ABSTRACT
Eukaryotic genomic DNA contains numerous high-affinity sites for transcription factors. Only a small fraction of these sites directly regulates target genes. Other high-affinity sites can serve as naturally present decoys that sequester transcription factors. Such natural decoys in genomic DNA may provide novel regulatory mechanisms for transcription factors.
Keywords: decoy, regulatory elements, sequestration, target search, transcription factors
Many transcription factors recognize particular DNA sequences and bind to them with high affinity, consequently playing crucial roles in gene regulation at a transcriptional level. The ChIP-on-chip and ChIP-seq methods have allowed for genome-wide studies on the binding sites of eukaryotic transcription factors in vivo.1 In particular, the Encyclopedia of DNA Elements (ENCODE) project provides extensive information about transcription factor association on genomic DNA.2 Such genome-wide studies showed that transcription factors bind to not only functional sites in the cis-regulatory elements of the genes but also many other apparently non-functional sites,3,4 though definitions of “functional” and “non-functional” are somewhat arbitrary and controversial.5,6 Transcription factors may also exhibit significantly strong affinities for quasi-specific sequences that are similar but not identical to target sequences. In fact, such non-cognate sites are known to play some roles in development.7 We refer to non-functional high-affinity sites (either identical or similar to target sequences) on genomic DNA as natural decoys (NDs). This term is defined as opposed to synthetic decoys, which are short DNA oligonucleotides designed to inhibit particular transcription factors for therapeutic purposes.8-11 In this Point-of-View article, we review how NDs influence transcription factors and gene regulation.
High abundance of natural decoys in genomic DNA
Eukaryotic transcription factors recognize a limited number of relatively short (typically <10 bp) DNA sequences.12,13 Simple probabilistic estimation implies that NDs are highly abundant in the nuclei. If a transcription factor recognizes an n-bp sequence of DNA, the transcription factor may also exhibit strong affinity for sequences with an m-bp match (m < n) to an n-bp recognition sequence. Given a pool of random sequences, the probability of finding an m-bp match in a window of n bps is given by 2(1/4)m(3/4)n-m nCm, where nCm represents the combinations, and a factor of 2 accounts for the complementary sequence match. Although somewhat simplistic, this calculation estimates that the total number of NDs (m ≥ 7) is ∼107 in 3 × 109 base pairs of the human genome for a transcription factor that recognizes 9-bp targets (n = 9). While ∼90% of the genome is inaccessible due to histones in chromatin,14 the estimated number of accessible NDs is still ∼106 sites on genomic DNA. In fact, biophysical studies on the inducible transcription factor Egr-1 (for which n = 9) suggested that genomic DNA contains ∼106–107 NDs, considerably impeding the Egr-1 target search on DNA.15,16 Compared to these numbers, functionally important target sites are far fewer (Fig. 1A). A typical transcription factor only targets ∼102–103 genes.14 Therefore, the total number of functional target sites for each transcription factor is approximately ∼102–104 sites in the genome, as cis-regulatory elements of each target gene typically involve only several binding sites of the same transcription factor. Thus, NDs overwhelmingly exceed target sites in number.
Figure 1.
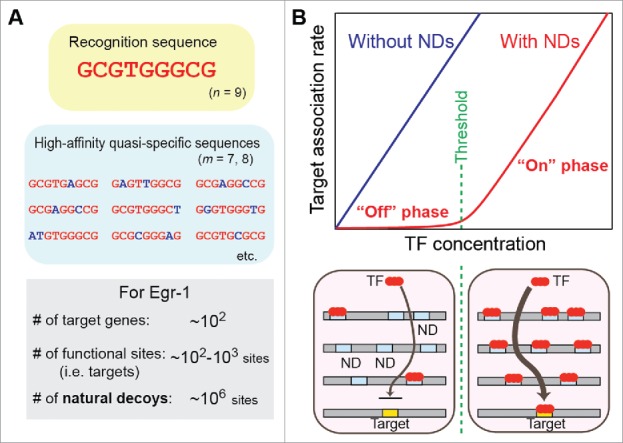
Natural decoys (NDs) for a transcription factor. (A) The recognition sequence and high-affinity quasi-specific sequences for Egr-1. (B) The influence of NDs on the target association kinetics of a transcription factor (TF). NDs preclude TFs from their targets. When all NDs are occupied by proteins, target association is drastically increased because the inhibitory effects of the NDs are eliminated. This phenomenon creates an on-off switch-like response.16,20
Functional sequestration by natural decoys
Because of their abundance, NDs should substantially influence transcription factors in vivo. Assuming that a nucleus is a sphere with a diameter of ∼6 μm,14 a quantity of ∼106 NDs corresponds to a concentration of ∼10−4 M in the nucleus. Even if only 1–10% of NDs are accessible, their concentration is as high as ∼10−5–10−6 M, which is far greater than the typical dissociation constants (10−10–10−7 M) for specific or quasi-specific DNA complexes of transcription factors. Consequently, binding to NDs would effectively sequester transcription factors and preclude them from binding to functional target sites. A well-characterized example is the ND for the transcription factor CCAAT/enhancer-binding protein α (C/EBPα). Because many NDs for this protein exist in tandem repeats (typically, thousands of copies) of 171-bp α-satellite DNA in the centromere regions of mammalian chromosomes, the C/EBPα molecules are effectively sequestered in the centromeres.17,18 This sequestration reduces the transcriptional capability of C/EBPα. An altered specificity mutation of C/EBPα, which reduces binding to α-satellite DNA but permits binding to the functional target sites, causes an elevation in the binding of C/EBPα to a promoter and an increase in transcriptional output from the promoter.17 This phenomenon suggests that NDs play a role in gene regulation via the functional sequestration of the transcription factor.
Sequestration in NDs may also have a positive impact on transcription factors. Burger et al. conducted a theoretical study on the potential role of NDs as protectants for transcription factors.19 If their DNA-bound states are less susceptible to proteolysis, NDs may prolong the mean lifetimes of transcription factors, partially offsetting the negative consequences of functional sequestration in non-functional regions.
Switch-like response via natural decoys
When NDs sequester a transcription factor, the regulation of its target genes requires a higher concentration of the transcription factor. More importantly, the genes' response to a change in the concentration of the transcription factor becomes non-linear and more like a binary on/off switch. Lee and Maheshri demonstrated this non-linear response in budding yeasts by quantitatively analyzing the effect of decoys in tandem repeats on target gene expression.20 Kemme et al. also showed kinetic data on the impact of NDs on the target search kinetics for Egr-1.16 Until the binding to the decoys is saturated, target association is not significant because the transcription factors are trapped at the NDs before reaching the targets (Fig. 1B). The concentration at which the saturation occurs corresponds to the threshold for the “on” state of the switch. At this point, the inhibitory effects of NDs are eliminated. When the level of a transcription factor exceeds this threshold, the target association of the transcription factor is drastically enhanced.
If the threshold exists between normal (100%) and 50% concentrations of a transcription factor, the heterozygous (+/−) and homozygous (−/−) knockouts of this transcription factor should result in almost equal changes in expression levels of its target genes. In fact, for example, a study using Dmp1+/+, Dmp1+/−, and Dmp1−/− mice showed such results for the transcription factor Dmp1 (Dmtf1).21 The on–off switch-like response via NDs may also be relevant to a sharp spatial boundary of expression in response to the gradient of the transcription factor Bicoid, a morphogen in Drosophila development.7 In the switch model, as long as a transition between the “on” and “off” phases is involved, even a relatively moderate up or downregulation of a transcription factor may result in drastic changes in expression levels of its target genes. This might be relevant to some diseases.
Cross-talk between transcription factors via natural decoys
Gene regulation by a transcription factor would be greatly enhanced when its NDs are blocked by other proteins, which weakens the functional sequestration (Fig. 2). Because most NDs are slightly different from the recognition sequence, a subset of NDs for a particular transcription factor may overlap with NDs for other transcription factors. Such overlaps would allow for indirect crosstalk between these transcription factors via NDs. For example, the sequestration of C/EBPα in the centromeres is reduced when another transcription factor, Pit-1, is co-expressed, causing an increase in the binding of C/EBPα to its functionally important sites to activate its target genes.17 This activation is probably because the recognition sequences of Pit-1 are similar but not identical to those of C/EBPα, which allows Pit-1 to selectively inhibit the NDs without blocking the functional target sites of C/EBPα. In this manner, Pit-1 enhances the function of C/EBPα through competitive interplay without involving direct protein–protein interactions.
Figure 2.
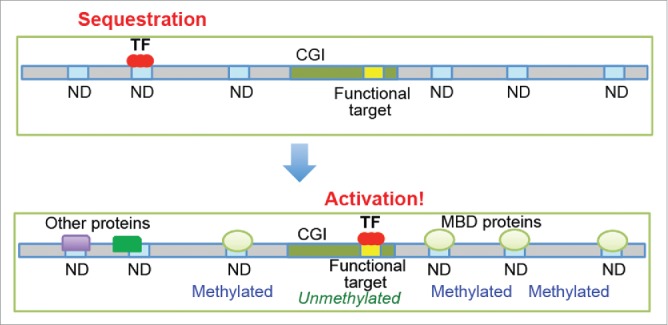
Potential mechanisms for the regulation of transcription factors via natural decoys (NDs). The accessibility of NDs may be modulated by other proteins, DNA methylation, and chromatin structures. When most NDs for a transcription factor become inaccessible, the transition from the “off” state to the “on” state (see Fig. 1B) may occur, which enhances the activity of the transcription factor.
Potential role of DNA methylation
DNA methylation may also control the sequestration of transcription factors in NDs. The methylation of CpG dinucleotides in DNA attracts methyl-CpG-binding proteins, such as MBD1, MBD2, and MeCP2.22 These proteins may block NDs for some transcription factors (e.g., ATF3, Egr-1, Elf1, E2F4, HIF1α, Nrf1, Sp1, and USF1)23 that recognize sequences that contain CpG dinucleotides. For example, the 9-bp recognition sequence of Egr-1 contains two CpG dinucleotides (see Fig. 1A), but their methylation does not affect the intrinsic affinity of Egr-1 for its target DNA.24 Many functionally important target sites of these transcription factors are located in CpG islands (CGIs). Because the total length of all CGIs is less than 1% of the genome size,22 the vast majority of NDs should be located outside CGIs. Interestingly, CpG methylation is rare (<10%) in CGIs, whereas the overall level of CpG methylation in genomic DNA is ∼85%.25 Therefore, the target sites in CGIs are likely unmethylated, whereas CpG dinucleotides within or near NDs are methylated. This distribution may enable methyl-CpG-binding proteins to selectively block NDs, assisting transcription activators in binding to functionally important sites within CGIs (Fig. 2). Further studies are required to examine this possibility.
Chromatin structure and natural decoys
The sequestration of transcription factors via NDs should also depend on chromatin structures. Janssen et al. studied drug-induced chromatin openings of DNA satellite V involving GAGAA repeats in Drosophila.26 They found that chromatin opening led to increased sequestration of the GAGA factor and reduced expression of its target genes.27 This finding suggests that the locations of NDs in the genome are important for functional sequestration of transcription factors. When NDs are located in accessible regions, they could sequester transcription factors to a greater degree. Because acetylation and methylation of histone tails are associated with the regulation of chromatin structure,28 the accessibility of NDs may, in principle, be assessed through the bioinformatics analysis of databases on nucleosome positions and histone modifications. Such investigations might allow for the prediction of the efficacy of NDs for each transcription factor.
Synthetic versus natural decoys for transcription factors
Abundant NDs may adversely impact the effectiveness of synthetic DNA decoys, which are short duplexes designed to inhibit particular transcription factors for therapeutic purposes. The synthetic decoy DNA strategy was first applied in 1995 by Morishita et al. to inhibit E2F, a transcription factor known to promote intimal hyperplasia after vascular injury.8 Since then, applications of this approach for various transcription factors have been examined. For example, STAT3, which is constitutively active in cancerous cells, has been targeted to abrogate the growth of head and neck cancer cells;9 Egr-1 to inhibit neointimal hyperplasia;10 and NF-κB to prevent myocardial infarction.11 Though this approach was successful in some applications, the synthetic decoys typically produced only modest inhibitory effects compared to other oligonucleotide-based gene suppression methods.29
This inadequate inhibition may be partially due to the presence of NDs in genomic DNA, although the short life span and poor delivery efficiency of the synthetic decoys may also be responsible. The synthetic decoys must compete with these NDs for transcription factors, and as mentioned above, the net concentration of the accessible NDs in the nuclei could be as high as ∼10−5–10−6 M. Because an uptake of the synthetic decoys at more than 10−6 M, in the nuclei of living cells, is very unlikely in practice, it is difficult for the synthetic decoys to competitively overcome the NDs unless the synthetic decoys exhibit a much higher affinity than the NDs. To achieve such conditions, the oxygen-to-sulfur substitution in the phosphate groups of DNA backbone that interact with the protein may be useful.30 Additionally, the inhibition of transcription factors by synthetic decoys should occur more effectively in the cytoplasm (i.e., pre-nuclear localization), due to the absence of NDs. Examples of transcription factors that exist in the cytoplasm before localizing to the nucleus include NF-κB and some nuclear hormone receptors. As a matter of fact, applications of the synthetic DNA decoy approach to NF-κB have been found to be successful. For the successful therapeutic application of synthetic decoys, it may be necessary to consider competition with NDs in genomic DNA.
Concluding remarks
NDs can be regarded as a novel class of regulatory DNA that controls the activities of sequence-specific transcription factors by precluding them from binding to their functional target sites on DNA. In the nuclei, NDs always exist in large quantities without the need for expression. However, NDs' inhibitory effects on transcription factors depend on various factors, such as chromatin structure, CpG methylation, and competition with other proteins. The functional activity of a transcription factor may be greatly enhanced through blocking of its NDs by other proteins. When most NDs become inaccessible, the transition of target association may resemble the behavior of an on–off switch. Thus, the sequestration of transcription factors in NDs could serve as a controllable mechanism of gene regulation. Since the ENCODE project gave a statement that 80% of the human genome is functional,2 the role of so-called “junk DNA” has been controversial.5,6 Each ND may be junk in terms of primary sequence and non-functional compared to the target sites, but ensemble of abundant NDs in the genome may have profound effects on functions of transcription factors. Currently, very little is known about NDs, and further characterizations, including the analysis of ND distributions in the genome, are necessary. Integration of cell biology, biochemistry, biophysics, and bioinformatics is required to delineate the roles of NDs in the transcriptional regulation of genes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the grants R01-GM107590 and R01-GM105931 from the National Institutes of Health (to J.I.).
References
- [1].Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science 2007; 316:1497-1502. [DOI] [PubMed] [Google Scholar]
- [2].The ENCODE project consortium An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fisher WW, Li JJ, Hammonds AS, Brown JB, Pfeiffer BD, Weiszmann R, MacArthur S, Thomas S, Stamatoyannopoulos JA, Eisen MB, et al.. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc Natl Acad Sci U S A 2012; 109:21330-21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al.. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 2008; 6:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Doolittle WF. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci U S A 2013; 110:5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, Elhaik E. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol 2013; 5:578-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Crocker J, Noon EP, Stern DL. The Soft Touch: Low-Affinity Transcription Factor Binding Sites in Development and Evolution. Curr Top Dev Biol 2016; 117:455-469. [DOI] [PubMed] [Google Scholar]
- [8].Morishita R, Gibbons GH, Horiuchi M, Ellison KE, Nakajima M, Zhang L, Kaneda Y, Ogihara T, Dzau VJ. A Gene-Therapy Strategy Using a Transcription Factor Decoy of the E2f Binding-Site Inhibits Smooth-Muscle Proliferation in-Vivo. Proc Natl Acad Sci U S A 1995; 92:5855-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, Robbins PD, Gadiparthi S, Burke NA, Watkins SF, et al.. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci U S A 2003; 100:4138-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ohtani K, Egashira K, Usui M, Ishibashi M, Hiasa KI, Zhao Q, Aoki M, Kaneda Y, Morishita R, Takeshita A. Inhibition of neointimal hyperplasia after balloon injury by cis-element “decoy” of early growth response gene-1 in hypercholesterolemic rabbits. Gene Ther 2004; 11:126-132. [DOI] [PubMed] [Google Scholar]
- [11].Morishita R, Sugimoto T, Aoki M, Kida I, Tomita N, Moriguchi A, Maeda K, Sawa Y, Kaneda Y, Higaki J, et al.. In vivo transfection of cis element ”decoy“ against nuclear factor-kappaB binding site prevents myocardial infarction. Nat Med 1997; 3:894-899. [DOI] [PubMed] [Google Scholar]
- [12].Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al.. Diversity and complexity in DNA recognition by transcription factors. Science 2009; 324:1720-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wunderlich Z, Mirny LA. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet 2009; 25:434-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alberts B, Johnson A, Lewis J, Morgan D, Martin R, Roberts K, Walter P. Molecular biology of the cell. New York: Garland Science; 2014 [Google Scholar]
- [15].Esadze A, Kemme CA, Kolomeisky AB, Iwahara J. Positive and negative impacts of nonspecific sites during target location by a sequence-specific DNA-binding protein: origin of the optimal search at physiological ionic strength. Nucleic Acids Res 2014; 42:7039-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kemme CA, Esadze A, Iwahara J. Influence of quasi-specific sites on kinetics of target DNA search by a sequence-specific DNA-binding protein. Biochemistry 2015; 54:6684-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu X, Wu B, Szary J, Kofoed EM, Schaufele F. Functional sequestration of transcription factor activity by repetitive DNA. J Biol Chem 2007; 282:20868-20876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tang QQ, Lane MD. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev 1999; 13:2231-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Burger A, Walczak AM, Wolynes PG. Abduction and asylum in the lives of transcription factors. Proc Natl Acad Sci U S A 2010; 107:4016-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee TH, Maheshri N. A regulatory role for repeated decoy transcription factor binding sites in target gene expression. Mol Syst Biol 2012; 8:576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mallakin A, Sugiyama T, Kai F, Taneja P, Kendig RD, Frazier DP, Maglic D, Matise LA, Willingham MC, Inoue K. The Arf-inducing transcription factor Dmp1 encodes a transcriptional activator of amphiregulin, thrombospondin-1, JunB and Egr1. Int J Cancer 2010; 126:1403-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 2006; 31:89-97. [DOI] [PubMed] [Google Scholar]
- [23].Blattler A, Farnham PJ. Cross-talk between site-specific transcription factors and DNA methylation states. J Biol Chem 2013; 288:34287-34294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zandarashvili L, White MA, Esadze A, Iwahara J. Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1. FEBS Lett 2015; 589:1748-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol 2013; 20:274-281. [DOI] [PubMed] [Google Scholar]
- [26].Janssen S, Durussel T, Laemmli UK. Chromatin opening of DNA satellites by targeted sequence-specific drugs. Mol Cell 2000; 6:999-1011. [DOI] [PubMed] [Google Scholar]
- [27].Janssen S, Cuvier O, Muller M, Laemmli UK. Specific gain- and loss-of-function phenotypes induced by satellite-specific DNA-binding drugs fed to Drosophila melanogaster. Mol Cell 2000; 6:1013-1024. [DOI] [PubMed] [Google Scholar]
- [28].Kouzarides T. Chromatin modifications and their function. Cell 2007; 128:693-705. [DOI] [PubMed] [Google Scholar]
- [29].Goodchild J. Therapeutic oligonucleotides. New York: Springer; 2011 [DOI] [PubMed] [Google Scholar]
- [30].Zandarashvili L, Nguyen D, Anderson KM, White MA, Gorenstein DG, Iwahara J. Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate. Biophys J 2015; 109:1026-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]


