Abstract
Transient receptor potential vanilloid-1 (TRPV1) receptor and proteinase-activated receptor 2 (PAR2) have been implicated in the mechanism of acid-induced inflammation in gastroesophageal reflux disease (GERD). We aimed to evaluate TRPV1 and PAR2 mRNA expression levels in the GERD patients and their relationship with endoscopic findings and reflux symptoms.
Sixteen healthy controls, 45 patients with erosive reflux disease (ERD), and 14 nonerosive reflux disease (NERD) patients received endoscopy and completed questionnaires. Quantitative real-time polymerase chain reactions (qPCR) of TRPV1, glial cell line-derived neurotrophic factor (GDNF), nerve growth factor (NGF), PAR2, and interleukin (IL)-8 were performed in the distal esophagus specimen.
The levels of TRPV1, GDNF, NGF, PAR2, and IL-8 mRNA expression were highest in the ERD group followed by NERD and control groups and the differences between control and ERD groups were statistically significant. Within the ERD group, patients with grade B in Los Angeles (LA) classification showed significantly higher levels of TRPV1, GDNF, and NGF mRNA expression than those with grade A. Presence of reflux symptoms was associated with significant higher levels of TRPV1, PAR2, and IL-8. Notably not extraesophageal but esophageal reflux symptoms were significantly associated with them.
Upregulation of TRPV1 and PAR2 pathways might play a role in the development of distal esophageal inflammation and reflux symptoms. And extraesophageal reflux symptoms might not be associated with these processes.
Keywords: gastroesophageal reflux disease, glial cell-derived neurotrophic factor, nerve growth factor, proteinase-activated receptor 2, transient receptor potential vanilloid receptor 1
1. Introduction
Major progress has been recently made in the understanding of the molecular basis of gastroesophageal reflux disease (GERD), suggesting a far more complex and multifactorial pathogenesis. [1 2 3] According to recent studies, molecular signs of inflammation in the mucosa can be detected even before microscopic or macroscopic changes become apparent and inflammatory process has been suggested as a possible GERD pathogenesis. [4 5 6] For example, interleukin (IL)-8, IL-1β, and various other inflammatory cytokines have been found in mucosal biopsies from patients with GERD. [7 8 9 10] Also, the role of proteinase-activated receptor 2 (PAR2) in the pathogenesis of GERD has been studied. PAR2 is activated by serine proteases and induces proinflammatory and neuroinflammatory effects.[ 11 12 13] In in vitro experiments of esophageal squamous cell lines, PAR2 expression was induced by exposure to acid and weakly acidic solutions. [11] In consistent with this concept, in patients with GERD, several studies have shown that PAR2 activation led to increased secretion of IL-8 and contributed to immune-mediated inflammatory damages to the esophageal mucosa. [12]
Furthermore, not only PAR2 but also acid sensitive receptors—such as the capsaicin-sensitive transient receptor potential channel vanilloid subfamily member 1(TRPV1)—have been proposed as a possible mechanism involved in the manifestation of gastrointestinal symptoms. [14 15 16 17] PAR2-dependent sensitization of this acid sensitive receptor has been demonstrated in dorsal root ganglia. [18] TRPV1 activation in this primary afferent neurons has not only evoked the sensation of burning pain but also induced inflammatory and neuroinflammatory effects, thereby causing GERD. [19 20 21 22]
Even though there are several studies showing that upregulation of TRPV1 and PAR2 is associated with GERD, its clinical influences on the GERD symptoms were not clearly evaluated so far. Therefore, we conducted this study to evaluate TRPV1- and PAR2-mediated activities and its relationship with the expression of glial cell line-derived neurotrophic factor (GDNF), nerve growth factor (NGF), and IL-8 in GERD patients including both reflux esophageal disease and nonerosive reflux disease. In addition, the association of the upregulation of TRPV1 and PAR2 with each GERD symptom and endoscopic finding was also investigated.
2. Materials and methods
2.1. Subjects
The subjects were enrolled prospectively at the Department of Gastroenterology of Seoul National University Bundang Hospital (SNUBH), between March 2013 and May 2015. All the subjects received upper gastrointestinal endoscopy and completed questionnaires about GERD symptoms, including both esophageal and extraesophageal symptoms, under the supervision of a well-trained interviewer. Those enrolled subjects visited SNUBH mainly for evaluating the origin of GERD symptoms or for screening of gastric cancer. Subjects were excluded if there was a history of gastrointestinal surgery, Barrett's esophagus, esophageal motility disorder, duodenal ulcer, benign gastric ulcer or gastroduodenal cancer, and any history of systemic disease requiring chronic medication (except for hypertension and diabetes mellitus).
The subjects were classified into 3 groups: erosive reflux disease (ERD), nonerosive reflux disease (NERD), and control groups. ERD group was defined as the patients with mucosal breaks at gastroesophageal junction in endoscopic findings, according to the Los Angeles (LA) classification of esophagitis. [22] NERD group was defined as the patients who had more than one episode of heartburn or acid regurgitation per week with normal endoscopic findings, and a positive response of the proton-pump inhibitor (PPI) trial, meaning that more than 50% of symptom frequency was improved after 2-week PPI intake. NERD patients are defined only on PPI response without pH or multichannel intraluminal impedance study. Subjects with no symptom and normal endoscopic finding from health check-up were assigned as the control group.
The Institutional Review Board of Seoul National University Bundang Hospital approved this study (B-1211/180–003), and written informed consent was obtained from all participants. The ClinicalTrials.gov registration number is NCT 02114216.
2.2. Symptom assessment
Symptom assessment was made by face-to-face interview with a questionnaire which has been developed for gastroesophageal reflux before endoscopy procedures. The questionnaire has been validated in the nationwide survey regarding erosive esophagitis and NERD in the health check-up subjects, which consisted of questions on 7 reflux symptoms. Those symptoms include both 2 esophageal and 5 extraesophageal symptoms. [23 24 25] Esophageal symptoms were heartburn and acid regurgitation, and extraesophageal symptoms were chest pain, cough, globus symptoms, hoarseness, and epigastric soreness. [23 24 25]
2.3. Upper endoscopy
During the endoscopic examination, 2 biopsies using standard biopsy forceps at a fixed position 3 cm above the squamo-columnar junction in order to achieve sample consistency not only from all GERD but also from control subjects. The extent of mucosal damage was noted and assessed using the LA grading system.
2.4. RNA isolation and reverse transcription
In order to stabilize and protect RNA in fresh specimens, biopsy specimens were stored in RNAlater Solution (Ambion, Austin, TX) at 4°C after endoscopy. Total RNA was extracted from biopsy specimens using TRIzol reagent (Invitrogen, Carlsbad, CA) as recommended by the manufacturer and the collected RNA was purified using RNeasy mini kits (Qiagen, Valencia, CA). RNA samples were diluted to a final concentration of 0.5 mg/mL in RNase-free water and stored at −80°C until use. Synthesis of the cDNA was performed with 1 mg of total RNA with M-MLV reverse transcription reagents (Invitrogen). The reverse-transcription reaction consisted of 4 μl of first-strand buffer, 500 mM deoxynucleoside triphosphate mixture, 2.5 mM oligo(dT) 12–18 primer, 0.4 U/ml ribonuclease inhibitor, and 1.25 U/ml Moloney murine leukemia virus reverse transcriptase (Invitrogen). The thermal cycling parameters for the reverse transcription were 10 minutes at 65°C, 50 minutes at 37°C, and 15 minutes at 70°C.
2.5. Real-time quantitative polymerase chain reaction (qPCR)
The primers used in real-time qPCR were designed using PrimerExpress Software V2.0 (Applied Biosystems, Foster City, CA) based on sequence information from the National Center for Biotechnology Information database. Real-time qPCR was performed in triplicate by using a StepOnePlus Real-time PCR (Applied Biosystems) with SYBR Premix Ex TaqTM (Takara Bio, Shiga, Japan) according to manufacturers’ instructions and protocols. Thermal cycling was performed as follows: initial denaturation at 95°C for 10 seconds followed by 40 cycles of 95°C for 5 seconds, and 60°C for 33 seconds. Homo b-actin was used as a reference; i.e., each sample was normalized on the basis of its b-actin content. The relative change in all target genes expression was determined by the fold-change analysis.
2.6. Statistical analysis
Statistical analyses were performed using one way ANOVA, Fisher's exact, χ2, Student's t, Mann–Whitney U, and Kruskal-Wallis tests. A P value of less than 0.05 was accepted as statistically significant. Data were expressed as mean ± standard deviation (SD).
3. Results
3.1. General characteristics
A total of 75 subjects were included in this study. Among them, 45 patients were categorized as the ERD group. The number of subjects included in the NERD group was 14. The control group included 16 subjects who had neither endoscopic alteration nor reflux symptoms.
Their demographic characteristics are summarized in Table 1. Mean age was older in the NERD group (52.7 years old) than ERD (52.3 years old, P = 0.032) There were more male patients in the ERD group (51.1%) and NERD (50.0%) group than the control group (43.8%) (P = 0.014, 0.024, respectively). There were more smokers in the NERD (35.7%) group than the ERD (24.4%) group (P = 0.012). Body mass index and alcohol habit were not significantly different among them.
Table 1.
Characteristics of the participating subjects.
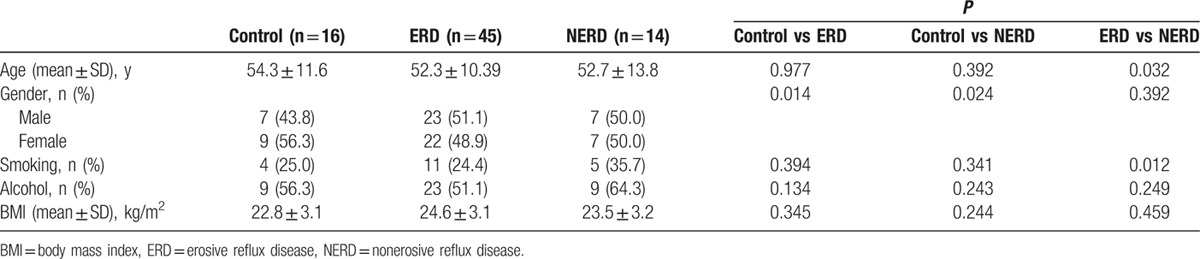
As Fig. 1 shows, the levels of TRPV1, GDNF, NGF, PAR2, and IL-8 mRNA expression were highest in the ERD patients (3.25 ± 0.10, 2.47 ± 0.11, 2.65 ± 0.29, 2.42 ± 0.17, 4.16 ± 0.32) followed by NERD (1.53 ± 0.10, 1.55 ± 0.14, 1.89 ± 0.07, 2.39 ± 0.36, 4.01 ± 0.35) and control (1.14 ± 0.12, 1.39 ± 0.06, 1.62 ± 0.08, 1.37 ± 0.20, 2.01 ± 0.60) groups. Especially, the differences between the control and ERD groups were statistically significant for all variables (P = 0.000, 0.030, 0.027, 0.002, 0.002, respectively). And between the control and NERD groups, only the levels of PAR2 and IL-8 were significantly different (P = 0.020, 0.031, respectively).
Figure 1.
Comparison of mRNA expression levels among three groups. ERD = erosive reflux disease, GDNF = glial cell line-derived neurotrophic factor, IL-8 = interleukin-8, NERD = nonerosive reflux disease, NGF = nerve growth factor, PAR2 = proteinase-activated receptor 2, TRPV1 = transient receptor potential vanilloid-1, ∗ P < 0.05.
3.2. Comparison of TRPV1 and PAR2 mRNA expression levels depending on the endoscopic grade of reflux esophagitis
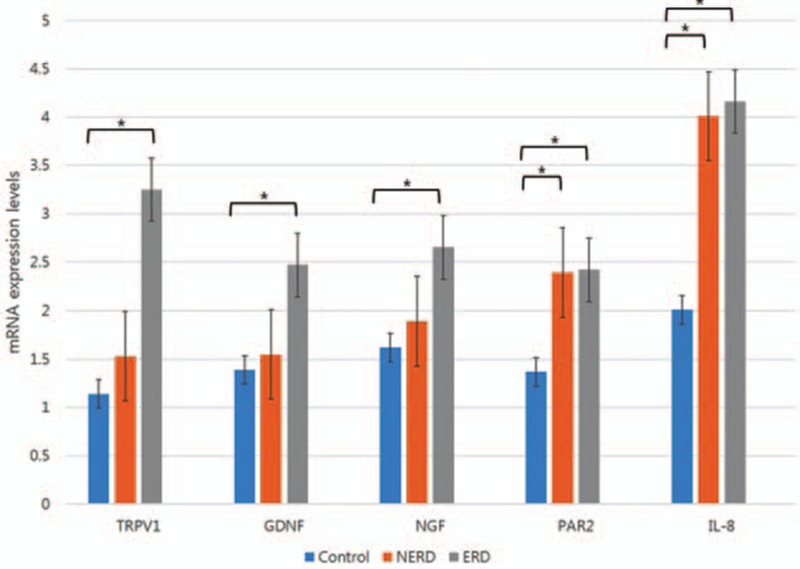
We analyzed whether endoscopic findings were associated with TRPV1 and PAR2 mRNA expression levels. Within the ERD group, subjects with grades A in the LA classification (3.05 ± 0.13, 1.66 ± 0.08, 3.66 ± 0.09, 1.69 ± 0.23, 4.29 ± 3.14) showed lower TRPV1, GDNF, NGF, PAR2, and IL-8 mRNA expression levels than those with grades B (4.10 ± 0.12, 2.90 ± 0.10, 4.01 ± 0.30, 1.91 ± 0.21, 4.57 ± 1.76) (Fig. 2). Especially, the differences between grade A and grade B were statistically significant only for TRPV1, GDNF, and NGF mRNA expression levels (P = 0.043, 0.024, 0.034, respectively).
Figure 2.
Comparison of mRNA expression levels depending on the Los Angeles classification among ERD subjects. ERD = erosive reflux disease, GDNF = glial cell line-derived neurotrophic factor, IL-8 = interleukin-8, NGF = nerve growth factor, PAR2 = proteinase-activated receptor 2, TRPV1 = transient receptor potential vanilloid-1, ∗ P < 0.05.
3.3. Comparison of TRPV1 and PAR2 mRNA expression levels in relation to the esophageal or extraesophageal reflux symptoms
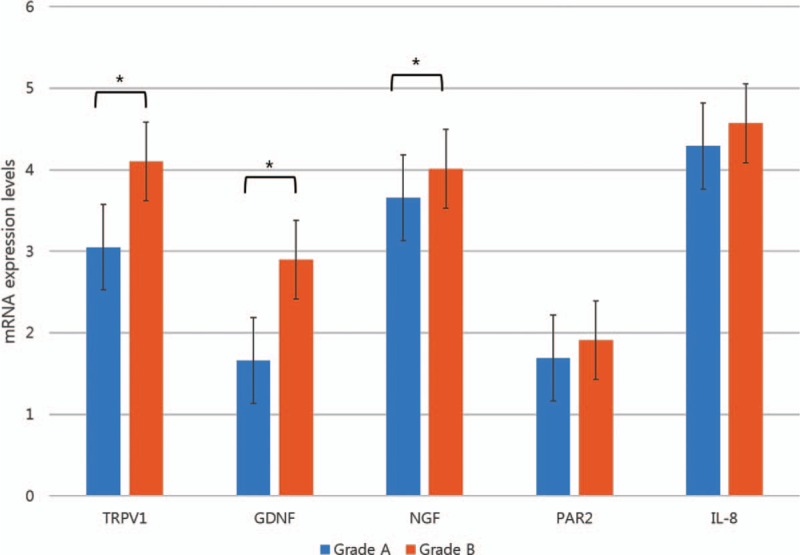
Each reflux symptom and its relationship with TRPV1 and PAR2 mRNA expression levels were analyzed within the ERD group. Eight subjects in the ERD group presented no reflux related symptoms and 37 subjects reported any reflux related symptoms irrespective of esophageal or extraesophageal reflux symptoms. Eleven subjects showed only extraesophageal reflux symptoms and 29 subjects showed only esophageal reflux symptoms. Subjects who complained both esophageal and extraesophageal reflux symptoms were 4.
Interestingly, those with esophageal reflux symptoms and those with both esophageal and extraesophageal reflux symptoms showed significantly higher levels of TRPV1, PAR2, and IL-8 mRNA expression than others (Fig. 3). Those with only extraesophageal reflux symptoms, such as chest pain, cough, globus symptoms, hoarseness, and epigastric soreness, did not show any relationship with upregulation of those genes.
Figure 3.
Comparison of mRNA expression levels in relation to the presence of reflux symptoms among ERD subjects. ERD = erosive reflux disease, GDNF = glial cell line-derived neurotrophic factor, IL-8 = interleukin-8, NGF = nerve growth factor, PAR2 = proteinase-activated receptor 2, TRPV1 = transient receptor potential vanilloid-1, ∗ P < 0.05. aExtraesophageal symptoms: chest pain, cough, globus symptoms, hoarseness, and epigastric soreness. bEsophageal symptoms: heartburn and acid regurgitation.
4. Discussion
We demonstrated that ERD was associated with the upregulations of TRPV1, GDNF, NGF, PAR2, and IL-8 gene expression levels. Also, endoscopic grade of reflux esophagitis and the presence of esophageal reflux symptoms showed upregulations of all those genes. This is the first study which showed comprehensively the relationship of pain-relating mediators (TRPV1, GDNF, NGF, PAR2, and IL-8) with esophageal or extraesophageal reflux symptoms in the ERD as well as NERD, so far.
ERD has been considered to originate from acid-induced inflammation. According to recent studies, not only TRPV1 receptors and neurotropic factors but also the interaction between PAR2 with TRPV1 seems to play an important role in this inflammatory process.[ 14 15] Similar to these previous studies, we demonstrated upregulation of TRPV1 and PAR2 in the ERD group. This result suggests downregulation of these genes can be a possible therapeutic target for the management of esophageal erosions.
Furthermore, we evaluated the relationship between those gene upregulation and endoscopic severities. Interestingly, endoscopic severity was associated only with TRPV1, GDNF, and NGF levels and not with PAR2 and IL-8 levels. This clinical implication has not been demonstrated so far, and which suggests that physical injury and inflammation is more closely associated with TRPV1 cascade reaction and PAR2 pathway might not be directly associated with physical inflammation.
So far, there have been no studies on the differences of symptomatic and asymptomatic ERD patients. In this study, we compared ERD patients with reflux symptoms to those without reflux symptoms. For the symptomatic ERD subjects, PAR2 pathway was significantly upregulated comparing to the asymptomatic ones. Moreover, NERD subjects showed higher expression levels in terms of PAR2 pathway only. Therefore, PAR2 pathway could be the important mediator for the manifestation of GERD symptoms and a promising candidate for the treatment of NERD patients.
Also, we further analyzed if there are any differences between esophageal and extraesophageal reflux symptoms, which has not been studied well yet. Notably, only esophageal reflux symptoms were relevant to the upregulation of these genes whereas extraesophageal reflux symptoms were not. This result suggests pathophysiology of extraesophageal reflux symptoms might be different from that of esophageal reflux symptoms. That is, not direct sensitization at the distal esophagus, but an indirect mechanism involving vagally mediated reflex from distal esophageal acid exposure would be more plausible for the extraesophageal reflux symptoms. Or, upregulation of inflammation-related genes might be located in other parts, such as proximal esophagus or pharynx.
Also, the data from this study are valuable for supporting the irrelevance between so-called extraesophageal reflux symptoms and esophageal inflammation from gastroesophageal acid reflux. Actually, the response rate to a proton pump inhibitor was lower in the patients with extraesophageal reflux symptoms than in those with typical esophageal reflux symptoms. [24 25 26 27 28 29 30 31 32 33] From this background, there has been a dispute whether extraesophageal reflux symptoms are truly related to the esophageal reflux. The present study can reinforce the suggestion that extraesophageal reflux symptoms might not be actually related to the gastroesophageal reflux and it could be entitled as a different disease spectrum.
This study has several limitations. First of all, we defined NERD group with using PPI-response and did not apply pH monitoring study. We have to concede that a positive or negative response to PPI therapy is only an empiric criterion and this definition can include heterogeneous subjects. For example, patients with normal esophageal acid exposure might respond to PPIs because of a possible placebo effect or the repair of subtle microscopic inflammatory damage. [34] Indeed, previous studies have underlined this important limitation that affects the diagnostic accuracy of the PPI test.[ 35 36] However, 24 hour pH monitoring could not be performed for most of the GERD suspicious patients because it is very cumbersome. For this reason, pH monitoring study could not be used in this study and this limitation should be considered for interpreting our results. In addition, the number of subjects who were enrolled in this study was relatively small. Actually, it was very difficult to get the consent for the biopsy from the esophageal mucosa. Thus, our results need to be reevaluated with more large number of subjects.
In conclusion, our study suggests that acid-induced inflammation in the esophagus seems to be associated with upregulation of TRPV1 and PAR2 and this upregulation might lead to the manifestation of esophageal reflux symptoms. This result implies that these genes can be a target for the therapeutic strategy of GERD. Extraesophageal reflux symptoms were not associated with upregulation of these genes and seem to originate from different pathophysiology. Thus, different therapeutic strategies might be needed for the patients with extraesophageal reflux symptoms.
Footnotes
Abbreviations: ERD = erosive reflux disease, GCRC = Global Core Research Center, GDNF = glial cell line-derived neurotrophic factor, GERD = gastroesophageal reflux disease, IL-8 = interleukin-8, LA = Los Angeles, NERD = nonerosive reflux disease, NGF = nerve growth factor, PAR2 = proteinase-activated receptor 2, qPCR = quantitative real-time polymerase chain reactions, SD = standard deviation, SNUBH = Seoul National University Bundang Hospital, TRPV1 = transient receptor potential vanilloid-1.
Funding/support: This work was supported by the National Research Foundation of Korea (NRF) grant for the Global Core Research Center (GCRC) funded by the Korean government (MSIP) (No. 2011–0030001).
The authors have no conflicts of interest to disclose.
References
- 1. Boeckxstaens G, El-Serag HB, Smout AJ, et al. Symptomatic reflux disease: the present, the past and the future. Gut 2014; 63:1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altomare A, Guarino MP, Cocca S, et al. Gastroesophageal reflux disease: Update on inflammation and symptom perception. World J Gastroenterol 2013; 21:6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barlow WJ, Orlando RC. The pathogenesis of heartburn in nonerosive reflux disease: a unifying hypothesis. Gastroenterology 2005; 128:771–778. [DOI] [PubMed] [Google Scholar]
- 4. Fitzgerald RC, Onwuegbusi BA, Bajaj-Elliott M, et al. Diversity in the oesophageal phenotypic response to gastro-oesophageal reflux: immunological determinants. Gut 2002; 50:451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fiocca R, Mastracci L, Engström C, et al. Longterm outcome of microscopic esophagitis in chronic GERD patients treated with esomeprazole or laparoscopic antireflux surgery in the LOTUS trial. Am J Gastroenterol 2010; 105:1015–1023. [DOI] [PubMed] [Google Scholar]
- 6. Mönkemüller K, Wex T, Kuester D, et al. Interleukin-1beta and interleukin-8 expression correlate with the histomorphological changes in esophageal mucosa of patients with erosive and non-erosive reflux disease. Digestion 2009; 79:186–195. [DOI] [PubMed] [Google Scholar]
- 7. Rafiee P, Nelson VM, Manley S, et al. Effect of curcumin on acidic pH-induced expression of IL-6 and IL-8 in human esophageal epithelial cells (HET-1A): role of PKC, MAPKs, and NF-kappaB. Am J Physiol Gastrointest Liver Physiol 2009; 296:388–398. [DOI] [PubMed] [Google Scholar]
- 8. Ma J, Altomare A, Guarino M, et al. HCl-induced and ATP-dependent upregulation of TRPV1 receptor expression and cytokine production by human esophageal epithelial cells. Am J Physiol Gastrointest Liver Physiol 2012; 303:635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng HH, Chang CS, Wang HJ, et al. Interleukin-1beta and -10 polymorphisms influence erosive reflux esophagitis and gastritis in Taiwanese patients. J Gastroenterol Hepatol 2010; 25:1443–1451. [DOI] [PubMed] [Google Scholar]
- 10. Queiroz DM1, Guerra JB, Rocha GA, et al. IL1B and IL1RN polymorphic genes and Helicobacter pylori cagA strains decrease the risk of reflux esophagitis. Gastroenterology 2004; 127:73–79. [DOI] [PubMed] [Google Scholar]
- 11. Yoshida N1, Katada K, Handa O, et al. Interleukin-8 production via protease-activated receptor 2 in human esophageal epithelial cells. Int J Mol Med 2007; 19:335–340. [DOI] [PubMed] [Google Scholar]
- 12. Huang SC. Protease-activated receptor-1 (PAR1) and PAR2 but not PAR4 mediate relaxations in lower esophageal sphincter. Regul Pept 2007; 142:37–43. [DOI] [PubMed] [Google Scholar]
- 13. Chen K, Zhang ZF, Liao MF, et al. Blocking PAR2 attenuates oxaliplatin-induced neuropathic pain via TRPV1 and releases of substance P and CGRP in superficial dorsal horn of spinal cord. J Neurol Sci 2015; 15:62–67. [DOI] [PubMed] [Google Scholar]
- 14. Ma J, Altomare A, Guarino M, et al. HCl-induced and ATP-dependent upregulation of TRPV1 receptor expression and cytokine production by human esophageal epithelial cells. Am J Physiol Gastrointest Liver Physiol 2012; 303:G635–G645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoshida N, Kuroda M, Suzuki T, et al. Role of nociceptors/neuropeptides in the pathogenesis of visceral hypersensitivity of nonerosive reflux disease. Dig Dis Sci 2013; 58:2237–2243. [DOI] [PubMed] [Google Scholar]
- 16. Choi YJ, Kim N, Kim J, et al. Upregulation of vanilloid receptor-1 in functional dyspepsia with or without Helicobacter pylori infection. Medicine (Baltimore) 2016; 95:e3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Y, Yang C, Wang ZJ. Proteinase-activated receptor 2 sensitizes transient receptor potential vanilloid 1, transient receptor potential vanilloid 4, and transient receptor potential ankyrin 1 in paclitaxel-induced neuropathic pain. Neuroscience 2011; 193:440–451. [DOI] [PubMed] [Google Scholar]
- 18. Huang ZJ, Li HC, Cowan AA, et al. Chronic compression or acute dissociation of dorsal root ganglion induces cAMP-dependent neuronal hyperexcitability through activation of PAR2. Pain 2012; 153:1426–1437. [DOI] [PubMed] [Google Scholar]
- 19. Guarino MP, Cheng L, Ma J, et al. Increased TRPV1 gene expression in esophageal mucosa of patients with non-erosive and erosive reflux disease. Neurogastroenterol Motil 2010; 22:746–751. [DOI] [PubMed] [Google Scholar]
- 20. Shan J, Oshima T, Chen X, et al. Trypsin impaired epithelial barrier function and induced IL-8 secretion through basolateral PAR-2: a lesson from a stratified squamous epithelial model. Am J Physiol Gastrointest Liver Physiol 2012; 15:G1105–G1112. [DOI] [PubMed] [Google Scholar]
- 21. Isomoto H, Nishi Y, Kanazawa Y, et al. Immune and Inflammatory Responses in GERD and Lansoprazole. J Clin Biochem Nutr 2007; 41:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Armstrong D, Bennett JR, Blum AL, et al. The endoscopic assessment of esophagitis: a progress report on observer agreement. Gastroenterology 1996; 111:85–92. [DOI] [PubMed] [Google Scholar]
- 23. Jo SY, Kim N, Lim JH, et al. Comparison of gastroesophageal reflux disease symptoms and proton pump inhibitor response using gastroesophageal reflux disease impact scale questionnaire. J Neurogastroenterol Motil 2013; 19:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim JY, Kim N, Seo PJ, et al. Association of sleep dysfunction and emotional status with gastroesophageal reflux disease in Korea. J Neurolgastroenterol Motil 2013; 19:344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim SE, Kim N, Oh S, et al. Predictive factors of response to proton pump inhibitors in Korean patients with gastroesophageal reflux disease. J Neurolgastroenterol Motil 2015; 21:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jung HK. Epidemiology of gastroesophageal reflux disease in Asia: a systematic review. J Neurogastroenterol Motil 2011; 17:14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fass R, Shapiro M, Dekel R, et al. Systematic review: proton-pump inhibitor failure in gastro-oesophageal reflux disease--where next? Aliment Pharmacol Ther 2005; 22:79–94. [DOI] [PubMed] [Google Scholar]
- 28. Lee ES, Kim N, Lee SH, et al. Comparison of risk factors and clinical responses to proton pump inhibitors in patients with erosive esophagitis and non-erosive reflux disease. Aliment Pharmacol Ther 2009; 30:154–164. [DOI] [PubMed] [Google Scholar]
- 29. Mohd H, Qua CS, Wong CH, et al. Non-cardiac chest pain: prevalence of reflux disease and response to acid suppression in an Asian population. J Gastroenterol Hepatol 2009; 24:288–293. [DOI] [PubMed] [Google Scholar]
- 30. Chunlertrith K, Boonsawat W, Zaeoue U. Prevalence of gastroesophageal reflux symptoms in asthma patients at Srinagarind Hospital. J Med Assoc Thai 2005; 88:668–671. [PubMed] [Google Scholar]
- 31. Qua CS, Wong CH, Gopala K, et al. Gastro-oesophageal reflux disease in chronic laryngitis: prevalence and response to acid-suppressive therapy. Aliment Pharmacol Ther 2007; 25:287–295. [DOI] [PubMed] [Google Scholar]
- 32. Heading RC, Monnikes H, Tholen A, et al. Prediction of response to PPI therapy and factors influencing treatment outcome in patients with GORD: a prospective pragmatic trial using pantoprazole. BMC Gastroenterol 2011; 11:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goh KL, Choi MG, Hsu WP, et al. Unmet treatment needs of gastroesophageal reflux disease in Asia: gastroesophageal reflux disease in Asia Pacific survey. J Gastroenterol Hepatol 2014; 29:1969–1975. [DOI] [PubMed] [Google Scholar]
- 34. Calabrese C, Bortolotti M, Fabbri A, et al. Reversibility of GERD ultrastructural alterations and relief of symptoms after omeprazole treatment. Am J Gastroenterol 2005; 100:537–542. [DOI] [PubMed] [Google Scholar]
- 35. Numans ME, Lau J, de Wit NJ, et al. Short-term treatment with proton-pump inhibitors as a test for gastroesophageal reflux disease: a meta-analysis of diagnostic test characteristics. Ann Intern Med 2004; 140:518–527. [DOI] [PubMed] [Google Scholar]
- 36. Dent J, Vakil N, Jones R, et al. Accuracy of the diagnosis of GORD by questionnaire, physicians and a trial of proton pump inhibitor treatment: the Diamond Study. Gut 2010; 59:714–721. [DOI] [PubMed] [Google Scholar]


