Abstract
Members of the genus Ranavirus (family Iridoviridae) are large double-stranded (ds) DNA viruses that are found world-wide infecting fish, amphibian and reptile ectothermic hosts. Ranavirus genomes range from 105 – 155 kbp in length and they are predicted to encode around 90–160 genes. Currently, our knowledge of the function of ~50% of these genes is known or inferred based on homology to orthologous genes characterized in other systems; however, the function of the remaining open reading frames (ORFS) is unknown. Therefore, in order to begin to uncover the function of unknown ORFs in ranaviruses we developed a standardized approach to generate a recombination cassette for any ORF in Ambystoma tigrinum virus (ATV). Our standardized approach quickly and efficiently assembles recombination cassettes and recombinant ATV. We have used this approach to identify two essential, one semi-essential and two non-essential genes in ATV.
Keywords: ranavirus, homologous recombination, gene knock-out, viral replication pathogenesis
1. Introduction
Ranaviruses (genus Ranavirus, family Iridoviridae) are large dsDNA viruses that infect a variety of ectothermic hosts world-wide including fish, amphibians and reptiles (Chinchar et al., 2009; Chinchar et al., 2011; Jancovich et al., 2015b). Members in this genus can be single or multi-host pathogens (Jancovich et al., 2001; Schock et al., 2008; Schock et al., 2009) and this group of viruses are now considered an emerging infectious disease (Chinchar, 2002; Tompkins et al., 2015). Ranaviruses have unit length genomes between 105 and 155 kbp in length and encode approximately 90–160 open reading frames (ORFs) or genes (Jancovich et al., 2015a). To that end, the function of about half of the ORFs encoded by ranaviruses are either known (e.g., the major capsid protein) or inferred (e.g., the viral subunits of RNA polymerase II) based on either experimental information or homology to other related viral proteins. The identity and function of the remaining ORFs, while highly conserved among iridoviruses, are unknown.
Identifying genes that are essential and non-essential for virus replication in cells in culture is an important step in determining the function of unknown ORFs or ORFs with a predicted function that has yet to be confirmed. In addition, non-essential genes in cells in culture may play a role in viral pathogenesis in the host. Therefore, understanding the repertoire of essential and non-essential viral genes in cells in culture will help in our understanding of the molecular determinants of viral replication, and potentially identify viral host-range and pathogenesis factors. There are a number of techniques used to identify essential and non-essential viral genes in cultured cells. For example, RNA interference or inhibition (RNAi) has been used to identify essential and non-essential ranavirus genes (Sample et al., 2007; Whitley et al., 2011; Xie et al., 2014). This approach is where ORFs are inhibited, or knocked-down, from being expressed at the mRNA level and viral growth can be used to measure the essentialness of the target ORF. This approach provides valuable information regarding the role of the ORF in viral replication in cells in culture; however, this technique is limited because you never achieve 100% knock-down of the target gene and the level or quantity of mRNA knock-down and viral growth used to classify ORFs as essential vs. non-essential can be arbitrary and difficult to interpret. In addition, it is difficult to quantitatively assess ORF knock-down at the protein level since there are limited molecular reagents available for every ranavirus ORF. In contrast, generating a recombinant virus, where an ORF is deleted, or knocked-out, from the virus directly identifies essential vs. non-essential genes as essential genes cannot be deleted from the virus whereas non-essential genes in cells in culture can be deleted from the virus. Therefore, the mutant virus generated by this knock-out approach can be used in further studies to characterize the function of that particular gene and its role in viral replication, host-range and pathogenesis.
Our lab is interested in identifying the essential and non-essential genes in Ambystoma tigrinum virus (ATV), a ranavirus that appears to be a host specific pathogen (Jancovich et al., 2001; Jancovich et al., 1997; Jancovich and Jacobs, 2011). Our ultimate goal is to understand which ATV genes, particularly those with unknown function, are essential or non-essential for viral replication in cells in culture as this will increase our understanding of ranavirus gene function, gain insight into the molecular mechanisms of viral replication and help in our understanding of viral pathogenesis. Therefore, we have developed a standardized procedure for creating recombination cassettes and recombinant ATV. Herein we describe this technique to generate recombination cassettes and our protocol to knock-out ORFs in ATV and show how this approach has successfully identified essential and non-essential genes in ATV.
2. Materials and Methods
2.1 Cells and Virus
Fathead minnow (FHM) cells were maintained in minimal essential medium with Hank’s salts (HMEM) supplemented with 5% fetal bovine serum (FBS) and 0.1 mM nonessential amino acids and vitamins. FHM cells were cultured at 20 – 22°C in the presence of 5% CO2. The Ambystoma tigrinum virus (ATV) isolated from the Sonoran tiger salamander in the San Rafael Valley, AZ was used in this study (Jancovich et al., 1997). To prepare virus stocks, wild-type (wt) ATV was amplified in FHM by infecting cells at a multiplicity of infection (MOI) of 0.01 pfu/cell. Cells and virus were harvested once cytopathic effects (CPE) reached 90 – 100%, frozen and thawed three times to release virus from the cell and clarified by centrifugation at 1,000 x g at 4°C for 10 minutes. The virus containing supernatant was then centrifuged at 25,000 x g at 4°C for one hour. The supernatant was removed and the pellet containing virus resuspended in 500 μl of 10 mM Tris, pH 8 before being layered onto a 20% (w/v) sucrose pad. Virus was purified by centrifugation at 31,000 x g at 4°C for 1.5 hours and virus was resuspened 100 μl of 10 mM Tris, pH. ATV was quantified by a standard plaque assay in FHM cells.
2.2 ATV ORFs Targeted
We targeted a total of 5 ATV ORFs (Table 1). Of the 5 ATV ORFs studied herein, two are found in all iridoviruses (i.e. an iridovirus core gene(Eaton et al., 2007); ORFs 11R and 25R) and one is found in all ranaviruses (40L). The remaining two ORFs (ORFs 53R and 54R) appear to be part of a multi-gene family found in all ranaviruses (Eaton et al., 2007; Jancovich et al., 2010). In addition, two ORFs, 25R and 40L, have a predicted function while the other three ORFs have an unknown predicted function (Table 1). All five ORFs tested in this study have orthologous ORFs predicted to be expressed as an early gene in frog virus 3 (FV3) (Majji et al., 2009).
Table 1.
ATV ORFs tested in this study.
| ORF #1 | Genome Location1 | Size (a.a.)2 | Predicted function1 | GenBank # | Category | Essential vs. Nonessential3 |
|---|---|---|---|---|---|---|
| 11R | 13,512–13,979 | 155 | unknown | AAP33188.1 | core gene | essential |
| 25R | 27,224–28,345 | 373 | RNase III | AAP33202.1 | core gene | semi-essential |
| 40L | 40,205–40,492 | 95 | CARD-domain | AAP33218.1 | ranavirus only | non-essential |
| 53R | 58,082–59,230 | 382 | unknown | AAP33232.1 | ranavirus multi-gene | essential |
| 54R | 59,613–60,710 | 365 | unknown | AAP33233.1 | ranavirus multi-gene | non-essential |
based on Jancovich et al., 2003
a.a. = amino acid
indentified in this study
2.3 Generating Recombination Cassettes
Recombination cassettes for all targets ORFs were generated by designing forward (for) and reverse (rev) primers to amplify the upstream (LA) and downstream (RA) flanking sequences. Primers were designed to initially amplify a PCR product around 1,000 nt up- and downstream from the start and end of the target sequence, respectively. These primers (ORF#_LA_for_1k and ORF#_RA_rev_1k, respectively) were paired with primers designed immediately before the start (ORF#_LA_rev) and after the end (ORF#_RA_rev) of the target gene. An adapter sequence (AF; 5’ GGTATAGGCGGAAGCGCC 3’) was added to the 3’ end of the LA reverse primer (AF_ORF#_LA_rev) and a second adapter (AR; 5’ GAACAGAAACTGATTAGCGAAGAAGAC 3’) was added to the 5’ end of the RA forward primer (AR_ORF#_RA_for). Each of these primers were designed to have a predicted melting temperature around 60°C. Pairing the ORF#_LA_for_1k primer with the AF_ORF#_LA_rev and the AR_ORF#_RA_for with ORF#_RA_1k_rev generated approximately 1 kb of sequence of both the left and right flanking homologous sequences with adapters at the 3’ end of the LA and the 5’ end of the RA. Using primers AF-CMV for and AR-NeoR rev, which target the cytomegalovirus (CMV)-green fluorescent protein (GFP)-neomycin resistance gene, which we will refer to as CMV-GNR, was PCR amplified using a pcDNA3.1 vector containing the GNR construct as a template. For each PCR reaction, 50 ng of plasmid or 100 ng of viral DNA was added to the High Fidelity PCR Master Mix according to the manufacturer’s instructions (Roche) and DNA was amplified with a single cycle of 94°C for 2 minutes, followed by 25 cycles of 94°C (30 seconds), 50°C (for primer sets seq for/rev and 500_for/rev) or 55°C (30 seconds) (for primer set 1k_for/rev), 72°C (90 seconds) and a final cycle of 72°C for 7 minutes. PCR products were visualized by 1% agarose gel electrophoresis and products were purified by Wizard® SV Gel and PCR Clean-Up System (Promega) system as described by the manufacturer after excision from 0.7% agarose gel. Purified PCR products were quantified by Nanodrop spectrophotometry. At this point we have three purified PCR products for each ATV ORF: the LA, RA and CMV-GNR.
To generate a recombination cassette by overlapping PCR, 50 ng of each PCR product (LA, RA and CMV-GNR) was added to 45 μl reaction (final volume) containing 1X iProof HF buffer, 200 μM of each dNTP, and 0.02 U/μl iProof DNA polymerase (BioRad). The recombination cassette assembly was initiated by a single cycle of 98°C (30 seconds), followed by 7 cycles of 98°C (10 seconds), 58°C (28 minutes), 72°C (150 seconds). After the completion of this program, 0.5 μM of the ORF#_LA_1k_for and ORF#_RA_1k_rev were added along with another 0.02 U/μl iProof DNA polymerase.
The reaction was then returned to the thermocycler and a second program consisting of a single cycle of 98°C (30 seconds), followed by 35 cycles of 98°C (10 seconds), 55°C (30 seconds), 72°C (150 seconds) and a final cycle of 72°C for 5 minutes was performed. PCR products were visualized and purified as described above. Purified recombination cassettes were then re-amplified using the ORF#_LA_500_for and ORF#_RA_500_rev primers using the High Fidelity PCR Master Mix as described above. PCR products were visualized and purified as described above and then cloned into pCR2.1®-TOPO® cloning vector as per the manufacturer’s instructions (Thermo Fisher Scientific). Colonies were screened for the recombination cassette using the seq for/rev primer set for each ORF (Table 2) and correctly constructed recombination cassettes were confirmed by sequencing. The recombination cassette was PCR amplified from the plasmid, agarose gel purified and quantified as described above for use in generating a knockout virus.
Table 2.
Primers used in this study.
| Name1 | Sequence (5′ – 3′) |
|---|---|
| AF-CMV for | GGTATAGGCGGAAGCGCCATGATGTACGGGCCAGATATACG |
| AR-NeoR rev | GTCTTCTTCGCTAATCAGTTTCTGTTCTCAGAAGAACTCGTCAAGAAGG |
| 11R seq for | TGTTATTCGTGGAGGCTG |
| 11R seq rev | ACAGAGACATTCCAGTCG |
| 11R LA for 1k | GTGATACGCACTCTCACTTGC |
| 11R LA for 500 | CGTCATGACCTGCACCG |
| 11R AF LA rev | GGCGCTTCCGCCTATACCTGCATTTTAAATGGACCCC |
| 11R RA rev 1k | GGCTGCAGACCCGGAACAG |
| 11R RA rev 500 | AACCCAATCGGACTTTGG |
| 11R AR RA for | GAACAGAAACTGATTAGCGAAGAAGACGGGATATAATCCAGAGATGAAC |
| 25R seq for | TCTCTCTGAAAAGACATTGC |
| 25R seq rev | GTTACACCTCATTCTCACG |
| 25R LA for 1k | AGTGTTTAGGGACTGGAAAGACG |
| 25R LA for 500 | GCAAGTGTAGGGTTCTCC |
| 25R AF LA rev | CATGGCGCTTCCGCCTATACCCTCGAGTGACTTGTTTTTC |
| 25R RA rev 1k | TCTCTCGGACTATGTGTCG |
| 25R RA rev 500 | CGAGTATAAATGTTGTACCAC |
| 25R AR RA for | GAACAGAAACTGATTAGCGAAGAAGACATAGAGTTTTAAACCTTTTCC |
| 40L seq for | GATGCACTGCCAATACAG |
| 40L seq rev | AAAACACTATTGCAAACACC |
| 40L LA for 1k | ACAAAATTGGATGGATGCATG |
| 40L LA for 500 | TAAAGCTCTCTGAGCACG |
| 40L AF LA rev | GGCGCTTCCGCCTATACCGGCTTATTGTGTAAAGCTGG |
| 40L RA rev 1k | AGTGCGGTCCAGTTTGCG |
| 40L RA rev 500 | ACGTGATACTTCCAGTCG |
| 40L AR RA for | GAACAGAAACTGATTAGCGAAGAAGACATAGAGATTAAGGACTTGTAGC |
| 53R seq for | TTTCGGGACCATTCACAG |
| 53R seq rev | GGTTGGAAACTAGCTAGC |
| 53R LA for 1k | ATGATCTTGGCGTAAGCC |
| 53R LA for 500 | CGATCGGCAACAGTCTAAG |
| 53R AF LA rev | GGCGCTTCCGCCTATACCTGTTGCGAGTTTAGTTTTGG |
| 53R RA rev 1k | TCCAGGATGGTGTACCTG |
| 53R RA rev 500 | TTACGGCGGCAACTGTCC |
| 53R AR RA for | GAACAGAAACTGATTAGCGAAGAAGACGCTGTAAAATAGTTTAGAGAC |
| 54R seq for | GATTCTATCACAACATTTTTAG |
| 54R seq rev | GAAAATATTTTATCACAAGATAC |
| 54R LA for 1k | GTTTTTTGCAACCGCACC |
| 54R LA for 500 | AAAACTGTGACGTTTGTGC |
| 54R AF LA rev | CATGGCGCTTCCGCCTATACC-TTGCTGAACCTGACGCAC |
| 54R RA rev 1k | GTGTCCTCTTTGGTCCTG |
| 54R RA rev 500 | CTAGAACAAGAATGAGGTG |
| 54R AR RA for | GAACAGAAACTGATTAGCGAAGAAGACGGCTTCTTAAACTGTTTCC |
GFP = green fluorescent protein; NeoR = neomycin phosphotransferase resistance gene; CMV = cytomegalovirus promoter; AF = adapter forward; AR = adapter reverse; for = forward; rev = reverse; seq = sequencing primer, ~50 bp from start/end of gene; 1k = ~1,000 bp from start/end of gene; 500 = ~500 bp from start/end of gene.
2.4 Generating Knockout ATV
Approximately 50% confluent monolayers of FHM cells in 35 mm dishes were infected with wtATV at a MOI of 0.01 for 1 hour at room temperature. While the virus was attaching, 500 ng of the target ATV ORF recombination cassette that had been PCR amplified and purified was added to FuGene® 6 transfection reagent according to the manufacturer’s instructions (Promega). This solution was incubated at room temperature for 20 minutes. After 1 hour, the virus inoculum was removed and replaced with the DNA-FuGene® 6 mixture. Cells were rocked with the transfection mixture for 1 hour at room temperature. After rocking, the infected/transfected cells were overlayed with 1X HMEM medium containing 5% FBS and incubated for 48 hours. Infections were then harvested and subjected to three rounds of freeze-thaw to release virus from the cell. The sample was then clarified by centrifugation at 1,000 x g for 10 minutes and recombinant viruses were selected by multiple blind passages in confluent monolayers of FHM cells in the presence of 1 mg/mL G418 (i.e. neomycin). wtATV, which is sensitive to G418, was used as a control. The presence of a GFP expressing, neomycin resistant virus plaque was indicative of the generation of a recombinant ATV with a knock-out of the target gene. GFP-neomycin resistant virus was then plaque purified up to four times in the presence of 1 mg/ml G418, grown to high titers as described above and viral DNA was isolated as previously described (Jancovich and Jacobs, 2011). PCR confirmation of the ORF knock-out virus and sequencing around the ATV gene of interest was performed using the seq for/rev primer pair described above.
3. Results
3.1 Generating Recombination Cassettes
To test ATV ORFs for being either essential or non-essential we first had to generate recombination cassettes for each targeted gene. A recombination cassette contains a screenable and selectable marker driven by a promoter and this construct is flanked by up- and downstream homologous sequences surrounding the target gene (Fig. 1A). We, and others making recombinant ranaviruses, have previously used an intrinsic ranavirus promoter consisting of the 200 nt of upstream sequence from the ICP-18 gene (Andino et al., 2015; Chen et al., 2011; Jancovich and Jacobs, 2011). However, upstream ranavirus flanking sequences also contain intrinsic viral promoters that may be similar in sequence to the ICP-18 promoter. As a result, this presented a challenge when performing overlapping PCR (data not shown). Therefore, we decided to use a cytomegalovirus (CMV) promoter to drive expression of the screenable/selectable marker GNR. This universal promoter, while around 800 nt in length, has limited homology to ranavirus promoters and therefore eliminated any technical complications with optimizing overlapping PCR. We used a green fluorescent protein (GFP) gene fused to neomycin resistance (NeoR) gene as our screenable and selectable marker since ATV is sensitive to neomycin (i.e. G418) and a neomycin phosphotransferase (i.e. G418 or neomycin resistance gene) has been used previously to generate recombinant ATV (Jancovich and Jacobs, 2011). This construct was termed CMV-GNR.
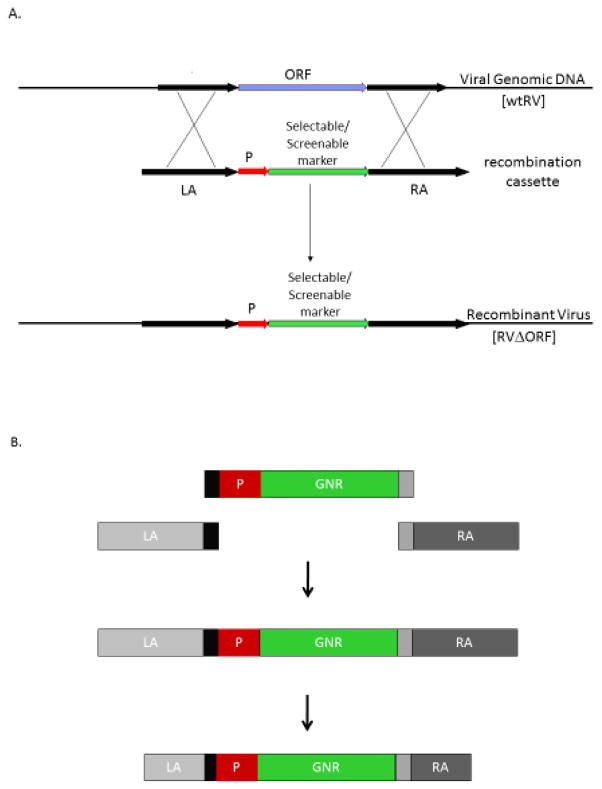
Figure 1.
Generating a knock-out ranavirus. (A.) Schematic representation of homologous recombination to knock-out a target gene or open reading frame (ORF). The process of generating a knock-out ranavirus (RV) deleted of the target gene requires the generation of a recombination cassette that contains homologous sequences (LA and RA) flanking a screenable and selectable reporter gene driven by a promoter (P). Cells are infected with wild-type virus and then transfected with the recombination cassette. Cells and virus are harvested after 48 hours and the recombinant virus deleted of the target ORF is selected by serial passaging in cells treated with selection specific components. Recombinant virus deleted of the target ORF will be resistant to the selection substance and produce easily observable plaques. Modified from Jancovich and Jacobs, 2011. (B.) Schematic representation of the standardized process to generate a recombination cassette. Primers are designed to amplify approximately 1,000 nt of the upstream (LA) and downstream (RA) flanking sequences for each target ORF and for the CMV-GNR cassette. The LA has an adapter sequence added to the 3’ end and the same adapter sequence is added to the 5’ end of CMV promoter. In addition, an different adapter is added to the 3’ end of GNR and the 5’ end of the RA. A standard overlapping PCR protocol assembles the recombination cassette. The recombination cassette is then agarose gel purified and re-amplified using primers that truncate the LA and RA sequences to around 500 nt. This PCR product is cloned, sequenced and used to generate a recombinant virus. P = promoter; LA = upstream flanking sequence; RA = downstream flanking sequence; GNR = GFP-neomycin resistant RV = ranavirus.
We then needed to develop a methodology that would allow us to standardize the process of generating a recombination cassette and knock-out ATV. While overlapping PCR is an efficient method for this process, each step to construct the recombination cassette must be individually optimized for each target ORF. In addition, using restriction enzymes to assemble recombination cassettes presents another challenge of having specific enzyme sites to clone each flanking sequence. This, however, will not work if the restriction site is found in the flanking sequences thereby requiring unique restriction sites and enzymes to construct each target ORF recombination cassette. Therefore, to standardize this process we utilized a forward (AF) and reverse (AR) adapter sequences. Adding these adapters to the 3’ end of the LA, the 5’ and 3’ end of the CMV-GNR construct and the 5’ end of RA sequences allows us to generate an overlapping PCR product for any target ORF using a standard overlapping PCR protocol (Fig. 1B).
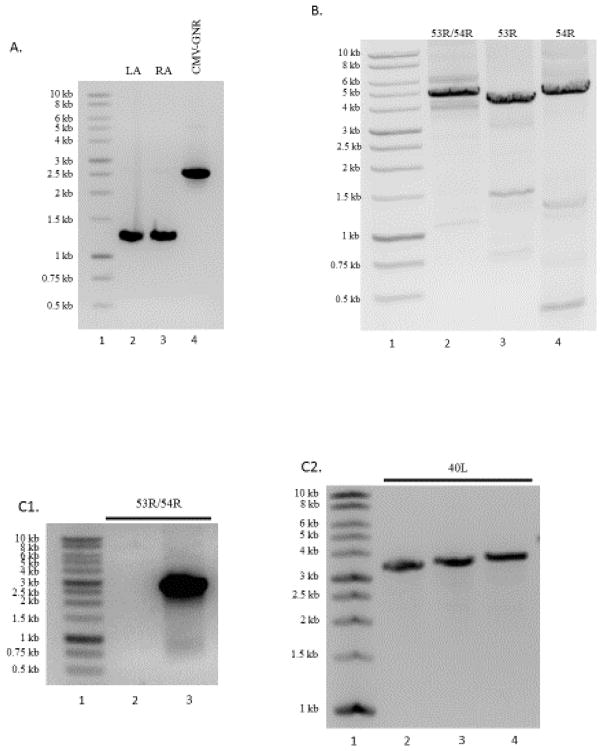
Recombination cassettes to knock-out 11R, 25R, 40L, 53R, 54R and 53/54R double deletion were assembled. We have included a representative agarose gel showing the PCR amplification of each individual piece of a recombination cassette that will be assembled (Fig. 2A) and the overlapping PCR assembly of recombination cassettes for 53R, 54R and a double deletion mutant 53/54R (Fig. 2B). The size of each LA/RA and overlapping PCR product varies based on where primers were designed to amplify the LA and RA homologous sequences. In addition to the recombination cassette, the prominent band around 4,500 nt in each lane, other non-specific bands can be observed in the gel (Fig. 2B). Our initial attempts to generate a recombinant ATV using this gel purified, overlapping PCR product proved difficult, as there was not enough correctly assembled DNA that could be extracted and used to efficiently recombine with viral DNA to generate a recombinant knock-out virus. In addition, re-amplification of the isolated PCR product using the ORF#_LA_1k_for and ORF#_RA_1k_rev primer set and the iProof DNA polymerase PCR assay repeatedly did not re-amplify this assembled PCR product (data not shown). As a result, we designed primers to re-amplify and truncate the LA and RA homologous sequences (i.e. ORF#_LA_500_for and ORF#_RA_500_rev). PCR was performed using the High Fidelity PCR Master Mix (Roche) resulting in a recombination cassette with truncated LA and RA sequences to around 500 nt in length. In addition, the use of the master mix allowed for TOPO® cloning of the PCR product. Colonies were screened for the correct insertion of the recombination cassette using the seq for/rev primer set for each target ORF (Fig. 2C1 and 2C2). We found that cloning this re-amplified recombination cassette was an important step in this process for the following reasons: (i) DNA can be sequenced using primers found on the plasmid as well as in the cloned PCR construct allowing for full sequence information to be obtained for each recombination cassette without designing more primers; (ii) the plasmid containing the recombination cassette is stable and can be stored as a glycerol stock in E. coli for future amplification (i.e. repeating experiments); (iii) the DNA molecule produced from re-amplification from the plasmid is pure and without any non-specific DNA allowing for efficient, repeatable construction of recombinant knock-out ATV. Re-amplified recombination cassette DNA was then agarose gel purified, quantified and used to generate a recombinant virus.
Figure 2.
Recombination cassette assembly. (A.) PCR amplification of the LA, RA and CMV-GNR products (lanes 2, 3 and 4, respectively). (B.) Assembly by overlapping PCR. Individual PCR products for the LA, RA and CMV-GNR are combined and assembled using a standard PCR protocol. Lane 1, molecular weight marker, and lanes 2 – 4 show the assembly of recombination cassettes to delete ORFs 53R and 54R (lane 2), 53R (lane 3) and 54R (lane 4) from ATV. (C.1). PCR screening of two colonies for the 53R/54R double mutant recombination cassette. A colony without the recombination cassette (lane 2) and one colony with the correctly inserted recombination cassette DNA (lane 3). (C.2). PCR colony screening of 3 colonies for the 40L recombination cassette. Each colony screened (lanes 2 – 4) contains the correct insertion of the 40L recombination cassette. LA = upstream flanking sequence; RA = downstream flanking sequence; CMV-GNR = cytomegalovirus promoter-GFP-neomycin resistance.
3.2 Generating Knockout ATV
We previously generated recombinant ATV using a MOI of 5 (Jancovich and Jacobs, 2011). While this amount of virus was adequate for creating a knock-out virus, the large amount of background wtATV required many rounds of selection and purification before a recombinant virus could be visualized and isolated. As a result, we reduced the MOI and therefore could visualize and purify a recombinant ATV in a much more timely and efficient manner. Since wtATV is sensitive to G418, each experiment included a wtATV as a control and we would consider the experiment a success if wtATV plaques were not present after 3 rounds of passage and selection using 1 mg/ml G418.
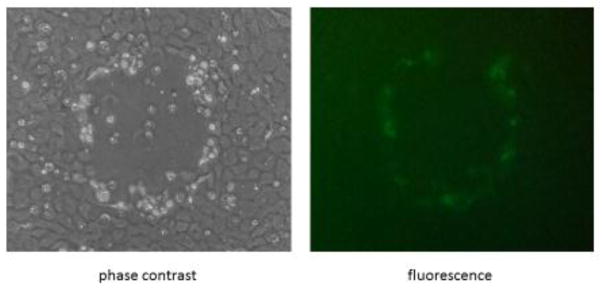
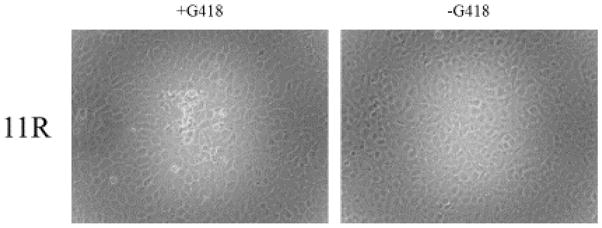
If the target gene is non-essential, GFP expressing, neomycin resistant plaques can be observed in as few as 2 to 3 blind passages (Fig. 3). Green plaques can then be plaque purified and the generation of a recombinant virus confirmed by PCR (Fig. 4). In contrast, semi-essential ORFs will show neomycin resistant green plaques in the presence of G418 but have an extremely small plaque phenotype when grown in the absence of G418 (Fig. 5, ATVΔ25R) or quickly revert back to wt virus once selection components are removed. ORFs that are essential cannot be deleted from the virus. In this situation, after multiple passages of the in vitro recombination experiment, either GFP expressing-neomycin resistant plaques are never observed (data not shown) or small, punctate regions on G418 treated cells can be observed (Fig. 6). These punctate clusters suggest some transfer of the neomycin resistant gene; however, GFP expressing cells or viral associated CPE was never observed when trying to knock-out an essential gene. Experiments were repeated multiple times to ensure the target gene was knocked-out (i.e. non-essential or semi-essential ORF) or unable to be knocked-out (i.e. essential ORF).
Figure 3.
Identification of a recombinant knock-out ranavirus. ATVΔ25R plaque under phase contrast and fluorescent microscopy. Recombinant ATV deleted of semi-essential or non-essential genes will be green and neomycin resistant as shown.
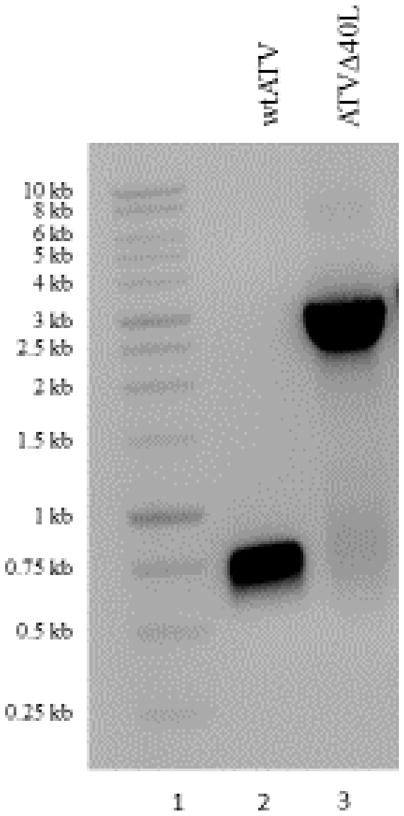
Figure 4.
PCR confirmation of a recombinant ranavirus. DNA isolated from wtATV shows a band with the molecular corresponding to the 40L ORF (lane 2) when using the sequence forward and reverse primers for that target ORF. In contrast, plaque purified ATVΔ40L shows a PCR product with a molecular weight consistent with the CMV-GNR construct (lane 3) thereby confirming knock-out of the target ORF.
Figure 5.
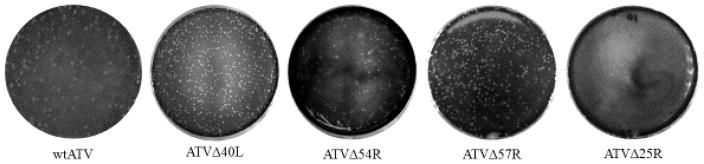
Plaque size comparison. Plaque morphology of wtATV as compared to ATVΔ40L, ATVΔ54R ATVΔ57R and ATVΔ25R. ATV ORFs 40L, 54R and 57R are non-essential gene in cells in culture (Jancovich and Jacobs, 2011). Each non-essentail ATV gene (i.e. ATVΔ40L, ATVΔ54R and ATVΔ57R) show a slight reduction in plaque size as compared to wtATV. In contrast, deletion of the ATV 25R ORF shows a much reduced plaque size compared to either the wtATV or any other recombinant virus.
Figure 6.
Essential ATV ORFs. ATV ORF 11R is essential as plaques expressing GFP in cells treated with or without neomycin are never observed.
ATV ORFs 40L and 54R were classified as non-essential genes as recombinant virus was easily isolated and purified. These ORF deletions show plaque morphologies that are similar to the deletion of another non-essential ATV gene, ORF 57R (Fig. 5) (Jancovich and Jacobs, 2011). ORFs 11R and 53R were classified as essential genes because recombinant viruses were never isolated and we could never generate a recombinant virus deleted of both the 53R and 54R ORFs because 53R is an essential gene and 54R is a non-essential gene. ATV ORF 25R was classified as a semi-essential gene as this recombinant could be generated and purified, but reverted back to wild-type once a selectable agent was removed during growth assays in cells in culture.
4. Conclusions
We have developed a standardized process and protocol to test ranavirus ORFs for being essential or non-essential in cells in culture by knock-out homologous recombination. This protocol requires the design of 6 unique primers (3 primer sets) for each target ORF and two universal primers (1 primer set) to assemble each recombination cassette, in addition to two other primers (1 primer set) used for diagnostics and sequencing across the target region. Once each piece of the recombination cassette has been generated by PCR, assembly of the recombination cassette can be performed using a standard PCR protocol that does not require optimization for each target ORF. Therefore, this unique assembly process can be used to screen any ORF in a ranavirus genome.
Other protocols to assemble and generate recombinant ranaviruses have been developed (Andino et al., 2015; Chen et al., 2011; He et al., 2012; Martin et al., 2015). While these protocols successfully generate recombinant ranaviruses deleted of the target ORF, our procedure does not require expensive DNA synthesis, the use of restriction enzymes or a complicated primer design to create a recombination cassette. Our protocol standardizes the process of recombination cassette assembly (i.e. overlapping PCR) in addition to maximizing the efficiency of knocking-out nonessential or semi-essential ranavirus genes (i.e. low MOI). In addition, our methodology utilizes a linear recombination cassette rather than a circular plasmid. Therefore, any recombinant virus generated is the result of knocking-out the target gene completely, as two recombination events are required for a mutant virus to be neomycin resistant and expressing GFP using a linear PCR product. In contrast, a single recombination event is possible when using circular DNA vectors for recombination cassettes resulting in a recombinant that can be selected and isolated but may not knock-out the target gene until two recombination events take place. Therefore, our protocol and procedure are advantageous over currently used methodologies.
Other technology to generate recombinant dsDNA viruses have been recently developed. For example, the CRISPR-Cas9 system has been optimized for generating recombinant poxviruses (Bi et al., 2014; Yuan et al., 2015a; Yuan et al., 2015b). However, at this time this technology has not been adapted for ranaviruses. That said, the protocol and procedure we have developed was used to identify two non-essential, one semi-essential and two essential genes in ATV. Therefore, our approach was successful identifying essential, semi-essential and non-essential genes. In addition, our standardized procedures will allow us to test other genes in ATV in hopes of learning more about the host-pathogen interactions.
This research has offered insight into ATV ORFs that play a role in viral replication in cells in culture as well as the genetic determinants of ATV pathogenesis. Generating ATVΔ40L was not surprising as an orthologue of this gene has been deleted from frog virus 3 (FV3) (Andino et al., 2015). Deletion of this gene showed reduced pathogenicity in Xenopus and therefore we predict ATVΔ40L will be attenuated in its natural host upon challenge as compared to wtATV. In addition, we predicted, based on other studies with RNase III-like genes (Hussain et al., 2010) that the ATV 25R would be a challenging, if not impossible, ORF to delete because this gene plays a significant role in viral replication. We were successful at generating an RNase III-like gene knock-out in ATV but this virus is very unstable and always reverts back to wtATV once neomycin is removed from the growth medium. Therefore this ATV gene plays a significant role in viral replication and therefore must influence viral pathogenesis. However, we were surprised to identify ATV ORF 53R as being essential and 54R non-essential. Previous studies in tiger frog virus suggest these multi-gene family ORFs were semi-essential based on RNAi (Xie et al., 2014). Perhaps since these two ORFs share sequence homology the RNAi assay knocked both genes down when using either the RNA inhibitory molecules developed for each ORF resulting in the classification of both ORFs as semi-essential (Xie et al., 2014). This provides a great example of the power of our technique and approach to identifying essential ORFs in ranaviruses. Finally, ATV ORF 11R has an unknown predicted function, although BLAST analysis shows homology with bacterial protein of unknown function. We were routinely unsuccessful at generating a knock-out 11R ATV. Therefore, this ATV gene plays an important role in ranavirus replication and further experimentation will be required to discover its function.
In summary, we have successfully developed and show the utility of a protocol to identify essential and non-essential genes in ATV. We are currently using this standardized process to continue to identify essential and non-essential genes in ATV. In addition, we are currently characterizing the function of the non-essential and semi-essential genes in ATV in hopes of better understanding the molecular mechanisms of viral pathogenesis in its natural host, tiger salamanders (Ambystoma tigrinum).
Highlights.
Develop a standardized protocol and procedure to generate recombination cassettes used to knock-out ranavirus genes
Optimized the recombination protocol to generate a knock-out ranavirus
Identified 2 essential, one semi-essential and 2 non-essential genes in Ambystoma tigrinum virus
Acknowledgments
This research was funded by a grant from the National Institutes of Health (1-R15AI101889-01).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andino FD, Grayfer L, Chen GC, Chinchar VG, Edholm ES, Robert J. Characterization of Frog Virus 3 knockout mutants lacking putative virulence genes. Virology. 2015;485:162–170. doi: 10.1016/j.virol.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Sun L, Gao D, Ding C, Li Z, Li Y, Cun W, Li Q. High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. Plos Pathog. 2014;10:e1004090. doi: 10.1371/journal.ppat.1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GC, Ward BM, Yu KH, Chinchar VG, Robert J. Improved Knockout Methodology Reveals That Frog Virus 3 Mutants Lacking either the 18K Immediate-Early Gene or the Truncated vIF-2 alpha Gene Are Defective for Replication and Growth In Vivo. Journal of Virology. 2011;85:11131–11138. doi: 10.1128/JVI.05589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchar VG. Ranaviruses (family Iridoviridae): emerging cold-blooded killers. Arch Virol. 2002;147:447–470. doi: 10.1007/s007050200000. [DOI] [PubMed] [Google Scholar]
- Chinchar VG, Hyatt A, Miyazaki T, Williams T. Family Iridoviridae: Poor Viral Relations No Longer. Curr Top Microbiol. 2009;328:123–170. doi: 10.1007/978-3-540-68618-7_4. [DOI] [PubMed] [Google Scholar]
- Chinchar VG, Yu KH, Jancovich JK. The Molecular Biology of Frog Virus 3 and other Iridoviruses Infecting Cold-Blooded Vertebrates. Viruses-Basel. 2011;3:1959–1985. doi: 10.3390/v3101959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton HE, Metcalf J, Penny E, Tcherepanov V, Upton C, Brunetti CR. Comparative genomic analysis of the family Iridoviridae: re-annotating and defining the core set of iridovirus genes. Virol J. 2007:4. doi: 10.1186/1743-422X-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He LB, Ke F, Zhang QY. Rana grylio virus as a vector for foreign gene expression in fish cells. Virus Research. 2012;163:66–73. doi: 10.1016/j.virusres.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Hussain M, Abraham AM, Asgari S. An Ascovirus-encoded RNase III autoregulates its expression and suppresses RNA interference-mediated gene silencing. J Virol. 2010;84:3624–3630. doi: 10.1128/JVI.02362-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancovich J, Qin Q, Zhang Q-Y, Chinchar VG. Ranavirus Replication: Molecular, Cellular, and Immunological Events. In: Gray MJ, Chinchar VG, editors. Ranaviruses. Springer International Publishing; 2015a. pp. 105–139. [Google Scholar]
- Jancovich J, Steckler N, Waltzek T. Ranavirus Taxonomy and Phylogeny. In: Gray MJ, Chinchar VG, editors. Ranaviruses. Springer International Publishing; 2015b. pp. 59–70. [Google Scholar]
- Jancovich JK, Bremont M, Touchman JW, Jacobs BL. Evidence for Multiple Recent Host Species Shifts among the Ranaviruses (Family Iridoviridae) Journal of Virology. 2010;84:2636–2647. doi: 10.1128/JVI.01991-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancovich JK, Davids EW, Seiler A, Jacobs BL, Collins JP. Transmission of the Ambystoma tigrinum virus to alternative hosts. Dis Aquat Organ. 2001;46:159–163. doi: 10.3354/dao046159. [DOI] [PubMed] [Google Scholar]
- Jancovich JK, Davidson EW, Morado JF, Jacobs BL, Collins JP. Isolation of a lethal virus from the endangered tiger salamander Ambystoma tigrinum stebbinsi. Diseases of Aquatic Organisms. 1997;31:161–167. [Google Scholar]
- Jancovich JK, Jacobs BL. Innate Immune Evasion Mediated by the Ambystoma tigrinum Virus Eukaryotic Translation Initiation Factor 2 alpha Homologue. Journal of Virology. 2011;85:5061–5069. doi: 10.1128/JVI.01488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majji S, Thodima V, Sample R, Whitley D, Deng Y, Mao J, Chinchar VG. Transcriptome analysis of Frog virus 3, the type species of the genus Ranavirus, family Iridoviridae. Virology. 2009;391:293–303. doi: 10.1016/j.virol.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V, Mavian C, Bueno AL, de Molina A, Diaz E, Andres G, Alcami A, Alejo A. Establishment of a Zebrafish Infection Model for the Study of Wild-Type and Recombinant European Sheatfish Virus. Journal of Virology. 2015;89:10702–10706. doi: 10.1128/JVI.01580-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample R, Bryan L, Long S, Majji S, Hoskins G, Sinning A, Olivier J, Chinchar VG. Inhibition of iridovirus protein synthesis and virus replication by antisense morpholino oligonucleotides targeted to the major capsid protein, the 18 kDa immediate-early protein, and a viral homolog of RNA polymerase II. Virology. 2007;358:311–320. doi: 10.1016/j.virol.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Schock DM, Bollinger TK, Chinchar VG, Jancovich JK, Collins JP. Experimental evidence that amphibian ranaviruses are multi-host pathogens. Copeia. 2008:133–143. [Google Scholar]
- Schock DM, Bollinger TK, Collins JP. Mortality Rates Differ Among Amphibian Populations Exposed to Three Strains of a Lethal Ranavirus. Ecohealth. 2009;6:438–448. doi: 10.1007/s10393-010-0279-0. [DOI] [PubMed] [Google Scholar]
- Tompkins DM, Carver S, Jones ME, Krkosek M, Skerratt LF. Emerging infectious diseases of wildlife: a critical perspective. Trends in parasitology. 2015;31:149–159. doi: 10.1016/j.pt.2015.01.007. [DOI] [PubMed] [Google Scholar]
- Whitley DS, Sample RC, Sinning AR, Henegar J, Chinchar VG. Antisense approaches for elucidating ranavirus gene function in an infected fish cell line. Dev Comp Immunol. 2011;35:937–948. doi: 10.1016/j.dci.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Xie JF, Lai YX, Huang LJ, Huang RQ, Yang SW, Shi Y, Weng SP, Zhang Y, He JG. Genome-wide analyses of proliferation-important genes of Iridovirus-tiger frog virus by RNAi. Virus Research. 2014;189:214–225. doi: 10.1016/j.virusres.2014.05.020. [DOI] [PubMed] [Google Scholar]
- Yuan M, Gao X, Chard LS, Ali Z, Ahmed J, Li Y, Liu P, Lemoine NR, Wang Y. A marker-free system for highly efficient construction of vaccinia virus vectors using CRISPR Cas9. Molecular therapy. Methods & clinical development. 2015a;2:15035. doi: 10.1038/mtm.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Zhang W, Wang J, Al Yaghchi C, Ahmed J, Chard L, Lemoine NR, Wang Y. Efficiently editing the vaccinia virus genome by using the CRISPR-Cas9 system. J Virol. 2015b;89:5176–5179. doi: 10.1128/JVI.00339-15. [DOI] [PMC free article] [PubMed] [Google Scholar]