ABSTRACT
UL36p (VP1/2) is the largest protein encoded by herpes simplex virus 1 (HSV-1) and resides in the innermost layer of tegument, the complex protein layer between the capsid and envelope. UL36p performs multiple functions in the HSV life cycle, including a critical but unknown role in capsid cytoplasmic envelopment. We tested whether UL36p is essential for envelopment because it is required to engage capsids with the cellular ESCRT/Vps4 apparatus. A green fluorescent protein (GFP)-fused form of the dominant negative ATPase Vps4-EQ was used to irreversibly tag ESCRT envelopment sites during infection by UL36p-expressing and UL36-null HSV strains. Using fluorescence microscopy and scanning electron microscopy, we quantitated capsid/Vps4-EQ colocalization and examined the ultrastructure of the corresponding viral assembly intermediates. We found that loss of UL36p resulted in a two-thirds reduction in the efficiency of capsid/Vps4-EQ association but that the remaining UL36p-null capsids were still able to engage the ESCRT envelopment apparatus. It appears that although UL36p helps to couple HSV capsids to the ESCRT pathway, this is likely not the sole reason for its absolute requirement for envelopment.
IMPORTANCE Envelopment of the HSV capsid is essential for the assembly of an infectious virion and requires the complex interplay of a large number of viral and cellular proteins. Critical to envelope assembly is the virally encoded protein UL36p, whose function is unknown. Here we test the hypothesis that UL36p is essential for the recruitment of cellular ESCRT complexes, which are also known to be required for envelopment.
INTRODUCTION
Herpesviruses replicate their genomes and assemble DNA-packaged capsids in the cell nucleus. It is generally accepted that capsids then bud into the inner nuclear membrane to generate primary enveloped perinuclear virions that subsequently fuse with the outer nuclear membrane, releasing mature nucleocapsids (“naked” capsids) into the cytoplasm (1–3). These capsids subsequently undergo secondary envelopment at a cytoplasmic organelle to assemble the mature, infectious virion (4–10).
Herpesvirus cytoplasmic envelopment is extremely complex, requiring the coordinated interaction of multiple viral and cellular polypeptides (8, 9, 11–16). An essential component of the envelopment apparatus is the conserved multifunctional protein UL36p (VP1/2) (17–21). UL36p is the largest structural polypeptide encoded by the members of the Herpesviridae (22, 23), and it forms the innermost layer (18, 24–32) of tegument, the complex protein scaffold between the capsid and envelope (8, 16, 33). UL36p attaches the capsid (18, 31, 32, 34) to multiple outer tegument components (24–30, 35, 36) that in turn bind integral membrane envelope proteins (1, 9, 37–40) and the lipid envelope (41–43). One important function of UL36p is to recruit UL37p (19, 20, 25, 29, 44, 45), a putative mimic of cellular multisubunit tethering complexes (28) that mediates capsid docking to organelles, including the trans-Golgi network (TGN) (27), perhaps via membrane anchors, such as the heterodimer gK/UL20p (46). In the absence of UL36p/UL37p, capsid/membrane docking can still occur, but the fidelity of capsid-organelle targeting appears to be impaired (47, 48). Stable and precise capsid/membrane association prior to envelopment may require cooperation between UL36p/UL37p and complexes containing the UL16p protein (21, 39, 49, 50).
UL36p appears to be critical for envelopment, since herpes simplex virus (HSV) and pseudorabies virus (PRV) strains lacking the UL36 gene are severely replication defective and accumulate nonenveloped cytoplasmic capsids, often in large paracrystalline arrays (17–20). A recent study identified two centrally positioned, conserved tryptophan-acidic motifs in HSV UL36p that are essential for secondary envelopment and plaque formation (51); a corresponding central region of PRV UL36p is also known to be essential for replication (52). Given the central role of UL36p in multiple protein-protein and protein-lipid interactions between capsid and envelope, it is reasonable to hypothesize that UL36p drives the budding process, or at least gathers essential structural components together at the site of envelopment; however, its specific function at this stage in assembly remains unknown.
In addition to requiring numerous virally encoded polypeptides, herpesvirus envelopment is dependent on the cellular ESCRT (endosomal sorting complex required for transport) apparatus (53–57), in common with retroviruses and many other virus families (15, 58). This is clearly the case for HSV (59, 60), PRV (61, 62), and Epstein-Barr virus (63), though conflicting results have been reported for cytomegalovirus (64, 65). The interplay between the ESCRT machinery and virally encoded structural proteins during capsid/membrane association and envelopment (8, 11, 21, 46, 66) is not understood, but it is an attractive hypothesis that UL36p recruits ESCRT components to the budding site, which is analogous to the function of retroviral Gag proteins (58, 67, 68). Consistent with this possibility, the amino terminus of HSV UL36p has been reported to interact with the ESCRT I component TSG101 (69); however, TSG101 expression is dispensable for the production of infectious virions (60).
In both normal and infected cells, the final step in the ESCRT pathway is ATP hydrolysis by the AAA ATPase Vps4 at the bud neck; this drives ESCRT III dissociation and membrane scission (70–73). Replacement of a glutamate by a glutamine in the Vps4 ATPase active site generates Vps4-EQ, an ATP-locked dominant negative allele (74, 75) that can block ESCRT III release and arrest viral envelopment (15, 59, 72, 76). We previously imaged green fluorescent protein (GFP)-tagged forms of Vps4 and Vps4-EQ in close association with fluorescently labeled HSV and PRV capsids by using fluorescence microscopy and infected-cell cytoplasmic extracts, and we postulated that these structures correspond to alphaherpesvirus capsids undergoing envelopment (62). Interestingly, these assembly intermediates could bind microtubules but were incapable of trafficking along them in an in vitro system, suggesting that envelopment must be completed before egress can occur (62).
If UL36p is essential for coupling of membrane-bound HSV capsids to the ESCRT apparatus, then deletion of the UL36 gene should result in a loss of capsid association with ESCRT components. Binding of the dominant negative Vps4-EQ-GFP protein provides a fluorescent marker that irreversibly tags sites of ESCRT recruitment and membrane budding; we therefore tested whether HSV capsids failed to colocalize with Vps4-EQ-labeled sites in the absence of UL36p. We found that loss of UL36p resulted in a two-thirds reduction in the efficiency of capsid/Vps4-EQ association. Nevertheless, the remaining ∼30% of UL36-null capsids were able to engage with the ESCRT apparatus and generate partially assembled envelopment intermediates resembling those seen in a UL36-expressing virus. We concluded that UL36p helps to couple HSV capsids to the ESCRT pathway but that this is not the sole reason for its absolute requirement for envelopment.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were maintained in Dulbecco modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum (NCS) and 1% penicillin-streptomycin (PS) (Gibco Laboratories). HS30 cells (17) were grown in DMEM supplemented with 1% PS, 10% fetal calf serum (FCS) (Gibco Laboratories), and 100 μg/ml Geneticin (Life Technologies). Cell lines expressing tetracycline-inducible GFP-tagged forms of Vps4 and Vps4-EQ were derived from T-REx HEK293 cells (Invitrogen) and have been described previously (62, 76). These cells were cultured in DMEM supplemented with 10% tetracycline-free FCS (Gemini-Bioproducts), 1% penicillin-streptomycin, and 100 μg/ml Zeocin (Gibco Laboratories) at 37°C. HSV-1 GS2822 expresses a monomeric red fluorescent protein (mRFP1)-tagged allele of the small capsid subunit (VP26) UL35 gene and was described previously (77–79). HSV-1 GS3494 was derived directly from the HSV-1 GS2822 parental bacterial artificial chromosome (BAC) by deletion of all UL36 sequences other than the start and stop codons (Gregory A. Smith, personal communication). All viruses were grown and titers determined by plaque assay on Vero or HS30 cell monolayers as previously described (80).
BAC transfection and PCR analysis.
A subconfluent monolayer of HS30 cells was cotransfected by use of Lipofectamine 2000 (Invitrogen) with GS3494 BAC DNA and a 16-fold molar excess of a Cre recombinase expression plasmid to catalyze excision of plasmid backbone sequences (79). At full cytopathic effect, at 72 h posttransfection, cells were harvested, subjected to 2 cycles of freeze-thawing, and sonicated, and virus yields were determined on HS30 cell monolayers. Individual plaques were picked from HS30 cell monolayers and amplified on HS30 cells to prepare stocks.
HS30 cells were mock infected or infected with a stock of plaque-purified HSV GS3494 (ΔUL36) or the UL36+ GS2822 parental control strain at a multiplicity of infection (MOI) of 5, and total infected-cell DNA was prepared after 18 h of infection. The DNA preparations were used as templates for PCRs as follows. A 403-bp region of the HSV strain F (79) UL19 gene was amplified using the forward primer 5′AGGACCGCTTTGTGACTGAG3′ and the reverse primer 5′GCGATAAACTCACACACGGC3′. A 177-bp region of the bacterial chloramphenicol acetyltransferase (CAT) gene (81) was amplified using the forward primer 5′GCCAATCCCTGGGTGAGTTT3′ and the reverse primer 5′AAGCATTCTGCCGACATGG3′.
Isolation of PNS.
Vero, HS30, or T-REx HEK293 cells expressing GFP-tagged Vps4 or Vps4-EQ were infected with GS3494 (ΔUL36) or GS2822 (UL36+) at an MOI of 20 for 1 h at 37°C. Cells were induced with 1 μM tetracycline at the time of infection. Infected cells were then overlaid with fresh prewarmed medium and incubated at 37°C for 16 h. Cells were then washed once with ice-cold MEPS buffer [5 mM MgSO4, 5 mM EGTA, 0.25 M sucrose, 35 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 7.1], scraped from the plate, and resuspended in MEPS buffer containing 2 mM phenylmethylsulfonyl fluoride (PMSF), 2% (vol/vol) protease inhibitor cocktail (Sigma), and 4 mM dithiothreitol (DTT). The postnuclear supernatant (PNS) was isolated following needle breakage of cells as previously described (47, 82).
Western blotting and antibodies.
Western blotting was performed as previously described (47). Primary antibodies were anti-VP5 (HA018-100; Virusys), anti-gD (sc-69802; Santa Cruz Biotechnologies), and anti-gB (sc-69798; Santa Cruz Biotechnologies). Anti-UL36p rabbit antiserum was a kind gift from Richard Courtney and John Wills. Secondary antibodies were alkaline phosphatase-conjugated goat anti-rabbit (Chemicon, Pittsburgh, PA) and alkaline phosphatase-conjugated goat anti-mouse (Antibodies Incorporated, Davis, CA) antibodies, as used previously (42).
Fluorescence microscopy.
Direct fluorescence microscopy studies were performed as previously described (48, 62) in the Analytical Imaging Facility of the Albert Einstein College of Medicine. PNS samples were attached to 35-mm glass-bottomed MatTek dishes (MatTek Corporation) precoated with 100 μg/ml poly-l-lysine (Sigma). Samples were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature and then washed with PBS. Images were taken using a Zeiss Axio Observer correlative light and electron microscope (CLEM) with a 63× oil immersion objective. Images were processed using Volocity software V 2.3 (PerkinElmer).
Correlative light and electron microscopy.
PNS samples were prepared and attached to MatTek dishes exactly as detailed above and then imaged using a Zeiss Axio Observer microscope and Axiovision Shuttle & Find software to mark regions of interest (ROIs). After fluorescence imaging, samples were fixed with 2.5% glutaraldehyde, dehydrated in ethanol, critical point dried using a Tousimis Samdri 790 apparatus, and then coated with chromium by use of an EMS 150T-ES sputter coater. The previously identified ROIs were automatically located in a Zeiss Supra 40 field emission scanning electron microscope (SEM) and imaged with a secondary electron detector. Fluorescence and SEM images were correlated by use of Zeiss AxioVision software (V 4.8).
Transmission electron microscopy.
Thin-section electron microscopy of intact infected cells was performed as described earlier (80). Dishes of infected cells were incubated with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, postfixed with 1% osmium tetroxide followed by 2% uranyl acetate, dehydrated through a graded series of ethanol, and embedded in LX112 resin (LADD Research Industries, Burlington, VT). Ultrathin sections were cut on a Reichert Ultracut UCT instrument, stained with uranyl acetate followed by lead citrate, and viewed on a JEOL 1200EX transmission electron microscope at 80 kV.
RESULTS
Preparation and properties of a UL36-null VP26-mRFP1 HSV strain.
The HSV-1 BAC GS3494 was a kind gift from Gregory A. Smith. This BAC encodes an in-frame fusion of monomeric red fluorescent protein (mRFP1) to the small capsid subunit VP26 and has all UL36 coding sequences deleted, other than the start and stop codons. Except for the UL36 deletion, GS3494 is identical to and directly derived from HSV-1 strain GS2822, which was described previously (77–79). In an earlier study, we used HSV-1 GS2822 to demonstrate colocalization of red fluorescent HSV capsids with Vps4-GFP- and Vps4-EQ-GFP-decorated organelles in infected-cell cytoplasmic extracts (62).
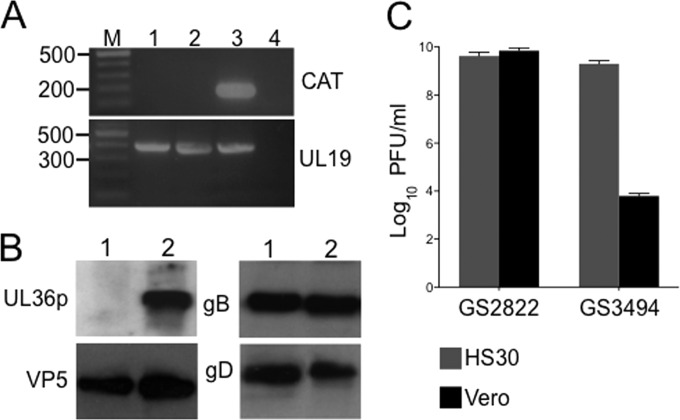
BAC DNA containing the UL36-null GS3494 genome was transfected into the UL36p-expressing Vero cell-derived line HS30 (17) in the presence of a 16-fold molar excess of a Cre recombinase-expressing plasmid. Infectious virus was recovered and propagated on HS30 cells, and individual plaques were picked and amplified. One plaque-purified isolate of GS3494 is characterized in Fig. 1. PCR analysis confirmed that this strain lacked the BAC-derived bacterial sequences expected to be excised by Cre-driven recombination; total DNA from GS3494-infected HS30 cells supported amplification of a region of the UL19 (VP5) gene but not a portion of the BAC vector-borne CAT gene (Fig. 1A). When replicating in Vero cells, GS3494 expressed the viral structural proteins VP5, gB, and gD at levels similar to those in the UL36+ parental strain, but it failed to express detectable levels of the UL36p protein (Fig. 1B). Loss of the UL36 gene correlated with an inability of the virus to replicate on Vero cells, but the replication defect was complemented by UL36p expression in the HS30 cell line (Fig. 1C).
FIG 1.
Preliminary characterization of UL36-null HSV strain GS3494. (A) PCR primers were used to amplify a 177-bp region of the bacterial chloramphenicol acetyltransferase (CAT) gene or a 403-bp region of the HSV UL19 gene, as indicated on the right. Lengths of DNA size markers (lane M), in base pairs, are shown on the left. Template DNA in each PCR was as follows: lane 1, total DNA from HS30 cells infected by UL36-null GS3494; lane 2, total DNA from HS30 cells infected by parental strain GS2822; lane 3, GS3494 BAC construct; lane 4, total DNA from uninfected HS30 cells. (B) Extracts of Vero cells infected by GS3494 (lanes 1) or the parental virus GS2822 (lanes 2) were subjected to SDS-PAGE and immunoblotted for UL36p, the major capsid protein VP5, and envelope proteins gB and gD, as indicated. (C) Vero (black bars) or HS30 (gray bars) cells were infected by GS2822 or GS3494 at an MOI of 0.01 and harvested after 72 h, and extracts were titrated on HS30 cell monolayers. Plaques formed by the UL36-null strain following replication on Vero cells likely correspond to residual input virus (propagated on HS30 cells).
UL36p-null HSV-1 GS3494 capsids associate with cytoplasmic organelles in intact cells.
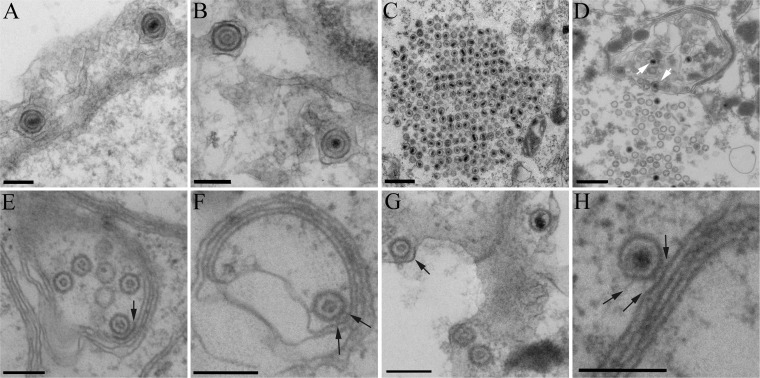
We showed earlier, by using infected-cell extracts and organelles prepared in vitro, that HSV capsids dock to cytoplasmic organelles in the absence of UL36p (48). To test whether capsid/membrane docking occurred in vivo in intact cells in the absence of UL36p, we infected Vero or UL36p-expressing HS30 cells with the UL36-null GS3494 strain and then fixed and sectioned cells for transmission electron microscopy (Fig. 2). As expected, in complementing HS30 cells, mature, enveloped GS3494 virions were assembled in the cytoplasm (Fig. 2A and B). In contrast, in Vero cells, loss of UL36p resulted in the characteristic accumulation of large paracrystalline arrays of cytoplasmic capsids (Fig. 2C), as others have previously observed (17, 19). Nevertheless, capsids were also frequently seen in intimate association with membranes (Fig. 2E to H), sometimes on the periphery of capsid aggregates (Fig. 2D, white arrows). Often, electron-dense material was visible as fibers reaching between the capsid shell and the lipid bilayer (Fig. 2F and H, black arrows) or lying along membranes adjacent to capsids, resembling sites of tegument deposition (Fig. 2E and G, black arrows). In some cases, capsids were partially enclosed by a membrane (Fig. 2D, right white arrow), but fully enveloped virions were never seen. We concluded that capsid/membrane docking can occur in the absence of UL36p in vivo, consistent with our earlier in vitro fractionation studies.
FIG 2.
A subset of HSV capsids are in close proximity to organellar membranes in intact cells in the absence of UL36p. Dishes of UL36p-expressing HS30 cells (A and B) or Vero cells (C to H) were infected by the UL36-null strain GS3494, fixed, and processed for thin-section electron microscopy. White arrows in panel D indicate examples of capsids on the edge of a large cytoplasmic cluster in close proximity to membranes. Black arrows indicate membrane-associated regions of electron density adjacent to capsids resembling tegument (E and G) or spokes (F and H) between the capsid and the lipid bilayer. Bars, 500 nm (C and D) and 200 nm (all other panels).
Testing the colocalization of HSV capsids with Vps4-EQ-GFP in the presence and absence of UL36p.
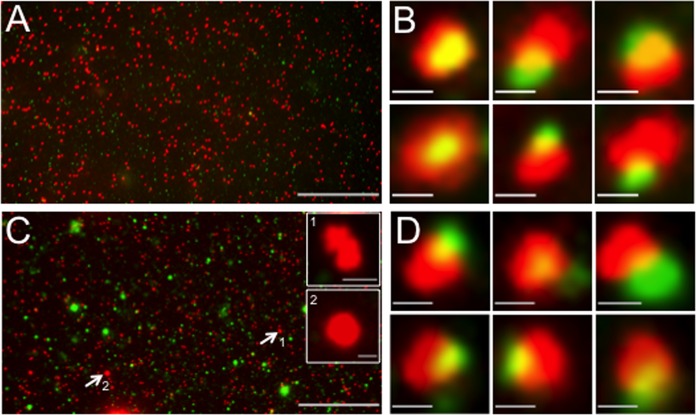
We prepared a postnuclear supernatant (PNS; cytoplasmic fraction) from GS2822- or GS3494-infected HEK293 cells expressing the dominant negative Vps4-EQ allele fused to GFP. The PNS was attached to polylysine-coated MatTek dishes, fixed, and imaged by fluorescence microscopy in the red (VP26-mRFP1 capsid subunit) or green (Vps4-EQ-GFP) channel, as we described earlier (48). Typical fluorescence fields and galleries of individual particles are shown for the parental strain GS2822 (Fig. 3A and B) and the UL36-null strain GS3494 (Fig. 3C and D). In the red (capsid) channel, microscopic fields of GS2822 and GS3494 extracts contained large numbers of discrete fluorescent particles of approximately uniform size. In a similar analysis of a GFP-VP26-expressing HSV strain, we used correlative light and electron microscopy to demonstrate that such puncta arise predominantly from individual capsids (48). In addition, larger structures were occasionally visible in the capsid-fluorescing channel of the GS3494 UL36-null strain (Fig. 3C). These were larger than 600 nm in (apparent) fluorescent length or diameter and not visible in extracts prepared from cells infected by UL36p-expressing GS2822 (Fig. 3A). We made similar observations using a VP26-GFP-expressing UL36-null HSV strain (48) and hypothesized that these structures might correspond to the large cytoplasmic capsid aggregates seen in the absence of UL36p; we tested this possibility as described below (see Fig. 5). We also observed a population of large green-fluorescing puncta in fields derived from GS3494-infected (Fig. 3C) but not GS2822-infected (Fig. 3A) cells. This implies greater heterogeneity in Vps4-EQ-GFP membrane/organelle association in the presence of the UL36-null virus, but we do not know the reasons for this at this time.
FIG 3.
Representative fields of HSV capsid-associated and Vps4-EQ-GFP-bound cytoplasmic particles. HEK293 cells were induced via tetracycline to express Vps4-EQ-GFP and then infected with HSV for 16 h, a PNS was prepared, and virions and organelles were attached to MatTek dishes. After fixation, fields of particles were imaged in the red (VP26-mRFP1) and green (Vps4-EQ-GFP) channels; merged images are shown. (A) Low-magnification field of a PNS from GS2822-infected cells. (B) Representative gallery of six individual particles from a field similar to that in panel A. (C) Low-magnification field of a PNS from GS3494-infected cells. Two large red fluorescent structures (indicated by white arrows and numbered 1 and 2) are shown as higher-magnification insets. (D) Representative gallery of six individual particles from a field similar to that in panel C. Bars, 30 μm (A and C), 1 μm (insets in panel C), and 0.5 μm (B and D).
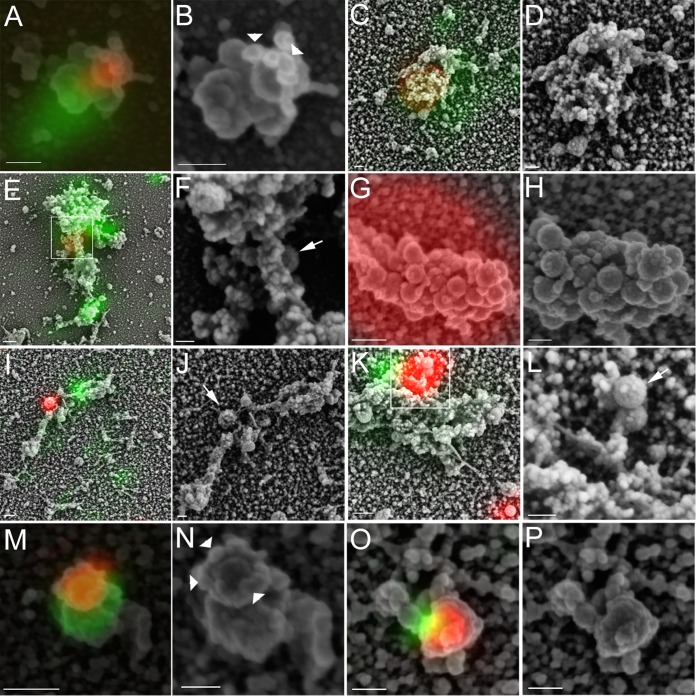
FIG 5.
Correlative light and electron microscopic analysis of HSV capsid/Vps4-EQ-GFP colocalization in the presence and absence of UL36p. Cytoplasmic particles from GS2822-infected (A to F) or GS3494-infected (G to P) Vps4-EQ-GFP-expressing HEK293 cells were prepared and imaged as described in the legend to Fig. 3. Samples were then processed for scanning electron microscopy (SEM) and images aligned. Images are shown in pairs; for each pair, the left panel shows an alignment of fluorescent red (capsid) and green (Vps4-EQ-GFP) images with the SEM image, and the right panel shows the SEM image alone, usually at a higher resolution. White boxes in panels E and K correspond to a region of the structure shown at a higher resolution in panels F and L, respectively. Bars for SEM/fluorescence-aligned images, 100 nm (A and O) and 200 nm (C, E, G, I, K, and M); bars for SEM-only images (B, D, F, H, J, L, N, and P), 100 nm. Arrows in panels F, J, and L label capsids docked to organelles but apparently not engaged with the ESCRT apparatus. Arrowheads (B and N) indicate bead-like structures around the leading edge of the membrane.
For both GS2822 and GS3494, we counted numbers of small and large (>600 nm) red fluorescent structures and scored how many colocalized with Vps4-EQ-GFP; we defined red (R) fluorescent particles as R+ G+ or R+ G− depending on whether they did or did not also fluoresce in the green (G) channel. We then calculated Pearson product-moment correlation coefficients (83) as detailed earlier (48) to identify the numbers of R+ G+ particles that showed substantial overlap of red and green fluorescence (we defined substantial fluorescence overlap as having a Pearson coefficient [ρ] of >0.6). These data are listed in Table 1. As a control, we performed similar studies using wild-type Vps4 fused to GFP. Wild-type Vps4 and Vps4-GFP do not interfere with HSV replication and envelopment (59, 62); we saw comparable numbers of total red fluorescent particles in the presence of Vps4-GFP and Vps4-EQ-GFP (Table 1), suggesting that the expression of viral structural proteins and capsid assembly were normal in the presence of Vps4-EQ-GFP, as we showed earlier (62). Unlike the dominant negative allele, wild-type Vps4-GFP associated only transiently with membranes, and small numbers of R+ G+ particles accumulated at the steady state (Table 1) (62).
TABLE 1.
Colocalization of GS3494 and GS2822 capsids with Vps4 and Vps4-EQ
| Virus/Vps4 allele | No. of particles/field (mean ± SD)a |
||||
|---|---|---|---|---|---|
| Total R+ particlesb | Total R+ G+ particles | R+ G+ particles with ρ value of >0.6 | R+ G− particles | R+ particles with >600-nm diam | |
| GS2822 (UL36+)/Vps4 | 344 ± 52 | 14 ± 6 | 5 ± 1 | 330 ± 48 | 0 |
| GS2822 (UL36+)/Vps4-EQ | 415 ± 157 | 73 ± 33 | 45 ± 20 | 341 ± 130 | 0 |
| GS3494 (ΔUL36)/Vps4 | 418 ± 60 | 14 ± 4 | 4 ± 1 | 403 ± 58 | 0 |
| GS3494 (ΔUL36)/Vps4-EQ | 452 ± 117 | 33 ± 13 | 16 ± 7 | 418 ± 107 | 3 ± 2 |
Numbers of individual microscopic fields examined were 16 (GS2822/Vps4), 20 (GS2822/Vps4-EQ), 12 (GS3494/Vps4), and 25 (GS3494/Vps4-EQ).
Number of mRFP1-associated particles in each field. Total numbers counted were 5,499 (GS2822/Vps4), 8,293 (GS2822/Vps4-EQ), 5,010 (GS3494/Vps4), and 11,290 (GS3494/Vps4-EQ).
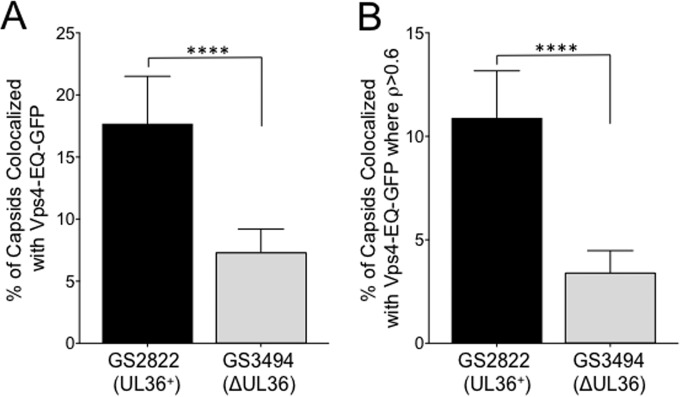
As summarized in Fig. 4, we found that a loss of UL36p resulted in a 60% (P < 0.0001) reduction in the number of Vps4-EQ-GFP-labeled capsid-associated structures (Fig. 4A). When we considered only particles with ρ values of >0.6, there were 68% fewer (P < 0.0001) in the absence of UL36p (Fig. 4B). No significant reduction was observed for the colocalization of capsids with Vps4-GFP, but the number of individual particles present, particularly those with ρ values of >0.6, was much smaller and reflects only those structures forming at the time of cell breakage (Table 1).
FIG 4.
Effect of UL36p loss on capsid/Vps4-EQ colocalization. Vps4-EQ-GFP-expressing cells were infected with HSV GS2822 (UL36+; black bars) or GS3494 (ΔUL36; gray bars). Postnuclear supernatants (PNS) were prepared, attached to MatTek dishes, fixed, and imaged. (A) Percentages of all mRFP1-fluorescing particles that colocalized with Vps4-EQ-GFP. (B) Percentages of all mRFP1-fluorescing particles that exhibited colocalization with Vps4-EQ-GFP with ρ values of >0.6. For both panels A and B, plotted values show means and standard deviations. Data were analyzed by the Student t test (****, P < 0.0001).
Ultrastructural analysis of capsid-associated Vps4-EQ-bound organelles by correlative light and electron microscopy.
To better understand the structure of each type of fluorescent particle in Table 1, we performed scanning electron microscopy (SEM) and aligned fluorescence and SEM images. For the UL36-expressing virus, R+ G+ particles with ρ values of >0.6 appeared in the SEM images as capsids partially encircled (Fig. 5A and B) or completely enclosed (Fig. 5C and D) by a Vps4-EQ-GFP-fluorescing membrane. Similar partially enveloped capsids were seen for the UL36p-null virus (Fig. 5M to P). R+ G+ particles exhibiting more limited fluorescence overlap appeared to arise from capsids docked to the surfaces of organelles, but at some distance from sites of ESCRT/Vps4-EQ recruitment. This was seen for both the UL36p-expressing virus (Fig. 5E and F) and the UL36-null strain (Fig. 5I to L). Often, capsids were attached to organelles via stalk-like structures (Fig. 5I to L). The large (>600-nm diameter) red fluorescent structures unique to the UL36-null strain (Fig. 3C; Table 1) aligned with large clusters of spheres that had the size and appearance of capsids (Fig. 5G and H), as we proposed earlier (48). The capsids in the aggregates often presented heterogeneous surface features overlying more regular radial bulges (Fig. 5H).
DISCUSSION
In earlier work, we identified membrane-bound capsids in fractions prepared from cells infected by HSV strains with partial (47) or complete (48) deletions of the UL36 gene. Although they were membrane associated, these viral particles were clearly noninfectious, consistent with the finding that HSV capsids fail to become enveloped when the UL36 gene is partially or completely deleted (17, 19). Our approach permitted ready quantitation of capsid-membrane association, but experiments were all performed in vitro using cytoplasmic extracts and buoyant density gradient fractions (47, 48). Here we show that in intact cells, in addition to cytoplasmic aggregates characteristic of UL36-null strains (17, 19), capsids also exist in intimate association with organellar surfaces (Fig. 2). We frequently observed electron-dense material reaching between the capsid and the bilayer (Fig. 2F and H) or lying along the membrane adjacent to the capsid (Fig. 2E and G). What was not clear from our earlier scanning electron microscopy studies (48) but is readily apparent in the thin sections is that docked capsids are frequently attached to curved stacks of membrane reminiscent of Golgi cisternae (Fig. 2D to F and H). This is consistent with our finding that the majority of membrane-bound capsids colocalized with the trans-Golgi network marker TGN46 in the presence and absence of UL36p (48).
In addition to C capsids (Fig. 2D, G, and H), we frequently observed UL36-null B capsids in close association with lipid membranes (Fig. 2E to G), though we have not yet performed a quantitative analysis. In an earlier study using a similar UL36-null HSV strain, we also observed numerous B capsids, along with C capsids, attached to isolated cytoplasmic organelles (48). Large numbers of cytoplasmic B capsids were unexpected, since it is generally accepted that C capsids are preferentially exported from the nucleus to the cytoplasm, at least during wild-type HSV infections. Trafficking of capsids from the nucleus involves interaction between the UL17p/UL25p-containing capsid vertex-specific component (CVSC), present on A, B, and C capsids (31, 84), and the UL31-nuclear export complex (85, 86). It is now clear that a fragment of the UL36p protein is also part of the mature CVSC on nuclear capsids (31), and it is possible that in a UL36-null strain the less stable proto-CVSC (31) does not effectively discriminate between C and B capsids for engagement with the nuclear export pathway. There is a precedent for such a result; loss of the UL15 or UL28 terminase subunit genes leads not only to a failure to package DNA but also to large numbers of B capsids reaching the cytoplasm and becoming enveloped (87, 88).
We previously suggested (48) that capsid-organelle docking might be mediated by cooperation between UL36p and the Golgi membrane-anchored UL11p-UL16p-UL21p complex characterized by the Wills laboratory (39, 49, 50, 89). Interestingly, cells infected by UL16p-null HSV strains accumulate membrane-docked capsid structures that are quite similar in appearance (90) to those we observed in the absence of UL36p (Fig. 2E and F), consistent with these proteins acting at a common step in assembly. We do not know whether the electron-dense layers and spokes lying between the capsid and the membrane (Fig. 2E to H) are involved in capsid attachment, but if so, they may represent the structure of this UL16p docking machinery.
If UL36p is essential for envelopment because it couples capsids to the ESCRT/Vps4 apparatus (59, 60, 62), then a loss of UL36p should disrupt this interaction. Using a GFP-fused version of Vps4-EQ as a marker for sites of ESCRT-driven envelopment, we quantitated the numbers of structures exhibiting capsid–Vps4-EQ-GFP colocalization in the presence and absence of UL36p (Table 1). As summarized in Fig. 4, we found that fewer capsids colocalized with Vps4-EQ-GFP when UL36p was absent. This was true for both capsid–Vps4-EQ-GFP structures in general (60% fewer) (Fig. 4A) and those that showed higher levels of colocalization of red/green fluorescence (ρ > 0.6), suggestive of close proximity (68% fewer) (Fig. 4B). Quantitative lipophilic dye staining and Western blotting of buoyant membrane fractions led us to conclude that capsid-membrane docking is normal in the absence of UL36p (48); the reduced colocalization with Vps4-EQ-GFP is therefore not because capsids fail to dock with organelles. Rather, they must be associating with organelles that are less likely to recruit ESCRT components. This is consistent with the organellar mistargeting of UL36-null capsids that we observed earlier (48) and with a role for the UL36p/UL37p complex in membrane tethering (27, 28). Alternatively, if capsids themselves stimulate ESCRT assembly upon docking, capsids lacking UL36p must have a reduced ability to do so.
Since capsids and Vps4-EQ-GFP/ESCRT complexes may exist in close proximity on the same organelle without physical association, and because we observed particles with various degrees of red/green fluorescence overlap (Table 1), we next examined these structures by correlative light and electron microscopy (CLEM). Regions of VP26-mRFP1 red fluorescence aligned with spherical structures of ∼140 nm in diameter; we previously saw identical structures emitting green fluorescence in the presence of VP26-GFP (48); this shape and size correspond well to the expected 125-nm diameter of the HSV capsid plus the approximately 16-nm-thick chromium sputter coating deposited during SEM preparation. Capsids were frequently in close association with pleomorphic vesiculotubular structures similar to those we earlier showed to be stained with lipophilic dyes (48) and that resemble isolated organelles and vesicles imaged by SEM (91, 92). CLEM analysis revealed that particles with high levels of red/green fluorescence colocalization correspond to capsids in intimate association with organellar envelopment sites. We observed capsids partially enclosed by Vps4-EQ-GFP-labeled membranes in the presence of UL36p (Fig. 5A and B) and also in its absence (Fig. 5M to P). These partially enveloped structures are exactly those expected to accumulate when neck closure and scission are inhibited by dominant negative Vps4-EQ-GFP (15, 59, 72, 73, 76, 93). Interestingly, the rim or neck of the unsealed envelope is decorated by a ring of structures resembling beads on a string (Fig. 5B and N, white arrowheads). Taking into account the thickness of the chromium sputter coating in these samples, the mean long-axis diameter of each “bead” is ∼12 nm. In size and appearance, these structures bear a striking resemblance to those formed by 10- to 16-nm Vps4-EQ-GFP oligomers (likely dodecamers) that align along underlying threads of membrane-bound ESCRT III polymers (94).
For the UL36p-expressing virus GS2822, capsids were also found to be obscured entirely by organellar membranes and were detectable only by their fluorescence emission (Fig. 5C and D). These structures may correspond to capsids that have undergone UL36p-dependent envelopment and become completely surrounded by membrane despite the presence of Vps4-EQ-GFP. The Vps4-EQ dominant negative phenotype is often incomplete (95, 96), as suppression of scission is a function of the relative copy numbers of Vps4-EQ and wild-type Vps4 in each Vps4 dodecamer and whether the Vps4-EQ subunits are among those that stochastically engage with ESCRT III during each cycle of scission (72, 73). Alternatively, though we did not observe them in the UL36-null strain, we cannot exclude the possibility that the structure in Fig. 5C and D corresponds to a docked or partially enveloped capsid lying underneath the organelle. Such capsids would be inaccessible to scanning electron microscopy.
R+ G+ particles (Table 1) that exhibited little or no overlap of the red and green fluorescence signals appeared to correspond to organelle-docked capsids lying some distance from Vps4/ESCRT assembly sites. These were the majority of particles seen in microscopic fields of PNS (Table 1) and were found in the presence of UL36p (Fig. 5E and F) and, to a lesser extent, also in its absence (Fig. 5I to L) (white arrows in these panels indicate capsids apparently docked to the surfaces of membranes). Such structures might correspond to capsids unable to engage the envelopment apparatus or possibly to a stage in HSV assembly downstream of docking but prior to ESCRT engagement/envelopment. However, at present, this is purely speculative.
Wild-type and UL36p-null capsids exhibit substantial surface features (Fig. 5) that presumably arise from capsomers, bound inner and outer teguments, and perhaps other cellular and viral proteins that are recruited to the cytoplasm during assembly. For wild-type cytoplasmic capsids, the VP5 penton vertices are decorated by UL25p, UL17p, and the UL36p/UL37p complex (31, 33, 86, 97–100), but we have not yet imaged sufficient numbers of particles to determine whether there are differences between UL36p+ and UL36p-null capsids. The UL36-null capsids present in large clusters do exhibit striking asymmetric and heterogeneous surface features (Fig. 5G and H), and it is an interesting question whether this may be the cause or result of their aggregation. Because of their structure, and since they are UL36-null specific, we propose that clusters such as these are the origin of the arrays of cytoplasmic capsids seen by thin-section EM (Fig. 2C) (17, 19).
Although statistically significant, the 60 to 68% reduction in capsid/ESCRT colocalization in the UL36-null strain (Table 1; Fig. 4) is rather modest. It is consistent with a role for UL36p in engagement with the ESCRT apparatus but appears to be inadequate to explain the almost complete failure of envelopment and the ∼4- to 5-log loss in infectivity typically seen for UL36-null strains (17, 19, 48). We also cannot yet discount the possibility that additional mutations acquired during construction of the GS3494 strain are responsible for the reduction in capsid/ESCRT colocalization. It remains clear that UL36p-null capsids can dock to ESCRT-bound organelles (Fig. 5I to L) and interact with the ESCRT machinery (Fig. 5M to P). It may be that UL36p-null capsids are structurally deficient in ESCRT III/Vps4 recruitment at a level of resolution beyond that used in our studies. Alternatively, UL36p or a UL36p-dependent factor(s) might act very late in envelopment, after Vps4 has been recruited to ESCRT III, to detach it from its membrane anchors (72, 93).
ACKNOWLEDGMENTS
We thank Gregory A. Smith for the HSV UL36-null BAC HSV-1 GS3494 and for helpful discussions. The HS30 cell line was generously provided by Prashant Desai. The Cre expression plasmid was a kind gift from Kartik Chandran. We thank Geoffrey Perumal, Leslie Gunther-Cummins, Frank Macaluso, and the Analytical Imaging Facility of the Albert Einstein College of Medicine for help with CLEM and Erik Snapp and the Liver Center Imaging and Cell Structure Core (P30 DK041296) for support and advice.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Mettenleiter TC, Muller F, Granzow H, Klupp BG. 2013. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol 15:170–178. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- 2.Bigalke JM, Heuser T, Nicastro D, Heldwein EE. 2014. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat Commun 5:4131. doi: 10.1038/ncomms5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bigalke JM, Heldwein EE. 2015. The great (nuclear) escape: new insights into the role of the nuclear egress complex of herpesviruses. J Virol 89:9150–9153. doi: 10.1128/JVI.02530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browne H, Bell S, Minson T, Wilson DW. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol 70:4311–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skepper JN, Whiteley A, Browne H, Minson A. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J Virol 75:5697–5702. doi: 10.1128/JVI.75.12.5697-5702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mettenleiter TC, Klupp BG, Granzow H. 2006. Herpesvirus assembly: a tale of two membranes. Curr Opin Microbiol 9:423–429. doi: 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Turcotte S, Letellier J, Lippe R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J Virol 79:8847–8860. doi: 10.1128/JVI.79.14.8847-8860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res 143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 10.Hollinshead M, Johns HL, Sayers CL, Gonzalez-Lopez C, Smith GL, Elliott G. 2012. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. EMBO J 31:4204–4220. doi: 10.1038/emboj.2012.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopp M, Granzow H, Fuchs W, Klupp B, Mettenleiter TC. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J Virol 78:3024–3034. doi: 10.1128/JVI.78.6.3024-3034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Browne H, Bell S, Minson T. 2004. Analysis of the requirement for glycoprotein M in herpes simplex virus type 1 morphogenesis. J Virol 78:1039–1041. doi: 10.1128/JVI.78.2.1039-1041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster TP, Chouljenko VN, Kousoulas KG. 2008. Functional and physical interactions of the herpes simplex virus type 1 UL20 membrane protein with glycoprotein K. J Virol 82:6310–6323. doi: 10.1128/JVI.00147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chouljenko DV, Kim IJ, Chouljenko VN, Subramanian R, Walker JD, Kousoulas KG. 2012. Functional hierarchy of herpes simplex virus 1 viral glycoproteins in cytoplasmic virion envelopment and egress. J Virol 86:4262–4270. doi: 10.1128/JVI.06766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owen DJ, Crump CM, Graham SC. 2015. Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses 7:5084–5114. doi: 10.3390/v7092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desai PJ. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol 74:11608–11618. doi: 10.1128/JVI.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schipke J, Pohlmann A, Diestel R, Binz A, Rudolph K, Nagel CH, Bauerfeind R, Sodeik B. 2012. The C terminus of the large tegument protein pUL36 contains multiple capsid binding sites that function differently during assembly and cell entry of herpes simplex virus. J Virol 86:3682–3700. doi: 10.1128/JVI.06432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandbaumhuter M, Dohner K, Schipke J, Binz A, Pohlmann A, Sodeik B, Bauerfeind R. 2013. Cytosolic herpes simplex virus capsids not only require binding inner tegument protein pUL36 but also pUL37 for active transport prior to secondary envelopment. Cell Microbiol 15:248–269. doi: 10.1111/cmi.12075. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs W, Klupp BG, Granzow H, Mettenleiter TC. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J Virol 78:11879–11889. doi: 10.1128/JVI.78.21.11879-11889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crump CM, Bruun B, Bell S, Pomeranz LE, Minson T, Browne HM. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J Gen Virol 85:3517–3527. doi: 10.1099/vir.0.80361-0. [DOI] [PubMed] [Google Scholar]
- 22.Mocarski ES., Jr 2007. Comparative analysis of herpesvirus-common proteins, p 44–58. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 23.Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ. 2009. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res 145:173–186. doi: 10.1016/j.virusres.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Ko DH, Cunningham AL, Diefenbach RJ. 2010. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2). J Virol 84:1397–1405. doi: 10.1128/JVI.01721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vittone V, Diefenbach E, Triffett D, Douglas MW, Cunningham AL, Diefenbach RJ. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J Virol 79:9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, Vittone V, Diefenbach E, Cunningham AL, Diefenbach RJ. 2008. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378:347–354. doi: 10.1016/j.virol.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 27.Pasdeloup D, Beilstein F, Roberts AP, McElwee M, McNab D, Rixon FJ. 2010. Inner tegument protein pUL37 of herpes simplex virus type 1 is involved in directing capsids to the trans-Golgi network for envelopment. J Gen Virol 91:2145–2151. doi: 10.1099/vir.0.022053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitts JD, Klabis J, Richards AL, Smith GA, Heldwein EE. 2014. Crystal structure of the herpesvirus inner tegument protein UL37 supports its essential role in control of viral trafficking. J Virol 88:5462–5473. doi: 10.1128/JVI.00163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly BJ, Bauerfeind R, Binz A, Sodeik B, Laimbacher AS, Fraefel C, Diefenbach RJ. 2014. The interaction of the HSV-1 tegument proteins pUL36 and pUL37 is essential for secondary envelopment during viral egress. Virology 454–455:67–77. doi: 10.1016/j.virol.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Mijatov B, Cunningham AL, Diefenbach RJ. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26–31. doi: 10.1016/j.virol.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Fan WH, Roberts AP, McElwee M, Bhella D, Rixon FJ, Lauder R. 2015. The large tegument protein pUL36 is essential for formation of the capsid vertex-specific component at the capsid-tegument interface of herpes simplex virus 1. J Virol 89:1502–1511. doi: 10.1128/JVI.02887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laine RF, Albecka A, van de Linde S, Rees EJ, Crump CM, Kaminski CF. 2015. Structural analysis of herpes simplex virus by optical super-resolution imaging. Nat Commun 6:5980. doi: 10.1038/ncomms6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grunewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 34.Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J Virol 83:6610–6623. doi: 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly BJ, Mijatov B, Fraefel C, Cunningham AL, Diefenbach RJ. 2012. Identification of a single amino acid residue which is critical for the interaction between HSV-1 inner tegument proteins pUL36 and pUL37. Virology 422:308–316. doi: 10.1016/j.virol.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 36.Svobodova S, Bell S, Crump CM. 2012. Analysis of the interaction between the essential herpes simplex virus 1 tegument proteins VP16 and VP1/2. J Virol 86:473–483. doi: 10.1128/JVI.05981-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gross ST, Harley CA, Wilson DW. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1–12. doi: 10.1016/j.virol.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 38.Chi JH, Harley CA, Mukhopadhyay A, Wilson DW. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J Gen Virol 86:253–261. doi: 10.1099/vir.0.80444-0. [DOI] [PubMed] [Google Scholar]
- 39.Loomis JS, Courtney RJ, Wills JW. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J Virol 77:11417–11424. doi: 10.1128/JVI.77.21.11417-11424.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Regan KJ, Bucks MA, Murphy MA, Wills JW, Courtney RJ. 2007. A conserved region of the herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE). Virology 358:192–200. doi: 10.1016/j.virol.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 41.Lee GE, Church GA, Wilson DW. 2003. A subpopulation of tegument protein Vhs localizes to detergent-insoluble lipid rafts in herpes simplex virus-infected cells. J Virol 77:2038–2045. doi: 10.1128/JVI.77.3.2038-2045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukhopadhyay A, Lee GE, Wilson DW. 2006. The amino terminus of the herpes simplex virus 1 protein Vhs mediates membrane association and tegument incorporation. J Virol 80:10117–10127. doi: 10.1128/JVI.00744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy MA, Bucks MA, O'Regan KJ, Courtney RJ. 2008. The HSV-1 tegument protein pUL46 associates with cellular membranes and viral capsids. Virology 376:279–289. doi: 10.1016/j.virol.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 44.Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J Virol 76:3065–3071. doi: 10.1128/JVI.76.6.3065-3071.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bottcher S, Granzow H, Maresch C, Mohl B, Klupp BG, Mettenleiter TC. 2007. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J Virol 81:13403–13411. doi: 10.1128/JVI.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jambunathan N, Chouljenko D, Desai P, Charles AS, Subramanian R, Chouljenko VN, Kousoulas KG. 2014. The herpes simplex virus type-1 UL37 protein interacts with viral glycoprotein gK and membrane protein UL20 and functions in cytoplasmic virion envelopment. J Virol 88:5927–5935. doi: 10.1128/JVI.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shanda SK, Wilson DW. 2008. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J Virol 82:7388–7394. doi: 10.1128/JVI.00225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kharkwal H, Furgiuele SS, Smith CG, Wilson DW. 2015. Herpes simplex virus capsid-organelle association in the absence of the large tegument protein UL36p. J Virol 89:11372–11382. doi: 10.1128/JVI.01893-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeh PC, Meckes DG Jr, Wills JW. 2008. Analysis of the interaction between the UL11 and UL16 tegument proteins of herpes simplex virus. J Virol 82:10693–10700. doi: 10.1128/JVI.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harper AL, Meckes DG Jr, Marsh JA, Ward MD, Yeh PC, Baird NL, Wilson CB, Semmes OJ, Wills JW. 2010. Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J Virol 84:2963–2971. doi: 10.1128/JVI.02015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivanova L, Buch A, Dohner K, Pohlmann A, Binz A, Prank U, Sandbaumhuter M, Bauerfeind R, Sodeik B. 2016. Conserved tryptophan motifs in the large tegument protein pUL36 are required for efficient secondary envelopment of herpes simplex virus capsids. J Virol 90:5368–5383. doi: 10.1128/JVI.03167-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JI, Luxton GW, Smith GA. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J Virol 80:12086–12094. doi: 10.1128/JVI.01184-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodman PG, Futter CE. 2008. Multivesicular bodies: co-ordinated progression to maturity. Curr Opin Cell Biol 20:408–414. doi: 10.1016/j.ceb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wollert T, Yang D, Ren X, Lee HH, Im YJ, Hurley JH. 2009. The ESCRT machinery at a glance. J Cell Sci 122:2163–2166. doi: 10.1242/jcs.029884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCullough J, Colf LA, Sundquist WI. 2013. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem 82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alam SL, Sundquist WI. 2007. Structural biology: ESCRT service. Nature 447:921–922. doi: 10.1038/447921a. [DOI] [PubMed] [Google Scholar]
- 57.Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. 2007. ESCRT-III recognition by VPS4 ATPases. Nature 449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 58.Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. 2011. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crump CM, Yates C, Minson T. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J Virol 81:7380–7387. doi: 10.1128/JVI.00222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pawliczek T, Crump CM. 2009. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J Virol 83:11254–11264. doi: 10.1128/JVI.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kramer T, Greco TM, Enquist LW, Cristea IM. 2011. Proteomic characterization of pseudorabies virus extracellular virions. J Virol 85:6427–6441. doi: 10.1128/JVI.02253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kharkwal H, Smith CG, Wilson DW. 2014. Blocking ESCRT-mediated envelopment inhibits microtubule-dependent trafficking of alphaherpesviruses in vitro. J Virol 88:14467–14478. doi: 10.1128/JVI.02777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee CP, Liu PT, Kung HN, Su MT, Chua HH, Chang YH, Chang CW, Tsai CH, Liu FT, Chen MR. 2012. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr virus. PLoS Pathog 8:e1002904. doi: 10.1371/journal.ppat.1002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tandon R, AuCoin DP, Mocarski ES. 2009. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J Virol 83:10797–10807. doi: 10.1128/JVI.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fraile-Ramos A, Pelchen-Matthews A, Risco C, Rejas MT, Emery VC, Hassan-Walker AF, Esteban M, Marsh M. 2007. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell Microbiol 9:2955–2967. doi: 10.1111/j.1462-5822.2007.01024.x. [DOI] [PubMed] [Google Scholar]
- 66.Han J, Chadha P, Meckes DG Jr, Baird NL, Wills JW. 2011. Interaction and interdependent packaging of tegument protein UL11 and glycoprotein E of herpes simplex virus. J Virol 85:9437–9446. doi: 10.1128/JVI.05207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol 13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. 2011. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J Virol 85:632–637. doi: 10.1128/JVI.01683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Calistri A, Munegato D, Toffoletto M, Celestino M, Franchin E, Comin A, Sartori E, Salata C, Parolin C, Palu G. 2015. Functional interaction between the ESCRT-I component TSG101 and the HSV-1 tegument ubiquitin specific protease. J Cell Physiol 230:1794–1806. doi: 10.1002/jcp.24890. [DOI] [PubMed] [Google Scholar]
- 70.Babst M, Wendland B, Estepa EJ, Emr SD. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J 17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Babst M, Sato TK, Banta LM, Emr SD. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J 16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shestakova A, Hanono A, Drosner S, Curtiss M, Davies BA, Katzmann DJ, Babst M. 2010. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol Biol Cell 21:1059–1071. doi: 10.1091/mbc.E09-07-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davies BA, Azmi IF, Payne J, Shestakova A, Horazdovsky BF, Babst M, Katzmann DJ. 2010. Coordination of substrate binding and ATP hydrolysis in Vps4-mediated ESCRT-III disassembly. Mol Biol Cell 21:3396–3408. doi: 10.1091/mbc.E10-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bishop N, Woodman P. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol Biol Cell 11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoshimori T, Yamagata F, Yamamoto A, Mizushima N, Kabeya Y, Nara A, Miwako I, Ohashi M, Ohsumi M, Ohsumi Y. 2000. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol Biol Cell 11:747–763. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor GM, Hanson PI, Kielian M. 2007. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J Virol 81:13631–13639. doi: 10.1128/JVI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith GA, Pomeranz L, Gross SP, Enquist LW. 2004. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc Natl Acad Sci U S A 101:16034–16039. doi: 10.1073/pnas.0404686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Antinone SE, Smith GA. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J Virol 84:1504–1512. doi: 10.1128/JVI.02029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J Virol 77:1382–1391. doi: 10.1128/JVI.77.2.1382-1391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Church GA, Wilson DW. 1997. Study of herpes simplex virus maturation during a synchronous wave of assembly. J Virol 71:3603–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marcoli R, Iida S, Bickle TA. 1980. The DNA sequence of an IS/-flanked transposon coding for resistance to chloramphenicol and fusidic acid. FEBS Lett 110:11–14. doi: 10.1016/0014-5793(80)80011-1. [DOI] [PubMed] [Google Scholar]
- 82.Harley CA, Dasgupta A, Wilson DW. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J Virol 75:1236–1251. doi: 10.1128/JVI.75.3.1236-1251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bolte S, Cordelieres FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 84.Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL. 2011. Residues of the UL25 protein of herpes simplex virus that are required for its stable interaction with capsids. J Virol 85:4875–4887. doi: 10.1128/JVI.00242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang K, Baines JD. 2011. Selection of HSV capsids for envelopment involves interaction between capsid surface components pUL31, pUL17, and pUL25. Proc Natl Acad Sci U S A 108:14276–14281. doi: 10.1073/pnas.1108564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol Cell 26:479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baines JD, Cunningham C, Nalwanga D, Davison A. 1997. The U(L)15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the U(L)15 gene product. J Virol 71:2666–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tengelsen LA, Pederson NE, Shaver PR, Wathen MW, Homa FL. 1993. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol 67:3470–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meckes DG Jr, Marsh JA, Wills JW. 2010. Complex mechanisms for the packaging of the UL16 tegument protein into herpes simplex virus. Virology 398:208–213. doi: 10.1016/j.virol.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Starkey JL, Han J, Chadha P, Marsh JA, Wills JW. 2014. Elucidation of the block to herpes simplex virus egress in the absence of tegument protein UL16 reveals a novel interaction with VP22. J Virol 88:110–119. doi: 10.1128/JVI.02555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morgado-Diaz JA, Nakamura CV, Agrellos OA, Dias WB, Previato JO, Mendonca-Previato L, De Souza W. 2001. Isolation and characterization of the Golgi complex of the protozoan Trypanosoma cruzi. Parasitology 123:33–43. [DOI] [PubMed] [Google Scholar]
- 92.Zhou J, Ghoroghi S, Benito-Martin A, Wu H, Unachukwu UJ, Einbond LS, Guariglia S, Peinado H, Redenti S. 2016. Characterization of induced pluripotent stem cell microvesicle genesis, morphology and pluripotent content. Sci Rep 6:19743. doi: 10.1038/srep19743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI, Hanson PI, Frost A. 2015. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350:1548–1551. doi: 10.1126/science.aad8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hanson PI, Roth R, Lin Y, Heuser JE. 2008. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol 180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fujita H, Yamanaka M, Imamura K, Tanaka Y, Nara A, Yoshimori T, Yokota S, Himeno M. 2003. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E Vps phenotype in mammalian cells. J Cell Sci 116:401–414. doi: 10.1242/jcs.00213. [DOI] [PubMed] [Google Scholar]
- 96.Hislop JN, Marley A, Von Zastrow M. 2004. Role of mammalian vacuolar protein-sorting proteins in endocytic trafficking of a non-ubiquitinated G protein-coupled receptor to lysosomes. J Biol Chem 279:22522–22531. doi: 10.1074/jbc.M311062200. [DOI] [PubMed] [Google Scholar]
- 97.Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol 73:3210–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Toropova K, Huffman JB, Homa FL, Conway JF. 2011. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J Virol 85:7513–7522. doi: 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Conway JF, Cockrell SK, Copeland AM, Newcomb WW, Brown JC, Homa FL. 2010. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J Mol Biol 397:575–586. doi: 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cardone G, Newcomb WW, Cheng N, Wingfield PT, Trus BL, Brown JC, Steven AC. 2012. The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J Virol 86:4058–4064. doi: 10.1128/JVI.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]