Abstract
Barrett's oesophagus (BE) is the premalignant condition associated with the development of oesophageal adenocarcinoma (OAC). Diagnostically, p53 immunohistochemistry remains the only biomarker recommended clinically to aid histopathological diagnosis. The emerging mutational profile of BE is one of highly heterogeneous lesions at the genomic level with many mutations already occurring in non-dysplastic tissue. As well as point mutations, larger scale copy-number changes appear to have a key role in the progression to OAC and clinically applicable assays for the reliable detection of aneuploidy will be important to incorporate into future clinical management of patients. For some patients, the transition to malignancy may occur rapidly through a genome-doubling event or chromosomal catastrophe, termed chromothripsis, and detecting these patients may prove especially difficult. Given the heterogeneous nature of this disease, sampling methods to overcome inherent bias from endoscopic biopsies coupled with the development of more objective biomarkers than the current reliance on histopathology will be required for risk stratification. The aim of this approach will be to spare low-risk patients unnecessary procedures, as well as to provide endoscopic therapy to the patients at highest risk, thereby avoiding the burden of incurable metastatic disease.
Keywords: Barrett's oesophagus, high-grade dysplasia, biomarkers, oesophageal adenocarcinoma, genome instability, aneuploidy, p53, SMAD4
There are two main subtypes of oesophageal cancer, squamous cell carcinoma and oesophageal adenocarcinoma (OAC), that are distinct from one another both in terms of the cell of origin and from an epidemiological perspective. OAC is the more common form in the West and showed a three-fold increase in incidence since 1971 in England and Wales (Lepage et al, 2008). The susceptibility of OAC has been linked to a history of chronic and severe reflux of acid and bile through development of the pre-cancerous condition Barrett's oesophagus (BE) (Lagergren et al, 1999). Other risk factors include Caucasian ethnicity, male sex, obesity (both in relation to a propensity for reflux and as an independent risk factor) and smoking. The survival rate for OAC remains poor, predominantly due to late clinical presentation with advanced disease.
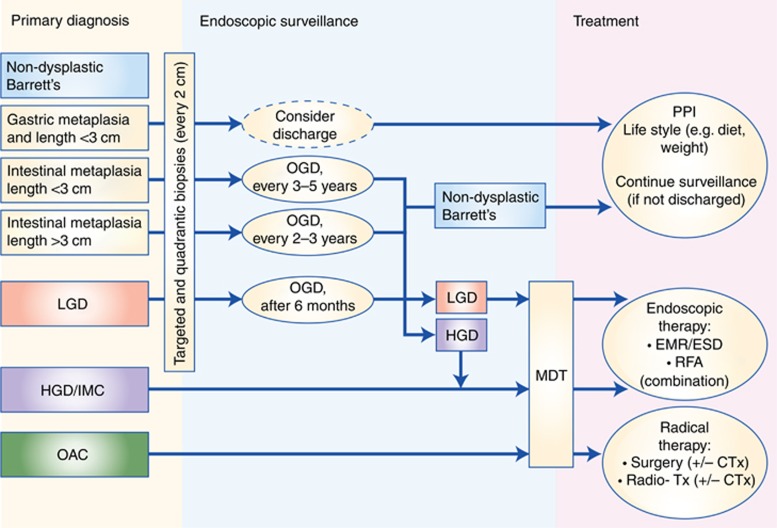
Upper gastrointestinal endoscopy combined with biopsy sampling of the distal oesophagus remains the current gold standard for diagnosing BE. According to the most recent British Society of Gastroenterology (BSG) guidelines (Fitzgerald et al, 2014), BE is characterised by the replacement of normal squamous epithelium of the distal oesophagus by columnar epithelium that is visible endoscopically and confirmed histologically. The guidelines of the American Gastroenterological Association (Spechler et al, 2011) require the presence of intestinal metaplasia (IM) in order to diagnose Barrett's. This is due to the body of research evidence that BE with IM is biologically more unstable than columnar epithelium without IM and, therefore, more likely to progress towards dysplastic or neoplastic lesions (Bhat et al, 2011). However, there is always the concern that IM may be missed due to sampling bias. For these reasons, the BSG guidelines do not require IM for Barrett's diagnosis but suggest that its presence or absence should be taken into consideration for patient management in terms of frequency of follow-up endoscopies (Figure 1) (Fitzgerald et al, 2014).
Figure 1.
Algorithm for the clinical management of patients with diagnosis of Barrett's oesophagus. Displayed is a summary of the currently recommended algorithm for surveillance and treatment of patients with Barrett's oesophagus according to the latest BSG guidelines. Patients with non-dysplastic Barrett's oesophagus should be included in endoscopic surveillance programmes at clearly defined intervals, apart from patients with short-segment Barrett's and only gastric metaplasia, which can be considered for discharge from surveillance. Patients with HGD and more advanced lesions should be discussed for therapeutic intervention at the local MDT. Patients with LGD need short-term follow up for confirmation. If the degree of dysplasia is confirmed by two independent pathologist treatment can also be discussed at the local MDT. EMR, endoscopic mucosal resection; ESD, endoscopic submucosal dissection; HGD, high-grade dysplasia; LGD, low-grade dysplasia; MDT, multidisciplinary team meeting; OAC, oesophageal adenocarcinoma; RFA, radio frequency ablation.
Overall, BE confers a low absolute risk of progression to OAC of 0.2–0.7% per patient per year (Hvid-Jensen et al, 2011; Desai et al, 2012). The risk of progression to cancer, however, increases with the diagnosis of epithelial dysplasia. Duits et al have reported an incidence rate of high-grade dysplasia (HGD) or invasive cancer of 9.1–13.4% per patient per year for patients confirmed to have low-grade dysplasia (LGD) by expert consensus; in contrast with 0.5–0.6% in patients who had no dysplasia at their initial biopsy (Curvers et al, 2010; Duits et al, 2015). However, the progression rate increases substantially to 25% when HGD is present (Kastelein et al, 2015).
National guidelines for the ongoing management of BE in the UK and the United States recommend repeat endoscopies in regular surveillance intervals (Figure 1) (Spechler et al, 2011; Fitzgerald et al, 2014). Surveillance aims to increase the proportion of patients in which neoplastic lesions are detected at early stages (HGD or intramucosal carcinoma) so that curative, endoscopic treatment can be given. Over the years, there has been contradictory evidence as to the benefits of surveillance. In a study reflecting everyday clinical practice, surveillance was not found to be associated with significantly reduced risk of death from OAC (Corley et al, 2013). On the other hand, a meta-analysis of 51 studies including more than 11 000 patients demonstrated that endoscopic surveillance of patients with non-dysplastic BE increases the likelihood for early detection of neoplastic lesions and therefore reduces mortality by more than 61% (mortality risk 0.386; 95% confidence interval (CI): 0.242–0.617) (Qiao et al, 2015). These data were supported by the results of a prospective multicentre cohort study from The Netherlands (Kastelein et al, 2016). It is, therefore, recommended that in all patients with BE, targeted biopsies should be taken from visible lesions suspicious for dysplastic changes of the mucosa, as well as four ‘random' quadrantic biopsies at 2 cm intervals over the entire extent of the Barrett's segment – the so-called Seattle protocol (Levine et al, 2000).
Endoscopic treatment entails mucosal resection of any visible lesion, followed by ablation of the entire Barrett's segment, which can be achieved using several methods including radio frequency ablation and argon plasma coagulation. This multi-modal approach has been demonstrated to be both effective and safe (Shaheen et al, 2009; Haidry et al, 2013). Whereas in the past, treatment was reserved for patients with HGD or intramucosal carcinoma, on the basis of new randomised controlled trial evidence treatment is now being offered to patients with LGD. It must be confirmed by two independent pathologists, given the subjectivity of this diagnosis and to avoid overtreatment in benign disease (Phoa et al, 2014). Patients diagnosed with invasive disease extending beyond the mucosa should be treated according to best practice guidelines, which generally involves peri-operative systemic chemotherapy (and/or radiotherapy) followed by surgery if it is deemed to be curative.
Genomic landscape of OAC
In order to understand the molecular genetic progression from BE to OAC, it is helpful to consider the genomic landscape of invasive disease. The genomic profile emerging from sequencing studies as part of the International Cancer Genome Consortium and The Cancer Genome Atlas is one of a highly mutated cancer with a mutation burden of around 10 single-nucleotide variations (SNVs) per megabase (Ross-Innes et al, 2015a). This rate is approaching as seen in those cancers with a well-defined carcinogen such as melanoma and lung cancer. Furthermore, it is a very heterogeneous disease with only a small number of genes that are recurrently mutated across multiple cases which are thus likely to be causative (so-called driver mutations). These recurrently mutated genes include tumour suppressors like TP53 and SMAD4, which had previously been identified as key in this disease, as well as MYO18B, SEMA5A, ARID1A and other members of the SWI/SNF chromatin remodelling complex (Dulak et al, 2013; Weaver et al, 2014). The lack of driver events in oncogenes is a particular challenge for identifying actionable targets to add to classical chemotherapy agents for molecular-targeted therapy.
The analysis of the pattern of base substitutions together with the exact base either side enables patterns or signatures to emerge. This analysis has aided identification of the causative mutagens, such as a signature associated with exposure to ultraviolet light in melanomas (Alexandrov et al, 2013). In OAC, a common mutational signature of T : A>G : C transversions in a CTT setting has been described, and it has been suggested that this could be a mutation pattern caused by acid exposure in the context of gastroesophageal reflux although this remains to be proven (Dulak et al, 2013; Nones et al, 2014; Weaver et al, 2014).
As well as singe-base substitutions Nones et al categorised 22 tumour samples based on larger chromosomal rearrangements termed structural variants (SV) into three classes: unstable genomes with ⩾450 SVs present across the genome (n=6), scattered alterations where <450 events were evenly distributed across the genome (n=2) and complex localised changes where SVs cluster only in a single or a few chromosomes (n=14) (Nones et al, 2014). The latter category could be explained by chromothripsis, a phenomenon of chromosomal shattering due to errors in chromosomal segregation during mitosis. So-called breakage-fusion-bridge (BFB) events define a further category of large-scale re-arrangements involving telomeric loss, chromosomal fusion and disrupted separation during anaphase. In OAC, a number of oncogenes were found to be amplified as a result of chromothripsis and BFB events including the oncogene myc (Nones et al, 2014). Such chromothripsis, or catastrophic events, occurring in a proportion of patients has a bearing on how we understand the evolution of OAC, which had hitherto been thought to be a gradual process.
Genomic landscape of Barrett's and progression to dysplasia and OAC
The overall mutation rate in non-dysplastic Barrett's is around 5.4–6.8 SNVs per megabase which, while being lower than in OAC, is higher than reported for many other invasive cancers including multiple myeloma and breast cancer (Stachler et al, 2015; Ross-Innes et al, 2015a). Stachler et al observed a significant difference in the mutation burden between non-dysplastic and dysplastic Barrett's when examining coding (exonic) mutations, which was not observed to the same degree by Ross-Innes et al where the pathology grade correlated poorly with the mutation rate observed across the entire genome (whole-genome sequencing). These findings underscore the difficulties in providing a histopathological grade that is a phenotypic readout from a complex genetic architecture, which becomes abnormal early on in the pathogenesis of this disease.
In terms of understanding the predilection of mutations for specific genes we have known for some time from candidate-gene studies that loss of the tumour suppressor p16 occurs commonly regardless of progression status, in comparison with p53 loss, which tends to occur later in the progression sequence (Reid et al, 2001; Leedham et al, 2008). The extent to which mutations can occur across a plethora of genes involved in cancer, even in patients with non-dysplastic Barrett's, has become apparent since the advent of genome-wide sequencing data. In one such study, whole-genome sequencing data in OAC was used to derive a custom panel of 26 genes and patients with stable, non-dysplastic Barrett's with many years of follow up (66 samples) were compared with cases with HGD and OAC (43 and 90 samples, respectively). They showed that similar mutation frequencies were observed across the disease states apart from the tumour suppressor genes TP53 and SMAD4. TP53 was recurrently mutated in HGD (72%) and OAC (69%) but only in one sample of non-dysplastic Barrett's in samples from patients who never progressed to HGD or OAC (P < 0.0001). SMAD4 was mutated at a lower frequency (13%) but was only found at the stage of invasive cancer (P=0.0061). Based on these observations, Weaver et al proposed that from the panel of genes examined, mutations in TP53 and SMAD4 mark the boundaries between non-dysplastic and dysplastic BE, and invasive cancer, respectively (Figure 2) (Weaver et al, 2014). Other sequencing studies observe p53 mutations in non-dysplastic tissue adjacent to cancer (Streppel et al, 2014; Stachler et al, 2015). Immunohistochemistry studies following patients over time also find aberrant p53 expression in non-dysplastic tissue in a low percentage of BE patients: 7% of non-progressors and 18% of progressors (Kastelein et al, 2013). Clinically, p53 is recognised as an adjunct to dysplasia diagnosis (Fitzgerald et al, 2014).
Figure 2.
Genetic events in the progression of Barrett's oesophagus. (A) Illustration of progression to oesophageal adenocarcinoma (OAC) from multiple clones with accumulation of mutations, with a predominance of tumour suppressor genes over time. CDKN2A loss of function is shown as an early event, p53 mutation likely to mark the boundary to dysplasia and SMAD4 mutations seen uniquely in the cancer. Copy-number changes, structural variants and genetic instability increase over time. A crisis event (denoted by *) may occur to promote rapid genomic instability and progression to cancer. Neutral clones may regress over time (e.g., grey and purple clones). (B) A diagram to show issues with sampling bias in a segment of Barrett's over time. Regions of the Barrett's lesion harbouring example mutations are highlighted. Heterogeneity within an individual segment of Barrett's means that endoscopic samples may be from tissue containing all, some or none of the mutations present in the entire lesion. No dysplasia is denoted by the blue dashed line and high-grade dysplasia (HGD) or oesophageal adenocarcinoma (OAC) in orange and pink, respectively. Even in regions described histologically as HGD and OAC, different combinations of mutations may be seen depending on sampling. For example, in panel 2, sequencing of sample 1 would show a CDKN2A mutation, sample 2 a p53 mutation and sample 3 would exhibit both mutations.
In a smaller study using the AmpliSeq Cancer Hotspot panel (which is a generic cancer panel), no mutations were seen at the baseline biopsies for the non-progressors (Del Portillo et al, 2015). This is likely due to the use of archival, formalin-fixed paraffin-embedded (FFPE) material and the difference in the respective gene panels as there were only three overlapping genes: TP53, SMAD4 and CDKN2A (Weaver et al, 2014). It is surprising that in the Del Portillo study CDKN2A wasn't found to be mutated in non-progressors, as this has previously been identified in a number of studies as an early event (Reid et al, 2001; Galipeau et al, 2007).
In addition to point mutations, chromosomal copy-number changes, called aneuploidy as well as gene-centric focal gains and deletions, have an important role in the progression to OAC (Dulak et al, 2012). Genome-wide sequencing data demonstrates that copy-number profiles of the tumour harbour significantly more gains and losses than the adjacent Barrett's (Ross-Innes et al, 2015a). Li et al previously investigated copy number using SNP arrays on biopsies from the index endoscopy (i.e. at first diagnosis of BE) and the penultimate endoscopy from OAC diagnosis for progressors. These were compared with similar time points for non-progressors (Li et al, 2014). While there were some focal changes seen in both patient cohorts, the copy-number profile of non-progressors remained relatively quiet over time, whereas the percentage of the genome harbouring somatic chromosomal alterations in progressor patients increased rapidly from 0 and 50% at baseline to approaching 100% within 48 months of the cancer diagnosis. These copy-number changes included genome doubling in some cases and increased genetic diversity (Li et al, 2014). The same authors have proposed a panel of 29 specific chromosomal alterations to predict the risk of BE progression (area under receiver operator curve=0.94) (Li et al, 2015). The samples from these retrospective studies were not histologically annotated; therefore, the relationship between these aberrations and the histological grade of dysplasia cannot be determined. The panel was tested in six OAC samples but has yet to be validated in an independent Barrett's cohort.
Clonal evolution
Inevitably, many of the studies in BE have relied on limited endoscopic biopsies and therefore the number of clones and their spatial relationship has not been forthcoming. Maley et al identified clones by differences in flow cytometric DNA content, pattern of loss of heterozygosity (LOH) as well as point mutations in CDKN2A (p16) and TP53 from multiple samples taken from within the same patient (Maley et al, 2006). They found that genetic diversity between the samples was strongly predictive of neoplastic progression of the Barrett's mucosa, and this was corroborated in 2010 in a follow-up study by the same group (Merlo et al, 2010; Li et al, 2014).
Maley et al proposed that a founder clone containing a p16 mutation providing a growth advantage to these cells leading to a clonal selective sweep across the Barrett's segment. Subsequent generation of dysplasia occurred via further clonal selective sweeps with loss of additional tumour suppressor genes conferring a selective advantage (Maley et al, 2006). Leedham et al in 2008 investigated regions of LOH as well as p16 and p53 point mutations on an individual crypt level. Their results suggested that heterogeneity in Barrett's lesions arose from multiple independent clones that evolved separately, resulting in a polyclonal mosaic of selective sweeps (Leedham et al, 2008). Ross-Innes et al assessed 1443 SNVs in 73 samples from a single patient's 10 cm BE segment, which had been collected over a period of 3 years (Ross-Innes et al, 2015a). In the non-dysplastic samples, six clones were identified with an initial clonal sweep but with just three SNVs common to all samples. These were three stochastic variants and not in known driver genes. Furthermore, dysplasia was derived from six distinct clones that did not correspond strictly to the dysplasia map generated by histopathological analysis (Ross-Innes et al, 2015a). Taken together, the polyclonal and thus heterogeneous nature of Barrett's explains the wide spectrum of the degree of mutational overlap between adjacent BE and OAC samples (Ross-Innes et al, 2015a). It should also be remembered that genetic mutations found in non-dysplastic Barrett's sampled adjacent to a cancer is not comparable to those found in non-dysplastic Barrett's in a patient who never progressed. This explains apparently conflicting evidence of the mutation spectrum observed between studies.
The prevailing view for the clonal evolution of OAC is that it occurs gradually through sequential loss of tumour suppressors culminating in loss of TP53 and cancer development. However, the whole-genome data suggests that in some cases TP53 mutation can lead to more rapid progression to cancer via chromosomal catastrophe (chromothripsis) or genome doubling and genetic instability (Figure 2) (Nones et al, 2014; Stachler et al, 2015).
Microenvironment
As well as characterising the epithelial cells of the tumour, it is also important to understand the contribution of interactions between different cellular components of the direct tumour microenvironment (He et al, 2013). Mechanisms that lead to an impairment of DNA damage repair mechanisms in the tumour can show a field effect on the surrounding mucosa, which is facilitated by the adjacent inflammatory processes (He et al, 2013). This is partly regulated by an increased population of regulatory T cells that can occur at the stage of reflux-related changes of the distal oesophagus, as well as by the activation of myeloid dendritic cells (Somja et al, 2013). The resulting cytokine milieu supports epithelial mesenchymal transition in the mucosa of the distal oesophagus, a process that is further maintained by cancer-associated fibroblasts and activation of PI3-Kinase/Akt signalling (Underwood et al, 2015). The cell-cell interaction network is also influenced by the formation and activation of cancer-associated adipocytes in the vicinity of tumour tissue (Trevellin et al, 2015). This is an area that requires further study.
Application of knowledge of somatic mutations for biomarkers of disease progression
Research is ongoing to identify individual markers, or marker panels that allow accurate detection of epithelial dysplasia and stratify patients for treatment, key studies are summarised in Table 1. This is particularly important given the rapid advances in endoscopic therapies, which have replaced oesophagectomy for patients with early disease. However, these therapies still have some risks and costs associated with them and hence it is important to stratify patients appropriately and avoid overtreatment.
Table 1. Biomarkers of progression of Barrett's oesophagus.
| Reference | Study design | Sample size | Baseline histology | Endpoint histology | Statistical findings | Validation cohort? | |
|---|---|---|---|---|---|---|---|
| P53 Immunohistochemistry | Murray et al, 2006 | Nested case-control study | 197 | ND | OAC | OR 11.7 (95% CI 2.37–30.0) | Yes – Kastelein et al, 2013 |
| Kaye et al, 2009 | Samples prospectively collected, retrospectively analysed | 143 | LGD | HGD/OAC | RR progression with consensus LGD: 1.56 | Yes – Kastelein et al, 2013 | |
| Kastelein et al, 2013 | Case-control study | 635 | ND | HGD/OAC | p53 overexpression: RR 5.6 (95% CI 3.1–10.3) loss of p53 expression: RR 14 (95% CI 5.3–37.2) | No | |
| 17p LOH | Reid et al, 2001 | Samples prospectively collected, retrospectively analysed | 256 | ND, ID and LGD | OAC | RR 16 (95% CI 6.2–39) | Yes – Galipeau et al, 2007 |
| Aneuploidy/tetraploidy | Reid, 2000 | Samples prospectively collected, retrospectively analysed | 322 | ND, ID and LGD | OAC | RR 11 (95% CI 5.5–21) | Yes – Galipeau et al, 2007; Bird-Lieberman et al, 2012a, 2012b |
| 17p LOH (p53), 9p LOH (p16) and abnormal DNA ploidy | Galipeau et al, 2007 | Samples prospectively collected, retrospectively analysed | 243 | ND | OAC | RR 38.7 (95% CI 10.8–138.5) | Yes – Di Pietro et al, 2015 |
| LGD, abnormal DNA ploidy and Aspergillus oryzae lectin | Bird-Lieberman et al, 2012a | Population-based nested case-control study | 380 | ND, ID and LGD | OAC | With baseline LGD: OR=3.90 (95% CI 2.39–6.37) Without baseline LGD: OR=3.31 (95% CI 1.81–6.05) | No |
| 8-gene methylation panel | Jin et al, 2009 | Retrospective double blinded | 195 | ND | HGD/OAC | AUC=0.843 at 2 years, AUC=0.829 at 4 years and AUC=0.840 | No |
| 4-gene methylation panel | Alvi et al, 2013 | Validated in retrospective and prospective cohorts | 46 | ND | OAC | AUC=0.988 97% specificity and 94% sensitivity Risk stratification: low risk: <2 genes, intermediate: 2 and high: >2 | Yes – Alvi et al, 2013 |
| 10 loci panel | Eluri et al, 2015 | Case-control study | 69 | ND | OAC | AUC=0.95 (95% CI 0.89–1) | No |
| 90 gene panel | Varghese et al, 2015 | Samples prospectively collected, retrospectively analysed | 150 | ND | OAC | Significantly higher rate of progression in patients regarded as high risk (P=0.047) | Yes – Varghese et al, 2015 |
Abbreviations: AUC=area under receiver-operating characteristic curve; CI=confidence interval; HGD=high-grade dysplasia; ID=indefinite for dysplasia; LGD=low-grade dysplasia; LOH = loss of heterozygosity; ND=non-dysplastic Barrett's; OAC=oesophageal adenocarcinoma; OR=odds ratio; RR=relative risk.
In the context of Barrett's, p53 alone and in combination with other markers has been investigated both as a diagnostic biomarker and a marker of progression (Reid et al, 2001; Weston et al, 2001; Murray et al, 2006; Kaye et al, 2009; Kastelein et al, 2013). As well as the possibility of sequencing, inactivating mutations of TP53 are also frequently detectable by immunohistochemistry providing a more clinically applicable test. Most mutations lead to stabilisation of the protein and hence increased the levels of expression, but a loss of staining can also be observed for truncating mutations (Kaye et al, 2009). In a nested case-control study of 197 patients with BE, the odds ratio (OR) for progression was 11.7 (95% CI: 1.93–71.4) in patients with higher p53 staining scores (Murray et al, 2006). A retrospective study of 635 patients confirmed this with a relative risk (RR) of 5.6 (95% CI: 3.1–10.3) (Kastelein et al, 2013). The risk for progression was even higher in cases with complete absence of p53 staining (RR 14.0; 95% CI: 5.3–37.2), and the risk of progression for patients with LGD increased from 15 to 33% if aberrant p53 staining was taken into account (Kastelein et al, 2013).
Assessment of p53 staining has also been shown to decrease the inter-observer variability between pathologists concerning the diagnosis of dysplastic lesions (Kaye et al, 2009, 2016). Therefore, taking all this evidence into account, immunohistochemistry for p53 has been suggested as an adjunct to aid the histological analysis of Barrett's biopsies in the recent BSG guidelines (Fitzgerald et al, 2014).
Marker panels
Galipeau et al built on previous work from their group (Reid et al, 2001) and reported that a panel of TP53 LOH, CDKN2A LOH and presence of tetraploidy indicated a RR for Barrett's progression of 38.7 (95% CI: 10.8–138.5) (Galipeau et al, 2007). In a more recent study, the LOH and microsatellite instability status of 10 specific gene loci were combined into a risk score to predict the progression towards HGD (Eluri et al, 2015).
In an approach designed to enable analysis of FFPE material, a combination of abnormal DNA ploidy and expression of the novel biomarker Aspergillus oryzae lectin resulted in a three-fold increased risk for progression (OR 3.31; 95% CI: 1.81–6.05), or nearly four-fold in patients with baseline LGD (OR 3.90; 95% CI 2.39–6.37) (Bird-Lieberman et al, 2012a, 2012b).
Varghese et al identified a predominantly MYC-regulated 90 gene signature to distinguish between HGD and non-dysplastic Barrett's (P<0.0001) using RNA microarray technology. In an independent validation cohort from the UK and The Netherlands, dysplastic samples were identified with an area under the curve of 0.87 (95% CI: 0.82−0.93). This panel is particularly useful given its ability to distinguish between patients with LGD who are likely to progress. Indeed, using this panel, the 64% of LGD categorised as being high risk had a significantly higher rate of progression (P=0.047) (Varghese et al, 2015).
Gene expression regulation by epigenetic factors has also been investigated in BE progression. Array data were used to generate a panel of four hypermethylated genes (SLC22A18, PIGR, GJA12 and RIN2), validated by pyrosequencing in an independent cohort, to risk stratify patients into high-, low- and intermediate-risk groups with 94% sensitivity and 97% specificity (Alvi et al, 2013). An 8-gene hypermethylation panel consisting of p16, RUNX3, HPP1, NELL1, TAC1, SST, AKAP12 and CDH13 has also been investigated in the context of BE progression (area under receiver-operating characteristic curve (AUC)=0.843 at 2 years, AUC=0.829 at 4 years and AUC=0.840 for the combined model) (Jin et al, 2009).
These biomarker studes have relied on the acquisition of endoscopically obtained biopsies. However, brush cytology techniques are increasingly being used to reduce the sampling error inherent from forceps biopsies and these methods can also be combined with molecular biomarkers to predict progression of non-dysplastic Barrett's (Timmer et al, 2015). Brush sampling, however, still requires direct endoscopic vision and non-endoscopic techniques like the Cytosponge are being developed. The Cytosponge is a densely folded sponge packed into a capsule that rapidly dissolves when entering the stomach. Once dissolved, the sponge can be retracted using the string. Cells of the oesophageal mucosal surface stick to the sponge that can then be assessed for biomarkers using immunohistochemistry and sequencing, such as p53 (Kadri et al, 2010; Ross-Innes et al, 2015b).
Combination of advanced endoscopic imaging modalities with molecular biomarkers
An alternative approach to reducing the number of random biopsies that are necessary according to the Seattle protocol is to perform a more sophisticated endoscopic assessment of the Barrett's mucosa using techniques such as narrow band imaging or auto-fluorescence imaging. This allows biopsies to be directed towards suspicious areas and then histopathological assessment can be combined with biomarker panels, which have the advantage of a more objective readout (e.g. p53, p16, cyclin A and altered ploidy); (Di Pietro et al, 2015). Similarly, confocal laser endomicroscopy allows the real-time detection of intestinalised epithelium and even dysplastic changes and this can also be combined with tissue-based biomarkers, resulting in excellent performance compared with standard histopathology (Tofteland et al, 2014; Di Pietro et al, 2015). An alternative approach to classical tissue-based assessment of biomarkers is the application of molecular probes that can be viewed during endoscopy in real time. One such approach takes advantage of the altered glycosylation patters of the mucosal surface during the progression towards cancer that can be imaged with a fluorescent lectin (Bird-Lieberman et al, 2012b). A similar approach using labelled peptides that bind specifically to dysplastic or neoplastic lesions in the oesophagus showed promising results and is under evaluation for clinical practice (Sturm et al, 2013).
Conclusions
Oesophageal adenocarcinoma is frequently precipitated by the loss of p53 and the resultant copy-number changes with a high frequency of heterogeneous genomic alterations. During its evolution, point mutations occur as early as non-dysplastic BE. It is, therefore, challenging to predict progression from Barrett's to OAC using biomarker approaches. However, the advent of increasingly cost-effective sequencing and array technologies mean that it is possible to develop predictive tests, for example copy-number alterations, without necessarily needing an approach limited to a small number of candidate genes. This is still an area for research. For clinical management of patients with BE undergoing surveillance, there is agreement by specialist societies that there should be consensus-based assessment of dysplasia assisted by p53 immunostaining and discussion of the management for individual cases within specialist multidisciplinary teams. The more that biomarkers are coupled with sampling methods that limit the bias inherent in random biopsies, the more accurate we can be at predicting progression and preventing OAC development.
Footnotes
RCF developed the Cytosponge technology and is named on related patents. This technology has been licensed by the MRC to Covidien GI Solutions, now Medtronic. The remaining authors declare no conflict of interest.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjörd JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jäger N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, López-Otín C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdés-Mas R, van Buuren MM, van't Veer L, Vincent-Salomon A, Waddell N, Yates LR. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR (2013) Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvi MA, Liu X, O'donovan M, Newton R, Wernisch L, Shannon NB, Shariff K, di Pietro M, Bergman JJ, Ragunath K, Fitzgerald RC (2013) DNA methylation as an adjunct to histopathology to detect prevalent, inconspicuous dysplasia and early-stage neoplasia in Barrett's esophagus. Clin Cancer Res 19: 878–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S, Coleman HG, Yousef F, Johnston BT, Mcmanus DT, Gavin AT, Murray LJ (2011) Risk of malignant progression in Barrett's esophagus patients: results from a large population-based study. J Natl Cancer Inst 103: 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird-Lieberman EL, Dunn JM, Coleman HG, Lao-Sirieix P, Oukrif D, Moore CE, Varghese S, Johnston BT, Arthur K, McManus DT, Novelli MR, O'Donovan M, Cardwell CR, Lovat LB, Murray LJ, Fitzgerald RC (2012. a) Population-based study reveals new risk-stratification biomarker panel for Barrett's esophagus. Gastroenterology 143: 927–35 e3. [DOI] [PubMed] [Google Scholar]
- Bird-Lieberman EL, Neves AA, Lao-Sirieix P, O'donovan M, Novelli M, Lovat LB, Eng WS, Mahal LK, Brindle KM, Fitzgerald RC (2012. b) Molecular imaging using fluorescent lectins permits rapid endoscopic identification of dysplasia in Barrett's esophagus. Nat Med 18: 315–321. [DOI] [PubMed] [Google Scholar]
- Corley DA, Mehtani K, Quesenberry C, Zhao W, De Boer J, Weiss NS (2013) Impact of endoscopic surveillance on mortality from Barrett's esophagus-associated esophageal adenocarcinomas. Gastroenterology 145: 312–9 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curvers WL, Ten Kate FJ, Krishnadath KK, Visser M, Elzer B, Baak LC, Bohmer C, Mallant-Hent RC, van Oijen A, Naber AH, Scholten P, Busch OR, Blaauwgeers HG, Meijer GA, Bergman JJ (2010) Low-grade dysplasia in Barrett's esophagus: overdiagnosed and underestimated. Am J Gastroenterol 105: 1523–1530. [DOI] [PubMed] [Google Scholar]
- Del Portillo A, Lagana SM, Yao Y, Uehara T, Jhala N, Ganguly T, Nagy P, Gutierrez J, Luna A, Abrams J, Liu Y, Brand R, Sepulveda JL, Falk GW, Sepulveda AR (2015) Evaluation of Mutational Testing of Preneoplastic Barrett's Mucosa by Next-Generation Sequencing of Formalin-Fixed, Paraffin-Embedded Endoscopic Samples for Detection of Concurrent Dysplasia and Adenocarcinoma in Barrett's Esophagus. J Mol Diagn 17: 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai TK, Krishnan K, Samala N, Singh J, Cluley J, Perla S, Howden CW (2012) The incidence of oesophageal adenocarcinoma in non-dysplastic Barrett's oesophagus: a meta-analysis. Gut 61: 970–976. [DOI] [PubMed] [Google Scholar]
- Di Pietro M, Bird-Lieberman EL, Liu X, Nuckcheddy-Grant T, Bertani H, O'donovan M, Fitzgerald RC (2015) Autofluorescence-directed confocal endomicroscopy in combination with a three-biomarker panel can inform management decisions in Barrett's esophagus. Am J Gastroenterol 110: 1549–1558. [DOI] [PubMed] [Google Scholar]
- Duits LC, Phoa KN, Curvers WL, Ten Kate FJ, Meijer GA, Seldenrijk CA, Offerhaus GJ, Visser M, Meijer SL, Krishnadath KK, Tijssen JG, Mallant-Hent RC, Bergman JJ (2015) Barrett's oesophagus patients with low-grade dysplasia can be accurately risk-stratified after histological review by an expert pathology panel. Gut 64: 700–706. [DOI] [PubMed] [Google Scholar]
- Dulak AM, Schumacher SE, Van Lieshout J, Imamura Y, Fox C, Shim B, Ramos AH, Saksena G, Baca SC, Baselga J, Tabernero J, Barretina J, Enzinger PC, Corso G, Roviello F, Lin L, Bandla S, Luketich JD, Pennathur A, Meyerson M, Ogino S, Shivdasani RA, Beer DG, Godfrey TE, Beroukhim R, Bass AJ (2012) Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res 72: 4383–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, McKenna A, Carter SL, Cibulskis K, Sivachenko A, Saksena G, Voet D, Ramos AH, Auclair D, Thompson K, Sougnez C, Onofrio RC, Guiducci C, Beroukhim R, Zhou Z, Lin L, Lin J, Reddy R, Chang A, Landrenau R, Pennathur A, Ogino S, Luketich JD, Golub TR, Gabriel SB, Lander ES, Beer DG, Godfrey TE, Getz G, Bass AJ (2013) Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet 45: 478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eluri S, Brugge WR, Daglilar ES, Jackson SA, Styn MA, Callenberg KM, Welch DC, Barr TM, Duits LC, Bergman JJ, Shaheen NJ (2015) The presence of genetic mutations at key loci predicts progression to esophageal adenocarcinoma in Barrett's esophagus. Am J Gastroenterol 110: 828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald RC, Di Pietro M, Ragunath K, Ang Y, Kang JY, Watson P, De Caestecker J (2014) British Society of Gastroenterology guidelines on the diagnosis and management of Barrett's oesophagus. Gut 63: 7–42. [DOI] [PubMed] [Google Scholar]
- Galipeau PC, Li X, Blount PL, Maley CC, Sanchez CA, Odze RD, Ayub K, Rabinovitch PS, Vaughan TL, Reid BJ (2007) NSAIDs modulate CDKN2A, TP53, and DNA content risk for progression to esophageal adenocarcinoma. PLoS Med 4: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidry RJ, Dunn JM, Butt MA, Burnell MG, Gupta A, Green S, Miah H, Smart HL, Bhandari P, Smith LA, Willert R, Fullarton G, Morris J, Di Pietro M, Gordon C, Penman I, Barr H, Patel P, Boger P, Kapoor N, Mahon B, Hoare J, Narayanasamy R, O'Toole D, Cheong E, Direkze NC, Ang Y, Novelli M, Banks MR, Lovat LB (2013) Radiofrequency ablation and endoscopic mucosal resection for dysplastic barrett's esophagus and early esophageal adenocarcinoma: outcomes of the UK National Halo RFA Registry. Gastroenterology 145: 87–95. [DOI] [PubMed] [Google Scholar]
- He H, Tian D, Guo J, Liu M, Chen Z, Hamdy FC, Helleday T, Su M, Ying S (2013) DNA damage response in peritumoral regions of oesophageal cancer microenvironment. Carcinogenesis 34: 139–145. [DOI] [PubMed] [Google Scholar]
- Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P (2011) Incidence of adenocarcinoma among patients with Barrett's esophagus. N Engl J Med 365: 1375–1383. [DOI] [PubMed] [Google Scholar]
- Jin Z, Cheng Y, Gu W, Zheng Y, Sato F, Mori Y, Olaru AV, Paun BC, Yang J, Kan T, Ito T, Hamilton JP, Selaru FM, Agarwal R, David S, Abraham JM, Wolfsen HC, Wallace MB, Shaheen NJ, Washington K, Wang J, Canto MI, Bhattacharyya A, Nelson MA, Wagner PD, Romero Y, Wang KK, Feng Z, Sampliner RE, Meltzer SJ (2009) A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett's esophagus. Cancer Res 69: 4112–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadri SR, Lao-Sirieix P, O'donovan M, Debiram I, Das M, Blazeby JM, Fitzgerald RC, Emery J, Boussioutas A, Morris H, Walter FM, Pharoah P, Hardwick RH (2010) Acceptability and accuracy of a non-endoscopic screening test for Barrett's oesophagus in primary care: cohort study. BMJ 341: c4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastelein F, Biermann K, Steyerberg EW, Verheij J, Kalisvaart M, Looijenga LH, Stoop HA, Walter L, Kuipers EJ, Spaander MC, Bruno MJ ProBar-study group (2013) Aberrant p53 protein expression is associated with an increased risk of neoplastic progression in patients with Barrett's oesophagus. Gut 62: 1676–1683. [DOI] [PubMed] [Google Scholar]
- Kastelein F, Van Olphen S, Steyerberg EW, Sikkema M, Spaander MC, Looman CW, Kuipers EJ, Siersema PD, Bruno MJ, De Bekker-Grob EW ProBar-study group (2015) Surveillance in patients with long-segment Barrett's oesophagus: a cost-effectiveness analysis. Gut 64: 864–871. [DOI] [PubMed] [Google Scholar]
- Kastelein F, Van Olphen SH, Steyerberg EW, Spaander MC, Bruno MJ (2016) Impact of surveillance for Barrett's oesophagus on tumour stage and survival of patients with neoplastic progression. Gut 65: 548–554. [DOI] [PubMed] [Google Scholar]
- Kaye PV, Haider SA, Ilyas M, James PD, Soomro I, Faisal W, Ragunath K (2009) Barrett's dysplasia and the Vienna classification: reproducibility, prediction of progression and impact of consensus reporting and p53 immunohistochemistry. Histopathology 54: 699–712. [DOI] [PubMed] [Google Scholar]
- Kaye PV, Ilyas M, Soomro I, Haider A, Atwal G, Menon S, Gill S, Richards C, Harrison R, West K, Ragunath K (2016) Dysplasia in Barrett's Oesophagus: p53 immunostaining is more reproducible then H&E diagnosis and improves overall reliability while grading is poorly reproducible. Histopathology; e-pub ahead of print 26 February 2016; doi:10.1111/his.12956. [DOI] [PubMed]
- Lagergren J, Bergstrom R, Lindgren A, Nyren O (1999) Symptomatic gastroesophageal reflux as a risk factor for esophageal adenocarcinoma. N Engl J Med 340: 825–831. [DOI] [PubMed] [Google Scholar]
- Leedham SJ, Preston SL, Mcdonald SA, Elia G, Bhandari P, Poller D, Harrison R, Novelli MR, Jankowski JA, Wright NA (2008) Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett's oesophagus. Gut 57: 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage C, Rachet B, Jooste V, Faivre J, Coleman MP (2008) Continuing rapid increase in esophageal adenocarcinoma in England and Wales. Am J Gastroenterol 103: 2694–2699. [DOI] [PubMed] [Google Scholar]
- Levine DS, Blount PL, Rudolph RE, Reid BJ (2000) Safety of a systematic endoscopic biopsy protocol in patients with Barrett's esophagus. Am J Gastroenterol 95: 1152–1157. [DOI] [PubMed] [Google Scholar]
- Li X, Galipeau PC, Paulson TG, Sanchez CA, Arnaudo J, Liu K, Sather CL, Kostadinov RL, Odze RD, Kuhner MK, Maley CC, Self SG, Vaughan TL, Blount PL, Reid BJ (2014) Temporal and spatial evolution of somatic chromosomal alterations: a case-cohort study of Barrett's esophagus. Cancer Prev Res (Phila) 7: 114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Paulson TG, Galipeau PC, Sanchez CA, Liu K, Kuhner MK, Maley CC, Self SG, Vaughan TL, Reid BJ, Blount PL (2015) Assessment of esophageal adenocarcinoma risk using somatic chromosome alterations in longitudinal samples in Barrett's esophagus. Cancer Prev Res (Phila) 8: 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, Paulson TG, Blount PL, Risques RA, Rabinovitch PS, Reid BJ (2006) Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 38: 468–473. [DOI] [PubMed] [Google Scholar]
- Merlo LM, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, Maley CC (2010) A comprehensive survey of clonal diversity measures in Barrett's esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res (Phila) 3: 1388–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray L, Sedo A, Scott M, Mcmanus D, Sloan JM, Hardie LJ, Forman D, Wild CP (2006) TP53 and progression from Barrett's metaplasia to oesophageal adenocarcinoma in a UK population cohort. Gut 55: 1390–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nones K, Waddell N, Wayte N, Patch AM, Bailey P, Newell F, Holmes O, Fink JL, Quinn MC, Tang YH, Lampe G, Quek K, Loffler KA, Manning S, Idrisoglu S, Miller D, Xu Q, Waddell N, Wilson PJ, Bruxner TJ, Christ AN, Harliwong I, Nourse C, Nourbakhsh E, Anderson M, Kazakoff S, Leonard C, Wood S, Simpson PT, Reid LE, Krause L, Hussey DJ, Watson DI, Lord RV, Nancarrow D, Phillips WA, Gotley D, Smithers BM, Whiteman DC, Hayward NK, Campbell PJ, Pearson JV, Grimmond SM, Barbour AP (2014) Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun 5: 5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoa KN, Van Vilsteren FG, Weusten BL, Bisschops R, Schoon EJ, Ragunath K, Fullarton G, Di Pietro M, Ravi N, Visser M, Offerhaus GJ, Seldenrijk CA, Meijer SL, ten Kate FJ, Tijssen JG, Bergman JJ (2014) Radiofrequency ablation vs endoscopic surveillance for patients with Barrett esophagus and low-grade dysplasia: a randomized clinical trial. JAMA 311: 1209–1217. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Hyder A, Bae SJ, Zarin W, O'neill TJ, Marcon NE, Stein L, Thein HH (2015) Surveillance in patients with Barrett's esophagus for early detection of esophageal adenocarcinoma: a systematic review and meta-analysis. Clin Transl Gastroenterol 6: e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BJ, Levine DS, Longton G, Blount PL, Rabinovitch PS (2000) Predictors of progression to cancer in Barrett's esophagus: baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol 95: 1669–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BJ, Prevo LJ, Galipeau PC, Sanchez CA, Longton G, Levine DS, Blount PL, Rabinovitch PS (2001) Predictors of progression in Barrett's esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol 96: 2839–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Becq J, Warren A, Cheetham RK, Northen H, O'donovan M, Malhotra S, di Pietro M, Ivakhno S, He M, Weaver JM, Lynch AG, Kingsbury Z, Ross M, Humphray S, Bentley D, Fitzgerald RC Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Study Group; Oesophageal Cancer Clinical and Molecular Stratification OCCAMS Study Group (2015. a) Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nat Genet 47: 1038–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Debiram-Beecham I, O'donovan M, Walker E, Varghese S, Lao-Sirieix P, Lovat L, Griffin M, Ragunath K, Haidry R, Sami SS, Kaye P, Novelli M, Disep B, Ostler R, Aigret B, North BV, Bhandari P, Haycock A, Morris D, Attwood S, Dhar A, Rees C, Rutter MD, Sasieni PD, Fitzgerald RC (2015. b) Evaluation of a minimally invasive cell sampling device coupled with assessment of trefoil factor 3 expression for diagnosing Barrett's esophagus: a multi-center case-control study. PLoS Med 12: e1001780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen NJ, Sharma P, Overholt BF, Wolfsen HC, Sampliner RE, Wang KK, Galanko JA, Bronner MP, Goldblum JR, Bennett AE, Jobe BA, Eisen GM, Fennerty MB, Hunter JG, Fleischer DE, Sharma VK, Hawes RH, Hoffman BJ, Rothstein RI, Gordon SR, Mashimo H, Chang KJ, Muthusamy VR, Edmundowicz SA, Spechler SJ, Siddiqui AA, Souza RF, Infantolino A, Falk GW, Kimmey MB, Madanick RD, Chak A, Lightdale CJ (2009) Radiofrequency ablation in Barrett's esophagus with dysplasia. N Engl J Med 360: 2277–2288. [DOI] [PubMed] [Google Scholar]
- Somja J, Demoulin S, Roncarati P, Herfs M, Bletard N, Delvenne P, Hubert P (2013) Dendritic cells in Barrett's esophagus carcinogenesis: an inadequate microenvironment for antitumor immunity? Am J Pathol 182: 2168–2179. [DOI] [PubMed] [Google Scholar]
- Spechler SJ, Sharma P, Souza RF, Inadomi JM, Shaheen NJ (2011) American Gastroenterological Association medical position statement on the management of Barrett's esophagus. Gastroenterology 140: 1084–1091. [DOI] [PubMed] [Google Scholar]
- Stachler MD, Taylor-Weiner A, Peng S, Mckenna A, Agoston AT, Odze RD, Davison JM, Nason KS, Loda M, Leshchiner I, Stewart C, Stojanov P, Seepo S, Lawrence MS, Ferrer-Torres D, Lin J, Chang AC, Gabriel SB, Lander ES, Beer DG, Getz G, Carter SL, Bass AJ (2015) Paired exome analysis of Barrett's esophagus and adenocarcinoma. Nat Genet 47(9): 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streppel MM, Lata S, Delabastide M, Montgomery EA, Wang JS, Canto MI, Macgregor-Das AM, Pai S, Morsink FH, Offerhaus GJ, Antoniou E, Maitra A, Mccombie WR (2014) Next-generation sequencing of endoscopic biopsies identifies ARID1A as a tumor-suppressor gene in Barrett's esophagus. Oncogene 33: 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm MB, Joshi BP, Lu S, Piraka C, Khondee S, Elmunzer BJ, Kwon RS, Beer DG, Appelman HD, Turgeon DK, Wang TD (2013) Targeted imaging of esophageal neoplasia with a fluorescently labeled peptide: first-in-human results. Sci Transl Med 5: 184ra61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer MR, Martinez P, Lau CT, Westra WM, Calpe S, Rygiel AM, Rosmolen WD, Meijer SL, Ten Kate FJ, Dijkgraaf MG, Mallant-Hent RC, Naber AH, van Oijen AH, Baak LC, Scholten P, Böhmer CJ, Fockens P, Maley CC, Graham TA, Bergman JJ, Krishnadath KK (2015) Derivation of genetic biomarkers for cancer risk stratification in Barrett's oesophagus: a prospective cohort study. Gut ; e-pub ahead of print 23 June 2015; doi:10.1136/gutjnl-2015-309642. [DOI] [PMC free article] [PubMed]
- Tofteland N, Singh M, Gaddam S, Wani SB, Gupta N, Rastogi A, Bansal A, Kanakadandi V, McGregor DH, Ulusarac O, Cherian R, Mathur SC, Sharma P (2014) Evaluation of the updated confocal laser endomicroscopy criteria for Barrett's esophagus among gastrointestinal pathologists. Dis Esophagus 27: 623–629. [DOI] [PubMed] [Google Scholar]
- Trevellin E, Scarpa M, Carraro A, Lunardi F, Kotsafti A, Porzionato A, Saadeh L, Cagol M, Alfieri R, Tedeschi U, Calabrese F, Castoro C, Vettor R (2015) Esophageal adenocarcinoma and obesity: peritumoral adipose tissue plays a role in lymph node invasion. Oncotarget 6: 11203–11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood TJ, Hayden AL, Derouet M, Garcia E, Noble F, White MJ, Thirdborough S, Mead A, Clemons N, Mellone M, Uzoho C, Primrose JN, Blaydes JP, Thomas GJ (2015) Cancer-associated fibroblasts predict poor outcome and promote periostin-dependent invasion in oesophageal adenocarcinoma. J Pathol 235: 466–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese S, Newton R, Ross-Innes CS, Lao-Sirieix P, Krishnadath KK, O'donovan M, Novelli M, Wernisch L, Bergman J, Fitzgerald RC (2015) Analysis of dysplasia in patients with Barrett's esophagus based on expression pattern of 90 genes. Gastroenterology 149: 1511–1518 e5. [DOI] [PubMed] [Google Scholar]
- Weaver JM, Ross-Innes CS, Shannon N, Lynch AG, Forshew T, Barbera M, Murtaza M, Ong CA, Lao-Sirieix P, Dunning MJ, Smith L, Smith ML, Anderson CL, Carvalho B, O'Donovan M, Underwood TJ, May AP, Grehan N, Hardwick R, Davies J, Oloumi A, Aparicio S, Caldas C, Eldridge MD, Edwards PA, Rosenfeld N, Tavaré S, Fitzgerald RC OCCAMS Consortium (2014) Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet 46: 837–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston AP, Banerjee SK, Sharma P, Tran TM, Richards R, Cherian R (2001) p53 protein overexpression in low grade dysplasia (LGD) in Barrett's esophagus: immunohistochemical marker predictive of progression. Am J Gastroenterol 96: 1355–1362. [DOI] [PubMed] [Google Scholar]