hartman K, Shintani AK, Walker CM, Kalams SA. 2004. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med 200:307–319. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, Brander C, Ogg G, Sullivan JS, Dyer W, Jones I, McMichael AJ, Rowland-Jones S, Phillips RE. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med 193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geels MJ, Dubey SA, Anderson K, Baan E, Bakker M, Pollakis G, Paxton WA, Shiver JW, Goudsmit J. 2005. Broad cross-clade T-cell responses to gag in individuals infected with human immunodeficiency virus type 1 non-B clades (A to G): importance of HLA anchor residue conservation. J Virol 79:11247–11258. doi: 10.1128/JVI.79.17.11247-11258.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leen AM, Bollard CM, Mendizabal AM, Shpall EJ, Szabolcs P, Antin JH, Kapoor N, Pai SY, Rowley SD, Kebriaei P, Dey BR, Grilley BJ, Gee AP, Brenner MK, Rooney CM, Heslop HE. 2013. Multicenter study of banked third-party virus-specific T cells to treat severe viral infections after hematopoietic stem cell transplantation. Blood 121:5113–5123. doi: 10.1182/blood-2013-02-486324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peggs KS, Verfuerth S, Pizzey A, Khan N, Guiver M, Moss PA, Mackinnon S. 2003. Adoptive cellular therapy for early cytomegalovirus infection after allogeneic stem-cell transplantation with virus-specific T-cell lines. Lancet 362:1375–1377. doi: 10.1016/S0140-6736(03)14634-X. [DOI] [PubMed] [Google Scholar]
- 37.Hill GR, Tey SK, Beagley L, Crough T, Morton JA, Clouston AD, Whiting P, Khanna R. 2010. Successful immunotherapy of HCMV disease using virus-specific T cells expanded from an allogeneic stem cell transplant recipient. Am J Transplant 10:173–179. doi: 10.1111/j.1600-6143.2009.02872.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.