ABSTRACT
Here, we discuss the overall architecture of the RNA polymerase I (Pol I) and III (Pol III) core enzymes and their associated general transcription factors in the context of models of the Pol I and Pol III pre-initiation complexes, thereby highlighting potential functional adaptations of the Pol I and Pol III enzymes to their respective transcription tasks. Several new insights demonstrate the great degree of specialization of each of the eukaryotic RNA polymerases that is only beginning to be revealed as the structural and functional characterization of all eukaryotic RNA polymerases and their pre-initiation complexes progresses.
KEYWORDS: pre-initiation complex, RNA polymerase I, RNA polymerase II, TFIIB, TFIIE, TFIIF, transcription
Introduction
Transcription is the first step of gene expression in which RNA is synthesized from a DNA template by a macromolecular machine called RNA polymerase (RNAP). In eukaryotes, transcription is carried out by three RNAPs. RNA polymerase I (Pol I) contains 14 subunits and solely transcribes the rRNA precursor gene that in yeast is consecutively cleaved into 25S, 18S, and 5.8S rRNA.1 S. cerevisiae Pol I only transcribes from one promoter that contains two regions to which upstream activity factor (UAF), TBP, and core factor (CF) bind and subsequently recruit Pol I. RNA polymerase III (Pol III) contains 17 subunits and produces all tRNAs, spliceosomal U6 snRNA, ribosomal 5S rRNA, but also other small, structured RNAs, generally not exceeding 400 nucleotides.2,3 Pol III uses three different promoter types. First, the 5S rRNA type I promoter that recruits TFIIIA, TFIIIB, and TFIIIC, second, the tRNA type II promoter that only recruits TFIIIC and TFIIIB, and third, the U6 snRNA type III promoter which in S. cerevisiae resembles the type II promoter. RNA polymerase II (Pol II) consists of 12 subunits and transcribes all mRNAs and most regulatory RNAs. Pol II is recruited to a large variety of different promoters that (like Pol I and Pol III) require multiple general transcription factors and chromatin modifying complexes, but in addition also co-activators such as the mediator complex that mediate the interaction with hundreds of activator proteins bound to specific enhancer elements. Together all these factors allow the precise regulation of Pol II's diverse transcriptome.
Despite this diversity, key steps in initiation follow a conserved pattern. During initiation of transcription, all RNAPs are recruited to a target gene with the help of general transcription factors that sequentially assemble on the promoter and form the pre-initiation complex (PIC). In a consecutive step, the DNA double strand in the closed complex is melted and the transcription bubble is formed in the open complex.4 Formation of the PIC is a key step during transcription that is tightly regulated and utilizes conserved mechanisms, especially during the recruitment and assembly of the core RNAP–PIC that comprises RNAP, TBP, TFIIA and TFIIB-, TFIIE- and TFIIF-like factors.5 In contrast to Pol I and Pol III, the transition from the closed to the open complex in Pol II also generally requires the additional general factor TFIIH, which harbors ATPase and helicase activities. The recent molecular structures of Pol I6,7 and Pol III8 contribute to the better understanding of general RNAP architecture and PIC assembly. At the same time they also illustrate functional adaptations as TFIIF- and TFIIE-like factors are incorporated as stable subcomplexes in the Pol I and Pol III enzymes in contrast to Pol II, where these factors are only recruited during PIC assembly.
Flexible arms of TFIIF- and TFIIE-like subcomplexes mark their specialization
Molecular structures of all three eukaryotic RNAPs are now available, which allows a broader view on the topology and function of their TFIIF- and TFIIE-like subcomplexes. Although these subcomplexes share a number of related domains, Pol I-, Pol II-, and Pol III-specific loops and extensions reach out towards the DNA, the DNA-binding cleft, the stalk and other subunits, thereby mediating their specific functionalities.6,9,10
In Pol II, TFIIF, and TFIIE subcomplexes are transiently recruited and form essential components of the Pol II–PIC,5 where they stabilize the DNA, directly recruit Pol II and facilitate formation of the open complex.11,12 TFIIE comprises the two subunits TFIIEα and TFIIEβ that span over the DNA binding cleft toward the Pol II protrusion and together contain three winged helix (WH) domains. TFIIFα/β also forms a heterodimer with its dimerization domains located on the lobe of the Pol II enzyme. Despite this peripheral position, several flexible extensions reach toward the cleft and the DNA duplex in the Pol II-PIC.10,12 TFIIFα and TFIIFβ both contain WH domains at their C-terminal ends which are important for transcription initiation and the recruitment of other Pol II-specific factors.10,12,13
In Pol I, the subcomplex A49/A34.5 is the structural homolog of TFIIF. Like TFIIF it associates to the lobe of Pol I via its dimerization domains, but also carries out related functions as subcomplex A49/A34.5 was reported to assist in Pol I initiation and elongation.14,15 Subunit A49 contains a tandem WH (tWH) domain at its C-terminus reminiscent of the two WH domains present in TFIIEβ (Fig. 1a).6 However, the TFIIF-like subcomplex A49/A34.5 contains additional features specific to the Pol I system. An extended C-terminal arm of A34.5 travels along the periphery of Pol I toward a small cavity where it anchors the subcomplex to three core subunits.6,7 The increased binding surface between this subcomplex and the Pol I core enzyme increases the stability of their interaction and might enhance processivity of Pol I as a functional adaptation toward the efficient transcription of long ribosomal precursor transcripts. In addition, the fusion of the tWH domain to A49 could be an evolutionary adaptation to the requirement of a single promoter in Pol I that would benefit from a more streamlined and specialized (but less regulated) recruitment machinery.
Figure 1.
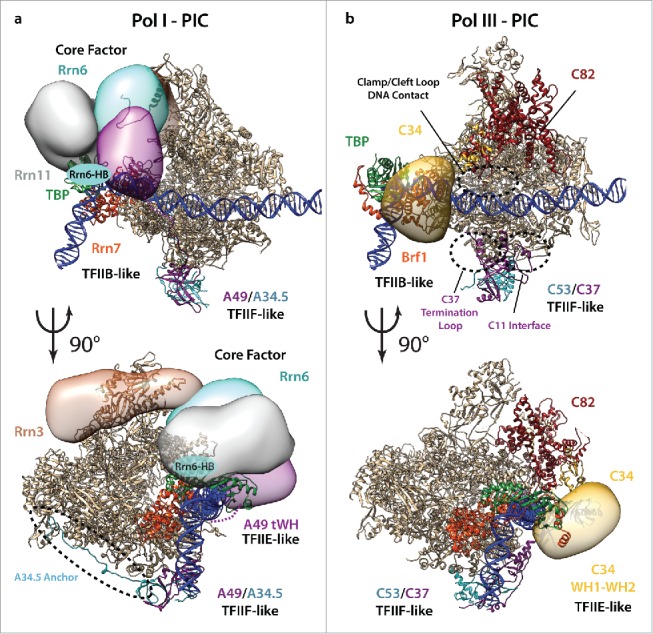
PIC—models for Pol I and Pol III. (a) Model of a Pol I–PIC depicting the TFIIB-, TFIIF-, and TFIIE-like factors together with core factor in a closed PIC. Pol I specific proteins except TBP are shown in color and adaptations are marked with black-dotted circles. The Pol I crystal structure (4c3i), the Pol II-TFIIB-TFIIF initial transcribing structure (4v1n), the Rrn3 crystal structure (3tj1) and the Pol II –PIC map (EMDB 3115) were used to model proteins and the DNA, and positioning of the CF proteins and Rrn3 was additionally aided by available crosslinking data24,32 and the described A43-Rrn3 and Rrn3-Rrn6 interactions.33 (b) Model of a Pol III—PIC highlighting Pol III specific TFIIB-, TFIIF-, and TFIIE-like factors. C82 is shown in red. Unique adaptations are depicted with black circles, the tentative position of C34 WH domains 1 and 2 is shown as yellow circles. The Pol III structure (5fja), the Brf2-TBP structure (4roc) and the Pol II –PIC map (EMDB 3115) were used to generate the model. Subunits and domains that could be positioned only approximately (Rrn3, 6, 11, C34-WH1 and WH2, and A49-tWH) are depicted schematically as low-resolution localization densities.
In Pol III, the TFIIE-, and TFIIF-like subcomplexes C82/C34/C31 and C53/C37 were shown to function in transcription initiation and termination9,16–18 and are similarly positioned on the RNAP core compared to Pol II and Pol I.19,20 Recent studies further support the view that C34 and TFIIE are structurally related,5,8,11 in agreement with their partially equivalent roles in transcription initiation during promoter opening and transition to the open complex.5,16 In contrast, the Pol III subunit C82 that positions four WH domains on the clamp domain of Pol III's largest subunit C160 does not contain direct structural counterparts in Pol I and Pol II. Indeed, in the elongating Pol III complex two WH domains of C82 reach toward downstream DNA, and the cleft loop from WH domain 2 protrudes through a cavity of the clamp, all of which are Pol III unique features. In the Pol III–PIC model (Fig. 1b), this cleft loop is contacting the closed DNA strand, and together with a small helix from the extended clamp head forms a positive patch.8 This patch could shift away from the DNA duplex by a conformational switch of the clamp head to the open clamp conformation. This feature could induce promoter melting during open complex formation and would constitute a Pol III-specific capability.
Strikingly, the recent structure of Pol III shows two remarkable adaptations of the C53/C37 subcomplex,8 the first being an extended contact surface of subunit C37 with C11 (the subunit that confers intrinsic RNA cleavage activity in Pol III), the second an extended C37 C-terminus that folds back to the dimerization domain over the Pol III lobe. This allows the positioning of residues required for transcription termination9,21 in close proximity to the putative path of the non-template DNA strand8 (Fig. 1b). In Pol III, transcription termination is a unique feature that unlike in other RNAPs only requires a terminator sequence of 5–7 thymidines in the non-template DNA strand.18 Both described features could mark adaptations toward the Pol III-specific transcription termination mechanism and indeed presumably function in a concerted manner.18,22 In summary, despite conserved positions and function of TFIIE- and TFIIF-like subcomplexes in the eukaryotic RNAPs, flexible N- and C-terminal extensions (some of which have not yet been structurally characterized) confer specialization and adaptation to their specific tasks in all three RNAPs.
C-terminal specialization in TFIIB-like factors in Pol I and Pol III modulate promoter-specific recruitment
All three eukaryotic RNAPs utilize TFIIB-like factors that together with TBP form essential components in assembling RNAP-PICs. TBP interacts with TFIIB in Pol II, with the TFIIB-like subunit Rrn7 but also with subunits Rrn6 and Rrn11 of the CF complex in Pol I and with TFIIIB subunit Brf1 in Pol III,23,24 thereby using an exceptional feature of TBP in PICs of all three eukaryotic RNAPs. TBP associates to TA-rich sequences in the upstream DNA (the TATA-box) and by doing so induces a ~90ᵒ bend in the DNA,25 which together with the TFIIB-like proteins sets the stage for RNAP recruitment.
Pol II-specific TFIIB contains a zinc ribbon domain in its very N-terminus followed by two short functional motifs, the B-linker and B-reader, and two cyclin domains.5,26 TFIIB is conserved among different species and recruits Pol II through its zinc ribbon domain and the first cyclin domain. Next, the B-reader motif contributes to transcription start site (TSS) selection and with the help of the B-linker, promoter melting is achieved.26,27 The two cyclin domains of TFIIB mediate the interaction with TBP throughout this process.
The two TFIIB-like paralogues in Pol I and Pol III, Rrn7 and Brf1, share a conserved N-terminal domain architecture with TFIIB.28 In fact, the zinc ribbon domains are interchangeable between Rrn7 and Brf1,28 and the B-linker is interchangeable among all three TFIIB-like proteins although its length varies between them.26,28 However, in contrast to TFIIB, both Rrn7 and Brf1 contain C-terminal extensions following the cyclin domains that mediate additional functionalities as discussed below.
In Pol I, the C-terminal extension of Rrn7 folds into several helices that interact with Pol I, TBP, and two additional components of the CF complex, Rrn6 and Rrn11.24 Therefore, the C-terminal extension of Rrn7 might serve as a central linker for the assembly and positioning of the Pol I PIC on the TSS. TAF1B, the human ortholog of Rrn7, contains a C-terminal domain with very weak sequence similarity to Rrn7, indicating independent interactions with transcription factors in yeast and human. Nevertheless, the N-terminal domain of TAF1B can replace the N-terminal domain of Rrn7, whereas a full TAF1B replacement led to cell lethality in Δrrn7 cells.29
In Pol III, the TFIIIB interaction between Brf1 and TBP is mediated by the cyclin folds and also by the extended C-terminus of Brf1, which additionally interacts with Bdp1 and Pol III.30 In the human system, additional functions of Pol III-specific TFIIB-like subcomplexes have evolved. Brf2, which is a vertebrate Brf1 ortholog, contains a C-terminal redox sensor that mediates an oxidative stress response thereby controlling the transcription of Brf2-dependent genes.31
In summary, despite the conservation of TFIIB-like factors in Pol I and Pol III, both Rrn7 and Brf1 show specific adaptations toward their respective RNAP and the associated GTFs. While in the Pol II system TFIIB seems to be mainly involved in TSS recognition and open complex formation, Rrn7 and Brf1 acquired additional functions in the regulation and recruitment of accompanying GTFs.
PIC models of Pol I and Pol III show potential promoter opening mechanisms
Structural studies on PICs have mainly been conducted on the Pol II system and although little is known about the PICs of Pol I and III, combining topological restraints and functional data have given good starting points for preliminary models of the Pol I and Pol III PICs in the past.4,5,19,24 In all structures and models of a closed PIC of Pol I, Pol II, and Pol III, the position of RNAP with respect to promoter DNA is conserved, flanked by upstream TFIIB (Pol II), TFIIIB component TBP/Brf1 (Pol III) or CF components TBP/Rrn7 (Pol I). However, increasing structural insight on Pol I, Pol II, and Pol III and associated GTFs allows expanding the PIC models of Pol I and Pol III (Fig. 1).
Pol I contains a unique transcription factor, Rrn3, which acts as a regulatory factor.32,33 Additionally, Rrn3 is needed to recruit Pol I and to establish the correct interactions between Pol I and CF.24 Interestingly, other studies have shown that CF alone is sufficient for recruitment of Pol I to the promoter in the absence of Rrn3, yet that this PIC is non-functional.28 The exact position of Rrn3 on the Pol I enzyme is still unclear and its role in the PIC is poorly understood. Nevertheless, the approximate position of Rrn3 in vicinity to the stalk could orchestrate the CF function in transcription initiation, thus stabilizing the PIC and ensuring reliability of Pol I-specific promoter recognition.
Interestingly, two CF subunits show structural similarity to TFIIIC. First, Rrn11 contains several tetra-trico peptide repeats (TPRs) similar to the τ131 subunit of TFIIIC, a motif often utilized as docking platform for additional factors. Second, CF subunit Rrn6 and TFIIIC subunit τ6034 both contain β-propellers as central cores. Additionally, the very C-terminus of Rrn6 contains many positively charged residues, which could interact with the promoter DNA in the vicinity of subunit A49 and the C-terminus of Rrn7. Interactions involving subunits Rrn3, Rrn6, and Rrn7 could promote the ATP-independent DNA melting in Pol I and together with the tWH domain of A49 stabilize open complex formation.
The Pol III–PIC extends to downstream TFIIIC, which was shown to interact with Pol III and TFIIIB.9,35 The displayed model shows much free space opposite of Pol III on the DNA (Fig. 1b) and possibly the TFIIIC subcomplex τA formed by subunits τ55, τ95, and τ131 could be located here. Notably, τA contacts the A-box, a 9-nucleotide DNA motif found close to the TSS.36 Furthermore, the two subunits τ55 and τ95 form a triple-β-barrel structure as also observed in TFIIF and similar to TFIIFβ, the C-terminus of τ95 contains a DNA-binding WH domain. Finally, the third τA subunit τ131 interacts with Bdp1 and TBP-Brf1,37 but also with Pol III subunits C53 and ABC10α,9,35 which positions τ131 in the close vicinity of TFIIIB and Pol III.
Summary
The PICs of Pol I and Pol III show several unique features that help them to fulfill their respective tasks. The stable incorporation of functional TFIIE- and TFIIF-like subcomplexes as additional subunits into Pol I and Pol III serves their specific transcriptional tasks by enabling increased processivity in Pol I and factor-reduced rapid transcription initiation and termination in Pol III. Therefore, it will be interesting to discover if the tWH domain of Pol I subunit A49 indeed functions as a TFIIE-like factor in Pol I. In Pol I, Rrn3 is essential for functional transcription and could stabilize open complex formation with a closed clamp. In Pol III, subunit C82 together with the extended clamp head might function in promoter melting induced by a transition between the closed clamp and open clamp state. Despite the conserved TFIIB-like architecture in the N-terminal part of Rrn7 in Pol I and Brf1 in Pol III, both proteins harbor additional functionalities in their C-terminal extensions. It will be interesting to reveal in greater detail the diverse interaction networks of Rrn7 and Brf1 and how they serve their distinct transcription tasks. In conclusion, integrated structural and functional analysis of all three RNAPs and their associated GTFs now allows studying the specialization of all three eukaryotic Pols and their PICs in greater detail giving insight into their specific adaptions in accordance with their distinct transcriptional activities.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
N.A.H. acknowledges support by the EMBL International PhD program.
Funding
Y.S., L.T., and C.W.M. acknowledge support by the ERC Advanced Grant (ERC-2013-AdG340964-POL1PIC) and J.K. by the EMBL Interdisciplinary Postdoc Program (EIPOD) under Marie Curie COFUND actions (PCOFUND-GA-2008-229597).
References
- [1].Russell J, Zomerdijk JC. The RNA polymerase I transcription machinery. Biochem Soc Symp 2006; 73:203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev 2002; 16:2593–2620. [DOI] [PubMed] [Google Scholar]
- [3].Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet 2007; 23:614–622. [DOI] [PubMed] [Google Scholar]
- [4].Sainsbury S, Bernecky C, Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol 2015; 16:129–143. [DOI] [PubMed] [Google Scholar]
- [5].Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell 2012; 45:439–446. [DOI] [PubMed] [Google Scholar]
- [6].Fernandez-Tornero C, Moreno-Morcillo M, Rashid UJ, Taylor NM, Ruiz FM, Gruene T, Legrand P, Steuerwald U, Müller CW. Crystal structure of the 14-subunit RNA polymerase I. Nature 2013; 502:644–649. [DOI] [PubMed] [Google Scholar]
- [7].Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. RNA polymerase I structure and transcription regulation. Nature 2013; 502:650–655. [DOI] [PubMed] [Google Scholar]
- [8].Hoffmann NA, Jakobi AJ, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen WJ, Sachse C, Müller CW. Molecular structures of unbound and transcribing RNA polymerase III. Nature 2015; 528:231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu CC, Lin YC, Chen HT. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol Cell Biol 2011; 31:2715–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Eichner J, Chen HT, Warfield L, Hahn S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J 2010; 29:706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature 2013; 495:481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Murakami K, Tsai KL, Kalisman N, Bushnell DA, Asturias FJ, Kornberg RD. Structure of an RNA polymerase II preinitiation complex. Proc Natl Acad Sci U S A 2015; 112:13543–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kilpatrick AM, Koharudin LM, Calero GA, Gronenborn AM. Structural and binding studies of the C-terminal domains of yeast TFIIF subunits Tfg1 and Tfg2. Proteins 2012; 80:519–529. [DOI] [PubMed] [Google Scholar]
- [14].Beckouet F, Labarre-Mariotte S, Albert B, Imazawa Y, Werner M, Gadal O, Nogi Y, Thuriaux P. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol Cell Biol 2008; 28:1596–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Albert B, Leger-Silvestre I, Normand C, Ostermaier MK, Perez-Fernandez J, Panov KI, Zomerdijk JC, Schultz P, Gadal O. RNA polymerase I-specific subunits promote polymerase clustering to enhance the rRNA gene transcription cycle. J Cell Biol 2011; 192:277–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Brun I, Sentenac A, Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J 1997; 16:5730–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kassavetis GA, Prakash P, Shim E. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J Biol Chem 2010; 285:2695–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Arimbasseri AG, Maraia RJ. Mechanism of Transcription Termination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element. Mol Cell 2015; 58:1124–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fernandez-Tornero C, Bottcher B, Rashid UJ, Muller CW. Analyzing RNA polymerase III by electron cryomicroscopy. RNA Biol 2011; 8:760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fernandez-Tornero C, Bottcher B, Rashid UJ, Steuerwald U, Florchinger B, Devos DP, Lindner D, Müller CW. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J 2010; 29:3762–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rijal K, Maraia RJ. RNA polymerase III mutants in TFIIFalpha-like C37 that cause terminator readthrough with no decrease in transcription output. Nucleic Acids Res 2013; 41:139–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Iben JR, Mazeika JK, Hasson S, Rijal K, Arimbasseri AG, Russo AN, Maraia RJ. Point mutations in the Rpb9-homologous domain of Rpc11 that impair transcription termination by RNA polymerase III. Nucleic Acids Res 2011; 39:6100–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bedwell GJ, Appling FD, Anderson SJ, Schneider DA. Efficient transcription by RNA polymerase I using recombinant core factor. Gene 2012; 492:94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Knutson BA, Luo J, Ranish J, Hahn S. Architecture of the Saccharomyces cerevisiae RNA polymerase I Core Factor complex. Nat Struct Mol Biol 2014; 21:810–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet 2007; 8:424–436. [DOI] [PubMed] [Google Scholar]
- [26].Sainsbury S, Niesser J, Cramer P. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 2013; 493:437–440. [DOI] [PubMed] [Google Scholar]
- [27].Chen BS, Hampsey M. Functional interaction between TFIIB and the Rpb2 subunit of RNA polymerase II: implications for the mechanism of transcription initiation. Mol Cell Biol 2004; 24:3983–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Knutson BA, Hahn S. TFIIB-related factors in RNA polymerase I transcription. Biochim Biophys Acta 2013; 1829:265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Knutson BA, Hahn S. Yeast Rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors. Science 2011; 333:1637–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Khoo B, Brophy B, Jackson SP. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev 1994; 8:2879–2890. [DOI] [PubMed] [Google Scholar]
- [31].Gouge J, Satia K, Guthertz N, Widya M, Thompson AJ, Cousin P, Dergai O, Hernandez N, Vannini A. Redox Signaling by the RNA Polymerase III TFIIB-Related Factor Brf2. Cell 2015; 163:1375–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Blattner C, Jennebach S, Herzog F, Mayer A, Cheung AC, Witte G, Lorenzen K, Hopfner KP, Heck AJ, Aebersold R, et al.. Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes Dev 2011; 25:2093–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J 2000; 19:5473–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mylona A, Fernandez-Tornero C, Legrand P, Haupt M, Sentenac A, Acker J, Müller CW. Structure of the tau60/Delta tau91 subcomplex of yeast transcription factor IIIC: insights into preinitiation complex assembly. Mol Cell 2006; 24:221–232. [DOI] [PubMed] [Google Scholar]
- [35].Dumay H, Rubbi L, Sentenac A, Marck C. Interaction between yeast RNA polymerase III and transcription factor TFIIIC via ABC10alpha and tau131 subunits. J Biol Chem 1999; 274:33462–33468. [DOI] [PubMed] [Google Scholar]
- [36].Taylor NM, Baudin F, von Scheven G, Muller CW. RNA polymerase III-specific general transcription factor IIIC contains a heterodimer resembling TFIIF Rap30/Rap74. Nucleic Acids Res 2013; 41:9183–9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Male G, von Appen A, Glatt S, Taylor NM, Cristovao M, Groetsch H, Beck M, Müller CW. Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly. Nat Commun 2015; 6:7387. [DOI] [PMC free article] [PubMed] [Google Scholar]


