Abstract
Background:
Gastric cancer (GC) is a highly aggressive cancer type associated with significant mortality owing to delayed diagnosis and non-specific symptoms observed in the early stages. Therefore, identification of novel specific GC serum biomarkers for screening purposes is an urgent clinical requirement.
Methods:
This study recruited a total of 432 serum samples from 296 GC patients split into the mining and testing sets. We aimed to screen for reliable protein biomarkers from matched serum samples based on mass spectrometry, followed by comparison with three representative conventional markers using receiver operating characteristic and survival curve analyses to ascertain their potential values as diagnostic and prognostic biomarkers for GC.
Results:
We identified an apoC-III fragment with confirmation in an independent test set from a second hospital. We found that the diagnostic ability of this fragment performed better than current standard GC diagnostic biomarkers both individually and in combination in distinguishing patients with GC from healthy individuals. Moreover, we found that this apoC-III protein fragment represents a more robust potential prognostic factor for GC than the three conventional markers.
Conclusions:
In view of these findings, we suggest that apoC-III protein fragment is a novel diagnostic and prognostic biomarker, a complement to conventional biomarkers in detecting GC.
Keywords: gastric adenocarcinoma, biomarker, serum
Gastric cancer (GC) is a highly aggressive cancer type associated with significant mortality, with high incidence worldwide (Jemal et al, 2011). The high mortality rates are attributable to delayed diagnosis and non-specific symptoms observed at the early GC stages (Roth, 2003; Krejs, 2010). The current prognosis for GC is poor, with <25% survival at 5 years after diagnosis (Ferlay et al, 2010; Siegel et al, 2013). Median survival for patients with advanced-stage GC remains at 8–10 months (Lorenzen and Lordick, 2011). Early detection has been shown to significantly improve the efficacy of cancer treatment, but diagnosis is often only possible after the appearance of the first clinical symptoms, which occurs too late for successful intervention. This is largely owing to the absence of specific and sensitive tests that would facilitate early screening and monitoring of cancerous states. Although serum pepsinogen (PG) tests, such as low PGI concentration and/or low PGI/II ratio, are suggestive screening tests in high-risk countries, such as Japan, they are good indicators of atrophic gastritis, rather than diagnostic markers of GC (Miki et al, 2003; Miki, 2006; Nasrollahzadeh et al, 2011).
Proteins secreted from tumour tissues have a greater likelihood of reaching the systemic circulation, and may therefore serve as a potential biomarkers for early detection (Kulasingam and Diamandis, 2008). Serum-based biomarkers are of considerable importance in early diagnosis of various diseases, including cancer. A proteome is the complete set of proteins found in a given cell type in any particular state. Proteomics is the systematic study of proteomes focusing on the large-scale identification and characterisation of proteins through measurement of protein expression and modifications in samples. Although serum is generated by coagulation, its proteome is vulnerable to the proteases involved in this cascade, as well as to those involved in the complement cascade, activated on blood clotting. Various pre-analytical parameters, such as the sampling device used, the clotting temperature and time, the storage duration and temperature, and the incubation temperature and handling, have proven the importance of uniform handling to exclude systematic pre-analytical inconsistency. Thus, false discovery can exert a distinct influence on the serum proteome, potentially leading to differences between results even when comparable patient populations and sample types are studied (Hsieh et al, 2006; Timms et al, 2007; Engwegen et al, 2008; Gast et al, 2009; Zeidan et al, 2009). New proteomic technologies are developing rapidly, which promote large-scale sample screening and facilitate identification of proteins associated with disease and treatment (Diamond et al, 2006; Smith et al, 2006). Mass spectrometry (MS), the most powerful proteomics tool, has evolved to a high-throughput level and improved to an extent that currently allows rapid and accurate analysis of several thousand proteins in a single study (Hirsch et al, 2004; Nilsson et al, 2010). Gel-free MS-based ‘shotgun' quantitative proteomics, a commonly used approach, is more sensitive and accurate relative to two-dimensional gel electrophoresis-based techniques (Tan et al, 2012). SDS–PAGE, followed by in-gel digestion, is a protein separation technique based on molecular weight (George et al, 2015). Surface-enhanced laser desorption/ionisation time-of-flight MS (SELDI-TOF-MS) based on the selective binding of proteins with different physicochemical properties on protein chip arrays has been successfully applied to uncover crucial molecular events in many tumour types, such as breast (Li et al, 2002; Rui et al, 2003; Laronga et al, 2003–2004; Clarke et al, 2005; Hu et al, 2005), liver (Kim et al, 2002; Feng et al, 2007; Zinkin et al, 2008), lung (Li et al, 2003; Xiao et al, 2003–2004; Au et al, 2008), prostate (Adam et al, 2002; Qu et al, 2002; Lin et al, 2007) and ovarian cancers (Gagnon et al, 2008). More recently, a new high-throughput workflow with matrix-assisted laser desorption/ionisation–time-of-flight/time-of-flight MS (MALDI-TOF/TOF MS) based on magnetic beads with different chemical chromatographic surfaces was established for the effective discovery and identification of serum peptides (Villanueva et al, 2004), whereby proteins selectively bound to magnetic beads are eluted and analysed with MALDI-TOF MS. Using this system, we identified novel biomarkers for papillary thyroid carcinoma, Wilms tumour and breast cancer (Fan et al, 2009, 2010; Wang et al, 2012), of which the utility has not been assessed in relation to the commonly used clinical markers.
Plasma-based tumour markers, including CEA (carcinoembryonic antigen), CA (carbohydrate antigen) 19-9 and CA125, are clinically applied to monitor GC (Sakamoto et al, 1987). These markers have shown utility in detecting disease recurrence after curative surgery (Pectasides et al, 1991), but display low sensitivity, lack specificity and often are not reproducible (Haglund et al, 1991; Kochi et al, 2000; Ychou et al, 2000; Oue et al, 2009). Although the combined use of these biomarkers has led to improved results to some extent, their value is still far from ideal (Aloe et al, 2003; Ucar et al, 2008). Several groups have employed quantitative proteomic approaches to identify novel secreted biomarkers of GC in the secretome and plasma (Chong et al, 2010; Yang et al, 2010; Loei et al, 2012; Marimuthu et al, 2013). Identification of novel and specific GC serum biomarkers for screening purposes is an urgent clinical requirement. The current study, with an independent test set from a second hospital, aimed to screen for reliable protein biomarkers from matched serum samples (preoperative, postoperative and relapsed) with SELDI-TOF-MS, followed by protein identification using MALDI-TOF/TOF MS, immunoassay-based protein confirmation and comparison with three representative conventional markers (CEA, CA19-9 and CA125) using receiver operating characteristic (ROC) and survival curve analyses to ascertain their potential value as diagnostic and prognostic biomarkers for GC.
Materials and methods
Patients and serum samples
Human serum samples were used based on Institutional Review Board-approved protocols, and written patient consent obtained where appropriate. A total of 432 serum samples from 296 individuals were included from the Division of General Surgery, the First Affiliated Hospital of Zhengzhou University, China, from June 2010 to March 2015 (mining set) and the Third Affiliated Hospital of Zhengzhou University, China, from May 2010 to January 2015 (testing set). The mining set consisted of 120 preoperative serum samples from 120 patients with GC, 106 corresponding postoperative serum samples (surgical treatment was either not conducted or abandoned due to financial constraints for 14 patients), 30 corresponding relapsed serum samples and 64 individuals, including 17 with gastritis and 47 healthy donors, as control. The testing set consisted of 60 preoperative serum samples from 60 patients with GC and 52 individuals, including 19 with gastritis and 33 healthy donors, as the control for the second study step. Meanwhile, fresh-frozen GC epithelium tissue samples from 75 patients were randomly obtained from the First and Third Affiliated Hospital of Zhengzhou University, whereas samples from 15 gastritis and 60 healthy donor fresh-frozen tissue were used as control. Patients were staged according to the World Health Organization, Lauren's classification and International Union against Cancer (UICC) TNM system. Details of clinicopathological data are summarised in Table 1. Patients subjected to curative intent resection were classified according to the following criteria: complete removal of primary gastric tumour, D2 dissection of regional lymph nodes, absence of macroscopic tumours remaining at the site of resection and absence of metastases in the liver, lungs or distant organs at the time of surgery. Postoperative follow-up visits were performed every 3 months for the first 2 years, followed by every 6 months up to 63 months or death. As of April 2015, complete periodic follow-up of all patients was recorded (Figure 2A).
Table 1. Demographics of GC patients enrolled in the study.
| Patient characteristics | Mining set | Testing set | χ2 or t value | P |
|---|---|---|---|---|
| No. of patients | 120 | 60 | — | — |
| Gender (male/female) | 77/43 | 36/24 | 0.297 | 0.586 |
| Age (year) | ||||
| ⩽50/>50 | 35/85 | 15/45 | 0.346 | 0.556 |
| Tumour size(cm) | ||||
| ⩽5/>5 | 33/87 | 18/42 | 0.123 | 0.726 |
| Stage | ||||
| I/II | 23/35 | 10/15 | 1.223 | 0.747 |
| III/IV | 53/9 | 28/7 | ||
| Depth of tumour invasion | ||||
| T1/T2 | 12/10 | 5/4 | 0.924 | 0.820 |
| T3/T4 | 33/65 | 14/37 | ||
| Lymph node metastasis | ||||
| N0/N1 | 26/23 | 11/10 | 0.575 | 0.902 |
| N2/N3 | 20/51 | 11/28 | ||
| Distant metastasis | ||||
| M0/M1 | 111/9 | 53/7 | 0.857 | 0.354 |
| Lauren classification | ||||
| Diffuse/intestinal type | 62/58 | 28/32 | 0.400 | 0.527 |
| Serosal invasion | ||||
| Negative/positive | 22/98 | 9/51 | 0.312 | 0.577 |
| Organ invasion | ||||
| Negative/positive | 88/32 | 42/18 | 0.222 | 0.638 |
| Vascular invasion | ||||
| Negative/positive | 36/84 | 20/40 | 0.207 | 0.649 |
| Peritoneal invasion | ||||
| Negative/positive | 103/17 | 49/11 | 0.529 | 0.467 |
| Tumour location | ||||
| Fundus | 12 | 5 | 1.271 | 0.736 |
| Body | 36 | 16 | ||
| Antrum | 62 | 31 | ||
| Diffuse | 10 | 8 | ||
| Median survival (months) | 32.5 | 37.8 | 1.065 | 0.302 |
Clinical marker(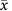 ±s) ±s) |
||||
| CEA (ng ml−1) | 5.500±1.843 | 6.014±1.662 | 1.819 | 0.071 |
| CA19-9 (U ml−1) | 50.003±20.886 | 45.161±18.808 | 1.514 | 0.132 |
| CA125 (U ml−1) | 42.002±18.100 | 46.454±19.320 | 1.521 | 0.130 |
Abbreviations: CA=carbohydrate antigen; CEA=carcinoembryonic antigen; GC=Gastric cancer.
For statistical analysis of CEA, CA19-9 and CA125, unpaired Student's t-test was used; for other variables, Pearson's χ2 test was used.
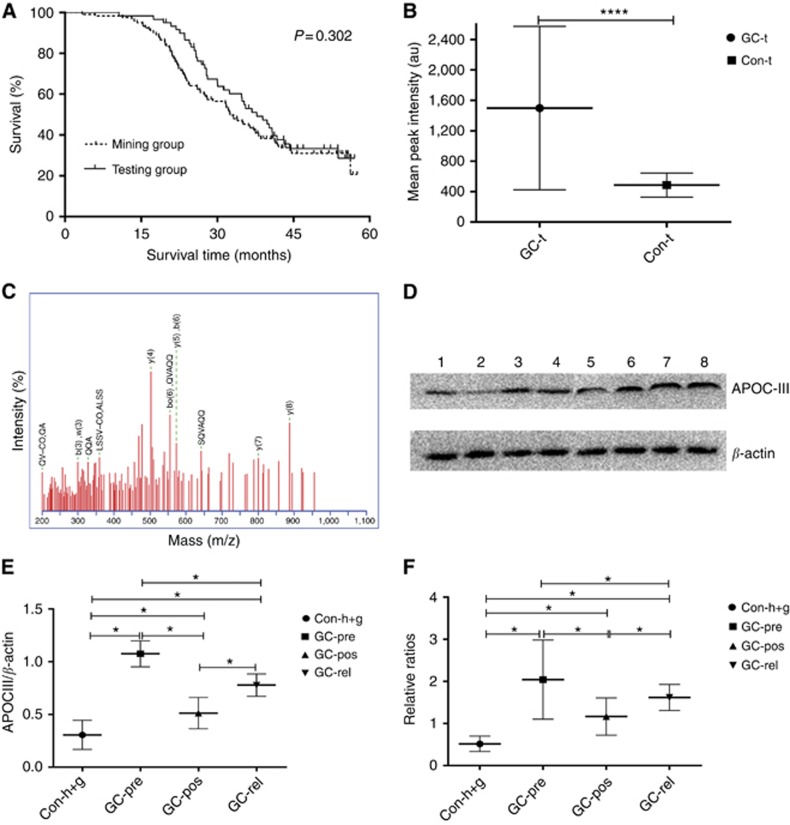
Figure 2.
Validation, identification and confirmation of the peak at 6449 Da. (A) Follow-up visits were performed for all included GC patients from the mining and blind testing sets (32.5 months vs 37.8 months; HR 1.225, 95% CI 0.838–1.771; χ2=1.065, P=0.302); (B) Validation of the peak at 6449 Da in the blind testing set. Intensity of the 6449 Da peak was higher in GC patients than that in the control (486.494±157.816 vs 1498.369±1075.437; t=6.719, P<0.0001), GC-t: GC patients in the blind testing set, Con-t: control samples in the blind testing set; (C) Representative MALDI-TOF/TOF-MS map of peptide segments obtained after enzymatic digestion of the protein and peptide segments with m/z of 6449 Da; (D) Representative WB analysis confirmed the SELDI-TOF-MS findings on the 6449 Da protein biomarker in 30 controls (15 with gastritis and 15 healthy donors), 30 matched GC-pre, GC-pos and GC-rel patients, lanes 1 and 2: healthy control, lanes 3 and 4: GC-pre patient, lane 5: GC-pos patient, lanes 6–8: GC-rel patient. β-Actin was used as a reference; (E) Grayscale scanning of WB bands, in which the ratio of grayscale value of apoC-III to β-actin was used as the analysed scale. Expression of the 6449 Da protein was significantly elevated in GC-pre sera, compared with control, decreased in GC-pos and upregulated again in GC-rel (0.306±0.138 vs 1.075±0.123 vs 0.512±0.148 vs 0.778±0.106; P<0.05); (F) Qualitative ELISA assay for apoC-III followed the trend of WB results, whereby optical density ratios of apoC-III following normalisation to the average of controls were calculated as the analysed scale (0.516±0.182 vs 2.043±0.940 vs 1.165±0.442 vs 1.518±0.309; P<0.05). *P<0.05, ****P⩽0.0001.
In addition, patients should have been clear of concomitant primary cancers and not received chemotherapy or radiotherapy before sample collection. All participants were pre- or postoperatively histologically verified with adenocarcinoma or gastritis via gastroscopy biopsy or histopathological examination by more than two senior pathologists. The benign lesion and healthy donor groups were age- and gender-matched with the GC group. In addition, serum levels of CEA, CA19-9 and CA125 were measured using commercial enzyme immunoassay kits with cut-off values set at 5 ng ml−1, 37 U ml−1 and 35 U ml−1, respectively. All overnight fasting serum samples from cancer and cancer-free individuals were collected in vacutainer tubes. Nutritional therapy of GC and control individuals at 72 h before blood sampling was based on lower insulin-secreting low-calorie, low-fat and high-fiber diet preparations. The duration of preoperative total parenteral nutrition preparatory phase ranged between 5 and 10 days (8 days on average). Enteral nutrition with elementary commercially available diet was begun 20 h after surgery, which was continued for 6 days; it was started at an 8 ml h−1 flow rate and increased gradually, with the volume doubled every 24 h, up to 100 ml h−1.During the initial 5 days after surgery, the patients were additionally supplemented parenterally via peripheral veins, similar to the preoperative period. As a routine pre-treatment examination at admission by our hospital laboratory, no significant differences were evident pertaining to the plasma lipid profile including high-density lipoprotein-cholesterol, low-density lipoprotein-cholesterol, cholesterol (CHO) and triglycerides (TAG) with cut-off values set at 0.91 mmol l−1, 3.61 mmol l−1, 5.20 mmol l−1 and 1.70 mmol l−1, respectively in GC vs control from both the mining and testing sets (P>0.05; Supplementary Table 1). Pre-analytical parameters, such as sampling device, clotting temperature and time, storage temperature and duration and, incubation temperature and handling, were controlled by following a standard protocol that was the same for cases and controls. Normal, benign gastric disease and GC serum samples were depleted of high-molecular weight proteins by acetonitrile precipitation. Sera were left at room temperature for 1 h, centrifuged at 3000 r.p.m. for 10 min, and stored at −80 °C. For immunological confirmation of apolipoprotein C-III (apoC-III) and β-actin, a similar procedure using rabbit anti-apoC-III and anti-β-actin antibodies (Cat. # sc-50377, Santa Cruz Biotechnology, Santa Cruz, CA, USA), one of the most authoritative antibody-producing suppliers at a dilution of 1 : 400 overnight at 4 °C, respectively, was employed. Details of SELDI-TOF-MS analysis, bioinformatic analysis, fractionation, identification and immunoassay-based confirmation of the candidate protein biomarkers are presented in online Supplementary File.
Statistical analysis
Data analysis was performed using the Zhejiang University Cancer Institute-ProteinChip Data Analysis System (ZUCIPDAS). The undecimated discrete wavelet transform method was applied to denoise the signals using Rice Wavelet Toolbox v. 2.4. Baseline correction was achieved by aligning spectra with a monotone local minimum curve, and mass calibration carried out by adjusting the intensity scale to the three peaks present in all the spectra. The peaks were filtered to maintain a signal-to-noise ratio (SNR) >3, whereby SNR was defined as the ratio of the height of the peak above baseline to the wavelet-defined noise. To distinguish between data from different groups, we used a non-linear support vector machine (SVM) classifier, originally developed by Vladimir Vapnik, with a radial-based function kernel, parameter Gamma of 0.6 and cost of constraint violation of 19. The leave-one-out crossing validation approach was applied to estimate the accuracy of this classifier. The capability of each peak in distinguishing data of different groups was estimated based on the P-value of Wilcoxon t-test.
Quantitative variables, presented as mean±s.d., were analysed with unpaired Student's t-test. Categorical variables were assessed using Pearson χ2 test. ROC curves were utilised to assess the diagnostic value of CEA, CA19-9, CA125 and the candidate protein biomarker. Survival curve analysis was performed using the Kaplan–Meier method, and significant levels assessed with the log-rank test. P-values were two-sided, and statistical significance set at P⩽0.05.
Results
Serum protein profiles and data processing
Serum samples from the mining set were analysed and compared using SELDI-TOF-MS with the WCX2 chip. All MS data were baseline-subtracted and normalised using total ion current, and peak clusters generated with Biomarker Wizard software. Wilcoxon rank sum tests to determine relative signal strength disclosed 15 differentially expressed proteins, including 9 upregulated and 6 downregulated protein peak intensities from samples of preoperative GC patients, compared with controls (Supplementary Table 2). Thirteen differentially expressed proteins, including 10 upregulated and 3 downregulated protein peak intensities, were observed for postoperative GC patients, compared with preoperative GC patients (Supplementary Table 3), and 15 differentially expressed proteins, including 9 upregulated and 6 downregulated protein peak intensities for relapsed GC patients, compared with postoperative GC patients (Supplementary Table 4). From the random combination of protein peaks with remarkable variation, the SVM screened out the model with maximum Youden index of the predicted value, leading to the identification of a marker positioned at 6449 Da with continuous dynamic presence in the control, preoperative, postoperative and relapsed GC patient sera. The 6449 Da protein was remarkably elevated in preoperative GC patient sera, compared with the control, but decreased in postoperative sera and upregulated again in relapsed GC samples (493.010±163.037 vs 1563.664±1080.217 vs 644.712±342.500 vs 1247.915±157.747; P<0.05; Figure 1A and B). In addition, the 6449 Da protein level progressively increased through clinical stages I, II, III and IV (649.724±620.964 vs 1506.884±1036.531 vs 2229.2995±2099.2703 vs 3081.431±2063.393; P<0.05; Figure 1C and D). Using leave-one-out cross-validation (LOOCV), the 6449 Da fragment was used to effectively distinguish GC from controls with an accuracy of 85.3% (157 out of 184), sensitivity of 85.0% (102 out of 120) and specificity of 85.9% (55 out of 64).
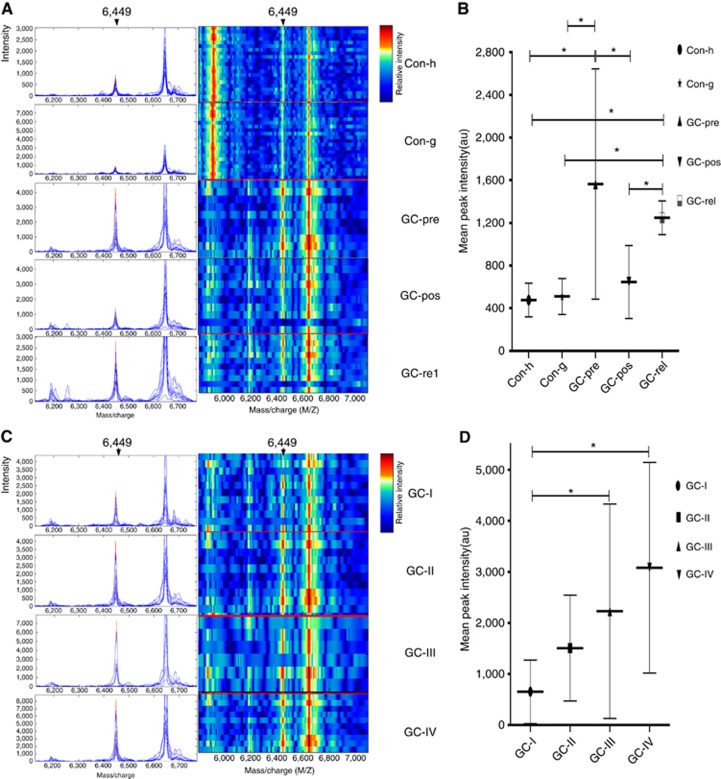
Figure 1.
Representative expression map of SELDI-TOF-MS analysis of sera from GC patients and healthy controls in the mining set. (A) Representative expression of the peak at 6449 Da (left panel, arrow) and gel views (right panel, arrow) from control (Con-h: healthy donors as control; Con-g: gastritis donors as control), preoperative GC (GC-pre), corresponding postoperative GC (GC-pos) and relapsed GC (GC-rel) patient sera; (B) The peak at 6449 Da was significantly elevated in preoperative GC patient sera, compared with the control, decreased in postoperative and upregulated again in relapsed GC samples, (493.010±163.037 vs 1563.664±1080.217 vs 644.712±342.500 vs 1247.915±157.747; P<0.05); (C) Typical expression of the 6449 Da peak (left panel, arrow) and gel views (right panel, arrow) at different stages of GC (GC-I: patient with stage I GC; GC-II: patient with stage II GC; GC-III: patient with stage III GC; GC-IV: patient with stage IV GC); (D) The level of the 6449 Da protein progressively increased with clinical stages I, II, III and IV (649.724±620.964 vs 1506.884±1036.531 vs 2229.2995±2099.2703 vs 3081.431±2063.393; P<0.05). *P<0.05.
Protein peak validation
The remaining 60 GC and 52 control serum samples (33 healthy donors and 19 with benign gastritis) were analysed as a blind testing set to validate the accuracy and validity of the 6449 Da protein marker identified from the mining set. The peak intensity of the 6449 Da protein was higher than that of control (486.494±157.816 vs 1498.369±1075.437; t=6.719, P<0.0001; Figure 2B). The 6449 Da marker distinguished GC samples from controls with an accuracy of 84.8% (95 out of 112), sensitivity of 86.7% (52 out of 60) and specificity of 82.7% (43 out of 52).
In-gel digestion and MALDI-TOF/TOF-MS identification of the candidate protein biomarker
Protein spots positioned at 6449 Da were excised from the gel with the Ettan Spot Picker, followed by digestion with trypsin and MALDI-TOF/TOF analysis (Figure 2C). The sequence of proteins and peptide segments with m/z of 6449 Da was E.MQPRVLALLASADASLLSFMYMKHATKTAKDVQVAQQAR.G (complete sequences are not listed owing to patent pending). Subsequent analysis using the MASCOT search programme and NCBI database led to identification of the peptide segment as apoC-III, with a matching rate of 59.3% and matching score of 104.9 points (Table 2).
Table 2. Peptide sequences obtained after enzymatic digestion of protein and peptide segments with m/z of 6449 Da. Peptide sequences identified using the NCBI database are in bold.
| m/z (Da) | Protein name | Peptides identified | Sequence |
|---|---|---|---|
| 6449 | APOC-III | R.VLLVVALLALLASAR.A | MQPRVLALLASADASLLSFMYMKHATKTAKDVQVAQQARGWVTDGFSSL |
| K.DALSSVQESQVAQQAR.G | |||
| R.ASEAEDASLLS FMQGYMK.H | |||
| K.HATKTAK.D | |||
| R.VLLVVALLALLASARASEAEDASLLSFMQGYMK.H |
Abbreviations: m/z=mass-to-charge ratio; NCBI=National Center for Biotechnology Information.
Confirmation of candidate protein biomarker expression using western blot (WB) and ELISA
As full-length human apoC-III is 10.9 kDa, the m/z 6449 Da biomarker peptide with total sequence coverage >59.3% represents the large fragment of apoC-III. Immunological analysis of apoC-III was performed using an available antibody purchased from authoritative antibody-producing supplier (Santa Cruz Biotechnology), specific for epitope corresponding to amino acids 25–99 mapping at the C-terminus of apoC-III of human origin covering >90% of the complete identified sequence. To further investigate the expression of apoC-III deduced from the results of SELDI-TOF-MS, WB was performed using serum specimens from 30 controls (15 with gastritis and 15 healthy donors), matched GC-pre, GC-pos and GC-rel patients (Figure 2D). Data were consistent with proteomic findings. The protein biomarker with β-actin used as the loading control was remarkably elevated in GC-pre sera, compared with control, but decreased in GC-pos and upregulated again in GC-rel (0.306±0.138 vs 1.075±0.123 vs 0.512±0.148 vs 0.778±0.106; P<0.05; Figure 2E). ELISA data on apoC-III levels in the same serum samples were consistent with WB findings (0.516±0.182 vs 2.043±0.940 vs 1.165±0.442 vs 1.518±0.309; P<0.05; Figure 2F), whereby optical density ratios of apoC-III following normalisation to corresponding control were calculated and used as the vertical scale. As apoC-III expression is confined to the liver and the intestine (and possibly adipose tissue), apoC-III expression in the gastric epithelium from 75 GC and 75 control individuals (15 with gastritis and 60 healthy donors) was confirmed via WB and ELISA, in which no significant differences were detected between GC and control individuals, respectively (WB: control, 0.175±0.027 vs. GC, 0.182±0.078; t=0.089, P=0.934; Supplementary Figure 1A and B and ELISA: control, 0.267±0.031 vs. GC, 0.368±0.196; t=1.789, P=0.076; Supplementary Figure 1C).
Diagnostic and prognostic value of the candidate protein biomarker, compared with CEA, CA19-9 and CA125
Relative intensities of the 6449 Da peak, together with CEA, CA19-9 and CA125 data in the testing set were used to determine diagnostic and prognostic value. Sensitivities were determined from the results of 60 patients with GC and specificities from 52 controls (19 with gastritis and 33 healthy donors). Taking the histologically verified results in patients as the golden standard, the diagnostic value of CEA was assessed by means of an ROC curve, the area under the curve value of which was calculated as 0.695 (95% CI: 0.597–0.793; P<0.0001; Figure 3A), compared with the control. ROC curves of CA19-9, CA125 and the 6449 Da peak were 0.610 (95% CI: 0.505–0.714; P=0.046; Figure 3A), 0.617 (95% CI: 0.513–0.721; P=0.033; Figure 3A) and 0.914 (95% CI: 0.870–0.957; P<0.0001; Figure 3A), respectively. ROC curves of the CEA, CA19-9 and CA125 combination and the CEA, CA19-9, CA125 and 6449 Da peak combination were 0.780 (95% CI: 0.696-0.863; P<0.0001; Figure 3B) and 0.959 (95% CI: 0.921–0.997; P<0.0001; Figure 3B), respectively. The cut-off level (1498.369 a.u.) for the 6449 Da peak was set at mean of data obtained in 60 patients with GC. CEA levels were <5 ng ml−1 in 27 cases, CA19-9 levels were <37 U ml−1 in 24 cases and CA125 levels were <35 U ml−1 in 24 cases. In 15 cases, CEA, CA19-9 and CA125 levels were not significantly elevated. Notably, however, the 6449 Da peak was above the cut-off level in 9 of the 15 cases. Thus, measurement of the peak may be used as a complement to CEA, CA19-9 and CA125 assessment in nine cases where the conventional tumour markers were not diagnostic. Sensitivity and specificity of combined CEA, CA19-9 and CA125 were 36.7% (22 out of 60) and 48.1% (25 out of 52), while a combination of CEA, CA19-9, CA125 and 6449 Da presented 88.3% (53 out of 60) sensitivity and 84.6% (44 out of 52) specificity, respectively.
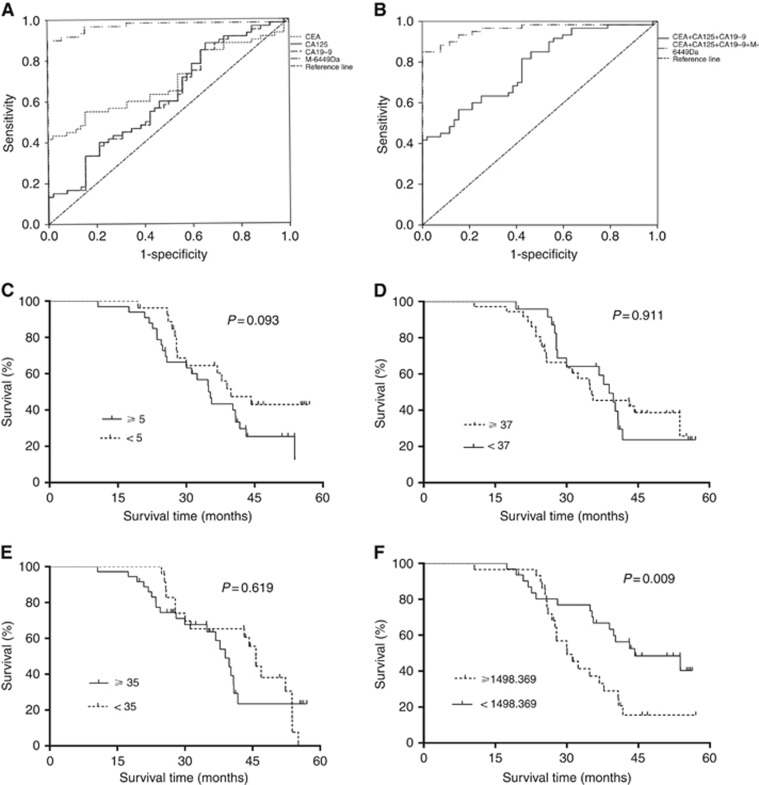
Figure 3.
Diagnostic and prognostic value of the candidate protein biomarker, compared with CEA, CA19-9 and CA125, based on ROC and Kaplan–Meier survival curve analyses. (A) Diagnostic values of CEA, CA19-9, CA125 and 6449 Da determined using ROC curves (area under the curve (AUC)=0.695, 95% CI: 0.597–0.793; P<0.0001 vs AUC=0.610, 95% CI: 0.505–0.714; P=0.046 vs AUC=0.617, 95% CI: 0.513–0.721; P=0.033 vs AUC=0.914, 95% CI: 0.870–0.957; P<0.0001) compared with controls; (B) Diagnostic value of the CEA, CA19-9 and CA125 combination and CEA, CA19-9, CA125 and 6449 Da peak combination, determined using ROC curves (AUC=0.780, 95% CI: 0.696–0.863; P<0.0001 vs AUC=0.959, 95% CI: 0.921–0.997; P<0.0001), compared with the controls; (C) Kaplan–Meier survival curve for CEA in GC patients from the testing set; (D) Kaplan–Meier survival curve for CA19-9 in GC patients from the testing set; (E) Kaplan–Meier survival curve for CA125 in GC patients from the testing set; (F) Kaplan–Meier survival curve between apoC-III-lower and -higher GC patients from the testing set, supporting upregulated 6449 Da peak intensity as a poor prognostic risk factor. M-6449Da: marker positioned at 6449 Da.
No significant differences in survival times were evident between GC patients with CEA levels greater than or equal to and those with CEA lower than cut-off values (35.3 months vs 39.8 months; HR 1.731, 95% CI 0.912–3.285; χ2=2.815, P=0.093; Figure 3C) in Kaplan–Meier survival analysis. Similar results were obtained for CA19-9 (35.3 months vs 39.0 months; HR 0.964, 95% CI 0.505–1.838; χ2=0.013, P=0. 911; Figure 3D) and CA125 (38.9 months vs 45.8 months; HR 1.169, 95% CI 0.625-2.232; χ2=0.247, P=0.619; Figure 3E). In contrast, survival times for GC patients with 6449 Da peak intensities greater than or equal to average relative intensity values were shorter than those of GC patients with peak intensities lower than the average relative intensity values (30.0 months vs 44.3 months; HR 2.233, 95% CI 1.263–4.679; χ2=6.680, P=0.009; Figure 3F), clearly indicating that higher 6449 Da peak intensity is an unfavourable prognostic factor.
Discussion
Patient blood samples are an ideal source of disease biomarkers owing to their ease of access. Several studies have identified potential candidates, but few have overcome validation and reproducibility issues to achieve clinical application (Poste, 2011). To our knowledge, this is the first study to characterise an apoC-III fragment as a potential diagnostic and prognostic biomarker based on MS across a broad spectrum of GC sera with an independent test set from a second hospital. We used protein chip MS to identify a unique serum protein positioned at 6449 Da with continuous dynamic presence in control, preoperative, postoperative and relapsed GC sera that effectively discriminates between sera from GC patients and healthy volunteers with high sensitivity and specificity in the mining set. The 6449 Da protein panel screened using a SVM was significantly upregulated in preoperative GC, decreased in postoperative sera and upregulated again in relapsed GC, compared with control, as validated in an independent testing set. In one previous study by Cohen et al (2011), the 9.4 kDa protein, identified as apoC-III was decreased in pre-operation stomach cancer sera as compared with control groups. The inconsistency with Cohen research team may be caused by socio-geographical difference and the instability of this protein, leading to further truncation during prolonged storage. Gast et al (2009), found that over long storage duration the intensity of some vulnerable proteins would decrease because of fragmentation, while the intensities of the fragments of these proteins would increase. Samples of both cases and controls analysed in the study by Cohen et al, were collected respectively from an unknown time on whereas samples of both case and control groups from the mining and testing sets in our study were all collected within the same 5-year window. Another, possibly more likely explanation for the discrepant result is difference in pre-analytical sample handling to which some proteins are more vulnerable. Consistent overnight fasting samples of both cases and controls from both mining and test sets analysed in our study were pre-analytically handled identically following a standard protocol. Samples were allowed to clot for 1 h before they were centrifuged for 10 min at 3000 rpm at room temperature. All blood samples with significantly distinctive fasting data (overnight fasting vs unknown fasting) from two commercial sources analysed in the study by Cohen et al, were allowed to clot for 30 min before they were centrifuged for a variable duration at inconstant centrifugation speed. We did not find a convincing explanation for this discrepant result, but it probably illustrates the above-mentioned susceptibility of apoC-III to external circumstances. For future studies, more effort should be put into the collection of blood samples of cases and controls with the use of more rigorous standardised and higher quality procedures. Remarkably, a increase of this protein was also found in two other GC sample-sets analysed by our group (publication in preparation). Although the specificity of the candidate protein biomarker using the method of LOOCV was somewhat lower in the testing than mining set, sensitivity was slightly higher in the testing set. In addition to the above dynamic changes in preoperative, postoperative and relapsed GC sera, the peak intensity of 6449 Da progressively increased with higher clinical stages, supporting the theory that excessive levels contribute to tumour progression. Using a combination of MS and immunological methods, the protein was identified as a fragment of apoC-III, a small 99 amino acid protein that forms a component of TAG-rich very-low-density lipoproteins and high-density lipoproteins.
Recent reports have indicated that apolipoprotein levels in blood can be used as potential biomarkers for different cancers. Apo-A1, a regulator of tumour growth and metastasis, has been shown to be involved in antiproliferative and proapoptotic activities via regulation of cancer cell differentiation (Zamanian-Daryoush et al, 2013; Kim et al, 2014). Hsu et al (2013) recently demonstrated a correlation of dyslipidemia-associated apo-A1 minor allele with unfavourable baseline characteristics in Taiwanese breast cancer patients, and the 10-year follow-up revealed poorest survival in patients carrying both minor alleles in the lymph node-negative group. ApoC-I was identified as a potential serum biomarker for colorectal cancer, hormone-refractory prostate cancer and liver fibrosis (Engwegen et al, 2006; Yamamoto-Ishikawa et al, 2009). Emerging roles of apoC-III synthesised in the liver, and to a minor degree, the small intestine, include directing the atherogenicity of high-density lipoprotein, intestinal dietary TAG trafficking and modulating insulin-secreting pancreatic beta-cell survival and apoptosis via activation of the mitogen-activated protein kinases p38 and extracellular signal regulated protein kinases 1 and 2 (Sol et al, 2009; Kohan, 2015), resulting in ephemeral hyperglycaemia which is difficult to detect abnormal glucose levels through traditional blood glucose monitoring means, a positive effect on GC progress via aquaporin 3 and risk factor for GC posed by Helicobacter pylori infection due to hypoinsulinemia (Ikeda et al, 2009; Zhou et al, 2015). There is still confusion, however, about the precise role of apoC-III in pancreatic beta-cell failure. Other reports indicate that apoC-III is a potential biomarker in pancreatic and breast cancer (Chen et al, 2007; McComb et al, 2007). However, these earlier reports employed MALDI-based screening and did not verify results obtained with immunoassay-based experiments or comparisons with representative conventional markers. Our WB and ELISA data confirmed that apoC-III is dynamically regulated in matched GC-pre, GC-pos and GC-rel patient sera, consistent with proteomic findings, suggesting that lipoprotein metabolism is dysregulated in GC. In addition, apoC-III expression in the gastric epithelium in GC vs control was confirmed via WB and ELISA. Further studies to determine whether apoC-III is secreted from GC cells and the molecular mechanisms underlying apoC-III-mediated GC progression are warranted. Moreover, an earlier study using matrix-assisted laser desorption/ionisation, time-of-flight and tandem MS reported that apoC-III could be suppressed by the serine–threonine kinase receptor–associated protein (STRAP), which promotes growth, and enhances tumourigenicity (Anumanthan et al, 2006).
Diagnosis based on measurement of a panel of biomarkers is more reliable than a single marker test owing to the multifactorial nature of cancer. Comparison of the diagnostic ability of the apoC-III fragment, determined based on ROC analysis, with the current standard GC diagnostic biomarkers (CEA, CA19-9 and CA125) revealed that the candidate protein biomarker is distinctly superior to all three standards, either individually or in combination, in distinguishing patients with GC from healthy individuals. Notably, the 6449 Da peak was elevated in 9 of 15 cases in which CEA, CA19-9 and CA125 were below cut-off levels, suggesting that the 6449 Da peak may be used as a complement to CEA, CA19-9 and CA125 in detecting GC, consistent with two previous reports using MS methodologies (Qian et al, 2005; Su et al, 2007). Similar to diagnosis results, Kaplan–Meier survival analysis disclosed that this 6449 Da protein fragment represents a new stronger potential prognostic factor for GC, compared with the three representative conventional markers, indicating that measurement of peak intensity positioned at 6449 Da in serum should improve estimation of postoperative survival chances for these GC patients. However, the identity of only a small proportion of the detected peaks has been confirmed, and the roles of many of the identified proteins in GC development are not yet known. Moreover, reliable validation of the potential GC biomarker in larger sample cohorts is necessary for translation to clinical application. The issue of whether a causal link exists between GC and abnormal lipid metabolism is yet to be established, and complicated by the fact that the detected peak may be associated with the inflammation state and not specific for GC.
Acknowledgments
We are grateful to Professor Lijun Wang from the School of Foreign Languages of Zhengzhou University for language editing and polishing of this paper. This work was supported by a grant (no.81172085) from the National Natural Science Foundation of China.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc).
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Adam BL, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, Semmes OJ, Schellhammer PF, Yasui Y, Feng Z, Wright GL Jr (2002) Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res 62(13): 3609–3614. [PubMed] [Google Scholar]
- Aloe S, D'Alessandro R, Spila A, Ferroni P, Basili S, Palmirotta R, Carlini M, Graziano F, Mancini R, Mariotti S, Cosimelli M, Roselli M, Guadagni F (2003) Prognostic value of serum and tumor tissue CA72-4 content in gastric cancer. Int J Biol Markers 18(1): 21–27. [DOI] [PubMed] [Google Scholar]
- Anumanthan G, Halder SK, Friedman DB, Datta PK (2006) Oncogenic serine-threonine kinase receptor-associated protein modulates the function of Ewing sarcoma protein through a novel mechanism. Cancer Res 66(22): 10824–10832. [DOI] [PubMed] [Google Scholar]
- Au JS, Cho WC, Yip TT, Law SC (2008) Proteomic approach to biomarker discovery in cancer tissue from lung adenocarcinoma among nonsmoking Chinese women in Hong Kong. Cancer Invest 26(2): 128–135. [DOI] [PubMed] [Google Scholar]
- Chen J, Anderson M, Misek DE, Simeone DM, Lubman DM (2007) Characterization of apolipoprotein and apolipoprotein precursors in pancreatic cancer serum samples via two-dimensional liquid chromatography and mass spectrometry. J Chromatogr A 1162(2): 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong PK, Lee H, Loh MC, Choong LY, Lin Q, So JB, Lim KH, Soo RA, Yong WP, Chan SP, Smoot DT, Ashktorab H, Yeoh KG, Lim YP (2010) Upregulation of plasma C9 protein in gastric cancer patients. Proteomics 10(18): 3210–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CH, Buckley JA, Fung ET (2005) SELDI-TOF-MS proteomics of breast cancer. Clin Chem Lab Med 43(12): 1314–1320. [DOI] [PubMed] [Google Scholar]
- Cohen M, Yossef R, Erez T, Kugel A, Welt M, Karpasas MM, Bones J, Rudd PM, Taieb J, Boissin H, Harats D, Noy K, Tekoah Y, Lichtenstein RG, Rubin E, Porgador A (2011) Serum apolipoproteins C-I and C-III are reduced in stomach cancer patients: results from MALDI-based peptidome and immuno-based clinical assays. PLoS One 6(1): e14540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond DL, Proll SC, Jacobs JM, Chan EY, Camp DG 2nd, Smith RD, Katze MG (2006) HepatoProteomics: applying proteomic technologies to the study of liver function and disease. Hepatology 44(2): 299–308. [DOI] [PubMed] [Google Scholar]
- Engwegen JY, Alberts M, Knol JC, Jimenez CR, Depla AC, Tuynman H, van Heukelem HA, Snel P, Smits ME, Cats A, Schellens JH, Beijnen JH (2008) Influence of variations in sample handling on SELDI-TOF MS serum protein profiles for colorectal cancer. Proteomics Clin Appl 2(6): 936–945. [DOI] [PubMed] [Google Scholar]
- Engwegen JY, Helgason HH, Cats A, Harris N, Bonfrer JM, Schellens JH, Beijnen JH (2006) Identification of serum proteins discriminating colorectal cancer patients and healthy controls using surface-enhanced laser desorption ionisation-time of flight mass spectrometry. World J Gastroenterol 12(10): 1536–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Shi L, Liu Q, Dong R, Zhang Q, Yang S, Fan Y, Yang H, Wu P, Yu J, Zheng S, Yang F, Wang J (2009) Discovery and identification of potential biomarkers of papillary thyroid carcinoma. Mol Cancer 8: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Wang J, Yang Y, Liu Q, Fan Y, Yu J, Zheng S, Li M, Wang J (2010) Detection and identification of potential biomarkers of breast cancer. J Cancer Res Clin Oncol 136(8): 1243–1254. [DOI] [PubMed] [Google Scholar]
- Feng Y, Tian ZM, Wan MX, Zheng ZB (2007) Protein profile of human hepatocarcinoma cell line SMMC-7721: identification and functional analysis. World J Gastroenterol 13(18): 2608–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127(12): 2893–2917. [DOI] [PubMed] [Google Scholar]
- Gagnon A, Kim JH, Schorge JO, Ye B, Liu B, Hasselblatt K, Welch WR, Bandera CA, Mok SC (2008) Use of a combination of approaches to identify and validate relevant tumor associated antigens and their corresponding autoantibodies in ovarian cancer patients. Clin Cancer Res 14(3): 764–771. [DOI] [PubMed] [Google Scholar]
- Gast MC, van Gils CH, Wessels LF, Harris N, Bonfrer JM, Rutgers EJ, Schellens JH, Beijnen JH (2009) Influence of sample storage duration on serum protein profiles assessed by surface-enhanced laser desorption/ionisation time-of-flight mass spectrometry (SELDI-TOF MS). Clin Chem Lab Med 47(6): 694–705. [DOI] [PubMed] [Google Scholar]
- George IS, Fennell AY, Haynes PA (2015) Protein identification and quantification from riverbank grape, Vitis riparia: Comparing SDS-PAGE and FASP-GPF techniques for shotgun proteomic analysis. Proteomics 15(17): 3061–3065. [DOI] [PubMed] [Google Scholar]
- Haglund C, Kuusela P, Roberts P, Jalanko H (1991) Tumour marker CA 125 in patients with digestive tract malignancies. Scand J Clin Lab Invest 51(3): 265–270. [DOI] [PubMed] [Google Scholar]
- Hirsch J, Hansen KC, Burlingame AL, Matthay MA (2004) Proteomics:current techniques and potential applications to lung disease. Am J Physiol Lung Cell Mol Physiol 287(1): L1–L23. [DOI] [PubMed] [Google Scholar]
- Hsieh SY, Chen RK, Pan YH, Lee HL (2006) Systematical evaluation of the effects of sample collection procedures on low-molecular-weight serum/plasma proteome profiling. Proteomics 6(10): 3189–3198. [DOI] [PubMed] [Google Scholar]
- Hsu MC, Lee KT, Hsiao WC, Wu CH, Sun HY, Lin IL, Young KC (2013) The dyslipidemia-associated SNP on the APOA1/C3/A5 gene cluster predicts post-surgery poor outcome in Taiwanese breast cancer patients: a 10-year follow-up study. BMC Cancer 13: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhang S, Yu J, Liu J, Zheng S (2005) SELDI-TOF-MS: the proteomics and bioinformatics approaches in the diagnosis of breast cancer. Breast 14(4): 250–255. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Doi Y, Yonemoto K, Ninomiya T, Kubo M, Shikata K, Hata J, Tanizaki Y, Matsumoto T, Iida M, Kiyohara Y (2009) Hyperglycemia increases risk of gastric cancer posed by Helicobacter pylori infection: a population-based cohort study. Gastroenterology 136(4): 1234–1241. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2): 69–90. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim SH, Lee SU, Ha GH, Kang DG, Ha NY, Ahn JS, Cho HY, Kang SJ, Lee YJ, Hong SC, Ha WS, Bae JM, Lee CW, Kim JW (2002) Proteome analysis of human liver tumor tissue by two-dimensional gel electrophoresis and matrix assisted laser desorption/ionization-mass spectrometry for identification of disease-related proteins. Electrophoresis 23(24): 4142–4156. [DOI] [PubMed] [Google Scholar]
- Kim MY, Yoo BC, Cho JY (2014) Ginsenoside-Rp1-induced apolipoprotein A-1 expression in the LoVo human colon cancer cell line. J Ginseng Res 38(4): 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochi M, Fujii M, Kanamori N, Kaiga T, Kawakami T, Aizaki K, Kasahara M, Mochizuki F, Kasakura Y, Yamagata M (2000) Evaluation of serum CEA and CA19-9 levels as prognostic factors in patients with gastric cancer. Gastric Cancer 3(4): 177–186. [DOI] [PubMed] [Google Scholar]
- Kohan AB (2015) Apolipoprotein C-III: a potent modulator of hypertriglyceridemia and cardiovascular disease. Curr Opin Endocrinol Diabetes Obes 22(2): 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejs GJ (2010) Gastric cancer: epidemiology and risk factors. Dig Dis 28(4–5): 600–603. [DOI] [PubMed] [Google Scholar]
- Kulasingam V, Diamandis EP (2008) Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol 5(10): 588–599. [DOI] [PubMed] [Google Scholar]
- Laronga C, Becker S, Watson P, Gregory B, Cazares L, Lynch H, Perry RR, Wright GL Jr, Drake RR, Semmes OJ (2003–2004) SELDI-TOF serum profiling for prognostic and diagnostic classification of breast cancers. Dis Markers 19(4-5): 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chen Z, Xiao Z, Wu X, Zhan X, Zhang X, Li M, Li J, Feng X, Liang S, Chen P, Xie JY (2003) Comparative proteomics analysis of human lung squamous carcinoma. Biochem Biophys Res Commun 309(1): 253–260. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW (2002) Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem 48(8): 1296–1304. [PubMed] [Google Scholar]
- Lin JF, Xu J, Tian HY, Gao X, Chen QX, Gu Q, Xu GJ, Song JD, Zhao FK (2007) Identification of candidate prostate cancer biomarkers in prostate needle biopsy specimens using proteomic analysis. Int J Cancer 121(12): 2596–2605. [DOI] [PubMed] [Google Scholar]
- Loei H, Tan HT, Lim TK, Lim KH, So JB, Yeoh KG, Chung MC (2012) Mining the gastric cancer secretome: identification of GRN as a potential diagnostic marker for early gastric cancer. J Proteome Res 11(3): 1759–1772. [DOI] [PubMed] [Google Scholar]
- Lorenzen S, Lordick F (2011) How will human epidermal growth factor receptor 2-neu data impact clinical management of gastric cancer? Curr Opin Oncol 23(4): 396–402. [DOI] [PubMed] [Google Scholar]
- Marimuthu A, Subbannayya Y, Sahasrabuddhe NA, Balakrishnan L, Syed N, Sekhar NR, Katte TV, Pinto SM, Srikanth SM, Kumar P, Pawar H, Kashyap MK, Maharudraiah J, Ashktorab H, Smoot DT, Ramaswamy G, Kumar RV, Cheng Y, Meltzer SJ, Roa JC, Chaerkady R, Prasad TS, Harsha HC, Chatterjee A, Pandey A (2013) SILAC-based quantitative proteomic analysis of gastric cancer secretome. Proteomics Clin Appl 7(5–6): 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McComb ME, Perlman DH, Huang H, Costello CE (2007) Evaluation of an ontarget sample preparation system for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in conjunction with normal-flow peptide high-performance liquid chromatography for peptide mass fingerprint analyses. Rapid Commun Mass Spectrom 21(1): 44–58. [DOI] [PubMed] [Google Scholar]
- Miki K (2006) Gastric cancer screening using the serum pepsinogen test method. Gastric Cancer 9(4): 245–253. [DOI] [PubMed] [Google Scholar]
- Miki K, Morita M, Sasajima M, Hoshina R, Kanda E, Urita Y (2003) Usefulness of gastric cancer screening using the serum pepsinogen test method. Am J Gastroenterol 98(4): 735–739. [DOI] [PubMed] [Google Scholar]
- Nasrollahzadeh D, Aghcheli K, Sotoudeh M, Shakeri R, Persson EC, Islami F, Kamangar F, Abnet CC, Boffetta P, Engstrand L, Dawsey SM, Malekzadeh R, Ye W (2011) Accuracy and cut-off values of pepsinogens I, II and gastrin 17 for diagnosis of gastric fundic atrophy: influence of gastritis. PLoS One 6(10): e26957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Mann M, Aebersold R, Yates JR 3rd, Bairoch A, Bergeron JJ (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods 7(9): 681–685. [DOI] [PubMed] [Google Scholar]
- Oue N, Sentani K, Noguchi T, Ohara S, Sakamoto N, Hayashi T, Anami K, Motoshita J, Ito M, Tanaka S, Yoshida K, Yasui W (2009) Serum olfactomedin 4 (GW112, hGC-1) in combination with Reg IV is a sensitive biomarker for gastric cancer patients. Int J Cancer 125(10): 2383–2392. [DOI] [PubMed] [Google Scholar]
- Pectasides D, Mylonakis A, Kostopoulou M, Papadopoulou M, Triantafillis D, Varthalitis J, Dimitriades M, Athanassiou A (1991) CEA, CA 19-9, and CA-50 in monitoring gastric carcinoma. Am J Clin Oncol 20(4): 348–353. [DOI] [PubMed] [Google Scholar]
- Poste G (2011) Bring on the biomarkers. Nature 469(7329): 156–157. [DOI] [PubMed] [Google Scholar]
- Qian HG, Shen J, Ma H, Ma HC, Su YH, Hao CY, Xing BC, Huang XF, Shou CC (2005) Preliminary study on proteomics of gastric carcinoma and its clinical significance. World J Gastroenterol 11(40): 6249–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Adam BL, Yasui Y, Ward MD, Cazares LH, Schellhammer PF, Feng Z, Semmes OJ, Wright GL Jr (2002) Boosted decision tree analysis of surface-enhanced laser desorption/ionization mass spectral serum profiles discriminates prostate cancer from noncancer patients. Clin Chem 48(10): 1835–1843. [PubMed] [Google Scholar]
- Roth AD (2003) Curative treatment of gastric cancer: towards a multidisciplinary approach? Crit Rev Oncol Hematol 46(1): 59–100. [DOI] [PubMed] [Google Scholar]
- Rui Z, Jian-Guo J, Yuan-Peng T, Hai P, Bing-Gen R (2003) Use of serological proteomic methods to find biomarkers associated with breast cancer. Proteomics 3(4): 433–439. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Haga Y, Yoshimura R, Egami H, Yokoyama Y, Akagi M (1987) Comparative effectiveness of the tumour diagnostics, CA 19-9, CA 125 and carcinoembryonic antigen in patients with diseases of the digestive system. Gut 28(3): 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A (2013) Cancer statistics. CA Cancer J Clin 63(1): 11–30. [DOI] [PubMed] [Google Scholar]
- Smith L, Lind MJ, Welham KJ, Cawkwell L Cancer Biology Proteomics Group (2006) Cancer proteomics and its application to discovery of therapy response markers in human cancer. Cancer 107(2): 232–241. [DOI] [PubMed] [Google Scholar]
- Sol EM, Sundsten T, Bergsten P (2009) Role of MAPK in apolipoprotein CIII-induced apoptosis in INS-1E cells. Lipids Health Dis 8: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Shen J, Qian H, Ma H, Ji J, Ma H, Ma L, Zhang W, Meng L, Li Z, Wu J, Jin G, Zhang J, Shou C (2007) Diagnosis of gastric cancer using decision tree classification of mass spectral data. Cancer Sci 98(1): 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HT, Lee YH, Chung MC (2012) Cancer proteomics. Mass Spectrom Rev 31(5): 583–605. [DOI] [PubMed] [Google Scholar]
- Timms JF, Arslan-Low E, Gentry-Maharaj A, Luo Z, T'Jampens D, Podust VN, Ford J, Fung ET, Gammerman A, Jacobs I, Menon U (2007) Preanalytic influence of sample handling on SELDI-TOF serum protein profiles. Clin Chem 53(4): 645–656. [DOI] [PubMed] [Google Scholar]
- Ucar E, Semerci E, Ustun H, Yetim T, Huzmeli C, Gullu M (2008) Prognostic value of preoperative CEA, CA 19-9, CA 72-4, and AFP levels in gastric cancer. Adv Ther 25(10): 1075–1084. [DOI] [PubMed] [Google Scholar]
- Villanueva J, Philip J, Entenberg D, Chaparro CA, Tanwar MK, Holland EC, Tempst P (2004) Serum peptide profiling by magnetic particle-assisted, automated sample processing and MALDI-TOF mass spectrometry. Anal Chem 76(6): 1560–1570. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang L, Zhang D, Fan Y, Jia Z, Qin P, Yu J, Zheng S, Yang F (2012) Identification of potential serum biomarkers for Wilms tumor after excluding confounding effects of common systemic inflammatory factors. Mol Biol Rep 39(5): 5095–5104. [DOI] [PubMed] [Google Scholar]
- Xiao X, Liu D, Tang Y, Guo F, Xia L, Liu J, He D (2003-2004) Development of proteomic patterns for detecting lung cancer. Dis Markers 19(1): 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto-Ishikawa K, Suzuki H, Nezu M, Kamiya N, Imamoto T, Komiya A, Sogawa K, Tomonaga T, Nomura F, Ichikawa T (2009) The isolation and identification of apolipoprotein C-I in hormonerefractory prostate cancer using surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Asian J Androl 11(3): 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Lim SK, Choong LY, Lee H, Chen Y, Chong PK, Ashktorab H, Wang TT, Salto-Tellez M, Yeoh KG, Lim YP (2010) Cathepsin S mediates gastric cancer cell migration and invasion via a putative network of metastasis-associated proteins. J Proteome Res 9(9): 4767–4778. [DOI] [PubMed] [Google Scholar]
- Ychou M, Duffour J, Kramar A, Gourgou S, Grenier J (2000) Clinical significance and prognostic value of CA72-4 compared withCEA and CA19-9 in patients with gastric cancer. Dis Markers 16(3–4): 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian-Daryoush M, Lindner D, Tallant TC, Wang Z, Buffa J, Klipfell E, Parker Y, Hatala D, Parsons-Wingerter P, Rayman P, Yusufishaq MS, Fisher EA, Smith JD, Finke J, DiDonato JA, Hazen SL (2013) The cardioprotective protein apolipoprotein A1 promotes potent anti-tumorigenic effects. J Biol Chem 288(29): 21237–21252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidan BA, Cutress RI, Murray N, Coulton GR, Hastie C, Packham G, Townsend PA (2009) Proteomic analysis of archival breast cancer serum. Cancer Genomics Proteomics 6(3): 141–147. [PubMed] [Google Scholar]
- Zhou Y, Wang Y, Wang S, Shen L (2015) Hyperglycemia promotes human gastric carcinoma progression via aquaporin 3. Dig Dis Sci 60(8): 2338–2345. [DOI] [PubMed] [Google Scholar]
- Zinkin NT, Grall F, Bhaskar K, Otu HH, Spentzos D, Kalmowitz B, Wells M, Guerrero M, Asara JM, Libermann TA, Afdhal NH (2008) Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res 14(2): 470–477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.