SUMMARY
The endosomal sorting complex required for transport (ESCRT) is a conserved protein complex that facilitates budding and fission of membranes. It executes a key step in many cellular events, including cytokinesis and multi-vesicular body formation. The ESCRT-III protein Shrub in flies, or its homologs in yeast (Snf7) or humans (CHMP4B), is a critical polymerizing component of ESCRT-III needed to effect membrane fission. We report the structural basis for polymerization of Shrub and define a minimal region required for filament formation. The X-ray structure of the Shrub core shows that individual monomers in the lattice interact in a staggered arrangement using complementary electrostatic surfaces. Mutations that disrupt interface salt bridges interfere with Shrub polymerization and function. Despite substantial sequence divergence and differences in packing interactions, the arrangement of Shrub subunits in the polymer resembles that of Snf7 and other family homologs, suggesting that this intermolecular packing mechanism is shared among ESCRT-III proteins.
Graphical Abstract
INTRODUCTION
The endosomal sorting complex required for transport (ESCRT) pathway is essential for eukaryotic membrane remodeling, with a critical role in numerous cellular functions (recently reviewed by Hurley, 2015) including scission of the cytokinetic bridge, biogenesis of intraluminal vesicles, and reformation/maintenance of the nuclear envelope. In addition to endogenous functions, the ESCRT system is also co-opted by membrane-enveloped viruses to escape the host cell via budding (Carlton and Martin-Serrano, 2007).
The ESCRT pathway is comprised of four major complexes (0, I, II, and III) that assemble at the cytoplasmic face of the membrane and produce outward deformation, budding, and scission. The 0, I, and II complexes determine spatiotemporal assembly of the machinery as well as recruitment of ubiquitylated vesicle cargo (McCullough et al., 2013). The ESCRT-III complex is sufficient for vesicle biogenesis in vitro, suggesting that the proteins making up this complex provide the biophysical force for membrane scission (Chiaruttini et al., 2015; Wollert et al., 2009). Polymer disassembly by the Vps4 ATPase is needed for in vivo function of the ESCRT system, presumably to support recycling of system components for subsequent rounds of vesicle biogenesis or scission (Lata et al., 2008).
The canonical ESCRT-III complex, as established for multi-vesicular body (MVB) formation, has four evolutionarily related core members, each containing a “Snf7” domain of ~165 residues and a variable C-terminal region that mediates interaction with other factors. The complex is formed by sequential loading of vacuolar protein sorting 20 (Vps20), Shrub, Vps24, and Vps2, with Shrub (also known as Snf7 in yeast, CHMP4B in humans) assembling into a polymerized form that is critical for all ESCRT-III functions (Teis et al., 2008; Wollert et al., 2009). In addition to these canonical core components, several accessory Snf7 domain proteins are required for a subset of ESCRT functions, including charged multivesicular body protein 1B (CHMP1B) and increased sodium tolerance 1 (IST1), which are linked to cytokinetic abscission and recruitment/activation of the AAA-ATPase Vps4 to the assembled ESCRT-III complex (Hurley, 2015).
A defining feature of many ESCRT-III family proteins is their propensity to polymerize spontaneously in vitro. These polymers bear striking similarities to ESCRT filaments directly observed in living cells at the cell surface and cytokinetic bridge (Cashikar et al., 2014; Guizetti et al., 2011; Hanson et al., 2008). The working model in the field presumes that polymerization provides the biophysical force responsible for membrane deformation (Chiaruttini et al., 2015). A structural and mechanistic understanding of how ESCRT-III proteins polymerize, most notably the universally required Shrub/Snf7/CHMP4B protein, has therefore been a major goal in the ESCRT field.
Previous crystal structures of the late-acting core member Vps24 and accessory protein IST1 have shown that their Snf7 domains have a central helical hairpin followed by a shorter helical bundle that packs against the hairpin (Bajorek et al., 2009; Muzioł et al., 2006) (Figures 1 and S1A). Mutation and truncation studies suggest that a C-terminal helix in these proteins serves as an auto-inhibitory element (Bajorek et al., 2009; Henne et al., 2012). These structures, however, did not reveal the structural basis for assembly of Snf7-domain proteins into the polymers required for formation of vesicles.
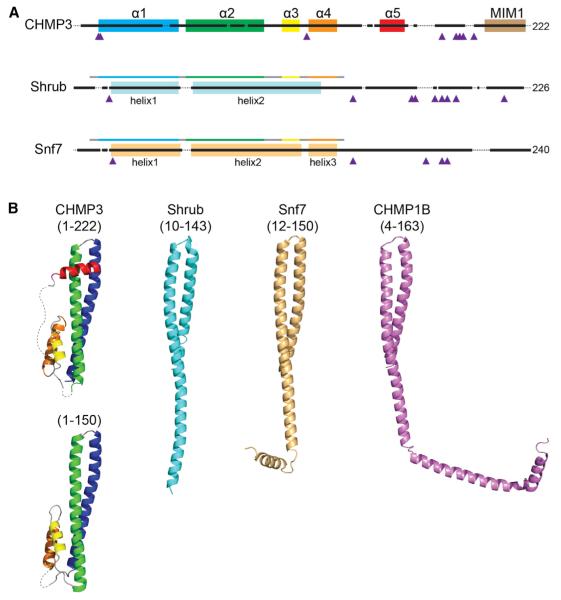
Figure 1. Structural Comparison of Shrub with CHMP3 and Snf7.
(A) Snf7 domain architecture aligned by primary sequence. Helices are indicated by colored boxes and prolines by purple triangles. For Shrub (light blue) and Snf7 (beige), helices are denoted based on the crystal structures, and the fragments used for crystallization of Shrub (10–143; this work) and Snf7 (12–150; Tang et al., 2015) are represented by bars above the structural schematic colored based on alignment to the canonical α1–α4 helices of CHMP3 shown above.
(B) Cartoon representations of CHMP3 (1–222, 3FRT; 1–150, 3FRV), Shrub (10–143, 5J45), Snf7 (12–150, 5FD9), and CHMP1B (4–163, 3JC1), colored using the scheme in (A).
See also Figures S1 and S3.
Here, we combine structural and biochemical studies to elucidate how Shrub, the core component of the ESCRT-III complex in flies, likely assembles into polymers at the atomic level. We report an X-ray structure of the core region of Shrub, which shows that individual Shrub subunits adopt an elongated conformation, and contact one another using complementary electrostatic surfaces in a staggered arrangement in the crystals. The core of the Shrub monomer bears striking similarity to that of the yeast ortholog Snf7, which also exhibits a nearly identical overall packing arrangement despite numerous individual differences in side-chain identities and their interactions (Tang et al., 2015). The structure of a co-polymer formed by distant paralogs of Shrub, the “accessory” factors CHMP1B and IST1, solved by cryo-electron microscopy, demonstrates a similar staggered, head-to-head interaction. Taken together, these findings argue for a universal mechanism of polymer assembly among ESCRT-III Snf7-family proteins.
RESULTS
Structure of the Shrub Polymerization Domain
To begin investigating Shrub structure-function relationships, we expressed a truncated form of the protein encompassing the Snf7 region (residues 10–143 from Drosophila melanogaster) aligned with the four-helix core of Vps24, (Figures 1A and S1) and solved its structure to 2.8 Å resolution (PDB: 5J45). Remarkably, however, this fragment of Shrub does not adopt the four-helix structure of the Vps24 core (Figure 1B), but instead forms a single elongated helical hairpin (Figure 1B), which closely resembles the recently determined structures of the Snf7 ESCRT-III subunit from yeast (Tang et al., 2015) and the CHMP1B subunit within a CHMP1B-IST1 co-polymer (Figure 1B) (McCullough et al., 2015).
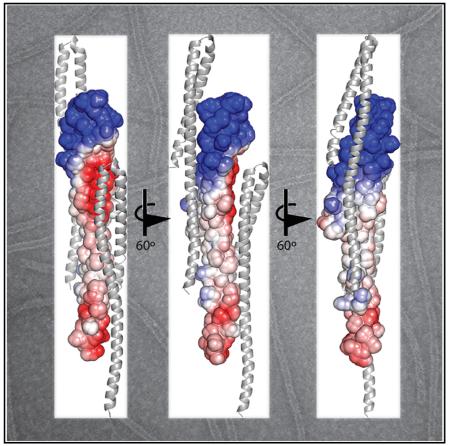
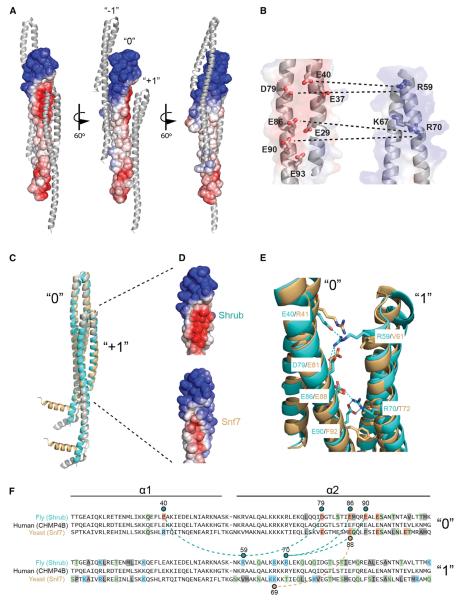
The packing of the monomers in the crystal lattice suggests a structural arrangement for the polymeric Shrub subunits in ESCRT-III complexes. Extensive crystal contacts between molecules in adjacent unit cells bury 760 Å2 of surface area at the subunit interface (Figure 2A). As the unit cell is translated, these adjacent monomers form an unbroken filament within the crystal. The smallest dimension of the crystal filament is ~5 nm, consistent with the range of widths of ESCRT-III polymers (4–15 nm) seen at the cell surface (Cashikar et al., 2014). The intersubunit contacts align so that the positively charged headpiece of one monomer (Figure 2A, subunit “0”) nestles into the negatively charged face of its preceding neighbor (subunit “−1”). The equivalent acidic face of this index monomer (subunit “0”) likewise serves as the acceptor surface for the basic face of the following monomer (subunit “+1”). An “open-book” representation showing the lattice contacts between adjacent subunits in the crystal highlights several salt bridge interactions present at this interface (Figure 2B). This overall charge distribution is highly conserved among metazoan Shrub orthologs (Figure S2). Key charged contacts in Shrub center on R59, which engages E40 and D79, and R70, which pairs with E86 and E90.
Figure 2. Key Lattice Interactions in the Shrub Crystal Structure.
(A) Three molecules from adjacent unit cells are shown and labeled as “−1,” “0,” and “+1.” The “0” subunit is represented as a surface, colored by charge (blue, positive to red, negative). Extensive crystal contacts formed by electrostatic interactions occur between molecules in adjacent unit cells.
(B) The electrostatic interface between adjacent molecules represented in “open book” form. Dashed lines indicate salt bridge interactions.
(C) Superposition of Shrub (cyan) onto the two conformations of the yeast ortholog Snf7 (tan, 5FD9; gray, 5FD7). Two subunits, and their observed crystal contacts, are shown for each of the independent crystal forms.
(D) Electrostatic surface representations (blue, positive to red, negative) for the indicated region of subunit “1.”
The packing arrangement of Shrub in the crystals reported here also bears a striking resemblance to the intermolecular packing of Snf7 in the structure reported recently by Tang et al. (2015). Despite analogous structural features at the global level (Figure 2C), the Shrub intersubunit interface relies heavily on electrostatic complementarity, whereas Snf7 contains a single salt bridge that is buttressed by hydrophobic interactions from nearby side chains (Figures 2D–2F). Of the six residues that form salt bridges at the interface between monomers in the fly structure, only one is conserved in yeast (Figure 2E). Overall, only 14 of the total 36 interface residues from the fly structure are conserved in yeast (Figure 2F). The electrostatic nature of the fly interface, however, appears to be conserved across multicellular organisms (Figure S2). Between flies and humans, for example, all six salt bridge forming residues are conserved (as well as 30 of 36 total interface residues overall; Figure 2F).
Shrub Polymerization Relies on Electrostatic Interactions
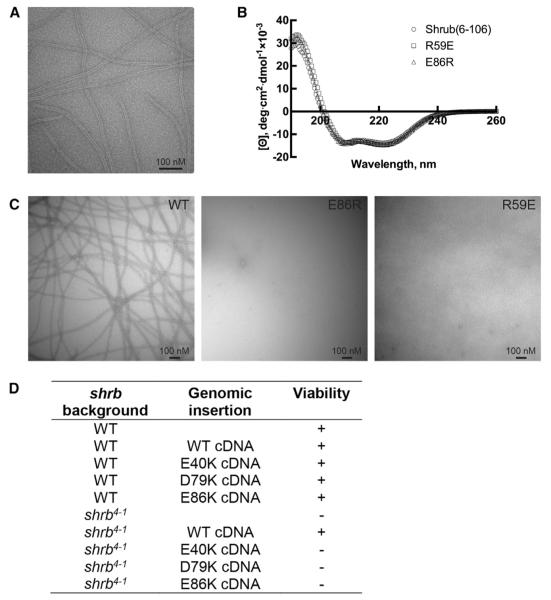
In order to test whether the electrostatic interface observed in the crystal is required for Shrub polymerization, we developed a polymer-formation assay using a fragment of Shrub containing only the electrostatic helical hairpin. A shortened Shrub protein containing residues 6–106 (see Figure S1) was purified in 1 M NaCl to maintain its monomeric state, dialyzed to 0.1 M NaCl, and then examined by negative stain electron microscopy (EM). The EM data show clear evidence for formation of Shrub polymers, which assemble as elongated, striated filaments (Figure 3A). The filament is ~10 nm and uniform in width as visualized by negative stain, dimensions consistent with formation of a polymer using the packing arrangement present in the larger fragment used for crystallization. The protein concentration required to generate observable polymers with the truncated construct is ~5-fold higher than that reported for a full-length Shrub homolog (Henne et al., 2012), likely reflecting an influence on polymer formation by conserved residues on the extended second helix (not present in the shorter Shrub molecule analyzed here). Nevertheless, these results show that the charged helical hairpin of Shrub forms filaments spontaneously and suggest that it functions as the “core” of the growing polymer.
Figure 3. Filamentation Is Driven by Electrostatic Interactions.
(A) The 6-106 fragment of Shrub at a concentration of 250 μM, was polymerized by dialysis into low salt buffer. Filaments were visualized by negative stain electron microscopy at a magnification of 68,000. Scale bars, 100 nM.
(B) Circular dichroism spectra of wild-type, R59E, and E86R variants of Shrub (6–106), acquired at 20°C.
(C) Wild-type Shrub and the indicated mutants were dialyzed to low salt and visualized by negative stain electron microscopy at a magnification of 30,000.
(D) Summary of the results from experiments attempting to rescue shrb4-1 (null) lethality using wild-type or mutant forms of Shrub delivered to the identical genomic landing site.
(E) Overlay of the observed crystal contact interfaces of Shrub (cyan) or Snf7 (light orange). The Shrub side-chains involved in direct salt bridge interactions (dotted lines) and the analogous yeast residues, are represented by sticks.
(F) Primary sequence alignment of the fly, human, and yeast orthologs. Residues at the crystal contact interfaces (within 5 Å of the neighboring protomer) are boxed and colored by class (acidic, red; basic, blue; polar, green; non-polar, gray). Residues involved in salt bridge interactions (dotted lines) are indicated by residue number and with a colored circle above (Shrub) or below (Snf7) the aligned sequences.
See also Figures S2 and S3.
We next disrupted the electrostatic complementarity of the subunit interface with the charge reversal mutations R59E or E86R and tested the ability of the mutated Shrub (6–106) proteins to polymerize spontaneously using the negative-stain EM assay. Both of these variants exhibit circular dichroism (CD) spectra indistinguishable from the wild-type protein, indicating that neither mutation disrupts the structure of the protein (Figure 3B), yet both charge reversal mutations of Shrub inhibit polymerization (Figure 3C). These results indicate that the electrostatic interfaces identified in the crystal lattice are also critical for polymerization in solution.
The putative electrostatic interface of the Shrub polymer was also evaluated using an in vivo rescue model for ESCRT function. Although Shrub-deficient flies (allele shrb4-1) die during embryogenesis and thus preclude cellular analysis, this genotype has been used to screen exogenously expressed Shrub molecules for proper function/phenotypic rescue (Sweeney et al., 2006). Consistent with this previously published work, shrb4-1 lethality is rescued by genomic insertion of the wild-type shrb cDNA sequence and its proximal upstream and downstream elements (Figure 3D). We then introduced single amino acid substitutions on the acidic face (E40K, D79K, or E86K) and tested whether these variants could also rescue the embryonic lethality. In contrast to wild-type Shrub, none of these point mutants were able to rescue the embryonically lethal phenotype (Figure 3D). (Note that wild-type and mutant shrb cDNA sequences were inserted into the same genomic landing site in order to match expression levels, but steady-state protein levels could not be directly analyzed. We made the decision to leave Shrub untagged because tagging at either terminus is likely to affect its functionality [Buchkovich et al., 2013; Sweeney et al., 2006; Teis et al., 2008]). These observations are also consistent with the interpretation that the electrostatic interface is required for Shrub polymerization and pathway function in vivo.
DISCUSSION
The ESCRT pathway is utilized in a diverse and critical set of cellular membrane remodeling functions. The fundamental event in all ESCRT-mediated remodeling is the assembly of filaments by the ESCRT-III polymerization component, called Shrub in flies, Snf7 in yeast, and CHMP4B in mammals.
In contrast to earlier structures of the related CHMP3 and IST1 proteins that form closed helical bundles (Bajorek et al., 2009; Muzioł et al., 2006; Xiao et al., 2009), but appear not to homopolymerize in vivo, Shrub and Snf7 crystallize as elongated helical hairpins. These structures show how the elongated forms of these proteins can serve as extended interaction surfaces for brick-like assembly of protein filaments. Shrub and Snf7 share a nearly identical protomer-protomer packing of subunits in their crystal lattices despite remarkable amino acid substitution along their interaction surfaces. Although the unicellular and multicellular interfaces are both clearly competent for polymerization, the interface in multicellular organisms has evolved to rely more heavily on electrostatic interactions at the interface. As the ESCRT pathway in animals contains an expanded number of homologs as well as regulatory chaperones, it is possible that the extensive electrostatic surface of animal orthologs may be essential for these additional heterotypic and/or regulatory adaptations. Because the fly and human orthologs share extensive sequence identity throughout the charged helical hairpin, the Shrub structure also offers a useful model for future studies of the human system.
One issue not yet fully resolved is whether the open conformation is constitutive for Shrub and its orthologs, or if both open (as observed in the crystals) and closed (as observed for CHMP3) forms exist in the cell. Cysteine cross-linking and pulse dipolar spectroscopy studies of Snf7 suggested a mixed conformational species in solution, but this work was necessarily performed in a context far removed from its normal cellular environment (Tang et al., 2015). If a structural transition is required for Shrub to convert from an inactive to an active conformation, it may be inhibited or triggered by a suite of regulatory factors.
Although required for all ESCRT-mediated functions, the Shrub/CHMP4B/Snf7 orthologs do not have a monopoly on filamentation. Recent work from McCullough et al. (2015) highlights many important similarities and differences between the assembly of a Shrub-type polymer and that of distant family members CHMP1B and IST1, which form large helical co-polymers with an open interior. As solved at ~4 Å resolution by cryo-EM, the copolymer consists of an inner CHMP1B strand, in an elongated form similar to Shrub (Figure 1B), and an exterior surface layer of IST1, present as a closed helical bundle (McCullough et al., 2015). Strikingly, the elongated CHMP1B protein exhibits an intermolecular, staggered head-to-head packing analogous to the crystal filament of Shrub (Figure S3) despite sharing only 14% sequence identity over the helical hairpin and no similarity in overall charge distribution. In addition, the co-polymer suggests an important structural role for the C-terminal portions of Shrub/Snf7, which were truncated in the fragments of these proteins used for the Shrub structure reported here and for the Snf7 structure from the Emr group (Tang et al., 2015). In the CHMP1B polymer, this helical element forms additional intermolecular contacts with distant monomers that stabilize the polymer as well as induce natural curvature (Figure S3).
Our studies provide strong support to an emerging model in which the staggered head-to-head interaction of elongated monomers is a general feature of polymerization of Snf7-family proteins from yeast to mammals. An additional key insight from our investigation of Shrub is the emergence of an important role for electrostatic interactions in driving polymerization of the metazoan Snf7 orthologs, a feature not necessarily evident from studies of Snf7 itself. Although the elongated conformation of Shrub/Snf7 is a prerequisite for this consensus polymerization model, the specification of interaction partners and filament geometry is also likely to depend on evolution of the residues at critical contact positions within this interface. Moreover, there appears to be a class of ESCRT-III proteins that is constitutively closed as a helical bundle, exemplified by Vps24 and IST1. In the case of the CHMP1B/IST1 co-polymer, the “closed” component IST1 alters the membrane-interacting exterior of the polymer. This general theme of heterotypic modulation may be replicated in the Shrub/Snf7 polymer to customize its membrane remodeling activity.
EXPERIMENTAL PROCEDURES
Protein Production
Shrub (residues 6–106 or 10–143) and Lgd (359–423) were cloned into pETHSUL vector using BamHI and XhoI restriction sites. Constructs were transformed into BL21(DE3)pLysS cells and expressed by IPTG induction at 18°C for 16 hr. “High salt” buffer (10 mM HEPES pH 7.4, 1 M NaCl) was used for cell lysis and protein isolation. Proteins were purified by affinity chromatography using HisPur Cobalt (Thermo Scientific) followed by cleavage of the His6-SUMO tag using Ulp1 peptidase. Untagged protein was then purified by size exclusion chromatography. Selenomethionine-labeled protein was produced as described (Molecular Dimensions).
Crystallization and Data Collection
To suppress oligomerization prior to crystallization, Shrub was co-purified with a minimal region of its regulatory chaperone Lethal (2) Giant Discs (Lgd, residues 359–423) (Martinelli et al., 2012; Troost et al., 2012). As previously reported for crystallization of a smaller fragment of human Shrub (Martinelli et al., 2012), the Lgd chaperone disassociated during the crystallization process to produce crystals containing only Shrub (data collection and refinement statistics, Table 1). To produce Lgd:Shrub complexes, each protein was independently isolated in “high salt” buffer, mixed to a molar excess of Lgd, and then dialyzed into “low salt” (10 mM HEPES pH 7.4, 0.1 M NaCl). The complex was then purified by size exclusion chromatography to remove free Lgd.
Table 1.
Crystallographic Data Collection and Refinement
| Data Collection | Shrub (MR-SAD) |
|---|---|
| Space group | P4(3) |
| Cell dimensions | |
| a, b, c (Å) | 29.77, 29.77, 174.31 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength | 0.9792 |
| Resolution (Å) | 50–2.76 (2.91–2.76) |
| Rmerge | 0.09 (1.09) |
| I/σI | 15.9 (1.5) |
| Completeness (%) | 98.1 (99.4) |
| Redundancy | 3.6 (3.7) |
| Wilson B-factor | 84.9 |
|
| |
| Refinement | |
|
| |
| Resolution (Å) | 43.58–2.76 |
| No. reflections | 3,792 |
| Rwork/Rfree | 0.216/0.269 |
| No. atoms | |
| Protein | 890 |
| B-factors | |
| Protein | 94.87 |
| RMSD | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.504 |
| Ramachandran (%) | |
| Favored | 98.2 |
| Allowed | 1.8 |
| Outlier | 0 |
| Rotamer outliers (%) | 4.1 |
| Clashscore | 1.1 |
The complex of selenomethione-labeled Shrub (10–143) and Lgd (359–423) was crystallized by vapor diffusion using the hanging drop method in 17% v/v PEG10K, 0.1 M NH4CH3CO2, and 100 mM Bis-Tris pH 5.5 at 20°C. For cryo-protection, crystals were dipped in the above solution also containing 15% glycerol and 15% ethylene glycol prior to plunging in liquid nitrogen. Data were obtained at the Advanced Photon Source, beam-lines 24-ID-C (NE-CAT).
Structure Determination
Diffraction images were indexed and integrated using XDS (Kabsch, 2010). Initial phases were produced by molecular replacement-single wavelength anomalous diffraction methods in Phenix (Adams et al., 2010) using PDB: 4ABM as a starting model. Refinement was performed in Phenix with experimental phase restraints and manual building/review in COOT (Emsley et al., 2010).
Structural Analysis
Electrostatic potential was determined using the Adaptive Poisson-Boltzmann Solver web server with a PARSE forcefield (Baker et al., 2001; Dolinsky et al., 2004). Surface accessible electrostatic potentials are shown at ±4 kT/e contouring. Homology models were generated using the Phyre2 server (Kelley and Sternberg, 2009) and energy minimization in Phenix. Sequence alignments were performed by Clustal Omega (Sievers and Higgins, 2014). Alignment of 3D structures was performed in Coot using secondary structure matching. Figures were produced using Pymol (Schrödinger).
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectra of wild-type, E86R, and R59E variants of Shrub (6–106) were acquired at protein concentrations between 7.5–15 μM on a Jasco J-815 instrument in phosphate buffer (10 mM, pH 7.4) containing potassium fluoride (1 M) at 20°C. For each protein, data were acquired in an 0.1 cm pathlength cell and represent the average of 10 scans taken at 50 nm/min with a 0.5-nm step size.
Electron Microscopy
Shrub proteins at 250 μM concentration were dialyzed from “high” to “low salt” buffer using 1 kDa cutoff mini-dialysis units (GE Healthcare). Dialyzed proteins were applied to glow-discharged carbon-coated grids, washed, and stained with 0.75% uranyl formate. Images were recorded using a 2-k CCD camera on a Tecnai G2 Spirit BioTWIN microscope operated at 120 kV.
Drosophila Expression and Phenotyping
The following fly stock was used for rescue: shrb4-1 FRTG13 (Sweeney et al., 2006). Rescue constructs were generated containing the shrb cDNA (Drosophila Genomics Resource Center, GH13992) flanked by the proximal shrb genomic elements (510 bp upstream and downstream, from BAC-shrb CH322-47O20; BACPAC Resources Center) and inserted into the genomic attP landing site at 86 Fb by BestGene (Bischof et al., 2007). Insertion chromosomes were balanced over SM6a-TM6B Tb, in order to allow recognition of experimental individuals during pupal stages by the loss of the dominant marker Tb.
Supplementary Material
In Brief.
The ESCRT complex facilitates budding and fission of cellular membranes. McMillan et al. report the X-ray structure of the fly ESCRT-III component Shrub. The packing of subunits in the structure, mirrored in the distant yeast homolog Snf7, suggests a general model for the subunit polymerization step required for membrane fission.
Highlights.
The X-ray structure of the metazoan ESCRT-III protein Shrub is resolved
Shrub polymerization relies on electrostatic interactions
Shrub monomers in the lattice interact in a staggered arrangement
Comparison with Snf7 suggests that the intermolecular packing mechanism is conserved
ACKNOWLEDGMENTS
This work was supported in part by NIH awards CA092433 and CA119070, project 3 (to S.C.B.), and by the Deutsche Forschungsgemeinschaft (DFG) through SFB 1208, project B01 (to T.K.).
Footnotes
ACCESSION NUMBERS
The accession number for the coordinates for the X-ray structure of the Shrub core (residues 10–143) reported in this paper is PDB: 5J45.
Supplemental Information includes three figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.06.093.
AUTHOR CONTRIBUTIONS
Conceptualization, B.J.M., S.C.B., and T.K.; Methodology, B.J.M., S.C.B., and T.K.; Investigation, B.J.M., C.T., H.J., and A.D.; Writing and Editing, B.J.M., S.C.B., T.K., and C.T.; Supervision, S.C.B., B.J.M., and T.K.; Funding, S.C.B. and T.K.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchkovich NJ, Henne WM, Tang S, Emr SD. Essential N-terminal insertion motif anchors the ESCRT-III filament during MVB vesicle formation. Dev. Cell. 2013;27:201–214. doi: 10.1016/j.devcel.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- Cashikar AG, Shim S, Roth R, Maldazys MR, Heuser JE, Hanson PI. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. eLife. 2014;3 doi: 10.7554/eLife.02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, Scheuring S, Roux A. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell. 2015;163:866–879. doi: 10.1016/j.cell.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mäntler J, Maar S, Poser I, Leonhardt H, Müller-Reichert T, Gerlich DW. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 2012;151:356–371. doi: 10.1016/j.cell.2012.08.039. [DOI] [PubMed] [Google Scholar]
- Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli N, Hartlieb B, Usami Y, Sabin C, Dordor A, Miguet N, Avilov SV, Ribeiro EA, Jr., Göttlinger H, Weissenhorn W. CC2D1A is a regulator of ESCRT-III CHMP4B. J. Mol. Biol. 2012;419:75–88. doi: 10.1016/j.jmb.2012.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI, et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 2015;350:1548–1551. doi: 10.1126/science.aad8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzioł T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Göttlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Sievers F, Higgins DG. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- Sweeney NT, Brenman JE, Jan YN, Gao FB. The coiled-coil protein shrub controls neuronal morphogenesis in Drosophila. Curr. Biol. 2006;16:1006–1011. doi: 10.1016/j.cub.2006.03.067. [DOI] [PubMed] [Google Scholar]
- Tang S, Henne WM, Borbat PP, Buchkovich NJ, Freed JH, Mao Y, Fromme JC, Emr SD. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. eLife. 2015;4:e12548. doi: 10.7554/eLife.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Troost T, Jaeckel S, Ohlenhard N, Klein T. The tumour suppressor Lethal (2) giant discs is required for the function of the ESCRT-III component Shrub/CHMP4. J. Cell Sci. 2012;125:763–776. doi: 10.1242/jcs.097261. [DOI] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Chen XW, Davies BA, Saltiel AR, Katzmann DJ, Xu Z. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol. Biol. Cell. 2009;20:3514–3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.