Abstract
Viruses have evolved diverse strategies to maximize the functional and coding capacities of their genetic material. Individual viral RNAs are often used as substrates for both replication and translation and can contain multiple, sometimes overlapping open reading frames. Further, viral RNAs engage in a wide variety of interactions with both host and viral proteins to modify the activities of important cellular factors and direct their own trafficking, packaging, localization, stability, and translation. However, adaptations increasing the information density of small viral genomes can have unintended consequences. In particular, viral RNAs have developed features that mark them as potential targets of host RNA quality control pathways. This minireview focuses on ways in which viral RNAs run afoul of the cellular mRNA quality control and decay machinery, as well as on strategies developed by viruses to circumvent or exploit cellular mRNA surveillance.
INTRODUCTION
Eukaryotic cells employ quality control mechanisms to monitor each step of mRNA metabolism, from transcription to translation. These mRNA surveillance and decay pathways are responsible for recognizing aberrant mRNAs caused by errors in the template genetic material or in steps of mRNA biogenesis, transport, or function. In addition to using quality control mechanisms to guard against production of deleterious proteins, it has become increasingly clear that cells also use them to regulate gene expression. mRNA surveillance begins immediately following RNA polymerase II initiation, with rapid degradation of pre-mRNAs lacking proper 5′ caps (1, 2). Efficient and accurate gene expression is further ensured by mechanisms to degrade RNAs in response to defects in transcription elongation, splicing, 3′-end formation, and nuclear export (3–10). Following export to the cytoplasm, ribosome-coupled quality control mechanisms such as nonsense-mediated decay (NMD) and no-go decay (NGD) monitor translation, removing mRNAs that cannot be productively translated or that contain premature termination codons (TCs) (reviewed in reference 11).
A general model for RNA quality control pathway target selection proposes that discrimination of decay substrates depends on kinetic competition between decay initiation and normal RNA processing or function. This model holds that defective RNAs take longer to complete processing or function than normal RNAs, giving a window in which quality control factors can act (12). In many cases, RNA defects are not directly detected; instead, the cell must monitor RNA features or function in order to compile a body of circumstantial evidence indicating that the RNA should be destroyed. Quality control pathways must therefore be carefully calibrated to efficiently detect potentially deleterious RNAs while avoiding promiscuous degradation of beneficial RNAs. While this is a problem for the cell, it represents an opportunity for so-called quality control pathways to also fulfill regulatory and antiviral roles. In parallel, the drive to develop new functions within the constraints of small genomes has resulted in viral RNAs that trigger quality control sensors. In this way, RNA quality control pathways can act as components of innate immune responses, with the key distinction that they do not generally signal to promote the activation of antiviral gene expression programs.
Just as studying the interactions between viral RNAs and cellular quality control proteins gives insight into the constraints governing viral evolution, viral RNAs have long been used to discover and dissect cellular gene expression mechanisms. Illustrating this point, multiple sections of this review refer to a sequence of steps that has been used repeatedly to wring insight from studies of viral RNAs: observation of a viral RNA exhibiting unusual behavior, identification of the RNA element responsible, careful structure-function studies, discovery of host factors positively or negatively affecting the RNA, and, finally, generalization of the phenomenon to include regulation of host RNAs. This template for discovery, combined with modern high-throughput sequencing and screening technologies, has revealed extensive interactions between viral RNAs and the host RNA quality control machinery, but much remains to be learned about both the physiological consequences and the underlying mechanisms of these complicated relationships.
FAILURE TO LAUNCH: NUCLEAR RNA QUALITY CONTROL
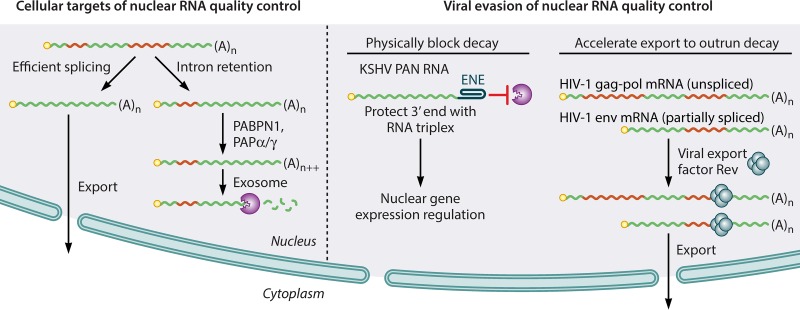
Herpesviruses express abundant unspliced, polyadenylated long noncoding RNAs (lncRNAs) that are retained in the nucleus (13). The prototypical member of this family is Kaposi's sarcoma herpesvirus (KSHV) polyadenylated nuclear (PAN) RNA (14–17), which regulates gene expression and is essential for KSHV replication (18–22). The extremely high levels of PAN RNA accumulation in the lytic phase of KSHV infection (up to 5 × 105 copies per cell) stimulated investigation of the RNA features responsible for PAN RNA abundance, leading to the identification an RNA element that protects PAN RNA from rapid nuclear deadenylation-dependent decay (23, 24). Structural studies of this “expression and nuclear retention element” (ENE) revealed a remarkable triple-helix RNA structure responsible for sequestering the RNA 3′ end from access by decay factors, a motif echoed in several viral and mammalian lncRNAs (25–28). Importantly, the ENE can be used to stabilize intronless β-globin mRNAs, which are usually retained and rapidly degraded in the nucleus (23).
In the absence of an ENE-like element, some intronless or inefficiently spliced mRNAs are subject to hyperadenylation by the canonical PAPα/γ poly(A) polymerases, in conjunction with the PABPN1 nuclear poly(A) binding protein (5, 6, 29–32). Such hyperadenylated transcripts are subsequently degraded by the nuclear exosome (Fig. 1). This process, now designated PABPN1- and PAPα/γ-mediated RNA decay (PPD), functions as a quality control mechanism to degrade several classes of mammalian RNAs, including spliced and unspliced lncRNAs, primary miRNA transcripts, and mRNAs with retained introns (5). Highlighting the extensive interactions between PPD and the viral life cycle, KSHV not only evades PPD using the ENE but also exploits the pathway. Specifically, the KSHV nuclease SOX, responsible for large-scale destruction of host mRNAs in lytic infection, enhances hyperadenylation and decay of nuclear mRNAs (33).
FIG 1.
Execution and evasion of host nuclear RNA decay. (Left) RNAs that are not efficiently exported from the nucleus (e.g., due to intron retention) undergo hyperadenylation and exosome-mediated decay. (Right) Viral RNAs can escape degradation by physically blocking access to the transcript 3′ end with specialized RNA structures such as the KSHV PAN ENE or by promoting export of unspliced or partially spliced mRNA. Red RNA segments indicate intronic sequences.
A CHANGE OF SCENERY: ESCAPING DECAY BY ACCELERATING EXPORT
PAN and related ncRNAs must remain in the nucleus to carry out their functions, but mRNAs and cytoplasmic ncRNAs can use a second option to avoid nuclear quality control: escape to the cytoplasm. Consistent with a mechanism in which PPD efficiency is determined by kinetic competition between nuclear export and mRNA decay, insertion of an intron into an otherwise unspliced mRNA promotes RNA export and stability (23, 34, 35). Alternatively, specific sequences in cellular intronless mRNAs can recruit the TREX mRNA export complex to speed export and avoid nuclear degradation (36). A similar paradigm appears to be at work in retroviruses, although a direct relationship between PPD and turnover of retroviral RNAs has not been demonstrated. Unspliced or singly spliced RNAs from retroviruses, including HIV, are retained in the nucleus and subject to rapid decay in the absence of specialized RNA export systems (37–39). Binding of viral export-promoting proteins such as HIV-1 Rev or direct recruitment of cellular mRNA export receptors by constitutive transport elements (CTEs) overcomes nuclear RNA instability by essentially ushering the RNAs away from danger (Fig. 1) (37, 40–46).
NMD
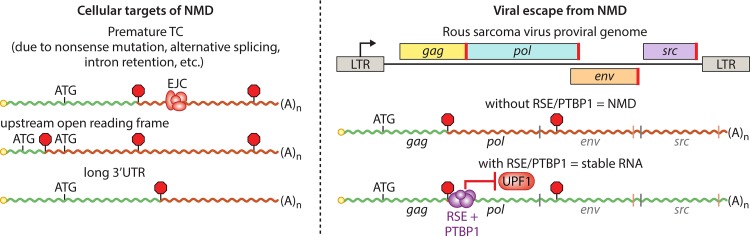
Nonsense-mediated mRNA decay (NMD) is the most extensively studied mRNA quality control pathway, and its target specificity makes it a threat to diverse viral RNAs. While NMD was originally identified as a means to downregulate mRNAs containing nonsense mutations, the pathway also has a continuous role in regulation of cellular gene expression, impacting an estimated 10% of all human genes (47). The work of many laboratories has resulted in a catalog of features that are associated with mRNA susceptibility to NMD, including premature stop codons, errors in splicing, upstream open reading frames (ORFs), 3′ untranscribed region (3′UTR) introns, long 3′UTRs, and others (48). In each of these cases, NMD is thought to act by determining whether a translation termination event is taking place at the correct position or whether it is in a location that indicates a problem with the mRNA (Fig. 2).
FIG 2.
Determinants of cellular and viral RNA susceptibility to NMD. (Left) RNA features leading to induction of NMD. In each case, translation termination occurs at sites distant from the 3′ end of the transcript. Decay is accelerated if termination takes place upstream of an EJC. (Right) RSV full-length mRNAs use the RSE to recruit host PTBP1 protein to the vicinity of the gag stop codon, preventing UPF1 association and inhibiting NMD. If the RSE is deleted or unable to bind PTBP1, RSV mRNAs are efficiently degraded by NMD. Positions of TCs typically used in full-length RNAs are indicated by stop signs, and positions of start and stop codons of additional ORFs are shown with light gray and red vertical lines, respectively. Red segments of RNAs indicate regions that may be recognized by UPF1 as aberrantly long 3′UTRs. LTR, long terminal repeat.
Presumably due to selective pressure to maximize coding capacity without increasing genome size, viral RNAs present several possible NMD-inducing features, including multiple open reading frames in a single mRNA, long 3′UTRs, retained introns, and translational recoding elements (49). Thus far, mRNAs from retroviruses and alphaviruses in mammals and positive-strand RNA viruses in plants have been found to be targets of NMD (50–53), but widespread investigations of viral RNA targeting by NMD have not been reported. Given the involvement of NMD in diverse viruses from plants to mammals, further studies are likely to yield more examples of either NMD-mediated viral restriction or viral strategies for NMD avoidance.
NMD is carried out by a complex network of protein factors that function together to determine whether an mRNA should be degraded or be allowed to continue producing protein. The highly conserved RNA helicase UPF1 is the central node in this process, engaging in interactions with target mRNAs, NMD pathway components, translation factors, and other components of mRNA-protein (mRNP) complexes (54). The initial phase of target discrimination by the NMD pathway involves nonspecific, ATPase-regulated RNA binding by UPF1, leading to preferential accumulation of the protein on long 3′UTRs and other mRNA segments not cleared by elongating ribosomes (55–59). In higher eukaryotes, the presence of an exon junction complex (EJC) downstream of the termination codon accelerates but is not essential for UPF1-mediated decay (60).
While the initial association of UPF1 with mRNAs can proceed in the absence of translation termination events, the NMD machinery relies on the ribosome for its indispensable ability to decode in-frame stop codons. The linkage between translation termination and NMD is direct: UPF1 interacts with eukaryotic release factors 1 and 3 (eRF1/3) at the terminating ribosome, in a complex that also contains the SMG-1 kinase (61). Activation of UPF1 phosphorylation and ATPase activity is promoted by several additional NMD proteins, including UPF2, UPF3, and DHX34 (62–66). SMG-1 phosphorylates UPF1 at multiple sites, which in turn leads to recruitment and activation of decay enzymes, including the NMD-specific endonuclease SMG6 (67–75).
NOW YOU SEE ME: NMD FACTORS RESTRICT ALPHAVIRUS REPLICATION
Alphaviruses are positive-strand single-stranded RNA (ssRNA) viruses that carry out their entire replication cycle in the cytoplasm. In an RNA interference (RNAi) screen for host protein regulators of Semliki Forest virus (SFV) replication, UPF1 was identified as a potential alphavirus restriction factor (51). In follow-up experiments, knockdown of UPF1 enhanced replication of both SFV and the related Sindbis virus. In addition to knockdown of UPF1, knockdown of NMD proteins SMG5 and SMG7 enhanced SFV replication, although depletion of several other core NMD factors, including SMG6, UPF2, and UPF3, had no apparent effect. Correspondingly, inactivation of NMD in cells infected with replication-defective SFV increased the half-life of genomic RNA. Further investigation of the mechanism of NMD-dependent alphavirus restriction revealed that the most obvious candidate NMD-sensitive SFV RNA feature, the ∼4,000-nucleotide (nt) 3′UTR downstream of the replicase ORF, was not required for increased translation of virus-derived reporter RNAs upon UPF1 knockdown. It remains unclear why SFV mRNAs are sensitive to NMD, but it is likely that UPF1-dependent destabilization of SFV RNAs is due to an unusual termination event that is in turn due to either placement or kinetics. An alternative possibility is that NMD can both directly degrade alphavirus RNAs (due to their long 3′UTRs or to another RNA feature) and indirectly inhibit replication by suppressing expression of proteins necessary for efficient viral replication.
NOW YOU DON′T: INHIBITION OF NMD AT SPECIFIC TERMINATION CODONS
The first indication that viral RNAs might be subject to decay in response to stop codon position dates to the very beginning of NMD investigations, even prior to the characterization of NMD proteins (76–78). In a series of studies, members of the Beemon laboratory discovered that mRNAs from the retrovirus Rous sarcoma virus (RSV) containing frameshifts or premature termination codons in the gag gene were destabilized in a UPF1-dependent manner (76, 77, 79). As the normal gag stop codon precedes a very long (∼7,000-nt) 3′UTR, it is also a potential NMD target, but wild-type full-length RSV mRNAs are not subject to NMD. With further investigation, it became clear that full-length RSV RNAs have evolved a mechanism that actively shields them from NMD, mediated by an ∼400-nt sequence found immediately downstream of the gag stop codon termed the RNA stability element (RSE) (80, 81). Studies of RSE structure and function revealed that it is a complex RNA element containing multiple functionally redundant segments (82, 83) which retains its protective activity when moved into proximity with NMD-inducing termination codons in RSV RNAs or reporter transcripts (81, 84).
Recently, several of us identified polypyrimidine tract binding protein 1 (PTBP1) as an essential cofactor for RSE-mediated stabilization of RSV mRNAs (Fig. 2) (84). Using RNA-based affinity purification and mass spectrometry, we found PTBP1 to be a prominent RSE-interacting protein, recruited via pyrimidine-rich binding sites found throughout the RSE. Mutation of these sequences abolished RSE-mediated protection of viral RNAs in chicken embryonic fibroblasts and reporter mRNAs in human cells, while addition of PTBP1 binding sites to NMD target mRNAs conferred stability in the absence of additional RSE sequence. PTBP1, which uses four RNA recognition motifs (RRMs) for high-affinity binding to pyrimidine-rich RNAs, is well known as a protein that binds intronic sequences to inhibit spliceosome assembly and induce exon skipping (85). Similarly, PTBP1 binding to potential NMD substrates prevents UPF1 association, inhibiting the early stages of NMD target selection (84). This mechanism represents another example of the journey from interesting viral RNA behavior to broad applicability for the cellular gene expression regulation outlined at the beginning of this review, as transcriptome sequencing (RNA-seq) studies and genome-wide profiling of PTBP1 RNA occupancy indicated that hundreds of human mRNAs may be stabilized by termination codon (TC)-proximal PTBP1 binding. Recruitment of PTBP1 to the vicinity of termination codons therefore appears to be a widespread strategy used by cellular and viral RNAs alike to signal to the NMD pathway that a particular stop codon should be ignored.
THE NUCLEAR OPTION: GLOBAL INHIBITION OF NMD BY VIRAL PROTEINS
An alternative approach for viruses to protect their RNAs from NMD is to globally inhibit the pathway. Thus far, representatives of two distinct virus families, flaviviruses (hepatitis C virus [HCV]) and retroviruses (human T cell leukemia virus [HTLV-1]), have been reported to disrupt cellular NMD activity (52, 53, 86). In the case of HCV, the viral core protein binds the EJC recycling factor PYM/WIBG, disrupting the interaction between PYM and the EJC. Through a mechanism that remains to be fully defined, this was in turn proposed to inhibit decay of multiple cellular NMD targets (86). Interestingly, two HTLV-1 proteins, Tax and Rex, have been proposed to be NMD inhibitors. First, Mocquet and colleagues identified interactions of HTLV-1 Tax, best characterized as a transcriptional transactivator, with the core NMD protein UPF1 and eIF3 translation initiation complex component INT6/eIF3E (53). A second report held that the viral RNA export cofactor Rex was responsible for inhibiting NMD, while Tax expression had only a minor effect (52). Further work will be required to fully dissect the relative contributions of HTLV-1 proteins to NMD inhibition and to understand the mechanisms underlying these observations. Both groups found that HTLV-1 RNAs remained somewhat susceptible to NMD in the presence of Tax and Rex, suggesting that these proteins may dial down NMD activity rather than abolishing it altogether (52, 53). Indeed, due to side effects of NMD inhibition such as cellular toxicity, alteration of gene expression, and activation of stress responses, it may not be advantageous for viruses to completely block NMD activity.
TRANSLATIONAL RECODING: A PROBLEM OR A SOLUTION?
Many viral RNAs contain elements that modulate the use of termination codons by promoting either translational readthrough or frameshifting. Generally, this is a mechanism to control the relative levels of production of two distinct forms of a viral polyprotein which are subsequently proteolytically processed to yield functional proteins (87). In most characterized cases, translational recoding elements contain pseudoknot RNA structures, although strong double-stranded RNA (dsRNA) hairpins have also been reported to mediate the activity (88). Structures that induce frameshifting or readthrough are typically found immediately downstream of a slippery sequence or a termination codon, respectively. While the mechanisms driving translational recoding are not completely understood, these structures are thought to put the ribosome under mechanical stress, which alters the process of stop codon recognition or disrupts reading frame maintenance (89, 90).
Translational recoding is an effective means to maximize the function of viral RNAs, but the placement of a partially suppressed stop codon upstream of a long ORF bears a strong resemblance to an mRNA containing a premature termination codon. Viral RNAs with this genome architecture must somehow avoid detection by NMD. One factor aiding viral RNA stability is that readthrough and frameshifting events can be inherently disruptive of NMD. Inefficient termination caused by small molecules, RNA elements, trans-acting proteins, and cellular conditions have all been demonstrated to inhibit NMD in organisms from yeast to humans (55, 91–98). The extent of induction of translational readthrough need not be large to have a substantial impact on RNA stability: readthrough efficiencies of approximately 1% have been observed to have a substantial impact on NMD susceptibility in multiple contexts (55, 97). Experiments with reporter mRNAs suggest that readthrough events can inhibit decay either by disrupting the association of UPF1 with sequences downstream of the suppressed termination codon or by disrupting a later step or steps required for initiation of decay, such as UPF1 phosphorylation or recruitment/activation of decay enzymes (55, 99).
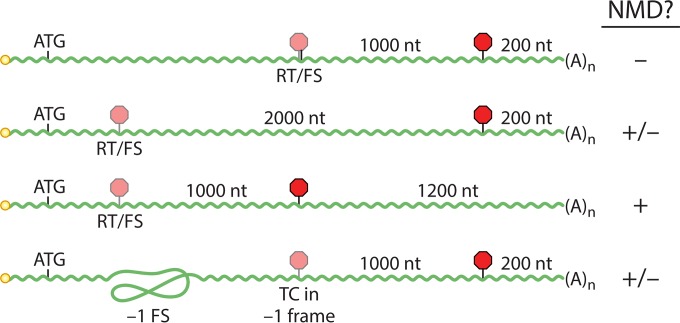
In addition to the absolute efficiency of frameshifting or readthrough, several factors likely combine to determine the decay susceptibility of a transcript undergoing programmed translational recoding (Fig. 3). Particular conditions must be met for recoding to be sufficient for protection from NMD. First, recoding must be efficient enough to compensate for the length of the conditionally translated downstream ORF, and, second, that ORF must end at a site that would not normally be recognized by NMD as a premature termination codon. Moreover, some recoding events can activate rather than inhibit NMD. For example, an efficient frameshifting element that causes use of a termination codon (TC) upstream of the normal TC can result in NMD (100). In sum, the position of the recoding event and the relative kinetics of elongation, termination, and decay induction all modulate the balance between stability and decay through NMD or, in some cases, NGD (101).
FIG 3.
Scenarios for NMD modulation by translational recoding events. Readthrough (RT) or frameshifting (FS) can protect transcripts from NMD, if the downstream TC is in a position not sensed by NMD (top [first] mRNA). This activity can be antagonized by increasing the length of the downstream ORF (second mRNA) or abolished if the downstream TC is itself followed by a long 3′UTR (third mRNA). In contrast, frameshifting events that cause usage of an upstream TC can induce decay (bottom mRNA). Hypothetical RNA lengths shown are for illustrative purposes only; lightly shaded stop signs indicate TCs that are not used constitutively. nt, nucleotide.
FISH OR CUT RNA: mRNA DECAY FOLLOWING RIBOSOME STALLING
Cells mount a concerted response to stalled ribosomes in which the stalled ribosomal subunits are recycled and the mRNA is degraded (11, 102, 103). Rescue of nonproductive ribosomes and the associated mRNA decay are mediated by homologs of eukaryotic release factors 1 and 3 (eRF1/3) PELO (Dom34 in yeast) and HBS1L (Hbs1 in yeast), respectively (104–106). Incorporation of PELO and HBS1L into an empty aminoacyl (A) site of a stalled ribosome is associated with endonucleolytic cleavage upstream of the ribosome by an as-yet-unidentified RNase and, in conjunction with ABCE1, ribosomal subunit splitting (106–108).
Two distinct decay pathways induced by stalled ribosomes, namely, no-go decay (NGD), which acts on mRNAs that have been damaged, that are highly structured, or that contain strings of rare codons, and non-stop decay (NSD), which acts on mRNAs lacking stop codons, have been previously described (104, 109, 110). In NSD, translation of an mRNA without a stop codon leads to translation elongation into the poly(A) tail, where the ribosome synthesizes polylysine and stalls (111, 112). As the protein factors required for NSD and NGD are the same, and as decay arises from the response to ribosome stalling in both cases, NSD can be considered a special case of NGD. The response to ribosome stalling performs two important functions: prevention of protein production from a potentially aberrant mRNA, and rescue of the ribosome from an unproductive state (106, 107, 113). While many viral RNAs contain highly structured elements within coding sequences and some, such as HIV, have unusual codon usage, it is not known whether such viral RNAs efficiently trigger NGD. Instead, the characterized examples of viral interactions with the NGD pathway feature more-complex routes to decay induction. For example, NGD has been observed to occur in response to depurination of viral RNAs by ectopic expression of the pokeweed antiviral protein, a member of a large class of bacterial and plant ribosome-inactivating proteins (114).
RNA QUALITY CONTROL BEGETS QUALITY VIRIONS?
Unexpectedly, recent findings suggest that HIV-1 may exploit no-go decay to fine-tune its own gene expression and ensure production of infectious virions. Mu et al. found that HIV-1 RNAs are susceptible to no-go decay through a mechanism dependent on translation of the Gag protein matrix domain (MA) (115). They proposed that a host factor, ATPase RuvB-like 2 (RVB2), interacts with the HIV-1 5′UTR and nascent MA peptide, impeding further translation of Gag or Gag-Pol protein. Ribosome stalling by this network of protein-RNA interactions causes destabilization of gag mRNA in a PELO-dependent manner, without affecting stability or translation of other viral mRNAs. The effect of RVB2 on HIV-1 Gag-Pol expression is counteracted by expression of an intact HIV-1 envelope protein, through a well-characterized interaction between the C-terminal tail of Env and MA (116). Together, these data suggest that HIV-1 uses NGD to suppress gag expression at early time points following initial proviral transcription, possibly to prevent production of virions that contain Gag and Gag-Pol but lack the Env protein necessary for infectivity (115). Once intracellular Env levels are high enough to efficiently compete with RVB2 for MA, the repression is relieved, and virions with a full complement of viral proteins can be produced. With this mechanism, HIV-1 thus may use a host RNA quality control pathway to maximize the quality of viral particles.
I JUST CAN′T QUIT YOU: VIRAL DEPENDENCE ON DECAY FACTORS
RNA quality control pathways are integral components of the cellular gene expression machinery, and their inactivation can cause widespread changes in cellular RNA and protein levels. This may explain why viruses sometimes depend on RNA decay factors for efficient replication, even as they attempt to shield viral RNAs from degradation. For example, HCV, in addition to suppressing NMD, requires the EJC recycling factor PYM (86). Likewise, the UPF1 protein has been described by two groups to be a positive regulator of HIV-1 replication at distinct steps: export and stability of viral mRNAs in late stages of infection and reverse transcription following entry (117–119). While both groups identified UPF1 as important for HIV-1 replication, Serquiña and colleagues did not observe the effect of UPF1 depletion on viral gene expression reported by Ajamian et al., for reasons that remain to be resolved. One possibility is that there are cell type-specific differences in the cellular effects of UPF1 depletion. Hinting at this possibility, UPF1 knockdown can severely impair viability of HeLa cells, used by Ajamian et al., while HEK-293 cells, used by Serquiña et al., appear to better tolerate loss of UPF1 (120; J. R. Hogg, unpublished observations). Interestingly, in both cases there are indications that enhancement of HIV-1 replication by UPF1 may operate through mechanisms distinct from its canonical NMD activities, as mutants of UPF1 inactive in NMD were reported to promote HIV-1 gene expression (117), while UPF2 was found to be dispensable for infectivity (119).
NSD/NGD factors have also been found to be essential for replication of diverse viruses, likely due to the enormous load viral replication can impose on the cellular translation machinery (121). Recent data from widely divergent systems suggest that high-level viral protein production may deplete the available pool of ribosomal subunits, leading to a requirement for efficient rescue of non-productively bound ribosomes (122, 123). This again creates a situation in which global inhibition of the pathway would preserve viral mRNA stability but would ultimately be deleterious to the virus. In Drosophila and plants, genetic screens for host factors promoting viral replication uncovered a dependence on the NGD factor PELO for ssRNA Drosophila C virus (DCV) and dsDNA tomato yellow leaf curl virus (TYLCV) replication, respectively (122, 123). In flies, PELO inactivation caused an increase in levels of 80S monosomes that did not appear to be productively engaged in translation, which corresponded to reduced synthesis of DCV capsid protein (122). These findings suggest that in the absence of efficient ribosome recycling, the available pool of free ribosomal subunits severely limits viral protein translation and replication. As with NMD, indirect effects due to the inactivation of an important component of the cellular gene expression machinery may also contribute to the lack of viral replication in cells deficient for NGD factors.
CONCLUSION
Viral interactions with host RNA quality control pathways can clearly have profound effects on both viruses and the cells in which they replicate. However, the underlying mechanisms by which viruses evade and manipulate RNA quality control are only beginning to be understood. Moreover, many more examples of viral restriction by RNA quality control or viral evasion of decay likely remain to be discovered. For viruses that efficiently counteract RNA quality control surveillance, these interactions can be particularly difficult to uncover. Whereas investigations of the impact of RNA decay pathways on viral replication and evolution have largely arisen from serendipity, the field may now benefit from systematic efforts to identify and characterize viral RNA elements that counteract quality control.
ACKNOWLEDGMENTS
I apologize to colleagues whose work I was unable to discuss due to space constraints. I express many thanks to Thomas Baird, Nazmul Haque, Aparna Kishor, Soumya Ranganathan, Zhiyun Ge, Lisa Postow, Stephen Goff, and Karen Beemon for helpful discussions and/or critical reading of the manuscript and to Patrick Lane for figure enhancement.
Funding Statement
This work was supported by HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI) | Intramural Research Program.
REFERENCES
- 1.Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M. 2010. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467:608–611. doi: 10.1038/nature09338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiao X, Chang JH, Kilic T, Tong L, Kiledjian M. 2013. A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol Cell 50:104–115. doi: 10.1016/j.molcel.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S, Nakamura M, Chen X, Zhang K, Meziane O, Boyer F, Parrinello H, Berkhout B, Terzian C, Benkirane M, Kiernan R. 2012. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell 150:1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson L, Kerr A, West S. 2012. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J 31:2566–2578. doi: 10.1038/emboj.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bresson SM, Hunter OV, Hunter AC, Conrad NK. 2015. Canonical poly(A) polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. PLoS Genet 11:e1005610. doi: 10.1371/journal.pgen.1005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemieux C, Marguerat S, Lafontaine J, Barbezier N, Bähler J, Bachand F. 2011. A pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Mol Cell 44:108–119. doi: 10.1016/j.molcel.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 7.Burgess SM, Guthrie C. 1993. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73:1377–1391. doi: 10.1016/0092-8674(93)90363-U. [DOI] [PubMed] [Google Scholar]
- 8.Bousquet-Antonelli C, Presutti C, Tollervey D. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102:765–775. doi: 10.1016/S0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 9.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 10.Burkard KT, Butler JS. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol 20:604–616. doi: 10.1128/MCB.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shoemaker CJ, Green R. 2012. Translation drives mRNA quality control. Nat Struct Mol Biol 19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doma MK, Parker R. 2007. RNA quality control in eukaryotes. Cell 131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 13.Conrad NK. 2016. New insights into the expression and functions of the Kaposi's sarcoma-associated herpesvirus long noncoding PAN RNA. Virus Res 212:53–63. doi: 10.1016/j.virusres.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun R, Lin SF, Gradoville L, Miller G. 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A 93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong W, Wang H, Herndier B, Ganem D. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci U S A 93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staskus KA, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson DJ, Ganem D, Haase AT. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol 71:715–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong W, Ganem D. 1997. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). J Virol 71:1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell M, Kim KY, Chang P-C, Huerta S, Shevchenko B, Wang D-H, Izumiya C, Kung H-J, Izumiya Y. 2014. A lytic viral long noncoding RNA modulates the function of a latent protein. J Virol 88:1843–1848. doi: 10.1128/JVI.03251-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossetto CC, Tarrant-Elorza M, Verma S, Purushothaman P, Pari GS. 2013. Regulation of viral and cellular gene expression by Kaposi's sarcoma-associated herpesvirus polyadenylated nuclear RNA. J Virol 87:5540–5553. doi: 10.1128/JVI.03111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossetto CC, Pari GS. 2011. Kaposi's sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J Virol 85:13290–13297. doi: 10.1128/JVI.05886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borah S, Darricarrère N, Darnell A, Myoung J, Steitz JA. 2011. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog 7:e1002300. doi: 10.1371/journal.ppat.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossetto CC, Pari G. 2012. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog 8:e1002680. doi: 10.1371/journal.ppat.1002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad NK, Steitz JA. 2005. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J 24:1831–1841. doi: 10.1038/sj.emboj.7600662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conrad NK, Mili S, Marshall EL, Shu M-D, Steitz JA. 2006. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell 24:943–953. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 25.Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. 2010. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science 330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tycowski KT, Shu M-D, Borah S, Shi M, Steitz JA. 2012. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep 2:26–32. doi: 10.1016/j.celrep.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilusz JE, JnBaptiste CK, Lu LY, Kuhn C-D, Joshua-Tor L, Sharp PA. 2012. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. 2012. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc Natl Acad Sci U S A 109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beaulieu YB, Kleinman CL, Landry-Voyer A-M, Majewski J, Bachand F. 2012. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein Nuclear 1. PLoS Genet 8:e1003078. doi: 10.1371/journal.pgen.1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bresson SM, Conrad NK. 2013. The human nuclear poly(a)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet 9:e1003893. doi: 10.1371/journal.pgen.1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goebels C, Thonn A, Gonzalez-Hilarion S, Rolland O, Moyrand F, Beilharz TH, Janbon G. 2013. Introns regulate gene expression in Cryptococcus neoformans in a Pab2p dependent pathway. PLoS Genet 9:e1003686. doi: 10.1371/journal.pgen.1003686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. 2010. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J 29:2173–2181. doi: 10.1038/emboj.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee YJ, Glaunsinger BA. 2009. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol 7:e1000107. doi: 10.1371/journal.pbio.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valencia P, Dias AP, Reed R. 2008. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc Natl Acad Sci U S A 105:3386–3391. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo MJ, Reed R. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci U S A 96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei H, Dias AP, Reed R. 2011. Export and stability of naturally intronless mRNAs require specific coding region sequences and the TREX mRNA export complex. Proc Natl Acad Sci U S A 108:17985–17990. doi: 10.1073/pnas.1113076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felber BKF, Hadzopoulou-Cladaras MH, Cladaras C, Copeland TC, Pavlakis GNP. 1989. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci U S A 86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz S, Felber BK, Pavlakis GN. 1992. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J Virol 66:150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zolotukhin AS, Michalowski D, Bear J, Smulevitch SV, Traish AM, Peng R, Patton J, Shatsky IN, Felber BK. 2003. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol Cell Biol 23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, Hammarskjöld ML. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci U S A 91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ernst RK, Bray M, Rekosh D, Hammarskjöld ML. 1997. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol 17:135–144. doi: 10.1128/MCB.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hidaka M, Inoue J, Yoshida M, Seiki M. 1988. Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J 7:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seiki M, Inoue J, Hidaka M, Yoshida M. 1988. Two cis-acting elements responsible for posttranscriptional trans-regulation of gene expression of human T-cell leukemia virus type I. Proc Natl Acad Sci U S A 85:7124–7128. doi: 10.1073/pnas.85.19.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenblatt JD, Cann AJ, Slamon DJ, Smalberg IS, Shah NP, Fujii J, Wachsman W, Chen IS. 1988. HTLV-II transactivation is regulated by the overlapping tax/rex nonstructural genes. Science 240:916–919. doi: 10.1126/science.2834826. [DOI] [PubMed] [Google Scholar]
- 45.Emerman M, Vazeux R, Peden K. 1989. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell 57:1155–1165. doi: 10.1016/0092-8674(89)90053-6. [DOI] [PubMed] [Google Scholar]
- 46.Hammarskjöld ML, Heimer J, Hammarskjöld B, Sangwan I, Albert L, Rekosh D. 1989. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol 63:1959–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mühlemann O, Jensen TH. 2012. mRNP quality control goes regulatory. Trends Genet 28:70–77. doi: 10.1016/j.tig.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Rebbapragada I, Lykke-Andersen J. 2009. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol 21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Quek BL, Beemon K. 2014. Retroviral strategy to stabilize viral RNA. Curr Opin Microbiol 18:78–82. doi: 10.1016/j.mib.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia D, Garcia S, Voinnet O. 2014. Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 16:391–402. doi: 10.1016/j.chom.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balistreri G, Horvath P, Schweingruber C, Zünd D, McInerney G, Merits A, Mühlemann O, Azzalin C, Helenius A. 2014. The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication. Cell Host Microbe 16:403–411. doi: 10.1016/j.chom.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 52.Nakano K, Ando T, Yamagishi M, Yokoyama K, Ishida T, Ohsugi T, Tanaka Y, Brighty DW, Watanabe T. 2013. Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: implications for retroviral replication. Microbes Infect 15:491–505. doi: 10.1016/j.micinf.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 53.Mocquet V, Neusiedler J, Rende F, Cluet D, Robin J-PP, Terme J-MM, Duc Dodon M, Wittmann J, Morris C, Le Hir H, Ciminale V, Jalinot P. 2012. The human T-lymphotropic virus type 1 tax protein inhibits nonsense-mediated mRNA decay by interacting with INT6/EIF3E and UPF1. J Virol 86:7530–7543. doi: 10.1128/JVI.07021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. 2013. Nonsense-mediated mRNA decay—mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta 1829:612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Hogg JR, Goff SP. 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143:379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kurosaki T, Maquat LE. 2013. Rules that govern UPF1 binding to mRNA 3′ UTRs. Proc Natl Acad Sci U S A 110:3357–3362. doi: 10.1073/pnas.1219908110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zünd D, Gruber AR, Zavolan M, Mühlemann O. 2013. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol 20:936–943. doi: 10.1038/nsmb.2635. [DOI] [PubMed] [Google Scholar]
- 58.Hurt JA, Robertson AD, Burge CB. 2013. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 23:1636–1650. doi: 10.1101/gr.157354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee SR, Pratt GA, Martinez FJ, Yeo GW, Lykke-Andersen J. 2015. Target discrimination in nonsense-mediated mRNA decay requires Upf1 ATPase activity. Mol Cell 59:413–425. doi: 10.1016/j.molcel.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. 2006. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melero R, Hug N, López-Perrote A, Yamashita A, Cáceres JF, Llorca O. 2016. The RNA helicase DHX34 functions as a scaffold for SMG1-mediated UPF1 phosphorylation. Nat Commun 7:10585. doi: 10.1038/ncomms10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hug N, Cáceres JF. 2014. The RNA helicase DHX34 activates NMD by promoting a transition from the surveillance to the decay-inducing complex. Cell Rep 8:1845–1856. doi: 10.1016/j.celrep.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Melero R, Uchiyama A, Castaño R, Kataoka N, Kurosawa H, Ohno S, Yamashita A, Llorca O. 2014. Structures of SMG1-UPFs complexes: SMG1 contributes to regulate UPF2-dependent activation of UPF1 in NMD. Structure 22:1105–1119. doi: 10.1016/j.str.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 65.López-Perrote A, Castaño R, Melero R, Zamarro T, Kurosawa H, Ohnishi T, Uchiyama A, Aoyagi K, Buchwald G, Kataoka N, Yamashita A, Llorca O. 15 January 2016. Human nonsense-mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res doi: 10.1093/nar/gkv1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chamieh H, Ballut L, Bonneau F, Le Hir H. 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 67.Loh B, Jonas S, Izaurralde E. 2013. The SMG5-SMG7 heterodimer directly recruits the CCR4-NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev 27:2125–2138. doi: 10.1101/gad.226951.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chakrabarti S, Bonneau F, Schüssler S, Eppinger E, Conti E. 2014. Phospho-dependent and phospho-independent interactions of the helicase UPF1 with the NMD factors SMG5-SMG7 and SMG6. Nucleic Acids Res 42:9447–9460. doi: 10.1093/nar/gku578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. 2006. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J 25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E. 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nicholson P, Josi C, Kurosawa H, Yamashita A, Mühlemann O. 22 July 2014. A novel phosphorylation-independent interaction between SMG6 and UPF1 is essential for human NMD. Nucleic Acids Res doi: 10.1093/nar/gku645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12:1187–1200. doi: 10.1016/S1097-2765(03)00443-X. [DOI] [PubMed] [Google Scholar]
- 73.Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. 29 September 2011. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 75.Unterholzner L, Izaurralde E. 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 76.Arrigo S, Beemon K. 1988. Regulation of Rous sarcoma virus RNA splicing and stability. Mol Cell Biol 8:4858–4867. doi: 10.1128/MCB.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barker GF, Beemon K. 1991. Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol Cell Biol 11:2760–2768. doi: 10.1128/MCB.11.5.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leeds P, Peltz SW, Jacobson A, Culbertson MR. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 79.LeBlanc JJL, Beemon KLB. 2004. Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. J Virol 78:5139–5146. doi: 10.1128/JVI.78.10.5139-5146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barker GF, Beemon K. 1994. Rous sarcoma virus RNA stability requires an open reading frame in the gag gene and sequences downstream of the gag-pol junction. Mol Cell Biol 14:1986–1996. doi: 10.1128/MCB.14.3.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weil JE, Beemon KL. 2006. A 3′ UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA 12:102–110. doi: 10.1261/rna.2129806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weil JE, Hadjithomas M, Beemon KL. 2009. Structural characterization of the Rous sarcoma virus RNA stability element. J Virol 83:2119–2129. doi: 10.1128/JVI.02113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Withers JB, Beemon KL. 2010. Structural features in the Rous sarcoma virus RNA stability element are necessary for sensing the correct termination codon. Retrovirology 7:65. doi: 10.1186/1742-4690-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ge Z, Quek BL, Beemon KL, Hogg JR. 8 January 2016. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. Elife doi: 10.7554/eLife.11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Keppetipola NK, Sharma S, Li QL, Black DLB. 2012. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol 47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramage HR, Kumar GR, Verschueren E, Johnson JR, Von Dollen J, Johnson T, Newton B, Shah P, Horner J, Krogan NJ, Ott M. 2015. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol Cell 57:329–340. doi: 10.1016/j.molcel.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bolinger C, Boris-Lawrie K. 2009. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology 6:8. doi: 10.1186/1742-4690-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dinman JD. 2012. Control of gene expression by translational recoding. Adv Protein Chem Struct Biol 86:129–149. doi: 10.1016/B978-0-12-386497-0.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Caliskan N, Peske F, Rodnina MV. 2015. Changed in translation: mRNA recoding by −1 programmed ribosomal frameshifting. Trends Biochem Sci 40:265–274. doi: 10.1016/j.tibs.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dunkle JA, Dunham CM. 2015. Mechanisms of mRNA frame maintenance and its subversion during translation of the genetic code. Biochimie 114:90–96. doi: 10.1016/j.biochi.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moriarty PM, Reddy CC, Maquat LE. 1998. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol 18:2932–2939. doi: 10.1128/MCB.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weiss SLW, Sunde RAS. 1998. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA 4:816–827. doi: 10.1017/S1355838298971990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Forge A, Schacht J. 2000. Aminoglycoside antibiotics. Audiol Neurootol 5:3–22. doi: 10.1159/000013861. [DOI] [PubMed] [Google Scholar]
- 94.Tork S, Hatin I, Rousset J, Fabret C. 2004. The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res 32:415–421. doi: 10.1093/nar/gkh201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Allamand V, Bidou L, Arakawa M, Floquet C, Shiozuka M, Paturneau-Jouas M, Gartioux C, Butler-Browne GS, Mouly V, Rousset J-P, Matsuda R, Ikeda D, Guicheney P. 2008. Drug-induced readthrough of premature stop codons leads to the stabilization of laminin alpha2 chain mRNA in CMD myotubes. J Gene Med 10:217–224. doi: 10.1002/jgm.1140. [DOI] [PubMed] [Google Scholar]
- 96.Sako Y, Usuki F, Suga H. 2006. A novel therapeutic approach for genetic diseases by introduction of suppressor tRNA. Nucleic Acids Symp Ser (Oxf) 2006:239–240. [DOI] [PubMed] [Google Scholar]
- 97.Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM. 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10:691–703. doi: 10.1261/rna.5147804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gonzalez-Hilarion S, Beghyn T, Jia J, Debreuck N, Berte G, Mamchaoui K, Mouly V, Gruenert DC, Déprez B, Lejeune F. 2012. Rescue of nonsense mutations by amlexanox in human cells. Orphanet J Rare Dis 7:58. doi: 10.1186/1750-1172-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hogg JR. 2011. This message was inspected by Upf1: 3′UTR length sensing in mRNA quality control. Cell Cycle 10:372–373. doi: 10.4161/cc.10.3.14735. [DOI] [PubMed] [Google Scholar]
- 100.Belew AT, Meskauskas A, Musalgaonkar S, Advani VM, Sulima SO, Kasprzak WK, Shapiro BA, Dinman JD. 2014. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature 512:265–269. doi: 10.1038/nature13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Belew AT, Advani VM, Dinman JD. 2011. Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast. Nucleic Acids Res 39:2799–2808. doi: 10.1093/nar/gkq1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Graille M, Séraphin B. 2012. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat Rev Mol Cell Biol 13:727–735. doi: 10.1038/nrm3457. [DOI] [PubMed] [Google Scholar]
- 103.Inada T. 2013. Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochim Biophys Acta 1829:634–642. doi: 10.1016/j.bbagrm.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 104.Doma MK, Parker R. 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Becker T, Armache J-PP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. 2011. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol 18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 106.Shoemaker CJ, Eyler DE, Green R. 2010. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pisareva VP, Skabkin MA, Hellen CUT, Pestova TV, Pisarev AV. 2011. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J 30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T. 11 April 2012. Dom34:Hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol Cell doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 109.Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC. 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 110.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 111.Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. 2007. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev 21:519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lu J, Deutsch C. 2008. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol 384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Saito S, Hosoda N, Hoshino S-I. 2013. The Hbs1-Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J Biol Chem 288:17832–17843. doi: 10.1074/jbc.M112.448977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gandhi R, Manzoor M, Hudak KA. 2008. Depurination of Brome mosaic virus RNA3 in vivo results in translation-dependent accelerated degradation of the viral RNA. J Biol Chem 283:32218–32228. doi: 10.1074/jbc.M803785200. [DOI] [PubMed] [Google Scholar]
- 115.Mu X, Fu Y, Zhu Y, Wang X, Xuan Y, Shang H, Goff SP, Gao G. 2015. HIV-1 Exploits the host factor RuvB-like 2 to balance viral protein expression. Cell Host Microbe 18:233–242. doi: 10.1016/j.chom.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 116.Tedbury PR, Freed EO. 2014. The role of matrix in HIV-1 envelope glycoprotein incorporation. Trends Microbiol 22:372–378. doi: 10.1016/j.tim.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, Gehring NH, Mouland AJ. 2008. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 14:914–927. doi: 10.1261/rna.829208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ajamian L, Abel K, Rao S, Vyboh K, García-de-Gracia F, Soto-Rifo R, Kulozik AE, Gehring NH, Mouland AJ. 2015. HIV-1 recruits UPF1 but excludes UPF2 to promote nucleocytoplasmic export of the genomic RNA. Biomolecules 5:2808–2839. doi: 10.3390/biom5042808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Serquiña AKPS, Das SRD, Popova EP, Ojelabi OA, Roy CKR, Göttlinger HGG. 2013. UPF1 is crucial for the infectivity of human immunodeficiency virus type 1 progeny virions. J Virol 87:8853–8861. doi: 10.1128/JVI.00925-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Azzalin CM, Lingner J. 2006. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol 16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 121.Li S, Kong L, Yu X. 2015. The expanding roles of endoplasmic reticulum stress in virus replication and pathogenesis. Crit Rev Microbiol 41:150–164. doi: 10.3109/1040841X.2013.813899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu X, He W-T, Tian S, Meng D, Li Y, Chen W, Li L, Tian L, Zhong C-Q, Han F, Chen J, Han J. 2014. pelo is required for high efficiency viral replication. PLoS Pathog 10:e1004034. doi: 10.1371/journal.ppat.1004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lapidot M, Karniel U, Gelbart D, Fogel D, Evenor D, Kutsher Y, Makhbash Z, Nahon S, Shlomo H, Chen L, Reuveni M, Levin I. 2015. A novel route controlling begomovirus resistance by the messenger RNA surveillance factor Pelota. PLoS Genet 11:e1005538. doi: 10.1371/journal.pgen.1005538. [DOI] [PMC free article] [PubMed] [Google Scholar]