Abstract
The nonenzymatic replication of RNA oligonucleotides is thought to have played a key role in the origin of life prior to the evolution of ribozyme-catalyzed RNA replication. Although the copying of oligo-C templates by 2-methylimidazole-activated G monomers can be quite efficient, the copying of mixed sequence templates, especially those containing A and U, is particularly slow and error-prone. The greater thermodynamic stability of the 2-thio-U(s2U):A base pair, relative to the canonical U:A base pair, suggests that replacing U with s2U might enhance the rate and fidelity of the nonenzymatic copying of RNA templates. Here we report that this single atom substitution in the activated monomer improves both the kinetics and the fidelity of nonenzymatic primer extension on mixed-sequence RNA templates. In addition, the mean lengths of primer extension products obtained with s2U is greater than those obtained with U, augmenting the potential for nonenzymatic replication of heritable function-rich sequences. We suggest that noncanonical nucleotides such as s2U may have played a role during the infancy of the RNA world by facilitating the nonenzymatic replication of genomic RNA oligonucleotides.
Introduction
The nonenzymatic replication and transmission of genetic information may have played an important role in the transition from prebiotic chemistry to cellular life by enabling the evolution of selectively advantageous ribozyme catalysts prior to the emergence of ribozyme RNA polymerases.1 The chemistry of nonenzymatic oligonucleotide replication was extensively explored by Orgel and his students and colleagues.2,3 Their most successful model system involved the use of 2-methylimidazole-activated mononucleotides to nonenzymatically copy short C-rich oligonucleotide templates.4 Considerable progress has since been achieved in demonstrating template-directed primer extension in the presence of activated nucleotide derivatives.5−8 However, several challenges still need to be addressed to show that nonenzymatic RNA replication could be sufficiently fast and accurate to allow for the evolution of new RNA-encoded functions within replicating protocells. Foremost among these problems is the slow rate of nonenzymatic primer extension on templates that contain A and U residues. The rate of primer extension varies by more than 2 orders of magnitude depending on the specific nucleotide being added to the growing chain. 2-Methylimidazole-activated AMP or UMP (2-MeImpA or 2-MeImpU)9 are added to the 3′-end of a primer (annealed to a complementary template) at a much slower rate than 2-MeImpG or 2-MeImpC.10−12 The rate of primer extension with 2-MeImpU is so slow as to be comparable to the rate of 2-MeImpU hydrolysis.1 A second major problem is that G:U and A:C wobble pairing lead to a very high error rate in nonenzymatic template copying,13 potentially precluding the emergence of functional RNAs such as ribozymes from diverse sequence populations.
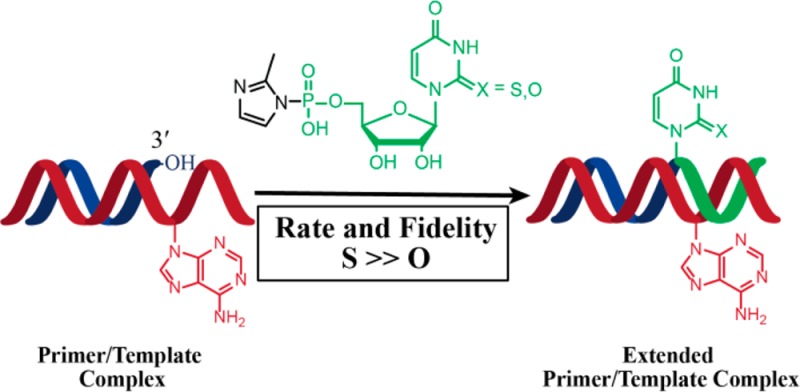
We sought to address these challenges by replacing U with s2U, a nucleobase known to significantly stabilize base pairing with A while modestly destabilizing wobble pairing with G.14,15 Remarkably, the thione-mediated stabilization of the s2U:A base pair within an RNA duplex is achieved without detectable structural perturbation,14 suggesting that the effect may be due at least in part to preorganization of the s2U-containing single strand.16 In addition the larger and more diffuse electron cloud of sulfur vs oxygen makes it more polarizable and hence may contribute to better stacking.15,17 Replacing thymine with 2-thiothymine in DNA is also stabilizing,18 and in RNA many occurrences of s2U also contain substitutions at the 5-position.19,20 We have therefore also explored the use of 2-thiothymine ribonucleotides (s2T) in RNA copying reactions.
In addition to the simple thermodynamic considerations, several independent arguments support the possibility that s2U or s2T might lead to improved nonenzymatic RNA copying. Nonenzymatic polymerization is most effective in the context of an A-form helix.13 s2U substitution in the anticodon loop of tRNA has been shown to increase the 3′-endo (N) conformer abundance of neighboring nucleotide sugars,21 and s2T stabilizes tRNAs from extreme thermophiles,22,23 most likely through stabilization of the 3′-endo sugar conformation. We have previously shown that monomers that are in the 2′-endo conformation in solution switch to the 3′-endo state upon binding to an RNA template.24 Since the s2U mononucleotide has been reported to exist in solution predominantly in the 3′-endo conformation,16,25 this preference should favor binding to the template. Finally, we have recently reported that nonenzymatic primer extension in another system, in which primer, template, and monomers are 3′-amino-2′,3′-dideoxynucleotides, exhibits enhanced rates and fidelity upon replacement of T with s2T.26 s2T also improves base pair discrimination in the context of PCR.27 Here we report the effects of s2U and s2T substitutions on both the rate and fidelity of nonenzymatic primer extension reactions in an all RNA system. We show that these substitutions in the activated monomer, but not in the template, contribute to an enhanced ability to copy mixed sequence templates and that the fidelity of copying is greatly increased.
Results
To examine the effect of 2-thio substitution on nonenzymatic RNA primer extension, we measured rates of primer extension using both 2-thio substituted U and ribo-T monomers and templates. We used two thiolated-nucleobase activated monomers, 2-thiouridine-5′-phosphor-(2-methyl)imidazolide (2-MeImps2U) and 2-thio-5-methyluridine-5′-phosphor-(2-methyl)imidazolide (2-MeImps2T), in nonenzymatic template-directed primer extension reactions with fluorophore-tagged RNA primers (Figure 1). We used two types of templates for this study, one type with six-nucleotide homopolymeric template regions U6 and A6, and a second type with a single U or A followed by five C residues, UC5 and AC5 (Figure 2; see also Supporting Information section 1 for a complete list of oligonucleotides used in this study). For pseudo-first-order rate determinations, primer extension reactions were studied for a maximum of 50 min, much less than the 2–5 day half-time of hydrolysis of the activated nucleotides. Pseudo-first-order rates and monomer–template dissociation constants were determined from plots of the fraction of unreacted primer as a function of time, for a series of activated nucleotide concentrations (Table 1; Figure 2; see also Supporting Information Figure 1).
Figure 1.
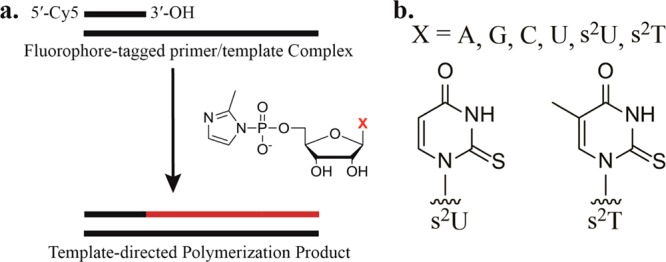
Template-directed primer extension system. (a) Primer extension reaction scheme. A 5′-Cy5-tagged RNA primer anneals to a complementary template. 2-MeImpX analogues form Watson–Crick base pairs on a complementary template and participate in template-directed primer extension. (b) Structure of thiolated uracil and thymine nucleobases in 2-methylimidazole-activated nucleotides.
Figure 2.
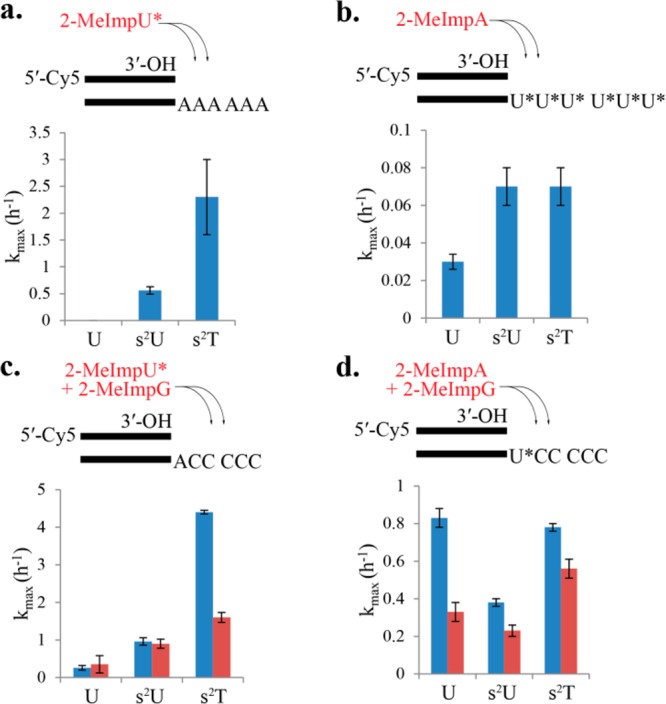
Kinetic studies of primer extension reactions. Pseudo-first-order rates were determined from the extent of primer disappearance as a function of time, and the resultant observed rates were determined as a function of activated nucleotide concentration to give kmax. Reaction conditions: 200 mM HEPES pH 7.0, 0.5 μM primer P1, 1.5 μM template, on ice. Buffer 1 (blue): 1.0 M NaCl, 200 mM MgCl2. Buffer 2 (red): 100 mM MgCl2. (a) Primer extension reaction on template T1 (A6) with 2-MeImpU* (U* = U, s2U, or s2T). (b) Primer extension reaction on templates T2 (U6), T3 (s2U6), or T4 (s2T6) with 2-MeImpA. (c) Primer extension reaction on template T8 (AC5) with 2-MeImpU* (U* = U, s2U, and s2T) and 40 mM 2-MeImpG. (d) Primer extension reaction on template T5 (UC5), T6 (s2UC5), and T7 (s2TC5) with 2-MeImpA and 40 mM 2-MeImpG.
Table 1. Kinetic Constants for Nonenzymatic Primer Extension Reactionsa.
| (1) N6 template |
(2) NC5 template |
(3) NC5 template |
|||||
|---|---|---|---|---|---|---|---|
| activated monomer | template nucleotide (N) | kmax (h–1) | Kd (mM) | kmax (h–1) | Kd (mM) | kmax (h–1) | Kd (mM) |
| 2-MeImpA | U | 0.027 ± 0.0036 | 31 ± 16 | 0.83 ± 0.045 | 9.2 ± 1.4 | 0.33 ± 0.051 | 8.0 ± 3.8 |
| 2-MeImpA | s2U | 0.067 ± 0.0081 | 15 ± 11 | 0.38 ± 0.019 | 3.3 ± 0.73 | 0.23 ± 0.026 | 1.6 ± 1.1 |
| 2-MeImpA | s2T | 0.071 ± 0.014 | 54 ± 31 | 0.78 ± 0.024 | 2.7 ± 0.38 | 0.56 ± 0.029 | 1.7 ± 0.51 |
| 2-MeImpU | A | <0.01 | ND | 0.26 ± 0.063 | 103.2 ± 41 | 0.35 ± 0.23 | 24 ± 32 |
| 2-MeImps2U | A | 0.56 ± 0.069 | 77 ± 23 | 0.96 ± 0.11 | 31 ± 7.6 | 0.90 ± 0.12 | 43 ± 9.4 |
| 2-MeImps2T | A | 2.3 ± 0.71 | 93 ± 62 | 4.4 ± 0.45 | 51 ± 8.4 | 1.6 ± 0.13 | 22 ± 3.8 |
Activated monomer and template nucleotides are indicated at left. 200 mM HEPES pH 7.0, 0.5 μM P1, 1.5 μM template, on ice and (1) 1.0 M NaCl, 200 mM MgCl2; (2) 1.0 M NaCl, 200 mM MgCl2, 40 mM 2-MeImpG; (3) 100 mM MgCl2, 40 mM 2-MeImpG.
We first considered the homopolymeric templates A6, U6, s2U6, and s2T6. While primer extension with 2-MeImpU on the A6 template was essentially undetectable at monomer concentrations up to 175 mM (kmax < 0.01 h–1), 2-MeImps2U and 2-MeImps2T resulted in maximal rates of primer extension (kmax) of 0.56 ± 0.069 and 2.3 ± 0.71 h–1, respectively (Table1; Figure 2a; see also Supporting Information Figure 1b(1)). It is notable that the single-atom oxygen to sulfur substitution resulted in readily observable primer extension; furthermore, methylation at the 5-position of 2-thiouracil increased the rate of primer extension an additional 4-fold. In contrast, primer extension with 2-MeImpA on U6, s2U6, and s2T6 templates approached the lower bounds of experimental measurement by this assay (kmax = 0.027 ± 0.0036, 0.067 ± 0.0081, and 0.071 ± 0.014 h–1, respectively) (Table 1; Figure 2b; see also Supporting Information Figure 1b(1)). These low rates and the modest effects of 2-thiolation and 5-methylation may reflect the poor stacking of U and modified U monomers and thus poor preorganization of templates consisting of multiple U (or s2U or s2T) residues in a row into an A-type helical conformation.
Although homopolymeric templates have been traditionally used to assess the reactivity of individual activated nucleotides, they are not directly relevant to potentially functional sequences, which are more likely to contain all four nucleotides. We therefore used a set of mixed sequence templates to examine primer extension in a more realistic setting. The template sequences followed the pattern 5′-C5N-(primer binding sequence)-3′ where N was A, U, s2U, or s2T. With the 5′-C5A-(primer binding sequence)-3′ template, the corresponding rates of monomer addition followed a similar pattern to those of the A6 template. The incorporation of either 2-MeImpU, 2-MeImps2U or 2-MeImps2T in the presence of 2-MeImpG led to kmax values of 0.26 ± 0.063, 0.96 ± 0.11, and 4.4 ± 0.45 h–1, respectively (Figure 2c, buffer 1 containing 200 mM HEPES pH 7.0, 200 mM MgCl2 and 1 M NaCl; also see Supporting Information Figure 1b(2)). Similarly, we found that the incorporation of 2-MeImpA in the presence of 2-MeImpG was approximately an order of magnitude faster on the single-U, s2U or s2T templates than on the corresponding homopolymeric templates, with observed rates of 0.83 ± 0.045, 0.38 ± 0.019, and 0.78 ± 0.024 h–1, respectively (Figure 2d, buffer 1; see also Supporting Information Figure 1b(2)). However, as with the homopolymer templates, thiolation of U or T in the template had little effect on the rate of primer extension. On the basis of the fact that base stacking plays a role in determining the melting temperature of complementary oligonucleotides,28 we suggest that the increased rates on the single-U or A templates, compared to the homopolymer templates, are best explained by improved stacking interactions of the incoming monomer (adjacent to the primer) with downstream G monomers base paired to the C5 portion of the template.
The above experiments were carried out using a high salt buffer (buffer 1, containing 200 mM MgCl2 and 1 M NaCl) that promotes base pairing by masking the interstrand repulsion of negatively charged phosphates. However, molar salt concentrations are incompatible with primitive fatty acid based vesicles,29 and we therefore examined primer extension without added NaCl and with a lower MgCl2 concentration. We observed that in buffer 2 (200 mM HEPES pH 7.0 and 100 mM MgCl2), kmax and Kd values were modestly reduced, by less than 2-fold in most cases (Table 1; Figure 2c,d; see also Supporting Information Figure 1b(3)).
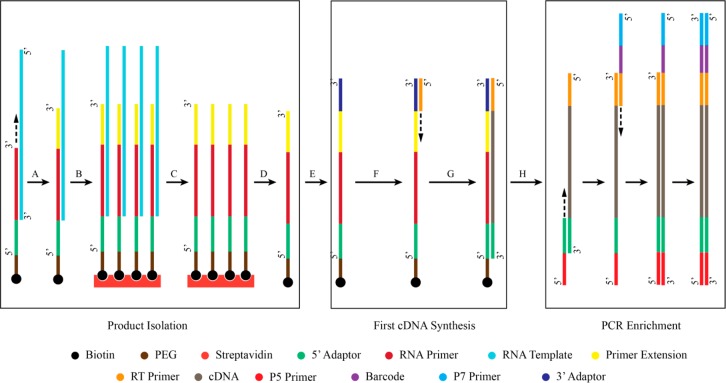
We used next generation sequencing (NGS) to assess the fidelity of the primer extension products obtained on mixed sequence templates, in the presence of competing activated monomers. We generated libraries of extended primers flanked by the necessary adaptor sequences at both the 5′ and 3′ ends, as illustrated in Figure 3. Briefly, nonenzymatic primer extension was carried out with a primer (P2) that included the 5′-adaptor sequence necessary for on-bead amplification during sequencing, and also included a 5′-biotin so that the template strand could be removed by a convenient bind and wash procedure using magnetic streptavidin beads. Following template removal, the nonenzymatically extended primer molecules were released from the streptavidin beads. An adaptor was then ligated to the 3′-end of each primer, followed by reverse-transcription and PCR to yield the library for sequence analysis.
Figure 3.
Schematic representation of MiSeq library assembly protocol. (A) Primer extension reaction. (B) Biotinylated-primer/streptavidin bead association. (C) Template removal. (D) Biotinylated primer/streptavidin dissociation. (E) 3′ Adaptor ligation. (F) Primer hybridization. (G) Reverse transcription: First strand cDNA synthesis. (H) PCR enrichment.
We compared primer extension across a 5′-GAGAGA-(primer binding sequence)-3′ template (T9) with U, s2U, and s2T as activated monomers (Figure 4a; see also Supporting Information Table 1a) in the presence of 2-MeImpC and sequenced the extended primers as described above to assess the extent and fidelity of the template copying reaction. Primer extension with 2-MeImpU and 2-MeImpC yielded largely products extended by only two or three nucleotides (Figure 4a, left panel). However, in an otherwise identical reaction with 2-MeImps2U and 2-MeImpC we observed a wider distribution of product lengths including full-length product, i.e., primer + six nucleotides (Figure 4a, middle panel). This effect was further enhanced when 2-MeImps2T replaced 2-MeImpU (Figure 4a, right panel).
Figure 4.
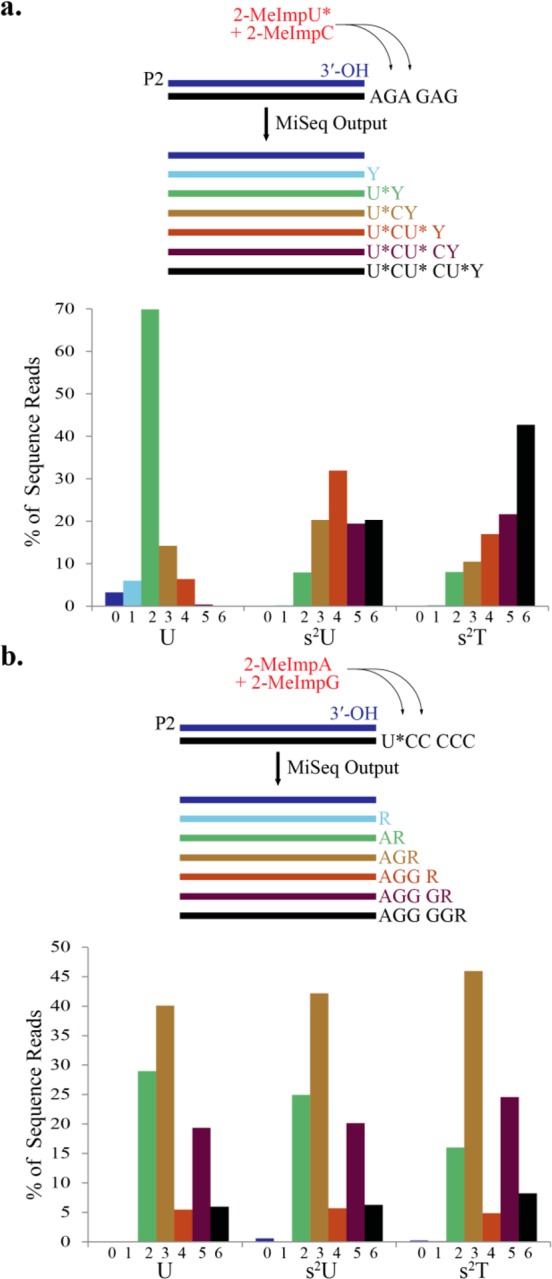
Sequence analysis of products of nonenzymatic primer-extension. (a) Top: Primer-extension was carried out on 2.0 μM template T9 (AGAGAG) in the presence of 40 mM 2-MeImpC and 50 mM 2-MeImpU* (U* = U, s2U, or s2T) on ice for 7 days. Bottom: Products were sequenced, and the sequence reads binned according to the number of nucleotides added to the primer; Y = C or U*. Reaction conditions: 2.0 μM primer P2, 200 mM HEPES pH 7.0, 100 mM MgCl2. (b) Top: Primer-extension was carried out on 2.0 μM template T5 (UC5), T6 (s2UC5), or T7 (s2TC5) in the presence of 40 mM 2-MeImpG and 50 mM 2-MeImpA on ice for 7 days. Bottom: Products were sequenced and the sequence reads binned according to the number of nucleotides added to the primer; R = A or G.
Examination of the sequences of the products of primer extension on the 5′-GAGAGA-(primer binding sequence)-3′ template revealed enhanced fidelity with the 2-MeImps2U and 2-MeImps2T monomers, compared to the standard 2-MeImpU monomer. In order to examine the effect of the modified nucleotides on G:U wobble mispairing, we examined all products in which the primer had extended by two or more bases and where the first nucleotide added was the correct U (or s2U or s2T). Correct primer extension would then result in incorporation of a C at position 2, while G:U mispairing would result in the incorporation of a U (or s2U or s2T). We found that G:U mispairing at the second position was diminished from 4.3% in the case of 2-MeImpU to 1.6% with 2-MeImps2U and 2.0% with 2-MeImps2T (Figure 5a; see also Supporting Information Table 2), corresponding to a 2–3 fold improvement in fidelity with the 2-thiolated U or T monomers. Because primer extension with 2-MeImpU was so inefficient, we were unable to compare the frequency of Watson–Crick vs wobble pairing at the next G residue in the template, which is at position 4.
Figure 5.
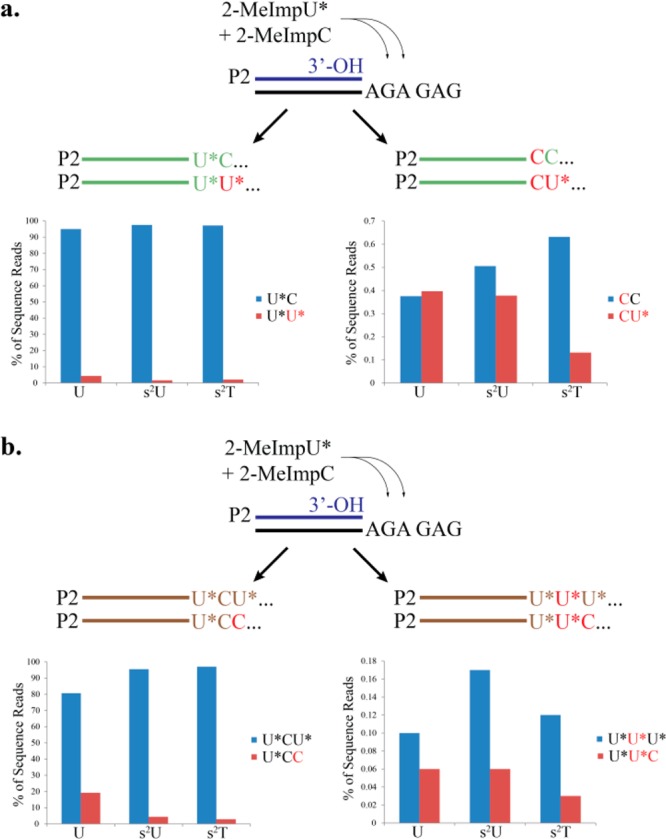
Fidelity of primer-extension reactions. Sequence reads obtained from the products of primer-extension on template T9 (AGAGAG), as described in Figure 4, were sorted into bins according to the number and identity of nucleotides added to the primer. (a) Left: products extended by at least two nucleotides, with correct incorporation of U* at position 1, were sorted according to whether the correct nucleotide C or the incorrect nucleotide U* was incorporated at position 2. Right: products extended by at least two nucleotides, with incorrect incorporation of C at position 1, were sorted according to whether the correct nucleotide C or the incorrect nucleotide U* was incorporated at position 2. (b) Left: products extended by at least three nucleotides, with correct incorporation at positions 1 and 2, were sorted according to whether the correct nucleotide U* or the incorrect nucleotide C was incorporated at position 3. Right: products extended by at least three nucleotides, with incorrect incorporation of U* at position 2, were sorted according to whether the correct nucleotide U*or the incorrect nucleotide C was incorporated at position 3. Misincorporated nucleotides are represented by red.
Surprisingly, we discovered that A:C mispairing was also diminished when U was replaced with s2U or s2T. The expected product of primer extension on the 5′-GAGAGA-(primer binding sequence)-3′ template at positions 1, 2, and 3 was primer-U*CU*. When we examined sequences where the first two nucleotides were correct, we found that an incorrect C was incorporated at position 3 over 19% of the time, following primer extension with 2-MeImpU and 2-MeImpC. We noted a 5–6 fold drop in the amount of primer-UCC when 2-thiolated U-derivatives were used, with only 4.3% and 2.9% C incorporation at position 3 when primer extension was carried out with 2-MeImps2U and 2-MeImps2U, respectively (Figure 5b; see also Supporting Information Table 2).
Finally, we observed that both G:U and A:C misincorporation largely terminated further primer extension and, furthermore, that the small amount of continued primer extension was highly error prone. We first consider the case of a G:U mismatch at position 2, corresponding to primer-UU products (see Supporting Information Table 2). Primer extension in the presence of 2-MeImpU and 2-MeImpC resulted in 2394 such sequences, but only 21 sequences corresponding to the addition of one more nucleotide to the growing primer, i.e., less than 1% continued extension. In contrast, we observed 53 134 sequences corresponding to the correct 2-nucleotide extension product primer-UC, and 13 077 sequences corresponding to continued primer extension by at least one more nucleotide, i.e., about 20% continued primer extension. Remarkably, primer extension following a G:U mismatch was highly error prone, with an error rate of almost 40%. Replacing 2-MeImpU with either 2-MeImps2U or 2-MeImps2T did not significantly increase the fidelity of post mismatch synthesis (25–50% error rate in both cases). Examination of sequences corresponding to an A:C mismatch at position +1 reveals similar poor fidelity at the following position.
To assess the capacity of s2U and s2T in the template to mitigate G:U mispairing, we performed nonenzymatic primer extension reactions with 5′-C5U*-3′ templates (where U* = U, s2U, or s2T) in the presence of both 2-MeImpA and 2-MeImpG. Interestingly, there was insignificant differentiation between U, s2U, or s2T when they were templating primer extension in this context, and the distributions of product length were comparable in all three cases (Figure 4b; see also Supporting Information Table 1b).
Discussion
A single-atom substitution of oxygen by sulfur at the 2-position of the activated uridine nucleotide 2-MeImpU significantly improves the rate and fidelity of nonenzymatic template-directed primer extension. Under different template and buffer conditions, the rate of primer extension consistently followed the order s2T > s2U > U. Surprisingly, this enhanced rate and fidelity of template copying was only observed with 2-thio-U or -T as the activated monomer, and no significant effect was observed when these modified nucleotides were present in the template strand. Although the reasons for this striking difference are not entirely clear, we suggest the following speculative explanation. We consider first an incoming U (or s2U or s2T) monomer. Although an s2U:A base pair at an internal position in a duplex stem is much more stable than a standard U:A base pair, little if any stabilization is seen for an s2U:A base pair at the end of a helix.15 This is consistent with the fact that we do not see a statistically significant decrease in the Kd of the 2-MeImps2U or 2-MeImps2T monomers relative to 2-MeImpU (although the Kd values for the very slow reactions on the A6 template have large standard errors). We do however see an increased maximal rate of reaction (at saturating monomer concentration) for the 2-thiolated monomers. We suggest that this stems from preorganization of the 2-thionucleotides in the 3′-endo conformation,16,25 which could help to properly position and orient the phosphate group of the incoming monomer for reaction with the attacking 3′-hydroxyl of the primer. Weaker hydrogen bonding between the sulfur of s2U(T) and the imino proton of G would weaken wobble pairing, favoring correct binding of C, and that together with a further distorted geometry may account for the decreased formation of U:G mismatches observed with s2U and s2T. We attribute the decreased frequency of A:C mismatches to the faster reaction rate of s2U or s2T (relative to U) when paired with A; effectively s2U and s2T outcompete C so that A:C mismatches do not have time to form. We now consider the substitution of U in the template with s2U or s2T. In this case, the binding of an incoming A monomer to the primer–template duplex is facilitated by the stronger stacking interactions of the purine nucleobase with flanking nucleotides, i.e., the 3′-nucleotide of the primer, and downstream monomers. As a result, the Kd of activated A is lower than that of the activated U monomers. In addition, template preorganization by an internal s2U or s2T may contribute to enhanced binding of 2-MeImpA, as observed for the 5′-CCCCCU* templates. However, once the activated A monomer is bound to the template, it has the same reactivity whether the template base is U, s2U, or s2T, and as a result, the rate at saturation (i.e., kmax) does not change. Accurate direct measurements of binding affinities and geometries may allow for experimental testing of the above hypotheses.
The stalling of primer extension following a mismatch has previously been shown, under certain circumstances, to lead to an enhanced effective fidelity of replication, because the first template copies to be completed tend to be the most accurate.30 However, this effect comes at the cost of significantly slowed overall rate of replication. Our data suggest that the formation of both U:G and C:A wobble pairs during nonenzymatic template-directed primer extension leads to a very strong stalling effect, i.e., a greatly decreased rate of primer extension following a wobble mismatch. The magnitude of this effect (ca. 20-fold, Figure 5) is surprising and should be examined in additional sequence contexts. Nevertheless, we now expect that replacing U with s2U or s2T should improve the overall rate of primer extension not only because of the increased rate at which the 2-thio monomers are incorporated but also because the increased accuracy of primer extension will lead to less stalling after mismatches. In addition, we note that the strongly decreased fidelity that we observed for primer extension following a mismatch is consistent with previous proposals that errors introduced during nonenzymatic RNA copying may be dominated by multiple sequential errors, as opposed to isolated single mutations.31 Such clustered mutations may speed the exploration of sequence space and the optimization of functions under selection. Enhancing the ability of RNA to make large jumps through sequence space may be particularly important in facilitating the emergence of novel RNA-coded functions.
Finally, our results raise the question of the prebiotic availability of s2U or s2T. It is conceivable that these 2-thio nucleotides could be generated spontaneously in a sulfur-rich early earth environment through a route analogous to that proposed for the synthesis of the canonical pyrimidine nucleotides.32 The experimental demonstration of an efficient pathway for the prebiotic synthesis of either s2U or s2T nucleotides would support their proposed role in facilitating nonenzymatic RNA replication during the origin of life.
Methods
Nonenzymatic Primer Extension
Representative reaction protocol: 4.0 μL of 1.0 M HEPES pH 7.0 buffer, 5.0 μL of nuclease-free water, 1.0 μL of primer P1, and 3.0 μL of template T5 were combined in a thin-walled PCR microtube. After being mixed well by pipetting up and down multiple times, the oligonucleotides were incubated at 90 °C for 5 min and annealed at 25 °C for 5 min. A 2.0 μL amount of 1.0 M MgCl2 was then added to the solution and mixed well. To initiate primer extension, 1.0 μL of 1.0 M 2-MeImpA and 4.0 μL of 200 mM 2-MeImpG were added to the lid of the microtube cap. The reaction was initiated when the nucleotides were spun down into the buffered primer/template solution. The solution was mixed well and incubated in a metal block in an ice bath for the duration of the experiment. Aliquots (4.0 μL each) were removed at 10, 20, 30, 40, and 50 min and were immediately quenched by addition to 26 μL of precipitation buffer [3.0 μL of 3.0 M NaOAc pH 5.5, 2.0 μL of 500 mM EDTA pH 8.0, 1.0 μL of 5 mg/mL glycogen, 2.0 μL of 10X TBE (1.0 M Tris, 1.0 M boric acid, and 20 mM EDTA pH 8.0), and 18.0 μL 8.0 M urea]. After the mixture was briefly vortexed to mix the contents, 75 μL of pure ethanol was added. The sample was mixed and kept at −25 °C for a minimum of 30 min and was then centrifuged at 15 000 RCF in an Eppendorf 5424R centrifuge at 4 °C. The RNA pellets were then taken up in 5.0 μL of 8.0 M urea in 1X TBE and incubated at 90 °C for 5 min before the RNA products were separated by 20% (19:1) denaturing PAGE. The gel was scanned with a Typhoon 9410 Variable Mode Imager, and the bands were quantified using the accompanying ImageQuant TL software package.
Illumina MiSeq Library Assembly
Nonenzymatic template-directed primer extension reactions were performed as described above except that 2.0 μM primer P2 and 2.0 μM template were used.
Bind and Wash Buffer
Ten millimolar Tris-HCl (pH 7.5), 10 mM EDTA (pH 8.0), 2.0 M NaCl; buffer A: 100 mM NaOH, 5 mM NaCl; buffer B: 10 mM NaCl; elution buffer: 95% formamide, 10 mM EDTA (pH 8.0); SSC buffer: 150 mM NaCl, 15 mM Na citrate, pH 7.0.
Bead Binding and Wash
A 100 μL amount of streptavidin MyOne C1 Dynabeads (10 mg/mL; Invitrogen) was separately decanted for each library assembly reaction. Gentle draw/expel pipetting was used as a bead mixing technique; vortexing was avoided. The beads were mixed and washed 3× with 200 μL “bind and wash” buffer and were allowed to sit on a magnetic rack for 1 min following each wash step. The supernatant was drawn off while the microtube remained on the magnetic rack. Beads were then washed 2 × 200 μL buffer A, followed by 2 × 200 μL buffer B. The RNA pellet from an extension reaction using primer P2 was diluted with 200 μL of nuclease-free water and 200 μL of “bind and wash” buffer. The RNA solution was added to the washed and decanted beads and then briefly mixed to suspend the beads in the RNA solution. The mixture was tumbled for 30 min at 25 °C. After the mixture was rested on the magnetic rack for 1 min, the supernatant was drawn off.
RNA Template Elution and Product Isolation
Beads with bound primer/template complex were mixed with 250 μL of SSC buffer and decanted after resting on the magnetic rack for 1 min. The beads were suspended in 100 μL of 150 mM NaOH and incubated at 25 °C for 10 min. The tube was placed on the magnetic rack for 1 min, and the supernatant was drawn off. The beads were then mixed with 250 μL of 100 mM NaOH and immediately placed on the magnetic rack for 1 min followed by removal of the supernatant. The biotinylated oligonucleotides were immediately eluted from the beads by mixing with 250 μL of elution buffer at 65 °C for 5 min followed by 1 min on the magnetic rack and removal of the supernatant. The RNA from this final supernatant was precipitated upon addition of 1120 μL of ethanol and 30 μL of 3.0 M NaOAc pH 5.5 and incubation for 30 min at −25 °C. The pellets were then used directly in subsequent steps.
3′-Adaptor Ligation and Wash
A 20.0 μL amount of 100 μM 3′-adaptor (5′-phosphate-AGA TCG GAA GAG CAC ACG TCT3′-3′-T-5′; DNA) and 6.0 μL of nuclease-free water were added to the biotinylated RNA pellet and placed in a thin-walled PCR microtube. After dissolution, the mixture was incubated at 65 °C for 2 min then placed on ice, and 4.0 μL of 10X T4 RNA Ligase I buffer, 4.0 μL of 50% PEG 8000, 4.0 μL of DMSO (molecular biology grade), and 2.0 μL of T4 RNA Ligase I (10,000 u/mL) were added. After mixing, the solution was incubated at 37 °C for 2 h. The samples were cleaned up with a QIAquick PCR Purification Kit (eluted with H2O not EB buffer from kit; Qiagen) to remove excess 3′-adaptor (22-mer) and retain the ligated product (92-mer). The samples were then lyophilized to dryness and taken up in 15 μL of nuclease-free water.
Reverse-Transcription and PCR
The following sequences used a combination of materials from SuperScript III First-Strand Synthesis SuperMix (Invitrogen) and NEBNext Multiplex Small RNA Library Prep Set 1 for Illumina (NEB). A 1.0 μL amount of SR RT Primer for Illumina (diluted 1:2; NEB) and 2.0 μL of annealing buffer (Invitrogen) were added to the 15 μL solution of RNA. The mixture was incubated in a thin-walled PCR microtube at 75 °C using a thermal cycler for 5 min, 37 °C for 15 min, and finally 25 °C for 15 min. At 25 °C, 20 μL of 2X First-Strand Reaction Mix (Invitrogen) and 2.0 μL of SuperScript III/RNaseOUT Enzyme Mix (Invitrogen) were added to the RNA solution, mixed, and incubated at 50 °C for 1 h and 85 °C for 5 min and then kept on ice. A 2.5 μL amount of SR Primer for Illumina (NEB), 2.5 μL of Indexed Prime (NEB), 5.0 μL of nuclease-free water, and 50 μL of LongAmp Taq 2X Master Mix (NEB) were added to the reverse-transcribed mixture and were cycled 12 times (30 s of initial denaturation, 94 °C; 15 s, 94 °C; 30 s, 62 °C; 15 s, 70 °C; 5 min of final extension at 70 °C; hold at 4 °C). The samples were desalted by QIAquick PCR Purification Kit.
Gel Purification and Quantification
The desalted and enzyme-free DNA was purified on a 20% TBE precast gel cassette (Invitrogen) with Quick-Load pBR322 DNA-MspI Digest (NEB) as a marker. The target band of 140 bp was sliced out, crushed with a disposable plastic RNase-free pestle (Fisher), and eluted with 250 μL of DNA Gel Elution buffer (NEB). The dsDNA was desalted with a QIAquick PCR Purification Kit and quantified by qPCR with a KAPA SYBR Fast Universal qPCR Kit for Illumina (KAPA Biosystems).
Sequencing and Sequence Analysis
Sequencing was performed on an Illumina MiSeq instrument. Samples were prepared as per the manufacturer’s recommendations with a 30% PhiX spike to bring an appropriate level of initial sequence diversity to the libraries. A multiplexed paired-end protocol was used: 25 nucleotides for Read-1, Index Read, 25 nucleotides for Read-2. Upon completion of sequencing, the data files were subjected to the following Python script. The script takes two FASTQ files, one for the forward read and one for the reverse read, and filters out reads that either lack the exact adapter sequence in the reverse read, lack the adapter in the forward read (with a maximum edit distance tolerance of 1), or have forward and reverse reads that are not identical in the region of nonenzymatic primer extension. It then outputs all retained sequences and their corresponding total read counts, as well as the number of reads discarded for the reasons mentioned above.
Acknowledgments
We thank A. Engelhart, T. Jia, A. Larsen, T. Olsen, T. Walton, A. Fahrenbach, and A. Björkbom for helpful discussions and U. Kim for help with next generation sequencing. This work was supported in part by grant CHE-0809413 from the National Science Foundation and by grant 290363 from the Simons Foundation to J.W.S. who is an investigator from the Howard Hughes Medical Institute.
Supporting Information Available
The complete list of oligonucleotides used in this study; rate plots and fits for all the kinetics studies; sequence reads and product distribution for fidelity studies; the Python script to generate the primer extension sequences and read counts. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
† Lathrop & Gage, LLP, 28 State St., Boston, MA 02109.
Author Contributions
§ B.D.H. and A.P. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Szostak J. W. J. Syst. Chem. 2012, 3, 2. [Google Scholar]
- Inoue T.; Orgel L. E. J. Am. Chem. Soc. 1981, 103, 7666. [Google Scholar]
- Orgel L. E. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 99. [DOI] [PubMed] [Google Scholar]
- Inoue T.; Orgel L. E. Science 1983, 219, 859. [DOI] [PubMed] [Google Scholar]
- Lohrmann R.; Orgel L. E. J. Mol. Evol. 1979, 12, 237. [DOI] [PubMed] [Google Scholar]
- Fakhrai H.; van Roode J. H. G.; Orgel L. E. J. Mol. Evol. 1981, 17, 295. [DOI] [PubMed] [Google Scholar]
- Sleeper H. L.; Lohrmann R.; Orgel L. E. J. Mol. Evol. 1979, 13, 203. [DOI] [PubMed] [Google Scholar]
- Sulston J.; Lohrmann R.; Orgel L. E.; Miles H. T. Proc. Natl. Acad. Sci. U. S. A. 1968, 59, 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. A. R.; Orgel L. E.; Wu T. Origins Life Evol. Biospheres 1993, 23, 285. [DOI] [PubMed] [Google Scholar]
- Joyce G. F. Cold Spring Harbor Symp. Quant. Biol. 1987, 52, 41. [DOI] [PubMed] [Google Scholar]
- Kurz M.; Göbel K.; Hartel C.; Göbel M. W. Angew. Chem. 1997, 109, 873. [Google Scholar]
- Wu T.; Orgel L. E. J. Am. Chem. Soc. 1992, 114, 7963. [DOI] [PubMed] [Google Scholar]
- Schrum J. P.; Ricardo A.; Krishnamurthy M.; Blain J. C.; Szostak J. W. J. Am. Chem. Soc. 2009, 131, 14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. K.; Davis D. R. Nucleic Acids Res. 1997, 25, 1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa S. M.; Disney M. D.; Turner D. H.; Kierzek R. Biochemistry 1999, 38, 16655. [DOI] [PubMed] [Google Scholar]
- Larsen A. T.; Fahrenbach A. C.; Sheng J.; Pian J.; Szostak J. W. Unpublished.
- Saenger W.Principles of Nucleic Acid Structure; Springer-Verlag: New York, 1984. [Google Scholar]
- Sintim H. O.; Kool E. T. J. Am. Chem. Soc. 2006, 128, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris P. F. Nucleic Acids Res. 2004, 32, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka M. A.; Milanowska K.; Osman Oglou O.; Purta E.; Kurkowska M.; Olchowik A.; Januszewski W.; Kalinowski S.; Dunin-Horkawicz S.; Rother K. M.; Helm M.; Bujnicki J. M.; Grosjean H. Nucleic Acids Res. 2013, 41, D262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M.; Takeishi K.; Ukita T. Biochim. Biophys. Acta 1971, 228, 153. [DOI] [PubMed] [Google Scholar]
- Watanabe K.; Yokoyama S.; Hansske F.; Kasai H.; Miyazawa T. Biochem. Biophys. Res. Commun. 1979, 91, 671. [DOI] [PubMed] [Google Scholar]
- Davanloo P.; Sprinzl M.; Watanabe K.; Albani M.; Kersten H. Nucleic Acids Res. 1979, 6, 1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N.; Zhang S.; Szostak J. W. J. Am. Chem. Soc. 2012, 134, 3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S.; Watanabe T.; Murao K.; Ishikura H.; Yamaizumi Z.; Nishimura S.; Miyazawa T. Proc. Natl. Acad. Sci. U. S. A. 1985, 82, 4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Blain J. C.; Zielinska D.; Gryaznov S. M.; Szostak J. W. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 17732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sismour A. M.; Benner S. A. Nucleic Acids Res. 2005, 33, 5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T.; SantaLucia J. J.; Burkard M. E.; Kierzek R.; Schroeder S. J.; Jiao X.; Cox C.; Turner D. H. Biochemistry 1998, 37, 14719. [DOI] [PubMed] [Google Scholar]
- Adamala K.; Szostak J. W. Science 2013, 342, 1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamani S.; Ichida J. K.; Antal T.; Treco D. A.; Leu K.; Nowak M. A.; Szostak J. W.; Chen I. A. J. Am. Chem. Soc. 2010, 132, 5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu K.; Kervio E.; Obermayer B.; Turk-MacLeod R. M.; Yuan C.; Luevano J.; J.-M; Chen E.; Gerland U.; Richert C.; Chen I. A. J. Am. Chem. Soc. 2013, 135, 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powner M. W.; Gerland B.; Sutherland J. D. Nature 2009, 459, 239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.