ABSTRACT
Eukaryotic mRNAs possess a methylated 5′-guanosine cap that is required for RNA stability, efficient translation, and protection from cell-intrinsic defenses. Many viruses use 5′ caps or other mechanisms to mimic a cap structure to limit detection of viral RNAs by intracellular innate sensors and to direct efficient translation of viral proteins. The coronavirus (CoV) nonstructural protein 14 (nsp14) is a multifunctional protein with N7-methyltransferase (N7-MTase) activity. The highly conserved S-adenosyl-l-methionine (SAM)-binding residues of the DxG motif are required for nsp14 N7-MTase activity in vitro. However, the requirement for CoV N7-MTase activity and the importance of the SAM-binding residues during viral replication have not been determined. Here, we engineered mutations in murine hepatitis virus (MHV) nsp14 N7-MTase at residues D330 and G332 and determined the effects of these mutations on viral replication, sensitivity to mutagen, inhibition by type I interferon (IFN), and translation efficiency. Virus encoding a G332A substitution in nsp14 displayed delayed replication kinetics and decreased peak titers relative to wild-type (WT) MHV. In addition, replication of nsp14 G332A virus was diminished following treatment of cells with IFN-β, and nsp14 G332A genomes were translated less efficiently both in vitro and during viral infection. In contrast, substitution of alanine at MHV nsp14 D330 did not affect viral replication, sensitivity to mutagen, or inhibition by IFN-β compared to WT MHV. Our results demonstrate that the conserved MHV N7-MTase SAM-binding-site residues are not required for MHV viability and suggest that the determinants of CoV N7-MTase activity differ in vitro and during virus infection.
IMPORTANCE Human coronaviruses, most notably severe acute respiratory syndrome (SARS)-CoV and Middle East respiratory syndrome (MERS)-CoV, cause severe and lethal human disease. Since specific antiviral therapies are not available for the treatment of human coronavirus infections, it is essential to understand the functions of conserved CoV proteins in viral replication. Here, we show that substitution of alanine at G332 in the N7-MTase domain of nsp14 impairs viral replication, enhances sensitivity to the innate immune response, and reduces viral RNA translation efficiency. Our data support the idea that coronavirus RNA capping could be targeted for development of antiviral therapeutics.
INTRODUCTION
Eukaryotic mRNAs possess a methylated 5′ guanosine cap linked to the penultimate nucleotide by a 5′-5′ triphosphate bridge (1). The 5′ capping of cellular mRNAs functions in RNA stability, pre-mRNA splicing, mRNA export from the nucleus, translation, and protection against cellular antiviral defenses (2). The canonical cellular capping process involves three enzymes: (i) an RNA triphosphatase (RTPase), which is responsible for cleaving the γ-phosphate of the nascent transcript, (ii) a guanylyltransferase (GTase), which transfers a GMP moiety to the 5′ diphosphate RNA, and (iii) an N7-methyltransferase (N7-MTase), which is responsible for transferring a methyl group from the methyl donor, S-adenosyl-l-methionine (SAM), to the N7 position of the guanosine base (3). These sequential reactions lead to formation of a cap-0 (7-methyl-Gppp) structure, which is thought to be the minimal cap determinant required for eIF4E recognition and efficient translation (4–6). Higher eukaryotes express 2′O-methyltransferases (2′O-MTase), which add a methyl group to the ribose 2′O position of the penultimate nucleotide of the cap-0 RNA. This reaction results in the formation of a cap-1 structure that allows cells to differentiate self from nonself RNAs in the cytoplasm (7, 8).
Eukaryotic viruses use host translation machinery, and many of these viruses encode capping enzymes. The diversity of enzymes and mechanisms used by viruses to synthesize capped RNA products suggests that there is selective pressure on viruses to cap their RNAs (9). Coronaviruses (CoVs) encode several enzymes within their large, positive-sense RNA genomes (27 to 34 kb) that are implicated in viral RNA capping. The coronavirus genome possesses a 5′ terminal cap and a 3′ poly(A) tail (10–12). All data to date support the hypothesis that CoV genomes are capped using the canonical mRNA capping pathway (13). Severe acute respiratory syndrome coronavirus (SARS-CoV) nsp13 displays RTPase activity in vitro (14). The CoV guanylyltransferase has not been identified but, according to the current model, would function to add a GMP to the diphosphate RNA product of nsp13. The RNA-dependent RNA polymerase (RdRp) of equine arteritis virus and SARS-CoV displays nucleotidylation activity (15). While further study is required to define the function of this activity in viral replication, it is possible that the RdRp participates in CoV RNA capping. nsp16 of feline coronavirus functions independently as a 2′O-MTase (16–18), but SARS-CoV nsp16 requires nsp10 as a cofactor for 2′O-MTase activity. SARS-CoVs lacking 2′O-MTase activity are recognized and sequestered by IFIT1 (13, 19–23) due to the lack of a cap-1 structure.
CoV nsp14 is a multifunctional protein with 3′-5′ exoribonuclease activity and N7-MTase activity (24, 25). nsp14-mediated N7-methylation of Gppp-RNA to form a cap-0 structure is a prerequisite for nsp10/16-mediated 2′O-methylation in vitro (13). A conserved DxG motif within the MTase domain is required for SAM binding in vitro, and alteration of these residues abolishes MTase activity in vitro (13, 26). However, the requirements of the CoV nsp14 N7-MTase during viral replication are not known. Therefore, we assessed the effect of mutations in the DxG motif of the MHV nsp14 N7-MTase on viral replication. We show that substitution of alanine at nsp14 D330 does not alter viral replication kinetics or increase sensitivity to beta interferon (IFN-β) treatment relative to wild-type (WT) MHV. However, substitution of alanine at nsp14 G332 impaired virus replication, resulting in delayed replication kinetics and decreased peak titer relative to WT MHV. In addition, nsp14 G332A virus displayed increased sensitivity to treatment of cells with IFN-β, and nsp14 G332A genomes were translated less efficiently in vitro and during infection. These data suggest that residue G332, but not residue D330, is required for MHV nsp14 N7-MTase activity and, collectively, that the regulation of CoV capping is likely more complex in the context of replicating virus than during in vitro biochemical studies with isolated proteins.
MATERIALS AND METHODS
Cells and viruses.
Murine delayed brain tumor (DBT) cells (27) and baby hamster kidney 21 cells expressing the MHV receptor (BHK-R) (28) were maintained at 37°C in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented to contain 10% fetal bovine serum (FBS; Invitrogen), 100 U/ml penicillin and streptomycin (Gibco), and 0.25 μg/ml amphotericin B (Corning). BHK-R cells were further supplemented to contain 0.8 mg/ml of G418 (Mediatech). Bone marrow-derived dendritic cells (BMDCs) were maintained in R10 medium (RPMI 1640 [Gibco] supplemented to contain 10% FBS, 2 mM l-glutamine, 100 μg/ml gentamicin [MP Biomedicals], 0.25 μg/ml amphotericin B, 50 μM beta-mercaptoethanol, 20 ng/ml granulocyte-macrophage colony-stimulating factor [GM-CSF], and 10 ng/ml interleukin-4 [IL-4]). Recombinant MHV strain A59 (GenBank accession number AY910861) was propagated as described previously (28).
Cloning, recovery, and verification of mutant viruses.
Site-directed mutagenesis was used to engineer point mutations in individual MHV genome cDNA fragment plasmids using the MHV infectious clone reverse genetics system (28). Viruses encoding firefly luciferase (FFL) fused to nsp2 were recovered using MHV A frag-FFL2 (29). Mutant viruses were recovered using BHK-R cells following electroporation of in vitro-transcribed genomic RNA. All mutagenized plasmids were sequenced (GenHunter Corporation, Nashville, TN) to ensure that no additional mutations were introduced. Recovered viruses also were sequenced to verify the engineered mutations.
Virus replication kinetics.
Subconfluent DBT cell monolayers were infected at a multiplicity of infection (MOI) of 1 PFU per cell at 37°C for 45 min. Inocula were removed, cells were washed with 1× phosphate-buffered saline (PBS), and fresh medium was added. Aliquots were harvested at various times postinfection. Viral titer at various intervals was determined by plaque assay (30).
5-FU sensitivity assays.
5-Fluorouracil (5-FU; Sigma) was prepared as a 200 mM stock solution in dimethyl sulfoxide (DMSO). Subconfluent DBT cells were treated with DMEM supplemented to contain various concentrations of 5-FU or DMSO alone at 37°C for 30 min (31). Drug was removed, and cells were infected with virus at an MOI of 0.01 PFU/cell at 37°C for 1 h. Inocula were removed, and cells were incubated in medium containing 5-FU or DMSO. Cell culture supernatants were collected at 24 h postinfection, and viral titers were determined by plaque assay.
IFN-β sensitivity assays.
Subconfluent DBT cells were treated with various concentrations of mouse IFN-β (PBL Assay Science) for 18 h prior to infection with virus at an MOI of 1 PFU/cell at 37°C for 45 min. Inocula were removed, cells were washed with PBS, and fresh medium was added. Cell culture supernatants were collected at the indicated times postinfection, and viral titers were determined by plaque assay.
IFN-β induction assays.
Subconfluent DBT cells were treated with 10 U/ml mouse IFN-β for 18 h prior to infection with virus at an MOI of 0.1 PFU/cell at 37°C for 45 min. Inocula were removed, cells were washed with PBS, and fresh medium was added. At 12 h postinfection, cell culture supernatants were aspirated and cell lysates were harvested by adding TRIzol reagent. Total RNA present in lysates was purified using the phenol-chloroform method. cDNA was generated by reverse transcription (RT)-PCR using 1 μg of total RNA as described previously (31). Mouse IFN-β expression levels relative to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were determined by quantitative real-time PCR (qPCR) using the Applied Biosciences 7500 real-time PCR system with the Power SYBR green PCR master mix and the IFN-β primers FWD, 5′-TCCGCCCTGTAGGTGAGGTTGAT-3′, and REV, 5′-GTTCCTGCTGTGCTTCTCCACCA-3′ and the GAPDH primers previously reported (31).
Generation and infection of BMDCs.
Primary BMDCs were isolated from the hind limbs of WT and IFN-α/β receptor-deficient (IFNAR−/−) C57BL/6J mice. Mice were euthanized by isoflurane overdose, and hind limbs were resected. Bone marrow cells were collected by flushing the femurs and tibiae with medium. Cells were strained through a 70-μm cell strainer, and red blood cells were lysed. Cells were cultured at 37°C in R10 medium supplemented to contain 20 ng/ml GM-CSF and 10 ng/ml IL-4. At 3 days postplating, cell culture supernatants were removed and replaced with fresh R10 medium. Six days postplating, cells were lifted using Cellstripper (Corning) and replated with fresh R10 medium in 24-well plates at a density of 105 cells/well and incubated at 37°C overnight. WT and IFNAR−/− BMDCs were infected with virus at an MOI of 0.01 PFU/cell at 37°C for 45 min. Inocula were removed, and fresh medium was added. Cell culture supernatants were collected 24 h postinfection, and viral titers were determined by plaque assay. All experiments with animals were performed in accordance with Vanderbilt University School of Medicine Institutional Animal Care and Use Committee guidelines.
Purification of virions and extraction of RNA.
Virion RNA was purified from subconfluent T150 flasks of BHK-R cells infected with WT-FFL or nsp14 G332A-FFL viruses at an MOI of 0.001 PFU/cell. When cytopathic effect was apparent throughout the monolayer, cell culture supernatants were collected and pooled into 50-ml conical tubes (Corning), clarified by centrifugation at 1,000 × g for 10 min, and stored at −80°C. Upon thawing, virus particles in the clarified supernatants were collected by ultracentrifugation at 106,750 × g overnight through a 5-ml, 20% (wt/wt) sucrose cushion in an SW32Ti rotor. The pelleted particles were resuspended in 200 μl MSE buffer (10 mM MOPS [morpholinepropanesulfonic acid] [pH 6.8], 150 mM NaCl, 1 mM EDTA) and incubated at 4°C overnight prior to resuspension by gently pipetting several times. Viral RNA was isolated from purified viral particles using TRIzol reagent (Invitrogen) and phenol-chloroform extraction.
In vitro translation reactions.
Viral genomic RNAs containing an in-frame firefly luciferase-encoding sequence were translated at 30°C for various intervals in 10 μl of rabbit reticulocyte lysate (Promega) in the presence of both 10 μM amino acid mixture without leucine and 10 μM amino acid mixture without methionine.
Firefly luciferase assays.
Subconfluent DBT cells were infected with virus at an MOI of 0.1 PFU/cell. At various intervals, cell culture supernatants were removed, cells were washed with PBS, and 100 μl of reporter cell lysis buffer (Promega) was added to each well. Cells lysates were frozen at −80°C to promote lysis and thawed at room temperature prior to quantifying firefly luciferase activity. Luciferase activity from cell lysates or in vitro translation reactions was quantified using a Veritas luminometer (Turner Biosystems) and the firefly luciferase assay system (Promega).
Determination of specific infectivity.
Subconfluent monolayers of DBT-9 cells were infected with virus at an MOI of 0.1 PFU/cell at 37°C for 45 min. Inocula were removed, fresh medium was added, and cells were incubated at 37°C for 24 h. Cell culture supernatants were collected, and viral titers were determined by plaque assay. Supernatants also were used for RNA genome isolation by addition of 100 μl supernatant to 900 μl TRIzol reagent, chloroform extraction by phase separation, and final purification using the PureLink Mini RNA kit (Ambion). Genome RNA was quantified using one-step qRT-PCR, and the particle-to-PFU ratio was calculated.
Genome RNA stability assay.
Subconfluent monolayers of DBT-9 cells were infected with virus at an MOI of 0.01 PFU/cell at 37°C for 45 min in the presence of DMSO or 100 μg/ml cycloheximide (CHX; Sigma). Inocula were removed, medium containing DMSO or 100 μg/ml CHX was added, and cell lysates were harvested at the indicated times postinfection by removing the cell culture supernatant and adding TRIzol reagent. Lysates were spiked with a known amount of in vitro-transcribed Renilla luciferase RNA, and total RNA was obtained by phenol-chloroform extraction. cDNA was generated by RT-PCR, and the number of viral genome copies present relative to Renilla luciferase was determined by SYBR green qPCR using nsp10 (31) and Renilla luciferase-specific primers (22).
Quantification of viral genomic RNA by qRT-PCR.
An RNA standard was prepared using the MHV A fragment (28) to generate a 931-nucleotide RNA. First, cDNA was generated by PCR amplification using the following primers: forward, 5′-TAATACGACTCACTATAGGGGGCTATGTGGATTGTTGTGG-3′, which initiates with a T7 promoter, and reverse, 5′-AATTCTTGACAAGCTCAGGC-3′. RNA for the standard curve was prepared using an mMessage mMachine T7 kit (Ambion) and purified using an RNeasy minikit (Qiagen). A standard curve was generated using 10-fold dilutions from 103 to 108 copies. A 5′ 6-carboxyfluorescein (FAM)-labeled probe (5′-TTCTGACAACGGCTACACCCAACG-3′ [Biosearch Technologies]) was used with forward (5′-AGAAGGTTACTGGCAACTG-3′) and reverse (5′-TGTCCACGGCTAAATCAAAC-3′) primers. Reaction mixtures were incubated on ice with enzyme added last. The final volume for reaction mixtures was 20 μl, with 150 nM probe, 900 nM each primer, 2 μl sample RNA, and 10 μl 2× ToughMix, one-step, low ROX enzyme mix (Quantas) per reaction mixture. Samples were quantified in duplicate using an Applied Biosciences 7500 real-time PCR system under the following conditions: 55°C for 10 min, 95°C for 5 min, 95°C for 30 s, and 60°C for 1 min, with the last two steps repeated 40 times. The standard curve was plotted using GraphPad Prism 6 software, and the number of genomes per microliter was calculated.
Statistical analysis.
Statistical tests were conducted using GraphPad Prism 6 software (La Jolla, CA) as indicated in the respective figure legends.
RESULTS
Recovery and replication kinetics of MHV nsp14 N7-MTase mutants.
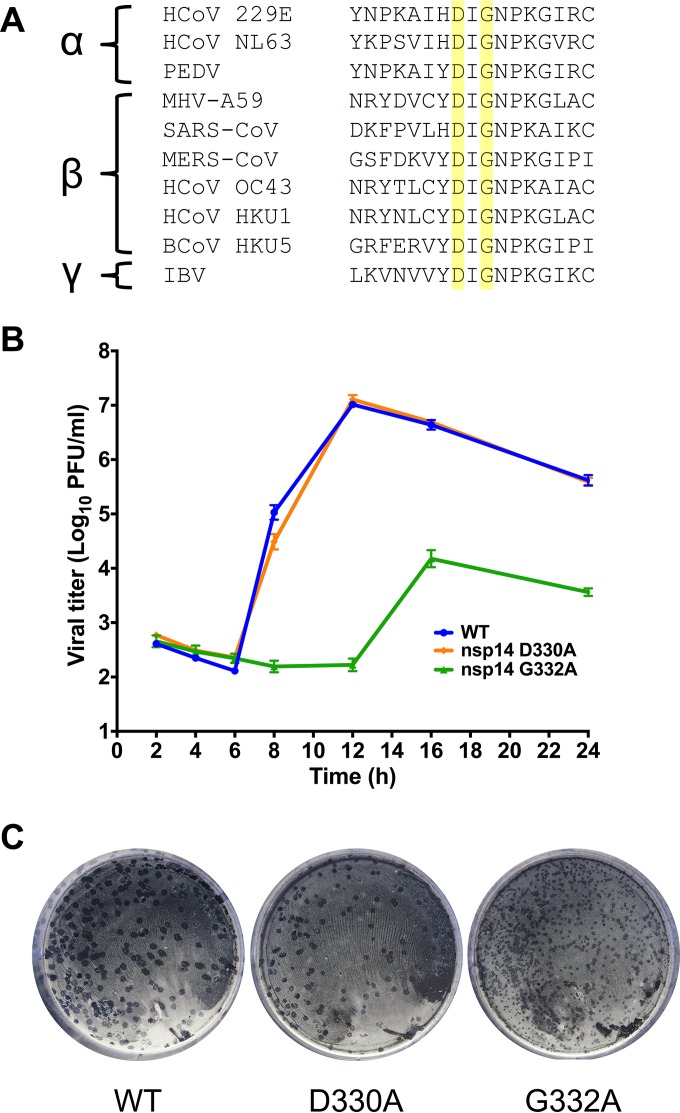
The DxG SAM-binding motif is conserved among the nsp14 N7-MTase domains of alpha-, beta-, and gammacoronaviruses (Fig. 1A). Mutations in this motif of SARS-CoV nsp14 ablate N7-MTase activity of purified proteins in vitro (13, 25, 26). To determine whether this motif is required for viral replication, we engineered alanine substitutions at the DxG SAM-binding motif in the MHV nsp14 N7-MTase domain. Virus containing either a D330A or G332A substitution in nsp14 was recovered, and the nucleotide sequence of each virus was confirmed across the nsp14-coding region. Following infection of DBT cells at an MOI of 1 PFU/cell, nsp14 D330A virus replicated with kinetics comparable to that of WT MHV (Fig. 1B). nsp14 D330A plaque morphology also was similar to that of WT MHV (Fig. 1C). In contrast, the nsp14 G332A virus began exponential replication 4 to 6 h later than WT MHV and reached a lower peak titer (1.5 × 104 PFU/ml) than that of WT MHV (107 PFU/ml) (Fig. 1B). The nsp14 G332A virus plaque size was also decreased relative to that of WT MHV (Fig. 1C). Thus, despite the requirement of D330 for nsp14 N7-MTase activity in vitro (13, 25, 26, 32), our data indicate that the D330A mutation has no detectable effect on MHV replication kinetics in cell culture.
FIG 1.
Replication kinetics of viruses with altered N7-MTase SAM-binding residues. (A) Alignment of GenBank ORF1b sequences of the α-, β-, and γ-CoVs shown demonstrates that SAM-binding residues (highlighted in yellow) are highly conserved. (B) DBT cells were infected with the viruses shown at an MOI of 1 PFU/cell. Cell culture supernatants were collected at the indicated times postinfection, and viral titers were determined by plaque assay. Error bars indicate standard errors of the means (SEM) (n = 6). (C) Plaque morphology of the viruses following agarose overlay plaque assay and fixation with 3.7% paraformaldehyde 24 h postinfection.
nsp14 D330A or G332A mutations do not significantly influence nsp14 ExoN activity.
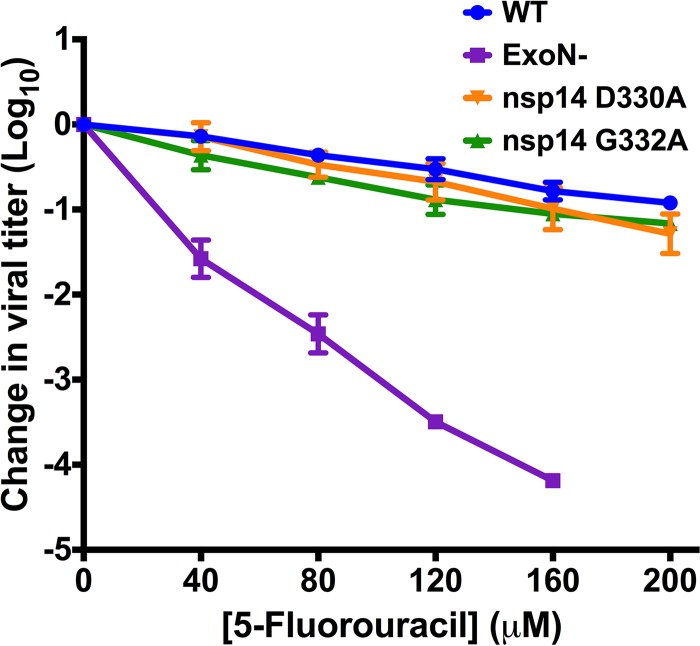
Coronavirus nsp14 is a multifunctional protein with two known enzymatic activities, a proofreading 3′-5′ exoribonuclease activity (ExoN) and N7-MTase activity (24, 25). Based on in vitro studies, the ExoN and N7-MTase domains of CoV nsp14 are interdependent (26). This conclusion is supported by the crystal structure of nsp14, demonstrating that the ExoN and N7-MTase domains interact through a large hydrophobic interface (32). In addition, disruption of ExoN (ExoN−) via mutations at two active-site residues decreases the replication fidelity of MHV and SARS-CoV and renders the viruses sensitive to the RNA mutagen 5-fluorouracil (5-FU) (30, 31, 33). Thus, 5-FU sensitivity has been shown to be an in vitro indicator of ExoN activity. Therefore, we tested whether the D330A or G332A mutations affect ExoN activity by treating cells with increasing concentrations of 5-FU or vehicle (DMSO) prior to infection with either nsp14 D330A or nsp14 G332A virus at an MOI of 0.01 PFU/cell (Fig. 2). The nsp14 D330A and nsp14 G332A viruses were not significantly altered in 5-FU sensitivity compared with WT MHV (no significant difference by one-way analysis of variance [ANOVA]). In contrast, the ExoN− virus displayed a concentration-dependent increase in 5-FU sensitivity. These results indicate that neither D330A nor G332A significantly alter ExoN activity during virus replication.
FIG 2.
N7-MTase mutants display WT-like sensitivity to the RNA mutagen 5-FU. DBT cells were treated with the indicated concentrations of 5-FU for 30 min prior to infection with the viruses shown at an MOI of 0.01 PFU/cell. Medium containing 5-FU or vehicle was added 30 min postinfection. After 24 h, cell culture supernatants were collected, and viral titers were determined by plaque assay. For each virus, titers were normalized to those following infection of DMSO-treated controls. Changes in viral titers for nsp14 D330A and nsp14 G332A viruses were not statistically significant relative to WT MHV by one-way ANOVA. Error bars indicate SEM (n = 4).
MHV nsp14 G332A is detected by and sensitive to the type I interferon-mediated innate immune response.
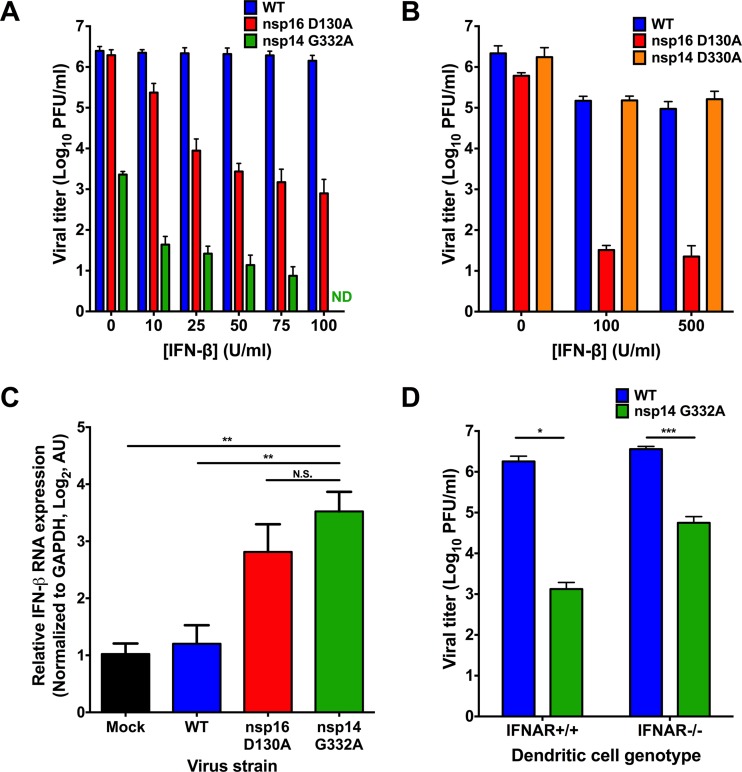
Coronavirus RNA capping likely follows the conventional capping pathway, with nsp14 N7-methylation being a prerequisite for 2′O-methylation in vitro (13). Therefore, decreased nsp14 N7-MTase activity should reduce overall 2′O-methylation, thereby increasing virus sensitivity to exogenous type I IFN due to recognition by IFIT1 and MDA5 (22, 23). To test this hypothesis, we pretreated DBT cells with murine IFN-β prior to infection with WT MHV, nsp16 D130A, which is an IFN-sensitive positive-control mutant virus due to ablated 2′O-MTase activity (19, 22, 23, 34), or nsp14 D330A or nsp14 G332A N7-MTase mutant viruses at an MOI of 1 PFU/cell. Cell culture supernatants were collected at either 12 or 24 h postinfection, and viral titers were determined by plaque assay. As expected, the nsp16 D130A virus was sensitive to IFN-β pretreatment (Fig. 3A). The nsp14 G332A virus demonstrated a dose-dependent increase in IFN-β sensitivity, which became undetectable by plaque assay at IFN-β concentrations greater than 75 U/ml (Fig. 3A). In contrast, nsp14 D330A virus displayed sensitivity to IFN-β comparable to WT MHV (Fig. 3B). Because nsp14 D330A displayed replication kinetics and resistance to IFN-β pretreatment indistinguishable from those of WT MHV, it is likely that the D330A substitution does not significantly affect N7-MTase activity. Therefore, we focused solely on the nsp14 G332A mutant for the remainder of the experiments in this study.
FIG 3.
nsp14 G332A virus exhibits increased induction of and sensitivity to IFN-β. DBT cells were treated for 18 h with the indicated concentrations of mouse IFN-β. Cells were infected with WT, nsp16 D130A, or nsp14 G332A virus and incubated for 24 h (A) or infected with WT, nsp16 D130A, and nsp14 D330A virus and incubated for 12 h (B). Cell culture supernatants were collected, and viral titers were determined by plaque assay. For each panel, error bars represent SEM (n = 4). ND, not detectable. (C) DBT cells were treated for 18 h with 10 U/ml mouse IFN-β. Cells were mock infected or infected with WT, nsp16 D130A, or nsp14 G332A virus at an MOI of 0.1 PFU/cell. At 12 h postinfection, cell lysates were harvested, total RNA was extracted, cDNA was generated, and IFN-β expression relative to GAPDH was determined by qPCR. Error bars indicate SEM (n = 9). N.S., not significant; **, P < 0.01 by Student's t test. (D) BMDCs were infected with either WT or nsp14 G332A virus at an MOI of 0.01 PFU/cell. At 24 h postinfection, cell culture supernatants were collected, and viral titers were determined by plaque assay. Error bars indicate SEM (n = 6). *, P < 0.05; ***, P < 0.001 by Student's t test.
In addition to an increased sensitivity to the effects of type I interferon pretreatment, coronaviruses lacking 2′O-MTase activity induce higher levels of IFN-β than do the WT viruses (8, 22, 23). Therefore, to determine whether nsp14 G332A is also recognized by innate sensors and subsequently induces type I interferon expression, we pretreated DBT cells with 10 U/ml murine IFN-β for 18 h prior to infection with WT MHV, nsp16 D130A, or nsp14 G332A viruses at an MOI of 0.1 PFU/cell. At 12 h postinfection, cell lysates were collected and the relative expression of IFN-β was determined by qPCR (Fig. 3C). As previously reported, infection with WT MHV marginally induced the expression of IFN-β (35) and infection with nsp16 D130A led to an upregulation of IFN-β relative to mock-infected cells (8, 22, 23). Furthermore, infection with nsp14 G332A led to a significant increase in the expression of IFN-β relative to mock and WT MHV-infected cells. These data further suggest that nsp14 N7-MTase activity precedes nsp16 2′O-MTase activity and the absence of either activity results in innate detection of the virus leading to the induction of type I interferon gene expression.
To determine the effect that increased sensitivity to IFN-β has on nsp14 G332A replication, we tested whether nsp14 G332A virus replication could be rescued in BMDCs lacking the IFN-α/β receptor (IFNAR−/−). IFNAR−/− cells lack the capacity to respond to type I IFNs and are thus incapable of mounting an effective IFN-dependent antiviral response (36). WT or IFNAR−/− BMDCs were infected with WT MHV or nsp14 G332A virus at an MOI of 0.01 PFU/cell, cell culture supernatants were collected 24 h postinfection, and viral titers were determined by plaque assay. Similar to experiments using DBT cells, nsp14 G332A virus replicated poorly in WT BMDCs relative to WT MHV (Fig. 3D). Titers of nsp14 G332A virus were increased approximately 40-fold in IFNAR−/− BMDCs (5.6 × 104 PFU/ml) compared with the titers of this virus in WT BMDCs (1.3 × 103 PFU/ml). However, despite the increase in viral titers of nsp14 G332A in IFNAR−/− BMDCs, titers were not restored to the level of WT MHV in IFNAR−/− BMDCs (3.6 × 106 PFU/ml). These data suggest that the impaired replication capacity of nsp14 G332A virus is only in part attributable to IFN sensitivity and, instead, this virus may manifest a more general replication defect.
nsp14 G332A genome translation is delayed during infection.
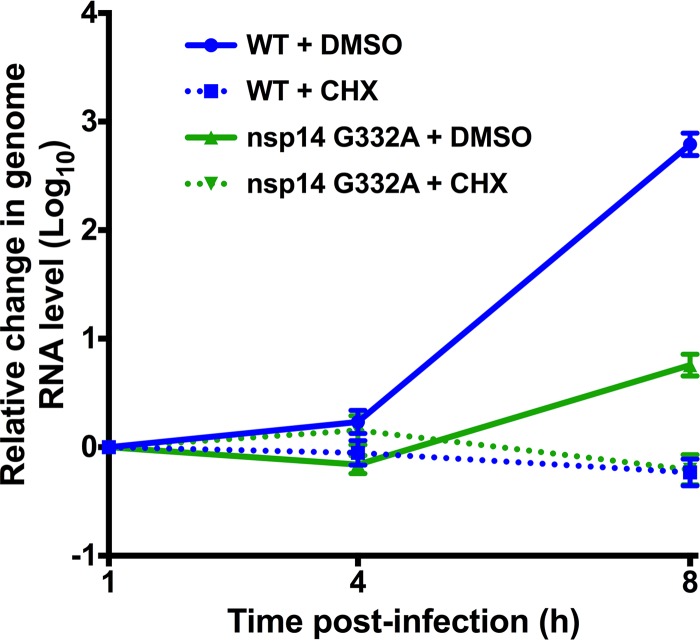
Since the absence of the IFNAR was insufficient to restore nsp14 G332A replication, other mechanisms, such as decreased genome RNA stability or decreased viral genome translation, may contribute to the replication defect of this virus. The 5′ capping of cellular mRNAs serves several important functions, one of which is to increase RNA stability (2, 9). To test the stability of the nsp14 G332A genome upon entry into the cell, we infected DBT cells with WT MHV or nsp14 G332A virus at an MOI of 0.01 PFU/cell in the presence of vehicle (DMSO) or 100 μg/ml cycloheximide (CHX). CHX inhibits the translation of input viral genomes and prevents expression of the viral RNA-dependent RNA polymerase, thereby allowing us to quantify the amount of coronavirus RNA present at later time points relative to input. At the indicated times postinfection, cell lysates were collected and spiked with a known amount of in vitro-transcribed Renilla luciferase, and the amount of viral RNA present relative to Renilla luciferase was determined by qPCR (Fig. 4). At each time point postinfection for CHX-treated samples, the level of nsp14 G332A RNA was similar to that of WT MHV, indicating that nsp14 G332A replication is not impaired due to decreased genome RNA stability.
FIG 4.
nsp14 G332A genomic RNAs are stable. DBT cells were infected with WT or nsp14 G332A virus at an MOI of 0.01 PFU/cell in the presence of vehicle (DMSO) or 100 μg/ml CHX. Cell lysates were harvested at the indicated times postinfection and spiked with a known amount of in vitro-transcribed Renilla luciferase RNA, and total RNA was obtained by phenol-chloroform extraction. cDNA was generated by RT-PCR, and the number of viral genome copies present relative to Renilla luciferase was determined by SYBR green qPCR using MHV nsp10 and Renilla luciferase-specific primers. Error bars indicate SEM (n = 6).
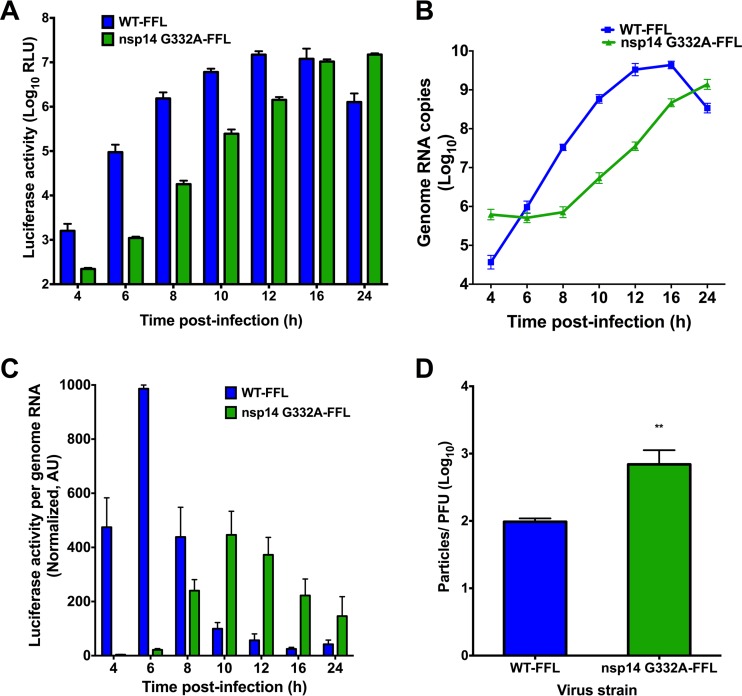
In addition to serving as a precursor for 2′O-methylation, N7-methylated guanosine 5′ caps are recognized by eIF4E and required for efficient translation of eukaryotic RNA (9, 37). To determine whether the nsp14 G332A mutation impairs viral translation efficiency, we first engineered virus encoding FFL as an in-frame N-terminal fusion with MHV nsp2 (29) in the open reading frame 1a (ORF1a) polyprotein coding sequence of the isogenic nsp14 G332A cloned genome. In this setting, FFL-nsp2 is the second protein translated from the input viral genome and becomes a reporter for viral protein translation. We infected DBT cells with either WT-FFL or nsp14 G332A-FFL virus at an MOI of 0.1 PFU/cell, and lysates were prepared at various intervals postinfection to quantify luciferase activity and viral genome RNA copy number. Luciferase activity accumulated more slowly following infection by nsp14 G332A-FFL virus relative to WT-FFL virus (Fig. 5A). The WT-FFL signal began to decline after 16 h due to destruction of the cell monolayer. In addition, levels of nsp14 G332A-FFL genomic RNA increased more slowly than those of WT-FFL (Fig. 5B). By quantifying both luciferase activity and viral genome copies, we were able to calculate the kinetics of translation. To determine the rate of translation at each time point postinfection, the ratio of luciferase activity to genome copies was determined using data shown in Fig. 5A and B. The ratio of luciferase activity to genome copies for WT-FFL was highest at early times postinfection (Fig. 5C). In contrast, the ratio of luciferase activity to genome copies was substantially less for the nsp14 G332A-FFL virus at early time points postinfection than for WT-FFL and failed to reach peak WT-FFL levels. These data demonstrate that nsp14 G332A-FFL virus requires more genomic RNA to achieve the WT levels of FFL activity, consistent with decreased translation efficiency of the mutant virus genome. Therefore, we next determined whether nsp14 G332A-FFL and WT-FFL virions are equivalently infectious by measuring the specific infectivity of each virus from infected DBT cell culture supernatants. The ratio of nsp14 G332A-FFL particles per PFU was approximately 7-fold more than WT-FFL (Fig. 5D). Thus, packaged nsp14 G332A-FFL genomes were less efficient at establishing infection than the WT.
FIG 5.
nsp14 G332A genomic RNAs are translated with delayed kinetics during infection. DBT cells were infected with either WT-FFL or nsp14 G332A-FFL virus at an MOI of 0.1 PFU/cell. At the times shown postinfection, cell culture supernatants were collected, and lysates were harvested and divided equally into two samples. For the first lysate sample, luciferase activity was quantified (A). For the remaining lysate sample, RNA was extracted, and genome RNA copies were quantified using real-time qRT-PCR with a standard curve and CoV nsp2-specific primers (B). (C) Translation of WT-FFL or nsp14 G332A-FFL genomes at the times shown postinfection as determined by luciferase activity per genome RNA copy number. Values were normalized to WT-FFL at 6 h postinfection. Error bars indicate SEM (n = 4). (D) Viral titers in cell culture supernatants from DBT cells infected with either WT-FFL or nsp14 G332A-FFL were determined by plaque assay, and the number of genome RNA copies present in the input supernatant was determined by one-step real-time qRT-PCR. The particle-to-PFU ratio was calculated by dividing the number of genome RNA copies by the viral titers. Error bars represent SEM (n = 4). **, P < 0.01 by Student's t test.
nsp14 G332A-FFL genomes are translated less efficiently than WT-FFL genomes in vitro.
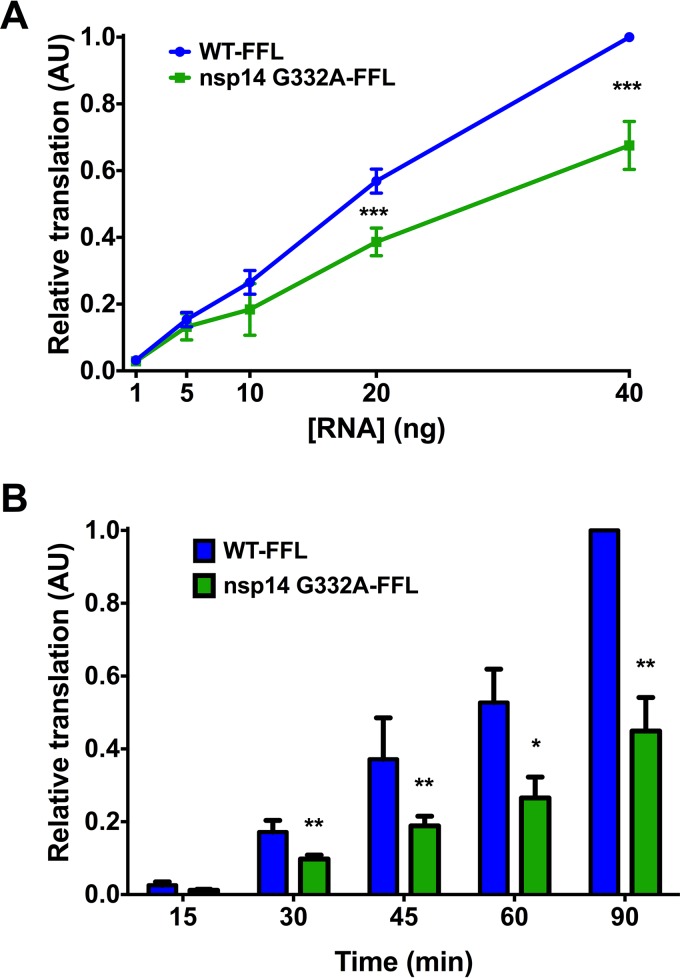
To directly assess the translation capacity of nsp14 G332A-FFL virus genomes, we isolated genome RNA from purified virions. Increasing concentrations of genome RNAs were incubated with rabbit reticulocyte lysates at 30°C for 1.5 h, and luciferase activity was quantified (Fig. 6A). Compared to WT-FFL genomes, FFL activity in the reticulocyte lysates was significantly reduced following incubation with nsp14 G332A-FFL genomes. In addition, we quantified the relative translation efficiency of equal amounts of WT-FFL and nsp14 G332A-FFL genomic RNA over time. At all time points tested after 15 min, FFL activity was significantly reduced following incubation of reticulocyte lysates with nsp14 G332A-FFL genomes relative to WT-FFL genomes (Fig. 6B). Taken together, our data indicate that the decreased replication capacity of the nsp14 G332A virus is attributable to IFN sensitivity and reduced translation efficiency.
FIG 6.
Purified nsp14 G332A genomic RNA is translated at lower efficiency in vitro. BHK-R cells were infected at an MOI of 0.001 PFU/cell with either WT-FFL or nsp14 G332A-FFL virus. Supernatants were harvested and clarified, and virions were collected by ultracentrifugation. Virion pellets were resuspended, TRIzol was added, and virion RNAs were purified using phenol-chloroform phase separation. Genome RNA copies were quantified using one-step real-time qRT-PCR with a standard curve and CoV nsp2-specific primers. (A) The concentrations of WT-FFL or nsp14 G332A-FFL genomic RNAs shown were translated in vitro at 30°C for 1.5 h, and luciferase activity was quantified. Translation values are relative to WT-FFL genomic RNA at 40 ng. Error bars represent SEM (n = 4). ***, P < 0.001 by Student's t test. (B) Equivalent numbers of either WT-FFL or nsp14 G332A-FFL genomic RNAs were translated in vitro for the times shown, and luciferase activity was quantified. Error bars represent SEM (n = 6). *, P < 0.05; **, P < 0.01 by Student's t test.
DISCUSSION
In this study, we engineered recombinant CoVs encoding alanine substitutions in the nsp14 N7-MTase at the SAM-binding-site residues, D330 and G332. We found that the N7-MTase SAM-binding-site mutants are viable and yield drastically different phenotypes during replication. Specifically, MHV nsp14 D330A virus replicates indistinguishably from WT MHV in all assays conducted, despite the requirement of this residue for SAM binding in vitro (26). There is precedent for such a contradiction. A previous study using vesicular stomatitis virus identified a SAM-binding residue within the L protein (G1674) that, when altered, does not affect viral replication or N7-MTase activity (38). The structure of the SARS-CoV nsp10-nsp14 complex reveals that D331 (D330 in MHV) is in close proximity to the SAM-binding site, but only G333 (G332 in MHV) directly contacts SAM (32). Since in vitro N7-MTase activity was assessed only for a SARS-CoV nsp14 D331A/G333A double mutant, it is not clear whether nsp14 D331 was required for N7-MTase activity in this study (32). However, a previous study using both in vitro functional assays and yeast complementation reported that SARS-CoV nsp14 D331 is essential for N7-MTase activity (26). Our study examined nsp14 N7-MTase in the context of viral replication. A potential difference between our work and previous studies of the CoV nsp14 N7-MTase is the use of MHV versus SARS-CoV proteins, respectively. Purified MHV nsp14 N7-MTase is not available in our lab for biochemical studies. However, our results will guide future experiments when such a system is established. During our study, we attempted to recover SARS-CoV nsp14 D331A, I332A, and G333A N7-MTase mutant viruses. However, viable viruses were not recovered after at least three attempts for each mutant. Nonetheless, the high conservation of the SAM-binding residues makes it unlikely that the differences observed between our work and previous biochemical studies are due to profoundly different N7-MTase catalytic mechanisms.
In contrast to nsp14 D330A virus, nsp14 G332A virus replicated with delayed kinetics and reached peak titers that were 1,000-fold less than those of WT MHV. CoV nsp14 has two domains: an N-terminal ExoN domain and a C-terminal N7-MTase domain. Mutations at D331 in SARS-CoV nsp14 do not affect ExoN activity in vitro (25, 26). However, the effect of altering residue G333 (G332 in MHV) on ExoN activity has not been reported using any system. It is unlikely that the G332A mutation in MHV nsp14 influences ExoN activity, as nsp14 G332A demonstrated WT-like sensitivity to the RNA mutagen 5-FU. Even a subtle alteration in ExoN activity should result in a detectable change in 5-FU sensitivity, particularly since we performed the assay using low-MOI conditions, which would increase mutagen incorporation during multistep replication (31, 34). The lack of enhanced 5-FU sensitivity for the nsp14 D330A and nsp14 G332A viruses indicates that mutations at these SAM-binding residues do not significantly dampen ExoN activity during virus replication. Additionally, since nsp14 G332A is resistant to 5-FU treatment, it is unlikely that the G332A phenotype is due to nsp14 instability or degradation.
Our data indicate that impaired replication of nsp14 G332A virus is likely due to a combination of factors, including increased detection by innate immune sensors and decreased translation efficiency of viral RNA. Binding of type I IFNs to the IFN receptor leads to expression of many IFN-stimulated genes and ultimately the establishment of an antiviral state (39). Coronavirus RNAs lacking 2′O-methylation are sensed by IFIT1, which is one of the most highly upregulated IFN-stimulated genes following IFN induction (40). While nsp14 D330A displayed WT-like sensitivity to pretreatment with IFN-β, nsp14 G332A virus did not replicate following IFN-β pretreatment with doses of >75 U/ml. However, initial titers were lower for nsp14 G332A. Thus, the concentration-dependent change in viral titer following IFN-β pretreatment was similar to that for the nsp16 D130A virus. The IFN-β sensitivity of nsp14 G332A likely results from a reduction in 2′O-methylation of viral RNA due to impaired N7-MTase activity. This hypothesis is supported by our data showing that infection with either nsp16 D130A or nsp14 G332A virus results in the induction of IFN-β gene expression. In addition, decreased N7-MTase activity due to the G332A mutation results in the delayed translation and decreased translation efficiency observed during viral replication and in vitro assays. Due to the highly impaired replication capacity of the nsp14 G332A virus, it has not been possible to directly determine the cap methylation status of nsp14 G332A virus genomes. Nevertheless, our results are consistent with functions of the N7-methylated 5′cap in promoting both viral and cellular translation (4–6). Decreased translation efficiency also could explain the lower specific infectivity observed for nsp14 G332A virus. Furthermore, it is possible that the delayed translation kinetics of nsp14 G332A genomic RNA increases innate sensing of the virus by delaying the early expression of multiple CoV IFN antagonists upon entry, resulting in decreased replication capacity.
Our data provide additional support for a sequential model of CoV RNA capping wherein N7-methylation precedes 2′O-methylation. In addition, our studies suggest that small-molecule inhibitors of the CoV nsp14 N7-MTase would impair virus replication and provide a pathogen-associated molecular pattern that would be quickly recognized by the innate immune response. Given the conservation of these enzymes, such inhibitors may have activity against diverse groups of coronaviruses.
ACKNOWLEDGMENTS
We thank Clint Smith for critical review of the manuscript and members of the Denison and Dermody laboratories for useful discussions.
REFERENCES
- 1.Shatkin AJ. 1976. Capping of eucaryotic mRNAs. Cell 9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Darnell JE. 1979. Transcription units for mRNA production in eukaryotic cells and their DNA viruses. Prog Nucleic Acid Res Mol Biol 22:327–353. doi: 10.1016/S0079-6603(08)60803-X. [DOI] [PubMed] [Google Scholar]
- 3.Furuichi Y, Shatkin AJ. 2000. Viral and cellular mRNA capping: past and prospects. Adv Virus Res 55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcotrigiano J, Gingras A-C, Sonenberg N, Burley SK. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89:951–961. doi: 10.1016/S0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 5.Filipowicz W, Furuichi Y, Sierra JM, Muthukrishnan S, Shatkin AJ, Ochoa S. 1976. A protein binding the methylated 5′-terminal sequence, m7GpppN, of eukaryotic messenger RNA. Proc Natl Acad Sci U S A 73:1559–1563. doi: 10.1073/pnas.73.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schibler U, Perry RP. 1977. The 5′-termini of heterogeneous nuclear RNA: a comparison among molecules of different sizes and ages. Nucleic Acids Res 4:4133–4150. doi: 10.1093/nar/4.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei CM, Gershowitz A, Moss B. 1975. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 8.Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decroly E, Ferron F, Lescar J, Canard B. 2012. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai MM, Stohlman SA. 1981. Comparative analysis of RNA genomes of mouse hepatitis viruses. J Virol 38:661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai MM, Patton CD, Stohlman SA. 1982. Further characterization of mRNAs of mouse hepatitis virus: presence of common 5-′end nucleotides. J Virol 41:557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masters PS. 2006. The molecular biology of coronaviruses. Adv Virus Res 66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B, Decroly E. 2010. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog 6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol 78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann KC, Gulyaeva A, Zevenhoven-Dobbe JC, Janssen GMC, Ruben M, Overkleeft HS, van Veelen PA, Samborskiy DV, Kravchenko AA, Leontovich AM, Sidorov IA, Snijder EJ, Posthuma CC, Gorbalenya AE. 2015. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res 43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LLM, Guan Y, Rozanov M, Spaan WJM, Gorbalenya AE. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Grotthuss M, Wyrwicz LS, Rychlewski L. 2003. mRNA cap-1 methyltransferase in the SARS genome. Cell 113:701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decroly E, Imbert I, Coutard B, Bouvet M, Selisko B, Alvarez K, Gorbalenya AE, Snijder EJ, Canard B. 2008. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J Virol 82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin T-Y, Schneller S, Züst R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Shi P-Y, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Su C, Ke M, Jin X, Xu L, Zhang Z, Wu A, Sun Y, Yang Z, Tien P, Ahola T, Liang Y, Liu X, Guo D. 2011. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog 7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Decroly E, Debarnot C, Ferron F, Bouvet M, Coutard B, Imbert I, Gluais L, Papageorgiou N, Sharff A, Bricogne G, Ortiz-Lombardia M, Lescar J, Canard B. 2011. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog 7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habjan M, Hubel P, Lacerda L, Benda C, Holze C, Eberl CH, Mann A, Kindler E, Gil-Cruz C, Ziebuhr J, Thiel V, Pichlmair A. 2013. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog 9:e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menachery VD, Yount BL Jr, Josset L, Gralinski LE, Scobey T, Agnihothram S, Katze MG, Baric RS. 2014. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-O-methyltransferase activity. J Virol 88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, Ziebuhr J. 2006. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A 103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, Guo D. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A 106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Tao J, Sun Y, Wu A, Su C, Gao G, Cai H, Qiu S, Wu Y, Ahola T, Guo D. 2013. Structure-function analysis of severe acute respiratory syndrome coronavirus RNA cap guanine-N7-methyltransferase. J Virol 87:6296–6305. doi: 10.1128/JVI.00061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W, Baric RS. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J Virol 70:3947–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yount B, Denison MR, Weiss SR, Baric RS. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol 76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freeman MC, Graham RL, Lu X, Peek CT, Denison MR. 2014. Coronavirus replicase-reporter fusions provide quantitative analysis of replication and replication complex formation. J Virol 88:5319–5327. doi: 10.1128/JVI.00021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol 81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith EC, Blanc H, Vignuzzi M, Denison MR. 2013. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog 9:e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y, Wu L, Shaw N, Gao Y, Wang J, Sun Y, Lou Z, Yan L, Zhang R, Rao Z. 2015. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc Natl Acad Sci U S A 112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, Lu X, Scherbakova S, Graham RL, Baric RS, Stockwell TB, Spiro DJ, Denison MR. 2010. Infidelity of SARS-CoV nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog 6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith EC, Case JB, Blanc H, Isakov O, Shomron N, Vignuzzi M, Denison MR. 2015. Mutations in coronavirus nonstructural protein 10 decrease virus replication fidelity. J Virol 89:6418–6426. doi: 10.1128/JVI.00110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth-Cross JK, Martínez-Sobrido L, Scott EP, García-Sastre A, Weiss SR. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J Virol 81:7189–7199. doi: 10.1128/JVI.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katze MG, He Y, Gale M. 2002. Viruses and interferon: a fight for supremacy. Nat Rev Immunol 2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 37.Gebauer FAT, Hentze MW. 2004. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5:827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Fontaine-Rodriguez EC, Whelan SPJ. 2005. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J Virol 79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diamond MS, Farzan M. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]