The importance of Abl kinase activity, the F-actin–binding site, and scaffolding ability in Abl’s many cell biological roles during Drosophila morphogenesis is examined. Abl is a robust multidomain scaffold with different protein motifs and activities contributing differentially to diverse cellular behaviors.
Abstract
Abelson family kinases (Abls) are key regulators of cell behavior and the cytoskeleton during development and in leukemia. Abl’s SH3, SH2, and tyrosine kinase domains are joined via a linker to an F-actin–binding domain (FABD). Research on Abl’s roles in cell culture led to several hypotheses for its mechanism of action: 1) Abl phosphorylates other proteins, modulating their activity, 2) Abl directly regulates the cytoskeleton via its cytoskeletal interaction domains, and/or 3) Abl is a scaffold for a signaling complex. The importance of these roles during normal development remains untested. We tested these mechanistic hypotheses during Drosophila morphogenesis using a series of mutants to examine Abl’s many cell biological roles. Strikingly, Abl lacking the FABD fully rescued morphogenesis, cell shape change, actin regulation, and viability, whereas kinase-dead Abl, although reduced in function, retained substantial rescuing ability in some but not all Abl functions. We also tested the function of four conserved motifs in the linker region, revealing a key role for a conserved PXXP motif known to bind Crk and Abi. We propose that Abl acts as a robust multidomain scaffold with different protein motifs and activities contributing differentially to diverse cellular behaviors.
INTRODUCTION
Biomedical science has twin goals: to define how cells and organisms work and to use this information to reveal what goes wrong in disease and develop better treatments. Collaboration between basic scientists and clinicians is key, with both sides informing the other. Few stories exemplify this better than that of Abelson kinase (Abl), one of the first identified human oncogenes. Chromosomal translocations activating Abl play a key role in chronic myelogenous leukemia (CML; Druker, 2008). The past 20 yr have seen parallel progress in understanding Abl’s normal function in development and revealing how its activation contributes to disease, thus improving treatment (Khatri et al., 2016).
Abl and its mammalian paralogue, Abl-related gene (Arg), are part of a kinase superfamily including Src that shares tandem N-terminal Src-homology domain 3 (SH3), Src-homology domain 2 (SH2), and tyrosine kinase domains (Figure 1A). In Abl family proteins, this is coupled to a C-terminal F-actin–binding domain (FABD; van Etten et al., 1994) via a long, less well-conserved linker. This structure suggested the hypothesis that Abl links cell signaling and cytoskeletal regulation, which was abundantly confirmed in vivo (Bradley and Koleske, 2009). For example, in immune cells, Abl regulates cytokine responses, immune synapse assembly, and T-cell migration in response to chemotactic cues (Huang et al., 2008). In the nervous system, Abl family members act downstream of axon guidance receptors to modulate growth cone guidance, synaptogenesis, and dendrite formation, regulating both actin and microtubules (Moresco and Koleske, 2003; Moresco et al., 2005; Lee et al., 2004; Li et al., 2005; Lin et al., 2009; O’Donnell and Bashaw, 2013). Finally, Abl family members modulate morphogenesis. The abl arg double-mutant mouse has neural tube closure defects (Koleske et al., 1998). Drosophila Abl regulates diverse events ranging from coordinated apical constriction in the invaginating mesoderm to cell shape changes in epidermal cells undergoing convergent elongation, collective cell migration, or wound repair, stem cell maintenance in the germline, and Golgi localization in neurons (Grevengoed et al., 2001, 2003; Fox and Peifer, 2007; Tamada et al., 2012; Kannan et al., 2014; Stine et al., 2014; Zulueta-Coarasa et al., 2014). Abl’s roles in cell migration are also important in cancer: deregulated Abl kinases can drive invadopodia formation and invasive behavior (Smith-Pearson et al., 2010; Mader et al., 2011).
FIGURE 1:
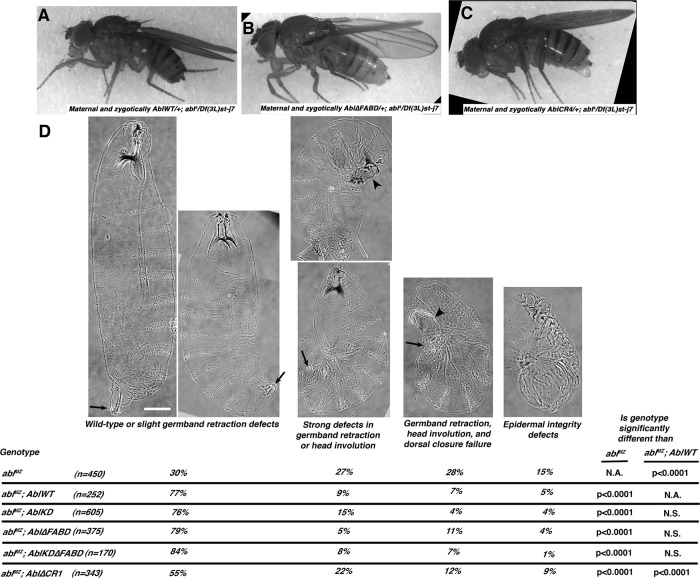
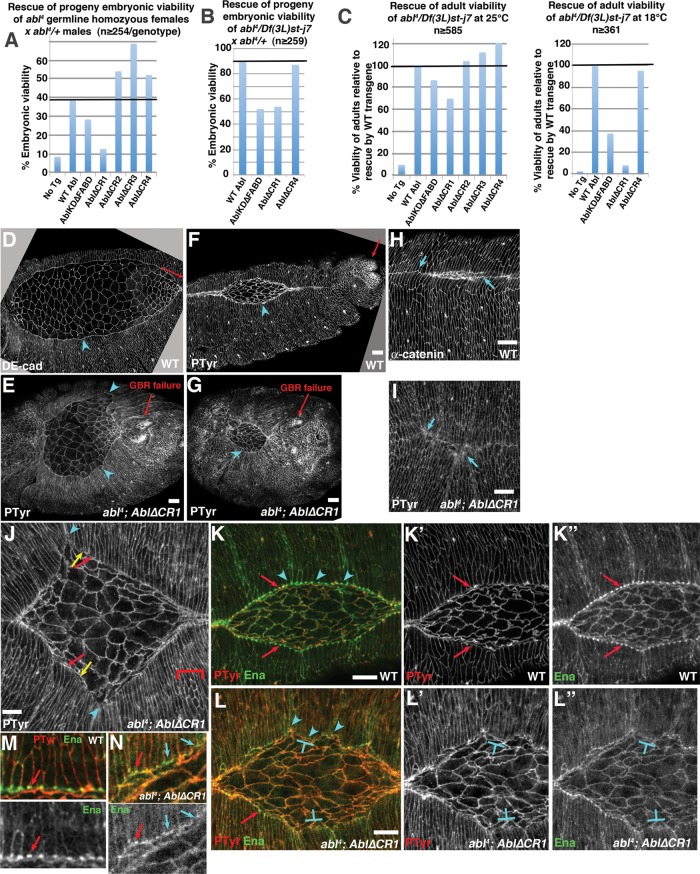
Wild-type human and Drosophila Abl proteins, mutants used, and rescue of viability. (A) Mammalian Abl and Arg and the single fly Abl share a core including highly conserved SH3, SH2, and tyrosine kinase domains and a C-terminal conserved region that binds actin in Abl and Arg (FABD). The linker is much less well conserved. In Arg, it contains a microtubule- and an additional actin-binding site, but the only shared motif is a PXXP SH3-binding site. (B) Mutants tested. All were C-terminally GFP-tagged, driven by the endogenous promoter (Fox and Peifer, 2007), and verified by Western blot to be expressed at levels similar to AblWT (Supplemental Figure S1). (C) Rescue of adult viability of abl4 hemizygous zygotic mutants at indicated temperature normalized to rescue by AblWT. (D, E) Rescue of embryonic viability of ablMZ mutants from abl4/ Df(3L)st-j7 (D) or abl4 germline homozygous mothers (E; 50% receive a wild-type abl gene paternally in both; crosses in Supplemental Figure S2). Full data for C–E are given in Table 1.
The importance of the Abl family in development and oncogenesis makes determining their mechanisms of action critical. Their structure and cell biological roles led to three models for function, which are not mutually exclusive. In one model, Abl/Arg, like other cytoplasmic kinases, phosphorylates protein targets that regulate cell behavior, thus altering their function. This model has substantial support. For example, Arg phosphorylates p190RhoGAP, regulating its subcellular localization and activity, with effects on cell contractility and migration. Contractility defects can be rescued by wild-type but not kinase-dead Arg (Hernandez et al., 2004; Bradley et al., 2006; Peacock et al., 2007). Arg also phosphorylates cortactin (Lapetina et al., 2009), altering actin assembly and regulating ruffling. Surprisingly, Arg’s effects on cell protrusions are largely rescued by kinase-dead Arg (Lapetina et al., 2009); inhibitor studies suggest that Abl kinase may take over in phosphorylating cortactin. Drosophila Abl’s best-characterized target is the actin regulator Enabled (Ena), which Abl negatively regulates (Gertler et al., 1995). Consistent with the kinase model, Abl phosphorylates Ena in vitro and in vivo. However, in contrast to this model’s simplest expectations, mutating all of the Abl phosphorylation sites on Ena does not hyperactivate Ena (Comer et al., 1998). Thus, whereas Abl family proteins phosphorylate many targets and can alter their activities, kinase activity is not the whole story.
In the second model, Abl family proteins directly regulate the cytoskeleton via their C-terminal FABD as well as, in mammalian Arg, other actin- and microtubule-binding sites in the linker. For example, microtubule-actin cross-linking by Arg’s C-terminal region is necessary and sufficient to modulate fibroblast lamellipodial dynamics (Miller et al., 2004), whereas the actin-binding domain can modulate cortactin (Lin et al., 2013). Intriguingly, Arg’s second actin- binding domain, which is not conserved in Abl, can stabilize actin filaments and activate the Arp2/3 complex (Courtemanche et al., 2015). The FABD also modulates kinase activity (Woodring et al., 2001) and targets Abl to certain substrates (Mitra and Radha, 2010).
In the third model, Abl family kinases act as scaffolds for assembling a signaling complex. In addition to the kinase domain and FABD, the Abl family shares protein-binding sites for other partners that mediate Abl/Arg localization or activity, position substrates for phosphorylation, or assemble multiprotein complexes. Ligand engagement by the SH2 and SH3 domains helps activate Abl (Hantschel and Superti-Furga, 2004) and modulates partner/substrate docking; for example, Arg’s SH3 domain mediates N-WASP interaction (Miller et al., 2010). The SH3 domain can also negatively regulate kinase activity—surprisingly, its ability to bind ligands is not essential for Bcr-Abl disease induction (Smith et al., 2003). Proline-rich motifs in the long, poorly conserved linker between the kinase domain and FABD bind other partners/substrates; for example, the most N-terminal PXXP motif binds the adaptors Crk and Nck (Feller, 2001) and the actin regulator Abi (Ibarra et al., 2005). These motifs play important roles in cell spreading (Antoku et al., 2008) and protrusive behavior (Lapetina et al., 2009). The sequence surrounding this N-terminal PXXP motif is the only linker region conserved between mammalian Abl, Arg, and Drosophila Abl (Figure 1A; we refer to it as conserved region 1 [CR1]).
Most studies probed individual properties conferred by Abl or Arg in cultured cells. In intact animals, the rules may be different, with essential roles for either kinase activity or the FABD in a subset of biological events or with greater redundancy. There have only been a handful of tests in whole animals, which probed only a small subset of Abl functions. In mice, kinase activity is important in rescuing abl-mutant postnatal lethality (Hardin et al., 1996), but most other murine Abl or Arg functions have not been similarly tested. Similarly, early experiments supported a role for the FABD in rescuing the partial lethality and immune defects of abl-null mice (Schwartzberg et al., 1991), but this mutant was assessed for only a few other tissues and developmental times (Qiu et al., 2010). Experiments in the 1990’s from Hoffmann’s lab began to test the importance of Abl kinase activity in Drosophila. Kinase-dead Abl restored adult viability of zygotic abl mutants, but tests in a sensitized genetic background suggested that it did not retain full function in this assay (Henkemeyer et al., 1990). In contrast, a severely truncated Abl retaining kinase activity but lacking both the linker and FABD could not rescue adult viability. However, this work was done when the array of known Abl functions was very limited, and the approach has not been substantially extended since then to explore Abl’s diverse roles in morphogenesis or the CNS. Similarly, although Abl family proteins clearly can act as scaffolds, the importance of the scaffolding role has been examined only in cultured cells.
Scientists also probed Abl’s mechanisms of action in leukemia. Kinase activity is clearly important for oncogenesis, such that the kinase inhibitor imatinib revolutionized CML treatment (O’Hare et al., 2005). Consistent with this, whereas Bcr-Abl induces a CML-like myeloproliferative disease in mice, kinase-dead Bcr-Abl does not (Zhang and Ren, 1998). However, although kinase activity is necessary for oncogenesis, it may not be sufficient. Roles for the FABD in cancer remain controversial, with the result differing, depending on the amount of Bcr sequence included in the Bcr-Abl fusion protein. p210Bcr-Abl does not require its FABD to induce leukemia (Wertheim et al., 2003), but deleting the FABD attenuates leukemogenesis by p190Bcr-Abl (Heisterkamp et al., 2000). Several effects of oncogenic Bcr-Abl or v-abl in cultured cells depend on the FABD, including the ability to transform fibroblasts, make lymphoblasts interleukin independent (McWhirter and Wang, 1993), modulate matrix adhesion (Wertheim et al., 2003), and suppress apoptosis and confer drug resistance to hematopoietic progenitors (Underhill-Day et al., 2006). Thus, although elegant experiments in cultured cells have revealed how kinase activity, cytoskeletal interactions, and scaffolding can influence individual cellular properties, the relative importance of these roles in whole animals is less clear. We thus tested the different hypotheses for Abl’s mechanism of action in morphogenesis, evaluating the importance of kinase activity, the FABD, and conserved linker motifs.
RESULTS
The F-actin–binding domain is not essential for rescuing embryonic or adult viability or fertility of abl-null mutants, whereas loss of kinase activity impairs but does not eliminate Abl function
To test the importance of kinase activity or direct regulation of the actin cytoskeleton in Abl function in vivo, we generated two mutants (Figure 1B): Abl kinase-dead (AblKD = AblK417N) and Abl∆FABD, which deletes the conserved FABD (van Etten et al., 1994). Fly Lys-417 corresponds to Lys-271 of human Abl, which the crystal structure reveals is part of the ATP-binding pocket (Schindler et al., 2000). This residue is highly conserved in tyrosine kinases, and its mutation eliminates their function. AblK417N eliminates kinase activity of fly Abl, as assessed by both kinase assays on Abl immunoprecipitated from Drosophila embryos (Henkemeyer et al., 1990) and failure to elevate phosphotyrosine levels after overexpression in vivo, in contrast to wild-type Abl (Stevens et al., 2007). Mutation of the analogous lysine in mouse Arg also eliminates kinase activity, and thus this mutant is the canonical kinase-dead mutant (e.g., Peacock et al., 2007).
Both mutants, as well as other mutants described later, were C-terminally green fluorescent protein (GFP) tagged and driven by the endogenous abl promoter. All transgenes were integrated into the same chromosomal location to minimize position effects, and transgenics were verified by PCR and/or sequencing (Supplemental Figure S1, A–H). GFP tagging does not interfere with the ability of Abl to rescue, and the endogenous promoter drives expression levels equivalent to that of endogenous Abl (Fox and Peifer, 2007). We confirmed that all of our transgenes are equivalently expressed (Supplemental Figure S1I).
We initially hypothesized that both kinase activity and the FABD would be essential for all Abl functions in morphogenesis or that each would be required for a subset of Abl’s functions. To test these hypotheses, we expressed each transgene in the absence of endogenous Abl and assessed their function. As a first functional test, we assessed whether they rescued the lethality caused by Abl loss. The abl zygotic mutants survive embryogenesis due to maternally contributed Abl, but most die as pupae (no transgene = no TG; Figure 1C, left, and Table 1). We first assessed whether our mutants rescued adult viability at 25°C. Abl∆FABD rescued adult viability as well as our wild-type abl transgene (AblWT; Figure 1C, left; full data and statistical tests in Table 1; rescue was normalized to that of AblWT). AblKD retained substantial rescuing ability only slightly reduced from that of AblWT (88%; Figure 1C, left, and Table 1). We further challenged the mutants by testing them at 18°C, which increases phenotypic consequences of Abl loss, perhaps by impairing cytoskeletal dynamics (Grevengoed et al., 2003). At 18°C, abl zygotic mutants are almost fully pupal lethal, but Abl∆FABD once again rescued as well or nearly as well as AblWT (91%; Figure 1C, right, and Table 1). In this assay, although AblKD retained substantial rescuing ability, it was significantly reduced from that of AblWT (51%) but did provide significant rescue over no transgene (2%; Figure 1C, right, and Table 1). We next tested whether AblKD and Abl∆FABD rescued embryonic viability of abl maternal and zygotic mutants (ablMZ), which are fully embryonic lethal (Grevengoed et al., 2001). To do this, we crossed adult females that lack zygotic Abl expression and carry one copy of specific Abl transgenes (abl4/Df(3L)st-j7; transgene/+ females; abl4 is a null allele (Fox and Peifer, 2007), and Df(3L)st-j7a is a chromosomal deletion removing abl and nearby genes) to abl4/+ heterozygous males that were homozygous for the transgene (Supplemental Figure S2A). Thus 50% of the progeny were maternally and zygotically abl null. By using two different alleles in these crosses, we removed issues associated with effects of other mutations on the chromosome during oogenesis of the abl4/Df(3L)st-j7 mothers. Surprisingly, Abl∆FABD fully rescued embryonic viability (97% viable; Figure 1D and Table 1); this is comparable to the viability of wild-type control flies and matches rescue by AblWT. AblKD once again was reduced in its function (embryonic viability of 62 vs. 89% for AblWT; Figure 1D and Table 1), but the level of rescue suggested that a subset of AblKD maternal/zygotic mutants survive embryogenesis.
TABLE 1:
Rescue of adult or embryonic viability of abl mutants by mutant transgenes.
| Percentage rescue of adult viability in abl4/Df(3L)st-j7 background (normalized to WT) | |||||
|---|---|---|---|---|---|
| Construct | Residues changed or deleted | 25°C | 18°C | Percentage rescue of progeny, embryonic viability of abl4/Df(3L)st-j7 female × abl4/+ male | Percentage rescue of progeny, embryonic viability of abl4 maternal germline female × abl4/+ male |
| No transgene | N/A | 9.4 (911)a | 2.1 (733) | N/A | 8.2 (527) |
| AblWT | N/A | 100 (1502)b | 100 (1335)b | 89.4 (901) | 38.9 (540)b |
| AblKD | K417N | 88.0 (1225)a,b | 50.9 (1009)a,b | 62.3 (705)a | 25.2 (726)a,b |
| Abl∆FABD | ∆1418–1520 | 102.6 (1238)b | 91.1 (1474)a,b | 97.1 (362) | 34.6 (515)b |
| AblKD∆FABD | K417N,∆1418–1520 | 86.4 (585)a,b | 37.1 (361)a,b | 52.1 (259)a | 28.2 (560)a,b |
| Abl∆CR1 | ∆734–789 | 69.6 (2149)a,b | 7.7 (972)a,b | 53.9 (267)a | 12.4 (606)a,c |
| Abl∆CR2 | ∆930–965 | 103.8 (56)b | ND | ND | 54.0 (303)b |
| Abl∆CR3 | ∆1003–1014 | 111.7 (110)b | ND | ND | 68.7 (256)b |
| Abl∆CR4 | ∆1063–1095 | 120.4 (906)b | 95.3 (797)a,b | 87.1 (340) | 52.0 (254)b |
In evaluating differences between genotypes, it is important to note that “statistically significant” differences using the test chosen may not always reflect important biological differences. We were thus cautious not to overinterpret small differences. Our more qualitative assessments of relevance are presented in Supplemental Figure S3.
Numbers in columns 5 and 6 (rescue of embryonic viability) are fractions of viable progeny and are not normalized to rescue by AblWT. Numbers in columns 3 and 4 (adult viability rescued) are reported as normalized to WTAbl. This was done because the cross (abl4/TM3 Sb × Df(3L)st-j7/TM3Sb) produced three classes of adults (abl4/TM3 Sb, Df(3L)st-j7/TM3Sb, and the relevant class abl4/Df(3L)st-j7). Because none of the genotypes is fully wild type, it would be problematic to compare progeny numbers to those expected from Mendelian segregation, as the mutations on the TM3 Balancer chromosome and other mutations on the Deficiency chromosome may also affect adult viability. We counted the number of adults of each genotype (distinguishable by linked markers Sb and Ki). For example, at 18°C, for AblWT, 488/1335 (36.6%) of the progeny were abl4/TM3 Sb, 397/1335 (29.7%) were Df(3L)st-j7/TM3Sb, and 450/1335 (33.7%) were the relevant class, abl4/Df(3L)st-j7. Thus, for other genotypes scored for adult viability at 18°C, the percentage of the progeny of the abl4/Df(3L)st-j7 genotype was divided by 33.7 to normalize to wild-type survival.
Numbers in parentheses are numbers of adults/embryos scored; N/A, not applicable; ND, not determined.
aSurvival worse than rescue by AblWT at p < 0.001 (all others not significantly worse or actually better than AblWT).
bSurvival better than no transgene at p < 0.01.
cSurvival better than no transgene at p < 0.05 (all others not significant).
Most striking, Abl∆FABD, like AblWT, allowed ablMZ mutants, which lack any endogenous Abl from the start of development, not only to survive embryogenesis, but also to survive to become fertile adults without obvious defects (Figure 2, A and B) that produce viable progeny. To confirm this surprising result, we PCR-genotyped adult progeny, verifying rescue (Supplemental Figure S3). In contrast, the subset of ablMZ mutants whose embryonic viability was rescued by AblKD did not survive to adulthood. Thus the FABD of Abl is not essential for successful embryonic or postembryonic development. In contrast, Abl kinase activity is critical for full Abl function, but AblKD retains significant function in promoting embryonic and postembryonic viability.
FIGURE 2:
Neither kinase activity nor the FABD is required to rescue adult viability and fertility of abl4 maternal/zygotic mutants or for many morphogenetic movements, as assessed by cuticle pattern. (A–C) Adults, anterior left, rescued by indicated transgene. All were otherwise maternally and zygotically abl-null mutant (abl4/Df(3L)st-j7 progeny of zygotically abl-null mutant mothers (maternal genotype abl4/Df(3L)st-j7; Supplemental Figure S2A; the altered bristles are caused by the Ki mutation on the Df(3L)st-j7 chromosome). (D) Rescue of embryonic morphogenesis, as assessed by larval cuticles, presented anterior up. Top, four classes of increasing severity. Arrows, correct germband retraction (left cuticle) or increasingly severe defects. Arrowheads indicate defects in head involution and dorsal closure. Scale bar, 50 μm. Bottom, fraction of cuticles in each category. All except Abl∆CR1 provide essentially wild-type levels of rescue. In D, embryos are progeny of a cross in which mothers had germlines homozygous for the null allele abl4 and fathers were abl4/+. Thus all embryos were maternally mutant for endogenous abl, and half were maternally and zygotically mutant.
Neither kinase activity nor the FABD is essential for cortical localization, localization to axons, or overall epidermal morphogenesis
One function of kinase activity or the FABD might be in ensuring correct subcellular localization. From gastrulation to germband retraction, Abl is found in a cytoplasmic pool and enriched at the cell cortex in cells of the ectoderm, a localization that is replicated by GFP-tagged AblWT (Fox and Peifer, 2007; Supplemental Figure S4A). AblKD and Abl∆FABD both retained cortical enrichment (Supplemental Figure S4, B and C). Abl is also enriched in CNS axons (Henkemeyer et al., 1990), as is GFP-tagged AblWT (Supplemental Figure S4G). AblKD and Abl∆FABD show no alterations in axonal enrichment (Supplemental Figure S4, H and I), even in the absence of wild-type Abl (Supplemental Figure S4, H and J; as we document later, however, AblKD did not rescue CNS defects). Thus neither kinase activity nor the FABD is essential for these aspects of Abl localization.
The lethality of abl mutants reflects Abl’s many roles in embryonic and adult morphogenesis and CNS development. The disadvantage of our abl4/Df(3L)st-j7 approach is that we could not directly compare embryos carrying our transgenes to comparable embryos without any transgene, since abl4/Df(3L)st-j7 females are essentially inviable (Figure 1C) and infertile. To circumvent this, we used the FLP/FRT/DFS approach (Chou and Perrimon, 1996) to generate females with germlines homozygous for abl4 either in the presence of a transgene or in the absence of any transgene as a control. We crossed them to males heterozygous for abl4—thus half of the progeny have no maternal or zygotic Abl, whereas half express paternally contributed Abl (Supplemental Figure S2B). This allowed direct comparison of ablMZ mutants rescued with our mutant transgenes to control ablMZ mutants without any transgene; the disadvantage is that our wild-type abl transgene (AblWT) does not fully rescue viability in this context, presumably due to homozygosity of other loci on the abl4 chromosome that enhance effects of Abl reduction. We thus used rescue by AblWT as our baseline. Abl∆FABD also significantly rescued embryonic viability, although it might be slightly diminished relative to AblWT (35% embryonic viability vs. 39% viability for AblWT and 9% viability for no transgene; Figure 1E and Table 1). AblKD also significantly rescued embryonic viability but was less effective than AblWT (25% viability vs. 39% viability for AblWT and 9% viability for no transgene; Figure 1E and Table 1).
Abl’s role in embryonic viability reflects important roles in cell behavior in many different tissues. To initially assess the requirement for kinase activity or the FABD in regulating morphogenetic movements, we examined cuticles of mutant embryos. The cuticle is the larva’s external skeleton and is secreted by the epidermis, and its features allow one to assess completion of morphogenetic movements such as germband retraction, dorsal closure, and head involution, all of which are defective in ablMZ mutants (Figure 2D; Grevengoed et al., 2001; half of the embryos receive a paternal wild-type abl gene and are partially rescued). Surprisingly, both AblKD and Abl∆FABD substantially rescued morphogenesis defects at rates similar to AblWT (Figure 2D, bottom). These data suggest that neither kinase activity nor the FABD is essential for completion of many morphogenetic movements, at least as assessed by cuticle patterning.
Abl∆FABD rescues cellularization, mesoderm invagination, and CNS development as effectively as AblWT, whereas AblKD has differential function in these events
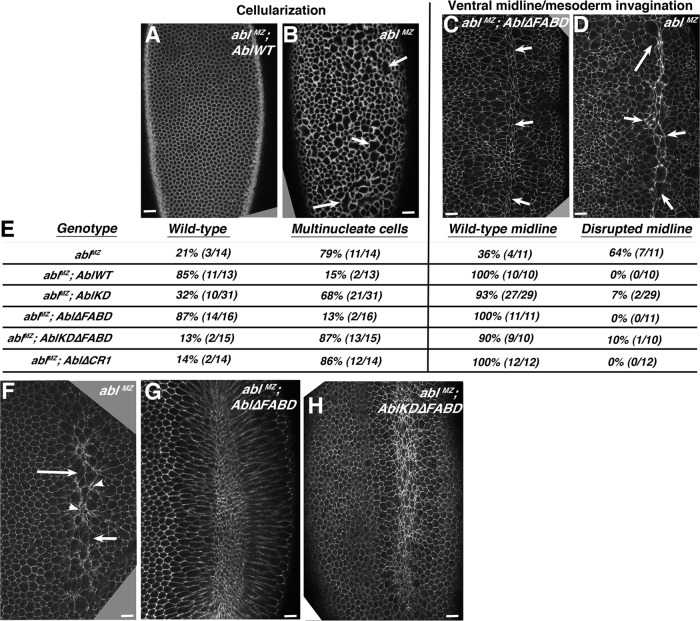
These data were surprising, demonstrating that neither kinase activity nor the conserved FABD is absolutely essential for viability or some aspects of normal morphogenesis. Thus we tested an alternate mechanistic hypothesis: that kinase activity and/or the FABD play specific roles in a subset of processes requiring Abl. The robustness of the embryonic program means embryos can survive despite defects in some cell biological events of morphogenesis. We thus examined cell shapes and cell movements underlying morphogenetic events depending on Abl: cellularization, coordinated mesoderm invagination, germband retraction, dorsal closure, and axon outgrowth (Grevengoed et al., 2001, 2003; Fox and Peifer, 2007). Abl loss during cellularization leads to excess Ena accumulation at the apical cortex, leading to aberrant actin accumulation, defects in actomyosin furrow ingression, and formation of multinucleate cells (Figure 3, A vs. B; Grevengoed et al., 2003). ablMZ mutants also have defects in coordinated apical constriction during gastrulation (Figure 3F; Fox and Peifer, 2007) and thus mesodermal cell invagination into the ventral furrow, once again due to aberrant Ena accumulation and apical actin abnormalities. This leads to defects at the ventral midline (Figure 3, C vs. D). Strikingly, Abl∆FABD rescued both cellularization and mesodermal invagination at rates similar to AblWT (Figure 3, C, E, and G; representative images of all genotypes are in Supplemental Figure S5). In contrast, AblKD provided only a subtle rescue of cellularization (Figure 3E and Supplemental Figure S5D; 32% cellularized normally vs. 21% for no transgene and 85% for AblWT). However, AblKD effectively rescued mesoderm invagination (Figure 3E and Supplemental Figure S5K).
FIGURE 3:
The FABD is not required for correct cellularization or mesoderm invagination, whereas AblKD and AblKD∆FABD display differential activity in these events. (A–D, F–H) Embryos, anterior at top. (A, B) abl4 maternal mutants have multinucleate cells at cellularization (B, arrows), which is effectively rescued by AblWT (A). (C, D) Defects in coordinated mesoderm invagination in abl4 maternal mutants lead to a very irregular ventral midline at stage 9 (D, arrows), which is rescued by Abl∆FABD (C, arrows). (E) Quantitation of degree of rescue by the indicated mutants. Left, multinucleate cells. Right, mesoderm invagination. Representative images of embryos of each genotype are shown in Supplemental Figure S5. (F–H) Embryos during mesoderm invagination. (F) ablMZ mutant showing lack of coordinated apical constriction (arrows vs. arrowheads). (G, H) Fixed images suggest that coordinated apical constriction is largely restored by Abl∆FABD or AblKD∆FABD. Scale bars, 15 μm. Embryos are progeny of a cross in which mothers had germlines homozygous for the null allele abl4 and fathers were abl4/+. Thus all embryos were maternally mutant for endogenous abl, and half were maternally and zygotically mutant.
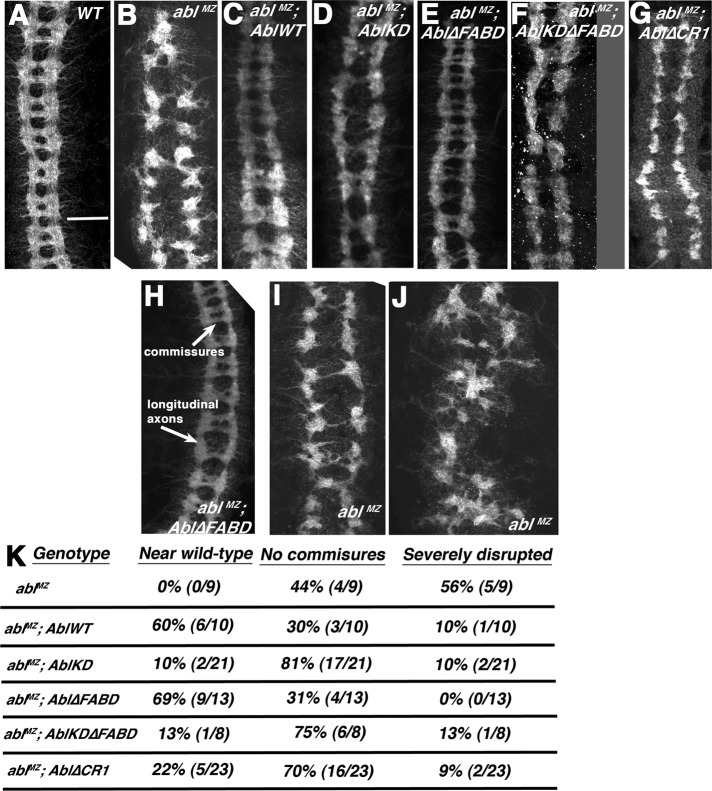
Abl also regulates CNS development via effects on axon path finding (Gertler et al., 1993; Grevengoed et al., 2001). Maternal/zygotic Abl loss leads to severe CNS defects—instead of the correct ladder of longitudinal and commissural axons, ablMZ mutants either have gaps in commissures (Figure 4, A vs. B and H vs. I, quantified in K) or more-severe axon disorganization (Figure 4, H vs. J, quantified in K). These defects are substantially but not fully rescued by AblWT—60% of embryos are fully rescued, whereas the others have commissural axon defects (Figure 4, C and K). Once again, Abl∆FABD (Figure 4E) rescued CNS defects as well as AblWT (Figure 4K). In contrast, AblKD retained only partial function (Figure 4D), rescuing the severe axon disorganization phenotype of some ablMZ mutants but providing little or no rescue of the commissureless phenotype (Figure 4K). Thus the FABD is not essential for completion of any of these three morphogenetic events, whereas kinase activity is more important for regulating some events (cellularization and CNS axon outgrowth) and less important for others (mesoderm invagination).
FIGURE 4:
Abl∆FABD rescues the CNS defects of ablMZ mutants, whereas AblKD, AblKD∆FABD, and Abl∆CR1 retain only partial activity. (A–G) Representative CNS pictures of wild type and indicated mutants, presented anterior up. (H–J) Examples of wild-type or near-wild-type CNS with normal longitudinal and commissural axons (H), a mutant lacking most commissures (I), and a mutant showing the severely disrupted phenotype (J). (K) Quantitation of defect frequency in the indicated mutants. Embryos are progeny of a cross in which mothers had germlines homozygous for the null allele abl4 and fathers were abl4/+. Thus all embryos were maternally mutant for endogenous abl, and half were maternally and zygotically mutant. However, for CNS scoring, mutants scored were selected to be maternally and zygotically mutant using a GFP-marked Balancer chromosome. Scale bar, 25 μm.
The FABD is not essential for cell shape changes or actin regulation during dorsal closure, whereas kinase activity plays a modulatory role
Among the processes most drastically affected by Abl loss is dorsal closure, in which epidermal cell sheets move dorsally to meet at the midline to enclose the embryo (Grevengoed et al., 2001). This involves coordinated cell shape change and migration in two tissues (Martin and Parkhurst, 2004) and thus offers an opportunity to look for subtle effects of loss of kinase activity or the FABD. The squamous amnioserosal cells cover the dorsal surface before closure (Figure 5, A and B, asterisks) and are attached laterally to the two epidermal epithelial sheets (Figure 5, A and B, arrowheads). Three forces drive closure, all of which require a well-organized actin cytoskeleton: 1) Epidermal leading edge (LE) cells assemble a contractile actomyosin cable, anchored cell to cell at cell junctions; this contracts, helping power closure. 2) Amnioserosal cells apically constrict, pulling the epidermal sheets dorsally. 3) Dorsal protrusions of LE cells guide zippering at the canthi as the sheets meet (Figure 5, A and B, arrows).
FIGURE 5:
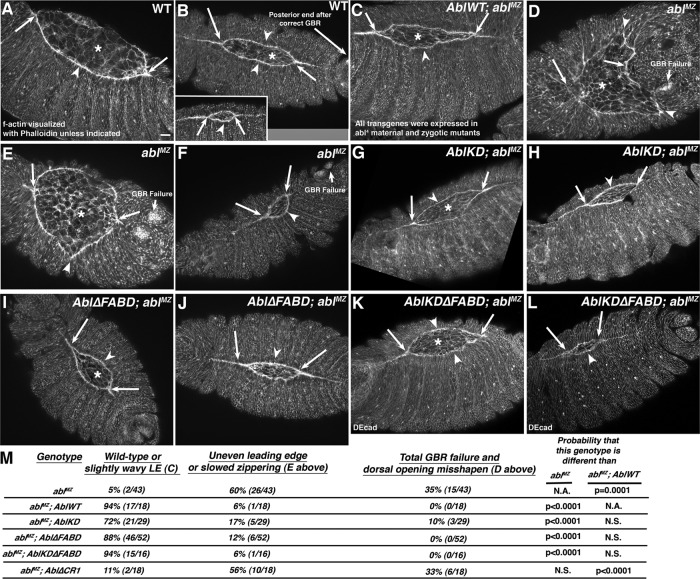
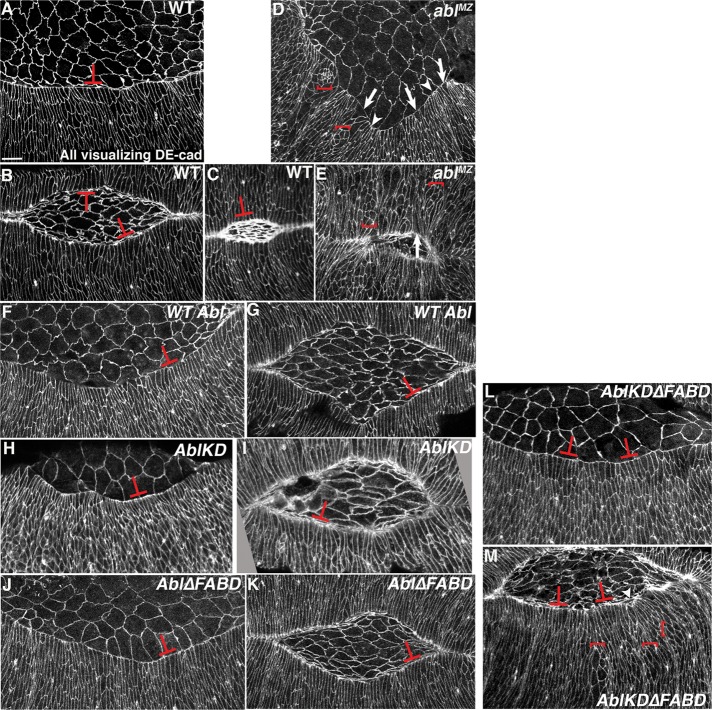
Abl proteins lacking kinase activity, the FABD, or both can restore a straight leading edge (LE) and effective zippering at the canthi during dorsal closure. Embryos, anterior left, dorsal toward viewer. Early (A, D), mid (B, C, E, G, I, K), or late dorsal closure (B, inset, F, H, J, L). Cell shapes and actin cable were visualized using phalloidin to detect F-actin. All mutants were selected to be maternally and zygotically mutant for endogenous Abl using a GFP-marked Balancer. (A, B) In wild type, amnioserosal contraction (asterisks) and the LE actin cable (arrowheads) drive closure, and epidermal sheets zip together at the canthi (arrows). Balanced forces leave the LE straight (arrowheads). Prior completion of germband retraction placed spiracles at the posterior end (beyond the right arrow in B). (C) AblWT rescues dorsal closure in abl4 maternal/zygotic mutants. (D–F) Almost all ablMZ mutants fail to fully retract the germband, so the spiracles are not at the posterior (germband retraction [GBR] failure). Most have severe LE defects (D, E, arrowheads), and zippering is delayed (D, E, arrows). Occasional mutants have a less severe phenotype (F) but still fail to fully retract their germband. (G–L) AblKD (G, H), Abl∆FABD (I, J), and AblKD∆FABD (K, L) each largely restore normal dorsal closure, with a relatively straight LE (arrowheads) and effective zippering (arrows). The mild LE waviness was also observed in embryos rescued with AblWT (C). Scale bar, 15 μm. (M) Quantitation of degree of rescue of dorsal closure in the different mutants. Embryos are progeny of a cross in which mothers had germlines homozygous for the null allele abl4 and fathers were abl4/+. Thus all embryos were maternally mutant for endogenous abl, and half were maternally and zygotically mutant. However, in this figure, mutants scored were selected to be maternally and zygotically mutant using a GFP-marked Balancer chromosome.
The ablMZ mutants have major defects in both dorsal closure and germband retraction, which precedes it (Figure 5, A and B vs. D and E, quantitated in M; Grevengoed et al., 2001). Thus most ablMZ mutants have very abnormally shaped “dorsal openings” (Figure 5, A–C vs. D–F, and M). We tested whether kinase activity or the FABD are essential for cell shape changes or collective cell migration during dorsal closure. Consistent with our cuticle data, we saw striking rescue of overall dorsal closure by both AblKD and Abl∆FABD (Figure 5, G–J, quantitated in M), with rescue by Abl∆FABD equivalent to that provided by AblWT (Figure 5C) and rescue by AblKD nearly as effective. In almost all embryos, germband retraction was successfully completed, the LE was relatively straight (Figure 5, G–J, arrowheads), and zippering at the canthi occurred normally (Figure 5, G–J, arrows). This suggests that kinase-dead and FABD-deleted mutants retain substantial function in regulating morphogenesis.
Dorsal closure is a robust process that compensates for loss of individual forces (Kiehart et al., 2000). Thus we tested whether our overall analysis missed subtle defects in cell biological events caused by absence of kinase activity or the FABD. Constriction of the LE actomyosin cable is one important driving force (Hutson et al., 2003). Normally, LE cells maintain even tension along the cable and thus have fairly similar widths at the leading edge (Figure 6, A–C, T-shapes). In ablMZ mutants, in contrast, some LE cells have splayed-open leading edges (Figure 6, D and E, arrows) whereas neighbors are hyperconstricted (Figure 6, D and E, arrowheads; Grevengoed et al., 2001), suggesting that actin cable integrity is reduced, and, when it fails, cells splay open while neighbors constrict. In addition, a subset of ablMZ epidermal cells completely fail to elongate along their dorsal-ventral axes, for unknown reasons (Figure 6, D and E, brackets). We hypothesized that although AblKD and Abl∆FABD rescued completion of closure, they might have defects in cell shape change. Strikingly, both AblKD (Figure 6, H and I, T-shapes) and Abl∆FABD (Figure 6, J and K, T-shapes) largely restored wild-type LE cell shapes, reducing the splayed-open and hyperconstricted leading edges seen in ablMZ mutants (Figure 6, D and E, arrows; rescue quantitated in Table 2) and rescuing uniform dorsal-ventral cell elongation and AblWT (Figure 6, F and G, and Table 2).
FIGURE 6:
AblKD, Abl∆FABD, and AblKD∆FABD each rescue the cell shapes of leading edge (LE) cells during dorsal closure. Embryos, anterior left and dorsal facing viewer, during early (A, D, F, H, J, L), mid (B, G, I, K, M), or late (C, E) dorsal closure, with cell shapes visualized using DE-cadherin. (A–C) Wild type. Even cable tension maintains relatively even-length LEs (T-shapes). (D, E) ablMZ. Some LE cells have splayed open LEs (arrows) while neighbors are hyperconstricted (arrowheads). In addition, some epidermal cells do not elongate (brackets). (F–M) AblWT (F, G), AblKD (H, I), Abl∆FABD (J, K), and AblKD∆FABD (L, M) each restores relatively even LE shapes (T-shapes; Table 2) and uniform epidermal cell elongation, although in AblKD∆FABD (L, M) occasional epidermal cells fail to elongate (M, brackets). Scale bar, 10 μm. Embryos are progeny of a cross in which mothers had germlines homozygous for the null allele abl4 and fathers were abl4/+. Thus all embryos were maternally mutant for endogenous abl, and half were maternally and zygotically mutant. However, in this figure, mutants scored were selected to be maternally and zygotically mutant using a GFP-marked Balancer chromosome.
TABLE 2:
Rescue of leading edge cell shape.
| Genotype | Number of “splayed open” leading edge cells per leading edge | Number of leading edges scored | Number of embryos scored |
|---|---|---|---|
| Wild type | 1.5 | 33 | 28 |
| abl4 MZ | 6.4 | 32 | 29 |
| abl4; AblWT | 2.3 | 26 | 18 |
| abl4; AblKD | 2.6 | 28 | 26 |
| abl4; AblΔFABD | 2 | 51 | 34 |
| abl4; AblKDΔFABD | 1.9 | 16 | 12 |
| abl4; AblΔCR1 | 4.8 | 18 | 12 |
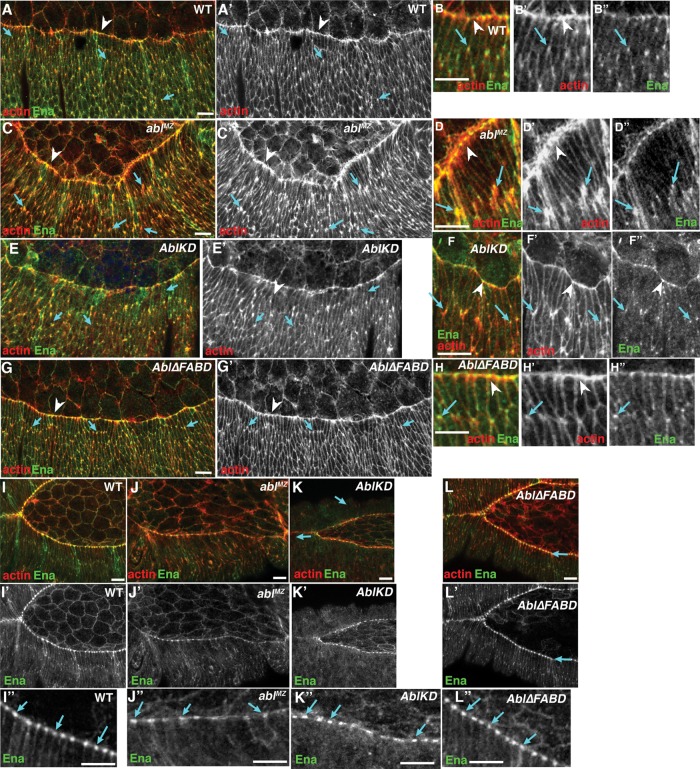
The cell shape defects in ablMZ mutants reflect underlying changes in Ena-mediated actin regulation (Grevengoed et al., 2001). Wild-type LE cells assemble a leading edge actomyosin cable (Figure 7, A–B′′, arrowheads), whereas more-ventral epidermal cells accumulate actin all around their cortex, with subtle enrichment at tricellular junctions (Figure 7, A–B′′, arrows). The actin regulator Ena is enriched at tricellular junctions of all epidermal cells during early dorsal closure (Figure 7, A–B′′, arrows; Gates et al., 2007); these coincide with and may promote the modest actin accumulation seen at tricellular junctions. During later dorsal closure, Ena accumulates in prominent “leading edge dots” at presumptive sites of cell–cell cable attachment (Figure 7, I–I′′, arrows). In ablMZ mutants, actin becomes highly elevated at dorsal-ventral cell boundaries of many epidermal cells during early dorsal closure, including those not at the LE (Figure 7, C–D′′, arrows). These “actin flares” overlap Ena’s normal tricellular junction localization, consistent with the possibility that they result from Ena misregulation. In later dorsal closure, LE Ena dots become irregular in ablMZ mutants (Figure 7, J–J′′, arrows), with some cells accumulating much less Ena. Abl∆FABD restored both normal low-level actin accumulation at tricellular junctions during early dorsal closure (Figure 7, G–H′′, arrows) and normal Ena LE dot localization later (Figure 7L′′, arrows). AblKD restored normal low-level actin accumulation at tricellular junctions during early dorsal closure, (Figure 7, E–F′′, arrows) but only partially restored Ena LE dot localization later (Figure 7, K′ and K′′, arrows). Together these data strongly suggest that the FABD is not essential for Abl to correctly regulate the cytoskeleton or cell shape changes during dorsal closure, whereas kinase activity plays a modulatory but not essential role.
FIGURE 7:
Abl regulation of actin levels and Ena localization does not require kinase activity or the FABD. (A–H) Lateral views of early stage 13 embryos, anterior to left. (B, D, F, H) Close-ups. (A, B) Wild type. The actin cable has formed at the LE (arrowheads), and Ena and actin also localize to tricellular junctions of more-ventral epidermal cells (arrows). (C, D) ablMZ. Elevated levels of actin accumulate both at LE (arrowheads) and tricellular junctions (arrows). (E–H) Defects in actin localization are rescued by both AblKD (E, F) and Abl∆FABD (G, H). (I–L) Stage 14. (I′′–L′′) Close-ups. (I) Wild type. Ena localizes to LE “dots” located near ends of the actin cable within each cell (arrows). (J) ablMZ. Ena localization to LE dots is reduced overall and much less regular (arrows). (K–L) AblKD (K) and Abl∆FABD (L) both rescue Ena localization. Scale bar, 10 μm. Embryos are progeny of a cross in which mothers had germlines homozygous for the null allele abl4 and fathers were abl4/+. Thus all embryos were maternally mutant for endogenous abl, and half were maternally and zygotically mutant. However, in this figure, mutants scored were selected to be maternally and zygotically mutant using a GFP-marked Balancer chromosome.
Deleting the FABD does not affect dorsal closure dynamics
Given the complete rescue of viability and morphogenesis by Abl∆FABD, we hypothesized that its role might only be apparent in effects on the dynamics of morphogenesis rather than their completion. Dorsal closure is robust (Kiehart et al., 2000), and some mutations, like that in the cadherin regulator p120catenin, do not affect completion of closure or accompanying cell shape changes but do slow the process (Fox et al., 2005). We thus used live imaging to examine whether FABD loss significantly slowed dorsal closure. We first measured the overall closure rate, assessing the rate of decrease in amnioserosal area in the last 60 min of dorsal closure. The wild-type closure rate is quite variable but averaged 63 μm2/min (Figure 8, A and E, and Supplemental Movie S1). The closure rate in ablMZ mutants (zygotically rescued embryos were eliminated using a GFP-marked Balancer chromosome) was significantly reduced (32 μm2/min; p = 0.03; Figure 8, B and E, and Supplemental Movie S2). Strikingly, AblWT (Figure 8C and Supplemental Movie S3) and Abl∆FABD (Figure 8D and Supplemental Movie S4) both rescued closure to rates statistically indistinguishable from wild type (Figure 8E). We also assessed the rate of zippering from the canthi, beginning when the opening was 120 μm—due to variability, even ablMZ-null mutants were not statistically distinguishable from wild type, but the trends were similar (Figure 8F)—with the slowest rate seen for ablMZ. Our quantitation of closure rates underestimates the rescue by AblWT and Abl∆FABD because almost half of unrescued ablMZ mutants completely failed to close (Figure 2D), and even in those that did, closure was qualitatively quite abnormal, with zippering defects (Figure 8B4, red arrow), abnormal leading edges, and failure to retract deep segmental grooves (Figure 8B6, blue arrows). Abl∆FABD also effectively rescued these qualitative aspects of closure. In summary, none of our assays revealed a significant diminution in the function of Abl lacking the FABD.
FIGURE 8:
Loss of the FABD does not impair the rate of dorsal closure. (A–D) Representative movie stills of embryos expressing moesin-GFP in the indicated mutant, dorsal up, anterior, as indicated (A, Supplemental Movie S1; B, Supplemental Movie S2; C, Supplemental Movie S3; D, Supplemental Movie S4). (E) Quantitation of rate of closure ± SE. Mean rate of change in amnioserosal area from 60 to 30 min and from 30 to 0 min before closure; n = 5–7 embryos. (F) Quantitation of mean rate of zippering ± SE. Rate of change in intercanthi distance beginning at 120 μm apart; n = 4–9 embryos. Scale bars, 50 μm. Embryos are progeny of a cross in which mothers had germlines homozygous for the null allele abl4 and fathers were abl4/+. Thus all embryos were maternally mutant for endogenous abl, and zygotically rescued mutants were eliminated using a GFP-marked Balancer chromosome.
Removing both kinase activity and the FABD severely reduces but does not eliminate Abl function
We considered two hypotheses explaining why both AblKD and Abl∆FABD retain substantial function: 1) both kinase activity and the FABD may be largely dispensable for some events, or 2) kinase activity and the FABD may function somewhat redundantly in modulating Abl’s scaffolding role, so that removing one does not totally disable Abl. To distinguish between these hypotheses, we generated a double mutant that is both kinase dead and lacks the FABD (AblKD∆FABD; Figure 1B).
AblKD∆FABD significantly rescued abl zygotic mutant adult viability at 25°C (Figure 1C, left;, and Table 1; 86% viable normalized to AblWT vs. 9% without a transgene). This rescuing activity was significantly reduced but not eliminated when we challenged AblKD∆FABD function further by assessing adult viability at 18°C (Figure 1C, right, and Table 1; 37% of the viability of AblWT vs. 2% without a transgene). Of interest, this was lower than that of both Abl∆FABD (91%) and AblKD (51%), suggesting that removing both kinase activity and the FABD may have additive effects on Abl function. AblKD∆FABD also retained some ability to rescue ablMZ mutant embryonic viability, but this was substantially reduced from AblWT (Figure 1, D and E, and Table 1); the 52% viable progeny of AblKD∆FABD; abl4/Df(3L)st-j7 + mothers (Figure 1D) likely relates only to those receiving paternal wild-type abl. Once again, the double mutant appeared more impaired than either single mutant (Abl∆FABD, 97% viability; AblKD, 62% viability). Finally, unlike AblWT or Abl∆FABD, AblKD∆FABD did not rescue adult viability of ablMZ mutants. These data are consistent with the possibility that whereas loss of the FABD alone does not substantially impair Abl function, removing it further reduces function of AblKD. However, Abl lacking both kinase activity and the FABD retained significant residual function.
We next explored whether AblKD∆FABD differentially rescued Abl’s embryonic roles. Of interest, morphogenesis (as assessed by cuticle patterning; Figure 2D) and coordinated mesoderm apical constriction (Figure 3, E, and F vs. H, and Supplemental Figure S5M) were rescued as well or nearly as well by AblKD∆FABD as by AblWT. Strikingly, however, Abl’s regulation of cellularization was not effectively rescued by AblKD∆FABD (Figure 3E and Supplemental Figure S5F), nor did it rescue correct midline axon guidance (Figure 4, F and K)–these paralleled roles in which AblKD also had defects. Finally, we examined dorsal closure. Consistent with the cuticle phenotype, overall dorsal closure was substantially rescued by AblKD∆FABD (Figure 5, K and L, quantitated in M), with an even LE (Figure 5, K and L, arrowheads) and sheet zippering (Figure 5, K and L, arrows) largely restored. AblKD∆FABD also largely rescued uniform LE cell shape (Figure 6, L and M, T-shapes, and Table 2), although we occasionally saw epidermal cell elongation failure and an uneven LE (Figure 6M, brackets and arrowhead). These data suggest that kinase activity and the FABD both contribute to full Abl function, but Abl lacking both still rescues many aspects of morphogenesis. These deficits in function are not solely due to a complete failure of cortical enrichment of AblKD∆FABD, as it remains cortically enriched (Supplemental Figure S4D). The differential rescuing ability of AblKD∆FABD could mean that different roles require quantitatively different thresholds of Abl activity or differ qualitatively, requiring particular combinations of Abl protein domains.
A conserved region in the Abl linker encoding a PXXP motif is critical for Abl function in vivo
The significant residual function of AblKD∆FABD suggested the hypothesis that other regions of Abl are critical for function. In addition to the SH3/SH2/kinase module and FABD, all Abl proteins contain a long linker, but its sequence is not well conserved from fly to mammal or even between mammalian Abl and Arg (Figure 1A). In Arg, the linker carries another actin-binding domain and a microtubule-binding domain (Miller et al., 2004), whereas fly Abl has a consensus binding site for the actin regulator Ena. To identify potential functional regions, we compared linkers of insect Abls, defining four short conserved regions of 12–56 amino acids, which we refer to as CR1–CR4 (Supplemental Figure S6). CR2 and CR3 do not contain any recognizable motifs. CR4 carries a perfect match to the “FP4 motif” that binds Ena’s EVH1 domain, as well as a second, less perfect match (LPPPP; Supplemental Figure S6). CR1 is the only motif with recognizable similarity to human Abl and Arg linkers—it includes the PXXP motif known to bind the adapters Crk and Nck and the actin regulator Abi (Figure 1A and Supplemental Figure S6; Feller, 2001; Ibarra et al., 2005).
To test whether these motifs contribute to Abl’s mechanism of action in development, we generated mutants deleting each motif individually. We first tested rescue of adult viability of abl zygotic mutants or embryonic viability of ablMZ mutants. Abl∆CR2 and Abl∆CR3 rescued both as well as AblWT (Figure 9, A and C, left, and Table 1). This suggests that CR2 and CR3 are not essential for Abl function in morphogenesis, and we did not analyze these mutants further. Owing to the presence of the FP4 motif, we examined Abl∆CR4 in more depth. Strikingly, Abl∆CR4 was as effective as AblWT at rescuing embryonic viability of ablMZ mutants (Figure 9, A and B; full data and statistical tests in Table 1), adult viability of abl4 zygotic mutants at 25°C (Figure 9C, left, and Table 1), and zygotic viability in our sensitized assay, which stresses cytoskeletal regulation by raising animals at 18°C (Figure 9C, right, and Table 1). Further, Abl∆CR4 rescued ablMZ mutants to adulthood (Figure 2C). Finally, Abl∆CR4 remained cortically enriched in vivo (Supplemental Figure S4F). Thus the putative Ena-binding motifs in CR4 are dispensable for Abl function, at least in our assays. Of course, Ena can also associate with Abl via its SH3 domain (Ahern-Djamali et al., 1999), so the linker FP4 motif may be redundant.
FIGURE 9:
CR1 with the PXXP motif is critical for Abl function in morphogenesis. (A, B) Rescue of embryonic viability of ablMZ mutants from abl4 germline homozygous (A) or abl4/Df(3L)st-j7 (B) mothers (50% of progeny are paternally rescued). (C) Rescue of adult viability of abl4 zygotic mutants at indicated temperatures, normalized to AblWT rescue. Full data for A–C are given in Table 1. (D–N) Stage 13–14 embryos, anterior left, dorsal up. (D–I) Embryos stained to visualize cell–cell junctions. Relative to wild type (D, F, H), Abl∆CR1 (E, G, I) fails to fully restore germband retraction (E, G, arrows) or a straight LE (D, F vs. E, G, arrowheads). (H, I) Even Abl∆CR1 mutants that do close have severe dorsal midline discontinuities (I, arrows) relative to the straight midline in wild type (H, arrows). (J) Abl∆CR1 mutants have the same large-scale defects at the LE (arrowheads), splayed open (red arrows; quantitated in Table 2) or hyperconstricted (yellow arrows) LE cells and failure of epidermal cells to elongate (brackets) seen in ablMZ mutants (Figure 6). (K–N) Abl∆CR1 fails to rescue LE Ena localization. (K, M) Close-ups. Wild type. The LE is straight (arrowheads), and Ena localizes uniformly to LE “dots” (arrows). (L, N) Close-ups. Abl∆CR1. The LE is wavy (arrowheads), and although Ena localizes to LE dots in occasional cells (red arrows), its localization is reduced and less well confined to the LE dots in most cells (T-shapes, blue arrows). Scale bars, 10 μm. In D–N, embryos are progeny of a cross in which mothers had germlines homozygous for the null allele abl4 and fathers were abl4/+. Thus all embryos were maternally mutant for endogenous abl, and half were maternally and zygotically mutant. However, in this figure, mutants scored were selected to be maternally and zygotically mutant using a GFP-marked Balancer chromosome.
In contrast, CR1 is critical for full Abl activity, although Abl∆CR1 retained residual function. Abl∆CR1 significantly rescued adult viability at 25°C (Figure 9C, left; full data and statistical tests in Table 1), although not as well as AblWT. However, at 18°C, Abl∆CR1’s ability to rescue adult viability was almost eliminated, although it did retain a small amount of residual activity (Figure 9C, right, and Table 1). Abl∆CR1 also exhibited substantially reduced rescue of embryonic viability of ablMZ mutants (Figure 9, A and B, and Table 1) and could not rescue ablMZ mutants to adulthood. Thus Abl∆CR1 was less functional than AblKD or Abl∆FABD and, in some assays, also appeared less functional than AblKD∆FABD (Figure 9A and Supplemental Figure S7).
Abl∆CR1 has substantially reduced ability to regulate morphogenesis, cell shape changes, and localization of the cytoskeletal regulator Ena
These data suggest that the CR1 region is critical for full Abl function. This could be due to a role in a single process critical for viability (e.g., CNS development) or reflect broader reduction in Abl’s diverse cell biological roles. To distinguish these hypotheses, we tested whether Abl∆CR1 rescued different embryonic events. We first used cuticles to assess completion of key morphogenetic movements. Abl∆CR1 failed to rescue two critical events—germband retraction and head involution (Figure 2D). This contrasted with all other mutants we tested, including AblKD∆FABD. However, Abl∆CR1 did provide some rescue of epithelial integrity relative to ablMZ mutants lacking any transgene, as fewer embryos had the most-severe epidermal integrity defects (Figure 2D).
To define the cell biological events in which Abl∆CR1 retained or lacked function, we examined Abl-regulated events in embryogenesis. Abl∆CR1 retained essentially wild-type activity in promoting mesoderm invagination (Figure 3E and Supplemental Figure S5N). In contrast, however, Abl∆CR1 failed to rescue cellularization (Figure 3E and Supplemental Figure S5G), and although Abl∆CR1 rescued the severe CNS disruption seen in some ablMZ mutants, it did not restore correct midline guidance (Figure 4, G and K). These data suggest a possible differential loss of function in Abl∆CR1. To further define the mechanistic functions retained or lost by Abl∆CR1, we examined dorsal closure, exploring whether Abl∆CR1 promoted correct cell shape change or regulated Ena localization. Consistent with cuticle analysis, most Abl∆CR1 mutants failed to fully retract their germbands (Figure 9, E and G vs. D and F, red arrows; 12 of 15 embryos scored failed). They also had the extremely uneven LE (Figure 9, E and J vs. D and F, arrowheads; quantitated in Figure 5M) seen in unrescued ablMZ mutants (Figure 5K). Even the subset that closed had significant discontinuities at the dorsal midline (Figure 9, H vs. I). Abl∆CR1 only partially rescued ablMZ mutant cell shape defects—LE cells had splayed open (Figure 9J, red arrows; quantitated in Table 2) and hyperconstricted LEs (Figure 9J, yellow arrows), suggesting defects in actin cable anchoring. Finally, Abl∆CR1 failed to restore localization of the actin regulator Ena—whereas Ena is highly and uniformly enriched at LE dots in wild type (Figure 9, K and M, arrows), in Abl∆CR1 mutants, Ena localization to LE dots was often reduced overall and less uniform (Figure 9, L, T-shapes, L′′, and N, blue arrows), as in ablMZ mutants. Thus Abl∆CR1 retains little if any function in dorsal closure. Together these data suggest that the CR1 motif plays a surprisingly important but not absolutely essential role in Abl function, with its loss differentially affecting some Abl-regulated events, like dorsal closure or CNS development, while not disrupting others, like mesoderm invagination. Once again, these deficits in function were not solely due to a failure of cortical enrichment, as Abl∆CR1 remained cortically enriched (Supplemental Figure S4E), although the degree of enrichment may be reduced from wild type and thus contribute to the deficit in function.
DISCUSSION
As a key regulator of cell behavior in development and an iconic example of targeted cancer therapy, Abl has attracted attention from scientists in many fields. Their studies defined the pathways that Abl regulates and revealed its capabilities as a kinase and a direct cytoskeletal regulator in cultured cells, but the relative importance of each role in morphogenesis was largely untested. Here we directly tested a series of mechanistic hypotheses about how kinase activity, the FABD, and other conserved protein-docking sites contribute to Abl’s diverse functions during morphogenesis in vivo.
The FABD is not essential for Abl’s diverse roles in morphogenesis, whereas kinase activity plays differential roles in different events
Two mechanistic models for Abl function in development and disease suggest that 1) Abl phosphorylates target proteins to modulate their activity or 2) Abl directly regulates cytoskeletal dynamics via its cytoskeletal binding sites. We tested these hypotheses using mutants lacking either kinase activity or the only predicted cytoskeletal-binding site in fly Abl, the FABD (Supplemental Figure S7). To our surprise, Abl∆FABD retained full activity in our assays—in the most dramatic demonstration, it rescued flies completely lacking endogenous maternal and zygotic Abl to fertile adults without morphological defects. Because some morphogenetic events regulated by Abl are robust, we looked carefully for cell biological defects that might not block viability, but even in those assays, Abl∆FABD rescued as well as AblWT. Thus, despite its sequence conservation, the FABD is not essential for many of Abl’s functions during normal development. It is interesting to note that O’Donnell and Bashaw (2013) carried out a detailed analysis of different roles played by Abl in the CNS, and their data suggest that although the FABD is dispensable for motor axon outgrowth, deleting it does lead to failure to rescue midline crossing of EW neurons, suggesting that Abl∆FABD is subtly impaired. We note that although the FABDs of mammalian Arg and Abl bind actin, this has not been experimentally demonstrated for fly Abl but is only predicted based on sequence conservation. It will be important to determine whether and how fly Abl interacts with actin. In contrast, Abl kinase activity is critical for full Abl function. In some events, such as cellularization, AblKD was similar to the abl-null mutant; previous data also suggest key roles for phosphorylation of β-catenin during convergent extension (Tamada et al., 2012) and Abl kinase activity in germline stem cells (Stine et al., 2014). In other events, such as rescue of zygotic adult or maternal/zygotic embryonic viability, AblKD retained partial function but was significantly less functional than wild-type Abl. Perhaps most surprising, in some events, including mesoderm invagination and completion of dorsal closure and germband retraction, it retained full or nearly full function. Consistent with this, analysis of Abl’s roles in axon outgrowth suggested both kinase-dependent and kinase-independent roles (O’Donnell and Bashaw, 2013). Thus kinase activity, although important for full Abl function, is not essential for all of its roles.
The conserved linker PXXP motif plays a surprisingly critical role in morphogenesis
In addition to the SH3:SH2:kinase module and FABD, all Abl proteins contain a long linker with a sequence that is divergent among mammalian paralogues or insect orthologues. Mammalian Arg has additional actin- and microtubule-binding sites in the linker, suggesting that family members may have distinct linker motifs contributing to function. Insect Abls share four short conserved motifs that may be partner-binding sites. Two, CR2 and CR3, do not contain known motifs, and neither proved critical for function. CR4 carries one perfect and one imperfect match for binding sites for Abl’s key regulatory target, Ena. Surprisingly, Abl∆CR4 was fully functional in all our assays. We hypothesize that this reflects redundancy/robustness of the Abl scaffold, as Abl’s SH3 domain can also bind Ena (Ahern-Djamali et al., 1999). Similarly, the second actin- and the microtubule-binding domains of Arg in the linker provide it with unique functions (e.g., Courtemanche et al., 2015). Perhaps variant Abl linkers provide raw material to create new binding motifs that modulate but are not critical for Abl function.
Although CR2–4 were dispensable, CR1, the only linker motif shared by fly and mammalian Abl, proved critical for Abl function. Abl∆CR1 failed to rescue embryonic viability of ablMZ mutants, and its activity in regulating individual morphogenetic events was severely impaired, although, like AblKD, Abl∆CR1 retained residual function in a subset of Abl’s roles. Thus CR1 is a critical and surprisingly less redundant part of Abl’s mechanism of action (Supplemental Figure S7). Several partners bind human Abl CR1, including Crk, Nck, and Abi (Hossain et al., 2012). These partners play strikingly different cell biological roles and can compete with one another for binding to Abl. Crk and Nck are signaling adapters, linking signals from receptor and nonreceptor tyrosine kinases, integrins, or cadherins to diverse outputs (Zandy and Pendergast, 2008; Bell and Park, 2012), whereas Abi is a multifunctional regulator of distinct actin polymerization machines (e.g., Ryu et al., 2009). It will be exciting to explore which contributes to different Abl functions in vivo. Existing mutational analysis may provide clues. Drosophila Abi is an excellent candidate, as it shares with Abl roles in cultured cell- protrusive behavior (Kunda et al., 2003) and embryonic axon guidance (Lin et al., 2009) and exhibits dose-sensitive genetic interactions with Abl (Lin et al., 2009). Nck is also a reasonable candidate, as maternal and zygotic Nck mutants (= Drosophila Dock) share significant embryonic CNS defects with abl, although these are not as severe as those of ablMZ mutants and in fact exhibit increased rather than decreased midline crossing (Desai et al., 1999)—effects on morphogenesis were not reported. Genetic analysis of Drosophila Crk was slowed by its location on the fourth chromosome and the lack of null alleles, but a cell-based RNA interference (RNAi) screen implicated both Abl and Crk in internalization of the pathogen Pseudomonas aeruginosa (Pielage et al., 2008), and analysis of both RNAi and a hypomorphic allele support Crk roles in pupal thoracic closure downstream of the receptor tyrosine kinase Pvr (Ishimaru et al., 2004). Of course, all three proteins likely have Abl-independent roles as well, complicating interpretation.
A model of Abl kinases as robust scaffolds with multiple semiredundant binding sites
Our understanding of receptor tyrosine kinases underwent a significant change 20 yr ago when it was realized that they are scaffolds that assemble signaling complexes, with kinase activity largely serving to create additional partner binding sites (Margolis and Skolnik, 1994). The surprising dispensability of the FABD and the significant function retained by both kinase-dead Abl and Abl∆CR1 in some events support a similar speculative model for Abl in which Abl assembles a multiprotein signaling/actin regulatory complex, with individual partners recruited by interactions with several proteins (Supplemental Figure S7). Thus, for example, a particular protein partner might bind both a tyrosine residue phosphorylated by Abl’s kinase and an actin-binding protein brought into the complex by Abl’s actin interaction or an SH3 domain protein that binds CR1. Eliminating one recruitment mechanism might only reduce but not eliminate inclusion of this partner from the complex, while eliminating both potential recruitment mechanisms would be more deleterious. Consistent with this, a double mutant lacking both kinase activity and the FABD (AblKD∆FABD) has, in some cases, phenotypes quantitatively more severe than AblKD or Abl∆FABD (Figure 9, A and C). However, AblKD∆FABD is not biologically dead—in fact, in some assays, it retained wild-type or nearly wild-type function, whereas in other events, it had residual activity. These data are consistent with a scaffolding model in which additional interaction sites exist, allowing AblKD∆FABD to retain residual scaffolding function by binding a subset of its partners. The SH2 and SH3 domains provide obvious examples. It will be important to explore their function in morphogenesis. In addition, in the absence of Abl kinase activity, other kinases may phosphorylate proteins in the complex, recreating SH2 binding sites normally created by Abl phosphorylation. Consistent with this, Abl is regulated by Src kinases in development and oncogenesis, with dual Abl/Src inhibitors emerging as effective treatments in CML resistant to the Abl-inhibitor Gleevec. Strikingly, overexpressing the Src-family kinase Hck confers Gleevec resistance; this requires Src kinase activity and correlates with Abl phosphorylation (Pene-Dumitrescu and Smithgall, 2010). Our data are reminiscent of work with platelet-derived growth factor receptors, for which mutating individual SH2-docking sites had complex, overlapping. or tissue-specific effects (Klinghoffer et al., 2002; Tallquist et al., 2003). This and recent work on phosphodependent protein complexes in yeast Hippo signaling (Rock et al., 2013) suggest that robust and complex scaffold assembly may be a broad feature of signaling.
As we further test our scaffolding model, it will be important to determine whether the differential loss of function of AblKD∆FABD or Abl∆CR1 in distinct morphogenetic events occurs because different roles require quantitatively different thresholds of Abl activity or because some roles require particular protein–protein interactions. Because deleting CR1 leads to defects in Ena localization, although Ena is not believed to bind this site, loss of particular interactions may have complex consequences.
Abl in mammalian development and oncogenesis
Mammalian Abl and Arg play important roles in events ranging from neural tube closure to immune system function to synaptogenesis. Cell-based studies identified cellular events regulated by Abl kinase activity or cytoskeletal interactions, respectively. Given our data, it will be exciting to explore whether mouse Abl is as robust a scaffold, by introducing mutants analogous to ours into abl and arg single- or double-mutant mice and analyzing the diverse Abl/Arg functions. Our results may also have implications for Bcr-Abl’s oncogenic role. Gleevec’s effectiveness in CML and the strong selection for mutations restoring kinase activity in relapsing Gleevec-treated patients clearly demonstrate that kinase activity is necessary for oncogenesis. How can we reconcile these results with ours? First, Bcr-Abl’s unregulated kinase likely phosphorylates targets outside Abl’s normal repertoire, some of which may drive oncogenesis. Second, the data do not demonstrate that kinase activity is sufficient for oncogenesis but instead that suggest kinase activity and direct cytoskeletal regulation cooperate in cell transformation. Experiments defining which aspects of Abl function are required for oncogenesis may now be warranted, using inducible Bcr-Abl mutants altering single “functions” and testing the robustness of this oncogenic kinase to perturbation (Huettner et al., 2000).
MATERIALS AND METHODS
Transgenic fly lines
The Xba/NotI fragment from pUASg-Abl::GFP (Fox and Peifer, 2007) containing 2 kb of upstream endogenous Abl promoter sequence and the Abl coding region was cloned into Xba/NotI sites of pUAStattP. Both missense and deletion mutants were made by PCR stitching of overlapping PCR products with mutations generated by primers in the overlapping segments. The forward mutagenic oligonucleotide primers for the Abl constructs are as follows:
Abl∆CR1: 5′CCAGCAGCAGGCCAGCACGCCCATGCCAGCCAACGCCAGATGCAATTTCATCG-3′
Abl∆CR2:
5′-GGTGACCAGTGCTCATCCCATCACTGAGGCTGCTCCTGCTCTTCCGGCAACTGC-3′
Abl∆CR3:
5′CGAAGGCCAGCCCCATTCCGCCACAGATGCAAAACAATGCGGCTGCCAGC-3′
Abl∆CR4:
5′GGAGTGCCTCGGGAGTGGCTTCAGGAGCGCCGGAGAGCGCTGTGCAGGCC-3′
Abl∆FABD: 5′AGCTCCGCCAGCTCCACACAGATATCAGGACTAGTGATTGGAGCTAGCATGGTG-3′
AblKD:
5′-TGGC AATACGGTGGCTGTTAACACGCTCAAGGA GGACACCAT-3′
The end of the boldface type in the primer sequences for the Abl deletions indicates the location of the start of the deletion. The underlined base indicates the location of the point mutation. The reverse primer for the respective constructs was the reverse complement of the forward primer. All constructs were sequenced to verify the desired mutational change and ensure that other changes had not been introduced. Transgenes were targeted to the left arm of the second chromosome by phiC31 integrase–mediated transgenesis (Bischof et al., 2007). Injections of transgenic constructs were performed by BestGene (Chino Hills, CA) into PBacyellow[+]-attP-3BVK00037 (cytogenetic map position, 22A3).
Genotyping assays
Fly genomic DNA was prepared as follows: one to five flies were frozen overnight at −20°C and then crushed with a barrier tip in 50 μl of 1× squishing buffer (10 mM Tris, pH 8.2, 1 mM EDTA, 25 mM NaCl) plus 1 μl of Proteinase K (10 mg/ml), incubated at 37°C for 30–60 min, and heat inactivated for 1–2 min at 95°C. Genomic DNA was used as a template for PCR. In Supplemental Figure S1, A and B, we verify the presence of the Abl kinase-dead mutation. Single flies were crushed as described. PCR was used to amplify a 238–base pair region around the kinase-dead mutation. Primer sequences used were, for KD forward, GTCTTTCCGCTGAGTCCCGAGCCG, and for KD reverse, CACCAATGAGCTGCACCAGA TTAGG.
The PCR product was sequenced using standard Sanger sequencing (University of North Carolina at Chapel Hill Genome Analysis Facility) using as primer KD-Seq, ACATCATGATGAAGCACAAGC. Note that the PCR primers used do not distinguish between the endogenous and the transgenic abl locus. Thus sequencing of this PCR product yields two peaks (an A for the endogenous locus and a C for the transgene) at the position of the point mutation, leading to an amino acid change that ablates Abl kinase activity.
In Supplemental Figure S1, D–H, we use PCR to verify the presence of the abl deletions in ∆FABD, ∆CR1, ∆CR2, ∆CR3, and ∆CR4. A 4-μl amount of each genomic DNA preparation was used for PCRs, following manufacturer’s instructions (Phusion polymerase; Thermo Scientific, Waltham, MA). Then 10–20 μl of the PCR was run on a 1.3–1.5% gel at 100 V. Primers sequences used were as follows:
∆FABD-F: GCACAAGCCAACAAAGCTAA
∆FABD-R: GAACTTCAGGGTCAGCTTGC
∆CR1-F: GGCGGTCAGGCCCTCACGCCGAACGCCCACCACAACGATCCGCACCAGCAGCAG
∆CR1-R: CCATTCGTGCTGAGGTCGTCGATGAAATTGCATCTGGCG
∆CR2-F: ACTTATCGCGAGGAGGATCC
∆CR2-R: GGCCTTCGGATTTAGTCTGG
∆CR3-F: GAGGCTGCTCCTGCTCTTC
∆CR3-R: GGAGGAGAACGTCATCATGG
∆CR4-F: CCACTACCGAAGGCACCATG
∆CR4-R: GTTGCCATTGTTTGGCAGCTG
For Supplemental Figures S1C and S3, PCR was done using two primers that flank the first intron of Drosophila abl (forward, 5′-CCTGGTCCGTGAAAGTGAAA-3′, and reverse, 5′-GGATCCTCTGAGATGCGGTA-3′). PCR products were resolved on a 3.5% NuSieve GTG agarose (Lonza, Basel, Switzerland) gel in 1× TBE (89 mM Tris base, 89 mM boric acid, 2 mM EDTA, pH 8.0). Wild-type endogenous abl yields a 166–base pair fragment, the abl4 mutant yields a 133–base pair fragment, and the abl transgenes yield a 93–base pair fragment due to the absence of an intron.
Fly stocks, viability, and phenotypic analysis of abl mutants and statistical tests
We used both y w and histone-GFP–expressing flies (Clarkson and Saint, 1999) as wild type. Zygotic abl mutants were generated by crossing Df(3L)st-7 Ki/TM3 Sb females to abl transgene (Tn[Abl])/Tn[Abl]; abl4/TM3 Sb males and selecting for Ki and against Sb (abl4/Df(3L)st-7 Ki). The fraction of progeny with this genotype seen when using the wild-type abl transgene (AblWT; 30.9% at 25°C and 33.7% at 18°C) was set as 100%, and other genotypes were normalized to this. Adult viabilities were compared by Fisher’s exact test (GraphPad, La Jolla, CA). Embryos and flies maternally and zygotically abl mutant (ablMZ) were generated in two ways: 1) using the dominant female sterile method (Chou and Perrimon, 1996) to make abl4 clones in the female germline and 2) using a deficiency spanning the abl locus transheterozygous to abl4 (cross schemes are shown in Supplemental Figure S2). Briefly, to generate abl germline clones, hs::Flp;;FRT 79 D-F ovoD/TM3 males were crossed with w;Tn[Abl]/Tn[Abl];FRT 79 D-F abl4/TM3 females. The 48- to 72-h-old progeny were heat shocked for 3 h at 37°C and allowed to develop to adulthood. Resulting virgin female flies of the genotype hs::FLP/+;Tn[Abl]/+; FRT 79 D-F abl4/ FRT 79 D-F ovoD were crossed with w;Tn[Abl]/Tn[Abl];FRT 79 D-F abl4/TM3, twi-GAL4,UAS-EGFP males and put into cups with apple juice/agar plates and yeast paste for embryo collection. The presence of a GFP-expressing Balancer chromosome allowed identification of paternally rescued embryos after germband extension. To generate embryos and flies maternally and zygotically mutant for abl using a deficiency, we used Df(3L) st-j7, Ki/TM6b (Bloomington Drosophila Stock Center #5416, Deletes73A2-73B2). For both generation of ablMZ mutants and assessment of rescue to adult viability by the abl transgenes, w; Df(3L) st-j7, Ki/ TM3, twi-GAL4,UAS-EGFP females were crossed with w;Tn[Abl]/Tn[Abl];FRT 79 D-F abl4/TM3 males. If the resulting w;Tn[Abl]/+;FRT 79 D-F abl4/Df(3L) st-j7, Ki females were viable, they were crossed to w;Tn[Abl]/Tn[Abl];FRT 79 D-F abl4/TM3, twi-GAL4,UAS-EGFP males and put into collection cups. Assessment of embryonic lethality and preparation of embryonic cuticles were done as in Wieschaus and Nüsslein-Volhard (1986). To compare cuticle phenotypes of abl4 mutants and embryos expressing different Abl transgenes in the abl4 mutant background, we used Fisher’s exact test (GraphPad). For each genotype, the numbers of cuticles falling into our three defective classes (strong defects in germband retraction, dorsal closure failure, and epidermal integrity defect) were grouped into a single defective category and compared with the number of cuticles in the wild-type category. Similarly, cuticle scores of each mutant transgene in the abl4 mutant background were compared with the wild-type transgene (AblWT) using the same approach. Fisher’s exact test was also used to compare the degree of rescue of dorsal closure in Figure 5M. To assess the completion of cellularization, we examined fixed embryos at cellularization, with cell outlines revealed by staining with phalloidin to mark either F-actin or DE-cadherin at a magnification that included ∼70% of the embryo surface. Embryos were scored as mutant if these images showed any cells in which cellularization failed, as assessed by counting cells with apical ends two or more times larger than the typical cell in the field (Figure 3B, arrows). All scored as mutant had >10 cells in which cellularization failed. To assess successful completion of ventral furrow invagination, we assessed fixed embryos after completion of germband extension. Successful completion of ventral furrow invagination was scored as embryos in which the ventral midline was straight and cells at the midline had relatively equal-sized apical ends (Figure 3C, arrows). In contrast, more than half of abl4 mutants had ventral midlines that were extremely irregular and in which many cells had failed to constrict apically (Figure 3D, arrows).
Embryo live imaging
Moesin-GFP–expressing embryos (Edwards et al., 1997) were dechorionated in 50% bleach and mounted in halocarbon oil (series 700; Halocarbon Products, River Edge, NJ) between a coverslip and a gas-permeable membrane (Petriperm; Sartorius, Edgewood, NJ). Imaging was performed with a PerkinElmer (Waltham, MA) UltraView spinning-disk confocal ORCA-ER camera (Hamamatsu, Hamamatsu City, Japan), Nikon (Melville, NY) 40× Plan Apo numerical aperture 1.3 objective, and MetaMorph (Molecular Devices, Sunnyvale, CA) software. The 0.5-μm z-stacks were maximum intensity projected to visualize the amnioserosa and leading edge, using the ImageJ 5-D plugin (National Institutes of Health, Bethesda, MD). Dorsal closure rates based on area were calculated by measuring the dorsal opening area at 60 and 30 min before closure, and the final rate was determined by averaging the closure rates from 60 to 30 min and from 30 min to completion. Data were analyzed using a Mann–Whitney U test in Matlab (MathWorks, Natick, MA).
Immunohistochemistry and immunoblotting
Flies were allowed to lay eggs for the appropriate time (1–24 h) on apple juice/agar plates with yeast paste. Embryos were dechorionated in 50% bleach, washed in 0.1% Triton-X, and fixed at room temperature in 1:1 heptane/3.7% formaldehyde diluted in phosphate-buffered saline (PBS) for 20 min. Embryos were devitillenized either by shaking in 1:1 heptane/methanol or by hand for phalloidin staining. Embryos were incubated in blocking solution (PBS/0.1% Triton-X and 1% normal goat serum) for ≥30 min, incubated overnight at 4°C in primary antibody diluted in blocking solution, and washed three times in blocking solution. Embryos were incubated for 2 h at room temperature in secondary antibody in blocking solution and washed three times in blocking solution. Embryos were mounted on glass slides in Aquapolymount (Polysciences, Warrington, PA). Imaging was done on a Zeiss (Oberkochen, Germany) LSM-5 Pascal or a Zeiss 710 scanning confocal microscope. Primary and secondary antibodies used were anti–D-cadherin (1:100), anti-Ena (1:500), anti-BP102 (1:200; all from the Developmental Studies Hybridoma Bank, Iowa City, IA), and anti-mouse and anti-rat immunoglobulin G (IgG) Alexa Fluor 568 and 647 (Molecular Probes, Waltham MA); some secondary antibodies were preabsorbed with fixed y w embryos. For F-actin staining, tetramethylrhodamine isothiocyanate–labeled phalloidin (Sigma-Aldrich, St. Louis, MO) was used at a dilution of 1:500–1:1000.
For immunoblotting, equal volumes of embryos were homogenized with a pestle in a microfuge tube in SDS–PAGE sample buffer, boiled for 5 min, run on a 7.5% SDS–PAGE gel, and transferred to a nitrocellulose membrane. For detection of the transgenic protein, anti-GFP (JL-8, 1:500 or 1:1000; Clontech, Mountain View, CA) was used. Anti–α-tubulin (1:10,000; Sigma-Aldrich), anti-γ–tubulin (clone GTU-88; 1:2500; Sigma-Aldrich), and anti-Pnut (1:30; Developmental Studies Hybridoma Bank) were loading controls. Secondary antibody was horseradish peroxidase–conjugated anti-mouse IgG (1:50,000; Pierce, Waltham, MA), and ECL Plus substrate kit (Pierce) was used for detection.
Supplementary Material
Acknowledgments
We are very grateful to Natalie McKeon for technical assistance and John Poulton for statistical advice. We thank the Developmental Studies Hybridoma Bank and the Bloomington Drosophila Stock Center for reagents and our lab members for thoughtful conversations. This work was supported by National Institutes of Health Grants R01GM47957 and R35 GM118096 to M.P., F32-GM106516 to K.D.S., and T32 CA009156 to A.J.S., Leukemia and Lymphoma Society Career Development Program Fellowship Grant 5339-08 to E.M.R., and a predoctoral fellowship from the American Heart Association to B.J.R.
Abbreviations used:
- Abl
Abelson kinase
- Abl∆FABD
Abl kinase lacking the conserved F-actin–binding domain
- AblKD
Abl kinase dead
- CR
conserved region
- Ena
Enabled
- LE
leading edge
- SH2
Src-homology domain 2
- SH3
Src-homology domain 3.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-05-0292) on July 6, 2016.
REFERENCES
- Ahern-Djamali SM, Bachmann C, Hua P, Reddy SK, Kastenmeier AS, Walter U, Hoffmann FM. Identification of profilin and src homology 3 domains as binding partners for Drosophila enabled. Proc Natl Acad Sci USA. 1999;96:4977–4982. doi: 10.1073/pnas.96.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoku S, Saksela K, Rivera GM, Mayer BJ. A crucial role in cell spreading for the interaction of Abl PxxP motifs with Crk and Nck adaptors. J Cell Sci. 2008;121:3071–3082. doi: 10.1242/jcs.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell ES, Park M. Models of crk adaptor proteins in cancer. Genes Cancer. 2012;3:341–352. doi: 10.1177/1947601912459951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley WD, Hernandez SE, Settleman J, Koleske AJ. Integrin signaling through Arg activates p190RhoGAP by promoting its binding to p120RasGAP and recruitment to the membrane. Mol Biol Cell. 2006;17:4827–4836. doi: 10.1091/mbc.E06-02-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley WD, Koleske AJ. Regulation of cell migration and morphogenesis by Abl-family kinases: emerging mechanisms and physiological contexts. J Cell Sci. 2009;122:3441–3454. doi: 10.1242/jcs.039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T-B, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating female germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson M, Saint R. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 1999;18:457–462. doi: 10.1089/104454999315178. [DOI] [PubMed] [Google Scholar]
- Comer AR, Ahern-Djamali SM, Juang J-L, Jackson PD, Hoffmann FM. Phosphorylation of Enabled by the Drosophila Abelson tyrosine kinase regulates the in vivo function and protein-protein interactions of Enabled. Mol Cell Biol. 1998;18:152–160. doi: 10.1128/mcb.18.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Gifford SM, Simpson MA, Pollard TD, Koleske AJ. Abl2/Abl-related gene stabilizes actin filaments, stimulates actin branching by actin-related protein 2/3 complex, and promotes actin filament severing by cofilin. J Biol Chem. 2015;290:4038–4046. doi: 10.1074/jbc.M114.608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai CJ, Garrity PA, Keshishian H, Zipursky SL, Zinn K. The Drosophila SH2-SH3 adapter protein Dock is expressed in embryonic axons and facilitates synapse formation by the RP3 motoneuron. Development. 1999;126:1527–1535. doi: 10.1242/dev.126.7.1527. [DOI] [PubMed] [Google Scholar]
- Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev Biol. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348–6371. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- Fox DT, Homem CC, Myster SH, Wang F, Bain EE, Peifer M. Rho1 regulates Drosophila adherens junctions independently of p120ctn. Development. 2005;132:4819–4831. doi: 10.1242/dev.02056. [DOI] [PubMed] [Google Scholar]
- Fox DT, Peifer M. Abelson kinase (Abl) and RhoGEF2 regulate actin organization during cell constriction in Drosophila. Development. 2007;134:567–578. doi: 10.1242/dev.02748. [DOI] [PubMed] [Google Scholar]
- Gates J, Mahaffey JP, Rogers SL, Emerson M, Rogers EM, Sottile SL, Van Vactor D, Gertler FB, Peifer M. Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development. 2007;134:2027–2039. doi: 10.1242/dev.02849. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Comer AR, Juang JL, Ahern SM, Clark MJ, Liebl EC, Hoffmann FM. enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev. 1995;9:521–533. doi: 10.1101/gad.9.5.521. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Hill KK, Clark MJ, Hoffmann FM. Dosage-sensitive modifiers of Drosophila abl tyrosine kinase function: prospero, a regulator of axonal outgrowth, and disabled, a novel tyrosine kinase substrate. Genes Dev. 1993;7:441–453. doi: 10.1101/gad.7.3.441. [DOI] [PubMed] [Google Scholar]
- Grevengoed EE, Fox DT, Gates J, Peifer M. Balancing different types of actin polymerization at distinct sites: roles for Abelson kinase and Enabled. J Cell Biol. 2003;163:1267–1279. doi: 10.1083/jcb.200307026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevengoed EE, Loureiro JJ, Jesse TL, Peifer M. Abelson kinase regulates epithelial morphogenesis in Drosophila. J Cell Biol. 2001;155:1185–1198. doi: 10.1083/jcb.200105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantschel O, Superti-Furga G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5:33–44. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- Hardin JD, Boast S, Mendelsohn M, de los Santos K, Goff SP. Transgenes encoding both type I and type IV c-abl proteins rescue the lethality of c-abl mutant mice. Oncogene. 1996;12:2669–2677. [PubMed] [Google Scholar]
- Heisterkamp N, Voncken JW, Senadheera D, Gonzalez-Gomez I, Reichert A, Haataja L, Reinikainen A, Pattengale PK, Groffen J. Reduced oncogenicity of p190 Bcr/Abl F-actin-binding domain mutants. Blood. 2000;96:2226–2232. [PubMed] [Google Scholar]
- Henkemeyer M, West S, Gertler F, Hoffmann F. A novel tyrosine kinase-independent function of Drosophila abl correlates with proper subcellular localization. Cell. 1990;63:949–960. doi: 10.1016/0092-8674(90)90498-4. [DOI] [PubMed] [Google Scholar]
- Hernandez SE, Settleman J, Koleske AJ. Adhesion-dependent regulation of p190RhoGAP in the developing brain by the Abl-related gene tyrosine kinase. Curr Biol. 2004;14:691–696. doi: 10.1016/j.cub.2004.03.062. [DOI] [PubMed] [Google Scholar]
- Hossain S, Dubielecka PM, Sikorski AF, Birge RB, Kotula L. Crk and ABI1: binary molecular switches that regulate abl tyrosine kinase and signaling to the cytoskeleton. Genes Cancer. 2012;3:402–413. doi: 10.1177/1947601912460051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YP, Comiskey EO, Dupree RS, Li SX, Koleske AJ, Burkhardt JK. The c-Abl tyrosine kinase regulates actin remodeling at the immune synapse. Blood. 2008;112:111–119. doi: 10.1182/blood-2007-10-118232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner CS, Zhang P, Van Etten RA, Tenen DG. Reversibility of acute B-cell leukaemia induced by BCR-ABL1. Nat Genet. 2000;24:57–60. doi: 10.1038/71691. [DOI] [PubMed] [Google Scholar]
- Hutson MS, Tokutake Y, Chang MS, Bloor JW, Venakides S, Kiehart DP, Edwards GS. Forces for morphogenesis investigated with laser microsurgery and quantitative modeling. Science. 2003;300:145–149. doi: 10.1126/science.1079552. [DOI] [PubMed] [Google Scholar]
- Ibarra N, Pollitt A, Insall RH. Regulation of actin assembly by SCAR/WAVE proteins. Biochem Soc Trans. 2005;33:1243–1246. doi: 10.1042/BST0331243. [DOI] [PubMed] [Google Scholar]
- Ishimaru S, Ueda R, Hinohara Y, Ohtani M, Hanafusa H. PVR plays a critical role via JNK activation in thorax closure during Drosophila metamorphosis. EMBO J. 2004;23:3984–3994. doi: 10.1038/sj.emboj.7600417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan R, Kuzina I, Wincovitch S, Nowotarski SH, Giniger E. The Abl/enabled signaling pathway regulates Golgi architecture in Drosophila photoreceptor neurons. Mol Biol Cell. 2014;25:2993–3005. doi: 10.1091/mbc.E14-02-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri A, Wang J, Pendergast AM. Multifunctional Abl kinases in health and disease. J Cell Sci. 2016;129:9–16. doi: 10.1242/jcs.175521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J Cell Biol. 2000;149:471–490. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinghoffer RA, Hamilton TG, Hoch R, Soriano P. An allelic series at the PDGFalphaR locus indicates unequal contributions of distinct signaling pathways during development. Dev Cell. 2002;2:103–113. doi: 10.1016/s1534-5807(01)00103-4. [DOI] [PubMed] [Google Scholar]
- Koleske AJ, Gifford AM, Scott ML, Nee M, Bronson RT, Miczek KA, Baltimore D. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron. 1998;21:1259–1272. doi: 10.1016/s0896-6273(00)80646-7. [DOI] [PubMed] [Google Scholar]
- Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Lapetina S, Mader CC, Machida K, Mayer BJ, Koleske AJ. Arg interacts with cortactin to promote adhesion-dependent cell edge protrusion. J Cell Biol. 2009;185:503–519. doi: 10.1083/jcb.200809085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Engel U, Rusch J, Scherrer S, Sheard K, Van Vactor D. The microtubule plus end tracking protein Orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron. 2004;42:913–926. doi: 10.1016/j.neuron.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Li W, Li Y, Gao FB. Abelson, enabled, and p120 catenin exert distinct effects on dendritic morphogenesis in Drosophila. Dev Dyn. 2005;234:512–522. doi: 10.1002/dvdy.20496. [DOI] [PubMed] [Google Scholar]
- Lin TY, Huang CH, Kao HH, Liou GG, Yeh SR, Cheng CM, Chen MH, Pan RL, Juang JL. Abi plays an opposing role to Abl in Drosophila axonogenesis and synaptogenesis. Development. 2009;136:3099–3107. doi: 10.1242/dev.033324. [DOI] [PubMed] [Google Scholar]
- Lin YC, Yeckel MF, Koleske AJ. Abl2/Arg controls dendritic spine and dendrite arbor stability via distinct cytoskeletal control pathways. J Neurosci. 2013;33:1846–1857. doi: 10.1523/JNEUROSCI.4284-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader CC, Oser M, Magalhaes MA, Bravo-Cordero JJ, Condeelis J, Koleske AJ, Gil-Henn H. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 2011;71:1730–1741. doi: 10.1158/0008-5472.CAN-10-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis B, Skolnik EY. Activation of Ras by receptor tyrosine kinases. J Am Soc Nephrol. 1994;5:1288–1299. doi: 10.1681/ASN.V561288. [DOI] [PubMed] [Google Scholar]
- Martin P, Parkhurst SM. Parallels between tissue repair and embryo morphogenesis. Development. 2004;131:3021–3034. doi: 10.1242/dev.01253. [DOI] [PubMed] [Google Scholar]
- McWhirter JR, Wang JY. An actin-binding function contributes to transformation by the Bcr-Abl oncoprotein of Philadelphia chromosome-positive human leukemias. EMBO J. 1993;12:1533–1546. doi: 10.1002/j.1460-2075.1993.tb05797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AL, Wang Y, Mooseker MS, Koleske AJ. The Abl-related gene (Arg) requires its F-actin-microtubule cross-linking activity to regulate lamellipodial dynamics during fibroblast adhesion. J Cell Biol. 2004;165:407–419. doi: 10.1083/jcb.200308055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MM, Lapetina S, MacGrath SM, Sfakianos MK, Pollard TD, Koleske AJ. Regulation of actin polymerization and adhesion-dependent cell edge protrusion by the Abl-related gene (Arg) tyrosine kinase and N-WASp. Biochemistry. 2010;49:2227–2234. doi: 10.1021/bi901721u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Radha V. F-actin-binding domain of c-Abl regulates localized phosphorylation of C3G: role of C3G in c-Abl-mediated cell death. Oncogene. 2010;29:4528–4542. doi: 10.1038/onc.2010.113. [DOI] [PubMed] [Google Scholar]
- Moresco EM, Donaldson S, Williamson A, Koleske AJ. Integrin-mediated dendrite branch maintenance requires Abelson (Abl) family kinases. J Neurosci. 2005;25:6105–6118. doi: 10.1523/JNEUROSCI.1432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco EM, Koleske AJ. Regulation of neuronal morphogenesis and synaptic function by Abl family kinases. Curr Opin Neurobiol. 2003;13:535–544. doi: 10.1016/j.conb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- O’Donnell MP, Bashaw GJ. Distinct functional domains of the Abelson tyrosine kinase control axon guidance responses to Netrin and Slit to regulate the assembly of neural circuits. Development. 2013;140:2724–2733. doi: 10.1242/dev.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hare T, Corbin AS, Druker BJ. Targeted CML therapy: controlling drug resistance, seeking cure. Curr Opin Genet Dev. 2005;16:92–99. doi: 10.1016/j.gde.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Peacock JG, Miller AL, Bradley WD, Rodriguez OC, Webb DJ, Koleske AJ. The abl-related gene tyrosine kinase acts through p190RhoGAP to inhibit actomyosin contractility and regulate focal adhesion dynamics upon adhesion to fibronectin. Mol Biol Cell. 2007;18:3860–3872. doi: 10.1091/mbc.E07-01-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pene-Dumitrescu T, Smithgall TE. Expression of a Src family kinase in chronic myelogenous leukemia cells induces resistance to imatinib in a kinase-dependent manner. J Biol Chem. 2010;285:21446–21457. doi: 10.1074/jbc.M109.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielage JF, Powell KR, Kalman D, Engel JN. RNAi screen reveals an AbI kinase-dependent host cell pathway involved in Pseudomonas aeruginosa internalization. PLoS Pathog. 2008;4:e1000031. doi: 10.1371/journal.ppat.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Cang Y, Goff SP. c-Abl tyrosine kinase regulates cardiac growth and development. Proc Natl Acad Sci USA. 2010;107:1136–1141. doi: 10.1073/pnas.0913131107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JM, Lim D, Stach L, Ogrodowicz RW, Keck JM, Jones MH, Wong CC, Yates JR, 3rd, Winey M, Smerdon SJ, et al. Activation of the yeast Hippo pathway by phosphorylation-dependent assembly of signaling complexes. Science. 2013;340:871–875. doi: 10.1126/science.1235822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JR, Echarri A, Li R, Pendergast AM. Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol Cell Biol. 2009;29:1735–1748. doi: 10.1128/MCB.01483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- Schwartzberg PL, Stall AM, Hardin JD, Bowdish KS, Humaran T, Boast S, Harbison ML, Robertson EJ, Goff SP. Mice homozygous for the ablm1 mutation show poor viability and depletion of selected B and T cell populations. Cell. 1991;65:1165–1175. doi: 10.1016/0092-8674(91)90012-n. [DOI] [PubMed] [Google Scholar]
- Smith KM, Yacobi R, Van Etten RA. Autoinhibition of Bcr-Abl through its SH3 domain. Mol Cell. 2003;12:27–37. doi: 10.1016/S1097-2765(03)00274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Pearson PS, Greuber EK, Yogalingam G, Pendergast AM. Abl kinases are required for invadopodia formation and chemokine-induced invasion. J Biol Chem. 2010;285:40201–40211. doi: 10.1074/jbc.M110.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan R, Gohl C, Fleige A, Klambt C, Bogdan S. Membrane-targeted WAVE mediates photoreceptor axon targeting in the absence of the WAVE complex in Drosophila. Mol Biol Cell. 2011;22:4079–4092. doi: 10.1091/mbc.E11-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens TL, Rogers EM, Koontz LM, Fox DT, Homem CC, Nowotarski SH, Artabazon NB, Peifer M. Using Bcr-Abl to examine mechanisms by which Abl kinase regulates morphogenesis in Drosophila. Mol Biol Cell. 2007;19:378–393. doi: 10.1091/mbc.E07-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine RR, Greenspan LJ, Ramachandran KV, Matunis EL. Coordinate regulation of stem cell competition by Slit-Robo and JAK-STAT signaling in the Drosophila testis. PLoS Genet. 2014;10:e1004713. doi: 10.1371/journal.pgen.1004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, French WJ, Soriano P. Additive effects of PDGF receptor beta signaling pathways in vascular smooth muscle cell development. PLoS Biol. 2003;1:E52. doi: 10.1371/journal.pbio.0000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada M, Farrell DL, Zallen JA. Abl regulates planar polarized junctional dynamics through beta-catenin tyrosine phosphorylation. Dev Cell. 2012;22:309–319. doi: 10.1016/j.devcel.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill-Day N, Pierce A, Thompson SE, Xenaki D, Whetton AD, Owen-Lynch PJ. Role of the C-terminal actin binding domain in BCR/ABL-mediated survival and drug resistance. Br J Haematol. 2006;132:774–783. doi: 10.1111/j.1365-2141.2005.05949.x. [DOI] [PubMed] [Google Scholar]
- van Etten RA, Jackson PK, Baltimore D, Sanders MC, Matsudeira PT, Janmey P. The COOH terminus of the c-Abl tyrosine kinase contains distinct F- and G-actin binding domains with bundling activity. J Cell Biol. 1994;124:325–340. doi: 10.1083/jcb.124.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JA, Perera SA, Hammer DA, Ren R, Boettiger D, Pear WS. Localization of BCR-ABL to F-actin regulates cell adhesion but does not attenuate CML development. Blood. 2003;102:2220–2228. doi: 10.1182/blood-2003-01-0062. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C. Looking at embryos. In: Roberts DB, editor. Drosophila, a Practical Approach. Oxford: UK: IRL Press; 1986. pp. 199–228. [Google Scholar]
- Woodring PJ, Hunter T, Wang JY. Inhibition of c-Abl tyrosine kinase activity by filamentous actin. J Biol Chem. 2001;276:27104–27110. doi: 10.1074/jbc.M100559200. [DOI] [PubMed] [Google Scholar]
- Zandy NL, Pendergast AM. Abl tyrosine kinases modulate cadherin-dependent adhesion upstream and downstream of Rho family GTPases. Cell Cycle. 2008;7:444–448. doi: 10.4161/cc.7.4.5452. [DOI] [PubMed] [Google Scholar]
- Zhang XW, Ren RB. Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin-3 and granulocyte-macrophage colony-stimulating factor in mice: A novel model for chronic myelogenous leukemia. Blood. 1998;92:3829–3840. [PubMed] [Google Scholar]
- Zulueta-Coarasa T, Tamada M, Lee EJ, Fernandez-Gonzalez R. Automated multidimensional image analysis reveals a role for Abl in embryonic wound repair. Development. 2014;141:2901–2911. doi: 10.1242/dev.106898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.