Abstract
Examining complete gene knockouts within a viable organism can inform on gene function. We sequenced the exomes of 3,222 British Pakistani-heritage adults with high parental relatedness, discovering 1,111 rare-variant homozygous genotypes with predicted loss of gene function (knockouts) in 781 genes. We observed 13.7% fewer than expected homozygous knockout genotypes, implying an average load of 1.6 recessive-lethal-equivalent LOF variants per adult. Linking genetic data to lifelong health records, knockouts were not associated with clinical consultation or prescription rate. In this dataset we identified a healthy PRDM9 knockout mother, and performed phased genome sequencing on her, her child and controls, which showed meiotic recombination sites localised away from PRDM9-dependent hotspots. Thus, natural LOF variants inform upon essential genetic loci, and demonstrate PRDM9 redundancy in humans.
Complete gene knockouts, typically caused by homozygous loss of function (LOF) genotypes, have helped identify the function of many genes, predominantly through studies in model organisms and of severe Mendelian-inherited diseases in humans. However, information on the consequences of knocking out most genes in humans is still missing. Naturally occurring complete gene knockouts offer the opportunity to study the effects of lifelong germline gene inactivation in a living human. A survey of LOF variants in adult humans demonstrated ~100 predicted LOF genotypes per individual, describing around ~20 genes carrying homozygous predicted LOF alleles and hence likely completely inactivated(1). Almost all these homozygous genotypes were at common variants with allele frequency >1%, in genes likely to have weak or neutral effects on fitness and health(1). In contrast, rare predicted LOF genotypes were usually heterozygous and thus of uncertain overall impact on gene function. A large exome sequencing aggregation study (ExAC), of predominantly outbred individuals, identified 1,775 genes with homozygous predicted LOF genotypes in 60,706 individuals(2). Furthermore, 1,171 genes with complete predicted LOF were identified in 104,220 Icelandic individuals (3), and modest enrichment for homozygous predicted LOF genotypes shown in Finnish individuals(4). However, even in these large samples, homozygous predicted LOF genotypes tend to be for variants at moderate (around 1%) allele frequency, and hence these approaches will not readily assess knockouts in most genes, which are lacking such variants.
Here, we identify knockouts created by rare homozygous predicted loss of function (rhLOF) variants by exome sequencing 3,222 Pakistani-heritage adults living in the UK who were ascertained as healthy, type 2 diabetic, or pregnant(5). These individuals have a high rate of parental relatedness (often with parents who are first cousins) and thus a substantial fraction of their autosomal genome occurs in long homozygous regions inferred to be identical by descent from a recent common ancestor (autozygous). We link the genotype to healthcare and epidemiological records, with the aims of i) describing the properties of, and assessing the health effects of, naturally occurring knockouts in an adult population, ii) understanding the architecture of gene essentiality in the human genome, through the characterization of the population genetics of LOF variants, and iii) studying in detail a knockout of the PRDM9 gene which plays a role in human meiotic recombination(6).
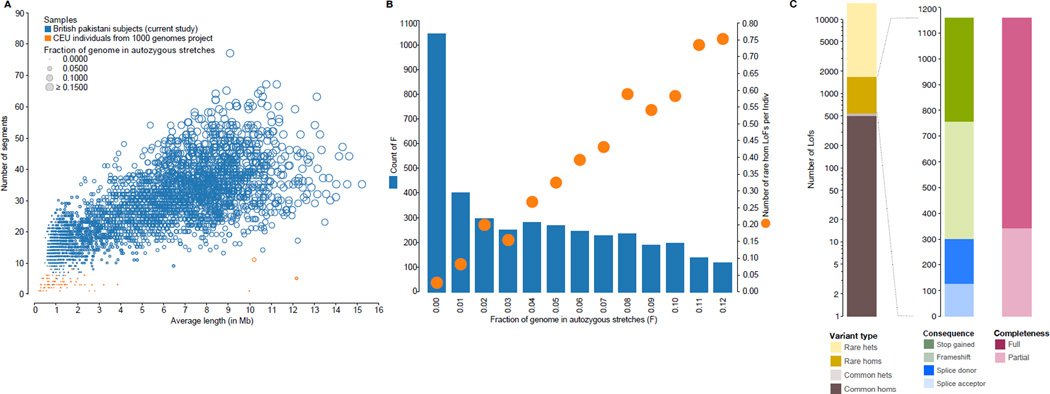
On average, 5.6% of the coding genome was autozygous, much higher than that in outbred European heritage populations (figs. 1A, S4). We identified, per subject, on average 140.3 non-reference predicted LOF genotypes comprising 16.1 rare (minor allele frequency <1%) heterozygotes, 0.34 rare homozygotes, 83.2 common heterozygotes and 40.6 common homozygotes. Nearly all rhLOF genotypes were found within autozygous segments (94.9%)(5), and the mean number of rhLOF per individual was proportional to autozygosity (fig. 1B). Overall we identified 1,111 rhLOF genotypes at 847 variants (575 annotated as LOF in all GENCODE-basic transcripts) in 781 different protein-coding genes (fig. 1C)(5) in 821 individuals. Autozygous segments were observed across all exomic sites with a density distribution not significantly different from random (5)(Shapiro-Wilks P=0.112). From these values we estimate that 41.5% of individuals with 6.25% autozygosity (expected mean for individual with first-cousin related but otherwise outbred parents) will have one or more rhLOF genotypes (fig. 1B).
Fig. 1. Discovery and annotation of rhLOF variants.
(A) Autozygous segment numbers and length for Pakistani heritage subjects in the UK, and 1000 Genomes project European (CEPH Utah residents with ancestry from northern and western Europe; CEU) individuals. (B) Autozygosity and rhLOF in 3,222 individuals. Count of number of individuals (left Y axis, blue columns) binned by fraction of autozygous genome (X axis, showing values from 0.00 to 0.12), with mean number of rhLOF genotypes per individual (right Y axis, orange circles). (C) Distribution of LOF variants by allele frequency, heterozygous or homozygous genotype, predicted protein consequence, and whether predicted for a full or partial set of GENCODE Basic transcripts for the gene.
The majority of identified genes with rhLOF genotypes (422) had not been previously reported, although 167 had been reported as containing homozygous or compound heterozygous LOF genotypes in Iceland, and 299 in ExAC. In total, 107 rhLOF genes were common to all three datasets (5) suggesting a subset of genes either tolerant of LOF and/or with higher rates of mutation. 89 rhLOF genotypes were homozygotes without observed heterozygotes, and we observed three subjects each with 5 rhLOF genotypes. On the basis of these observations we predict that in 100,000 subjects with first-cousin related parents of the same genetic ancestry we would expect at least one knockout in ~9,000 of the ~20,000 human protein-coding genes (fig. S3)(5).
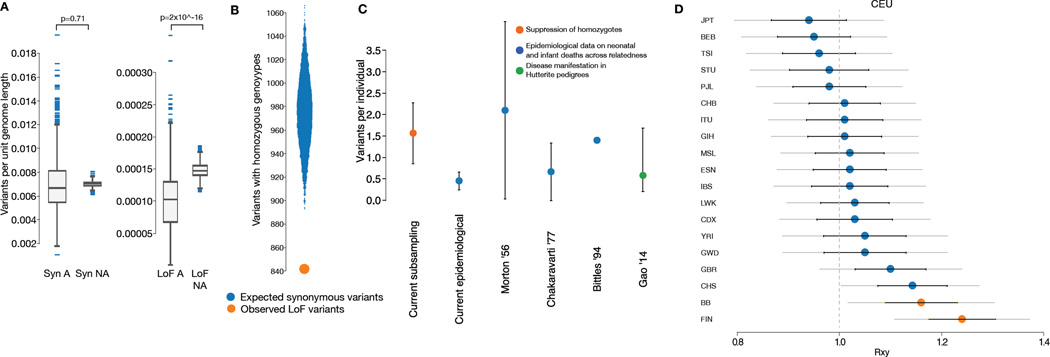
We observed a lower density of annotated rare LOF variants within autozygous tracts, where they are homozygous, compared to outside autozygous tracts, where they are typically heterozygous, indicative of direct negative selection on a fraction of homozygotes (fig. 2A). We matched each of the 16,708 rare annotated LOF (heterozygous and homozygous) variants to a randomly selected synonymous variant of the same allele frequency, and observed 842 rare LOF variants with >= 1 homozygous genotype versus an average of 975.5 rare synonymous variants with >= 1 homozygote, indicating a deficit of 13.7% (95% confidence interval 8–20%) of variants with rhLOF genotypes (fig. 2B)(5). We attribute this deficit to some rhLOF genotypes resulting in early lethality or severe disease and thus being incompatible with our selection criteria as healthier adults, although our data does not inform whether these are due to fewer high-penetrance, or more low-penetrance variants. This deficit is higher than in the Icelandic population (6.4%)(3), consistent with that analysis being biased towards more common variants already subject to selection.
Fig. 2. Population genetic analysis of rhLOF variants.
(A) Comparison of number of LOF variants per unit length in autozygous regions (LOF A) with expected rate from non-autozygous sections (LOF NA) showing suppression of rhLOFs (t-test). A similar analysis of synonymous (Syn) variants shows no significant differences. (B) Observed number of variants with homozygote genotypes in 16,708 rare LOF variants (orange circle) versus a frequency matched subsampling of synonymous variants (blue violin plot). (C) Quantification of the recessive lethal load carried on average by a single individual. Direct subsampling estimate for rhLOF variants from current study (orange circle); epidemiological estimates from correlating infant mortality rates to estimated autozygosity in current and published data (blue circles); direct estimate from large Hutterite pedigree (green circle). 95% confidence intervals as black bars. (D) Relative number of derived LOF alleles that are frequent in one population and not another (under neutrality the expectation is 1.0), calculated for 1000 Genomes Project populations and the current Birmingham/Bradford Pakistani heritage population (BB), compared to the CEU population. Error bars represent ±1 (black) or 2 (grey) standard errors, and significant differences (RA/B jackknife test) versus CEU population are highlighted in orange circles.
We then combined the calculated deficit rate with the observed number of heterozygous annotated LOF variants, integrating across allele frequencies, to obtain a direct estimate of the recessive lethal load per person. This suggests that a typical individual from the population we sampled carries 1.6 recessive annotated LOF lethal-equivalent variants in the heterozygous state(5). This is similar to previous estimates of the lethal load calculated by correlating the number of miscarriages, stillbirths and infant mortalities with the level of autozygosity (fig. 2C)(7, 8), and also similar to measurements in other species(9). Using epidemiological data from 13,586 mothers from the same Born In Bradford birth cohort studied in our genetic analysis, we estimated 0.5 lethal equivalents resulting in miscarriage, stillbirth or infant mortality per individual in our population(5). The difference between our two estimates can be accounted for by the fact that the first includes embryonic lethals, whereas the second only involves deaths after a registered pregnancy, suggesting that there are twice as many recessive mutations that are embryonically lethal as those that result in fetal or infant death. Controlling for other effects by comparing to synonymous mutations, we see a significant but moderate decrease (RA/B jackknife test P=0.04) in the rhLOF mutational load in our Pakistani heritage population dataset compared to outbred populations from the 1000 Genomes Project, although this is less than that caused by the historic bottleneck in the Finnish population (FIN in fig. 2D)(5).
We examined 215 genes with rhLOF in our dataset that have an exact 1:1 mouse:human gene ortholog. From mouse gene knockout data there were 52 genes where a lethal mouse phenotype had been reported on at least one genetic background(10). Whether or not the mouse ortholog knockout is lethal is not associated with alteration of protein function, duplication or changes in gene expression(5). Genes containing rhLOF showed 50% fewer molecular interactions compared to all genes in the STRING interactome dataset (Kruskal-Wallis P=3.4 × 10−9), predominantly driven by the Binding Interaction class (Kruskal-Wallis P=9.3 × 10−11). We saw a similar reduction in the Icelandic data (table S4), in contrast to both known pathogenic LOF variants and pathogenic gain-of-function (GOF) variants reported in Orphanet, which showed increased overall molecular interactions (P=1.1 × 10−6, 2 × 10−12 respectively)(5). Furthermore rhLOF genes that are drug targets have 11.4% phase I to approval rate versus 6.7% for all target-indication pairs (chi-squared P=0.046), although we observed no difference in the proportion of genes known or predicted to be druggable targets(11) for rhLOF genes (15%) compared to all genes (13%, P=0.098)(5).
In subjects from the Born In Bradford study, where full health record data was available, we observed 54 rhLOF genotypes in 52 individuals in Online Mendelian Inheritance in Man (OMIM) confirmed recessive disease genes. Our expectation was that these would be enriched for false positive observations(1). After a quality control analysis of the sequence-based genotype calls(5), we inspected the annotation of these variants(1). We considered 16 of 54 rhLOF genotypes to be possible genome annotation errors (i.e. incorrectly described as LOF) (5)(table S2). Only six of the remaining 38 rhLOF subjects had definite lifetime primary health record diagnoses recorded consistent with the OMIM phenotype, with a further three genotypes suggestively compatible (table S3). We suggest that the remaining 29 are due to a combination of incomplete penetrance[12–16], late onset of disease (i.e. not yet having occurred), individuals with mild symptoms not seeking medical attention, unrecognised technical issues with sequencing or annotation (e.g. tissue specific alternative splicing), or dubious evidence to support the gene-phenotype assignment (in table S3 we assess the available evidence for these possibilities).
We next assessed electronic health records in the Born In Bradford adults, focusing on the time since study recruitment(5). Drug prescription rate and clinical staff consultation rate have previously been shown to correlate strongly with health status(17). We compared individuals with one or more rhLOF (n=638) to individuals without rhLOF (n=1524), and found no association with prescription rate (logistic regression, OR 1.001, 95% CI 0.988 - 1.0144) or consultation rate (OR 1.017, 95% CI 0.996 - 1.038), nor any associations in rhLOF subgroups (5).
One of our subjects was a healthy adult mother with a predicted rare homozygous LOF mutation in PRDM9, which we confirmed experimentally(5)(fig. S7A, S7B). PRDM9 is the major known determinant of the genomic locations of meiotic recombination events in humans and mice through its DNA binding site zinc finger domain (6, 18, 19). We excluded that this rhLOF was from a somatic loss of heterozygosity event on the basis that this subject is heterozygous, not homozygous, on both sides of the 25Mb autozygous region(fig. S7C). Her lifetime primary and secondary care health records were unremarkable. Her genotype predicts protein truncation in the SET methyltransferase domain (thus lacking the DNA-binding zinc-finger domain) which we confirmed in an in vitro expression system (fig. S8A). We observed absence of increase in H3K4Me3 global methylation on transfection (20) of the truncation allele (fig. S8A), and that R345Ter specifically disrupted PRDM9-dependent H3K4Me3 methylation at hotspots (fig. S8B).
We determined the locations of meiotic recombination in the maternal gamete transmitted from the mother to her child by 10X Genomics long-range molecularly-phased whole-genome sequencing and identified 39 candidate crossovers(5). Using double strand break (DSB) maps and a maximum likelihood model to account for variability in region size and hotspot density(18), we estimated that only 5.9% (2 log unit confidence interval: 0 - 24%) of the observed PRDM9 knockout duo maternal gamete crossovers matched DSB sites from wild type PRDM9-A allele homozygotes(5). In comparison, in a control mother-child CEPH pedigree duo homozygous for PRDM9-A we estimated that 52.1% (confidence interval: 36 - 69%) of the crossovers occurred in known DSB sites. Using similar methods we saw that 18.5% of crossovers observed in the PRDM9 knockout duo (confidence interval: 1% - 42%) occurred in linkage disequilibrium based hotspots versus 75.7% in the control duo (confidence interval 57%-89% consistent with a previously published estimate of an average of 60% of crossovers occurring at hotspots(18))(5).
Prdm9 knockout mice demonstrate abnormal location of recombination hotspots with enrichment at gene promoters and enhancers, and also fail to properly repair double-stranded breaks and are infertile (both sexes sterile)(21, 22). Dogs, which lack Prdm9, retain recombination hotspots which unlike humans or knockout mice occur in high GC content regions (23). It has been speculated that dog recombination is controlled by an ancestral mammalian mechanism, and that PRDM9 competes and usurps these sites when active in non-canids(23, 24). However we did not see an increased overlap in our PRDM9-knockout duo crossover intervals with promoters and their flanking regions or enrichment in GC content, compared to the control duo(5). Thus the healthy and fertile PRDM9-deficient adult human suggests differences from both mice and dogs, and supports the possibility of alternative mechanisms of localizing human meiotic crossovers(25, 26).
Together these data suggest that apparent rhLOF genotypes identified by exome or genome sequencing from adult populations require cautious interpretation. Although this class of variants has the greatest predicted effect on protein function, loss of most proteins is relatively harmless to the individual, and even in previously annotated disease genes predicted rare LOF homozygotes may not always be as clinically relevant as often considered. This becomes of increasing importance now that exome and genome sequencing is rapidly expanding into healthier adults. We anticipate that further efforts to identify naturally occurring human knockouts, whether in bottlenecked populations, or more efficiently as here in subjects with related parents, will yield both new data relevant to clinical interpretation, and new biological insights, as exemplified by our investigation here of a PRDM9 deficient healthy and fertile woman.
Supplementary Material
Acknowledgments
The study was funded by the Wellcome Trust (WT102627 & WT098051), Barts Charity (845/1796), Medical Research Council (MR/M009017/1). This paper presents independent research funded by the National Institute for Health Research (NIHR) under its Collaboration for Applied Health Research and Care (CLAHRC) for Yorkshire and Humber. Core support for Born in Bradford is also provided by the Wellcome Trust (WT101597). VN was supported by the Wellcome Trust PhD Studentship (WT099769). DGM and KK were supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM104371. ERM is funded by NIHR Cambridge Biomedical Research Centre. HH is supported by awards to establish the Farr Institute of Health Informatics Research, London, from the Medical Research Council, Arthritis Research UK, British Heart Foundation, Cancer Research UK, Chief Scientist Office, Economic and Social Research Council, Engineering and Physical Sciences Research Council, NIHR, National Institute for Social Care and Health Research, and Wellcome Trust. Born in Bradford is only possible because of the enthusiasm and commitment of the Children and Parents in BiB. We are grateful to all the participants, health professionals and researchers who have made Born in Bradford happen. We thank B MacLaughlin (QMUL) for assistance, and J Rogers (HSCIC) for advice. We would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
Footnotes
RD declares his interests as a founder and non-executive director of Congenica Ltd., that he owns stock in Illumina Inc. from previous consulting and is a scientific advisory board member of Dovetail Inc. MSL and KG are employees of 10X Genomics Inc.
Data reported in the paper are presented in the Supplementary Materials, and are available under a Data Access Agreement at the European Genotype-phenome Archive (www.ebi.ac.uk/ega) under accession numbers EGAS00001000462, EGAS00001000511, EGAS00001000567, EGAS00001000717 and EGAS00001001301.
SUPPLEMENTARY MATERIALS
<URL www.sciencemag.org/>
Materials and Methods
Figs. S1 to S8
Tables S1 to S8
References (27–60)
Data Files S1 to S3
REFERENCES AND NOTES
- 1.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Exome Aggregation Consortium et al. Analysis of protein-coding genetic variation in 60,706 humans. 2015 doi: 10.1038/nature19057. http://biorxiv.org/content/early/2015/10/30/030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sulem P, et al. Identification of a large set of rare complete human knockouts. Nat. Genet. 2015;47:448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- 4.Lim ET, et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Supplementary Materials. [Google Scholar]
- 6.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty R, Chakravarti A. On consanguineous marriages and the genetic load. Hum. Genet. 1977;36:47–54. doi: 10.1007/BF00390435. [DOI] [PubMed] [Google Scholar]
- 8.Bittles AH, Neel JV. The costs of human inbreeding and their implications for variations at the DNA level. Nat. Genet. 1994;8:117–121. doi: 10.1038/ng1094-117. [DOI] [PubMed] [Google Scholar]
- 9.McCune AR, et al. A low genomic number of recessive lethals in natural populations of bluefin killifish and zebrafish. Science. 2002;296:2398–2401. doi: 10.1126/science.1071757. [DOI] [PubMed] [Google Scholar]
- 10.Liao B-Y, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 12.Flannick J, et al. Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes. Nat. Genet. 2013;45:1380–1385. doi: 10.1038/ng.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rehm HL, et al. ClinGen--the Clinical Genome Resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.UK10K Consortium et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston JJ, et al. Individualized iterative phenotyping for genome-wide analysis of loss-of-function mutations. Am. J. Hum. Genet. 2015;96:913–925. doi: 10.1016/j.ajhg.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Driest SL, et al. Association of Arrhythmia-Related Genetic Variants With Phenotypes Documented in Electronic Medical Records. JAMA. 2016;315:47–57. doi: 10.1001/jama.2015.17701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brilleman SL, et al. Implications of comorbidity for primary care costs in the UK: a retrospective observational study. Br. J. Gen. Pract. 2013;63:e274–e282. doi: 10.3399/bjgp13X665242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker CL, et al. Multimer Formation Explains Allelic Suppression of PRDM9 Recombination Hotspots. PLoS Genet. 2015;11:e1005512. doi: 10.1371/journal.pgen.1005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 22.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Axelsson E, et al. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 2012;22:51–63. doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berg IL, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 2010;42:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pratto F, et al. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichten M. Molecular biology. Putting the breaks on meiosis. Science. 2015;350:913. doi: 10.1126/science.aad5404. [DOI] [PubMed] [Google Scholar]
- 27.Bundey S, Alam H. A five-year prospective study of the health of children in different ethnic groups, with particular reference to the effect of inbreeding. Eur. J. Hum. Genet. 1993;1:206–219. doi: 10.1159/000472414. [DOI] [PubMed] [Google Scholar]
- 28.Rees SD, et al. Replication of 13 genome-wide association (GWA)-validated risk variants for type 2 diabetes in Pakistani populations. Diabetologia. 2011;54:1368–1374. doi: 10.1007/s00125-011-2063-2. [DOI] [PubMed] [Google Scholar]
- 29.Wright J, et al. Cohort Profile: the Born in Bradford multi-ethnic family cohort study. Int. J. Epidemiol. 2013;42:978–991. doi: 10.1093/ije/dys112. [DOI] [PubMed] [Google Scholar]
- 30.Raynor P. Born in Bradford Collaborative Group, Born in Bradford, a cohort study of babies born in Bradford, and their parents: protocol for the recruitment phase. BMC Public Health. 2008;8:327. doi: 10.1186/1471-2458-8-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jun G, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mills RE, et al. Natural genetic variation caused by small insertions and deletions in the human genome. Genome Res. 2011;21:830–839. doi: 10.1101/gr.115907.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunningham F, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue Y, et al. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science. 2015;348:242–245. doi: 10.1126/science.aaa3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 42.Morton NE, Crow JF, Muller HJ. AN ESTIMATE OF THE MUTATIONAL DAMAGE IN MAN FROM DATA ON CONSANGUINEOUS MARRIAGES. Proc. Natl. Acad. Sci. U. S. A. 1956;42:855–863. doi: 10.1073/pnas.42.11.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheridan E, et al. Risk factors for congenital anomaly in a multiethnic birth cohort: an analysis of the Born in Bradford study. Lancet. 2013;382:1350–1359. doi: 10.1016/S0140-6736(13)61132-0. [DOI] [PubMed] [Google Scholar]
- 44.Gao Z, Waggoner D, Stephens M, Ober C, Przeworski M. An estimate of the average number of recessive lethal mutations carried by humans. Genetics. 2015;199:1243–1254. doi: 10.1534/genetics.114.173351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Do R, et al. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat. Genet. 2015;47:126–131. doi: 10.1038/ng.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volkamer A, Kuhn D, Grombacher T, Rippmann F, Rarey M. Combining global and local measures for structurebased druggability predictions. J. Chem. Inf. Model. 2012;52:360–372. doi: 10.1021/ci200454v. [DOI] [PubMed] [Google Scholar]
- 47.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rivas MA, et al. Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015;348:666–669. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boomla K, Hull S, Robson J. GP funding formula masks major inequalities for practices in deprived areas. BMJ. 2014;349:g7648. doi: 10.1136/bmj.g7648. [DOI] [PubMed] [Google Scholar]
- 50.Kelaher M, Paul S, Lambert H, Ahmad W, Smith GD. The applicability of measures of socioeconomic position to different ethnic groups within the UK. Int. J. Equity Health. 2009;8:4. doi: 10.1186/1475-9276-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braveman PA, et al. Socioeconomic status in health research: one size does not fit all. JAMA. 2005;294:2879–2888. doi: 10.1001/jama.294.22.2879. [DOI] [PubMed] [Google Scholar]
- 52.Fairley L, et al. Using latent class analysis to develop a model of the relationship between socioeconomic position and ethnicity: cross-sectional analyses from a multi-ethnic birth cohort study. BMC Public Health. 2014;14:835. doi: 10.1186/1471-2458-14-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 54.Auton A, et al. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 2013;9:e1003984. doi: 10.1371/journal.pgen.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.al Singhal S E. Stable recombination hotspots in birds. 2015 doi: 10.1126/science.aad0843. http://biorxiv.org/content/early/2015/07/23/023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 57.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 58.Cleary JG, et al. Joint variant and de novo mutation identification on pedigrees from high-throughput sequencing data. J. Comput. Biol. 2014;21:405–419. doi: 10.1089/cmb.2014.0029. [DOI] [PubMed] [Google Scholar]
- 59.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 60.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.