Abstract
Recently, radiogenomics or imaging genomics has emerged as a novel high-throughput method of associating imaging features with genomic data. Radiogenomics has the potential to provide comprehensive intratumour, intertumour and peritumour information non-invasively. This review article summarizes the current state of radiogenomic research in tumour characterization, discusses some of its limitations and promises and projects its future directions. Semi-radiogenomic studies that relate specific gene expressions to imaging features will also be briefly reviewed.
INTRODUCTION
In recent years, a new direction in cancer research has emerged to address high-throughput methods of associating imaging features with genomic data.1–3 This approach is referred to as radiogenomics or imaging genomics. The imaging characteristics of a disease are also called its imaging phenotype or radiophenotype, while the genomic information defines the molecular phenotype or genotype of the disease. Research to uncover the underlying genetic causes of individual variation in sensitivity to radiation using high-throughput genomic methods has also been referred to as “radiogenomics” and is not discussed in this review.4
Much of the discussion of personalized medicine has focused on molecular characterization using genomic and proteomic technologies.5 However, a limitation of these approaches is the need to acquire tissue samples through invasive surgery or biopsy.6 Although some genetic analyses have been incorporated into clinical practice in recent years, large-scale genome-based cancer characterization is not routinely performed owing to its cost, turnaround time and technical complexity required for data analysis and interpretation.7 In addition, samples are often obtained from a small portion of a heterogeneous lesion and may not accurately represent the lesion's anatomic, functional and physiologic properties.8 Even more importantly, it is not feasible to obtain the tissue multiple times during treatment in order to monitor response. Consequently, it is still a challenge to incorporate genomics or proteomics into routine clinical practice.
Imaging has great potential for in vivo tumour characterization because it can provide a more comprehensive view of the entire tumour than biopsy samples alone.9 For example, imaging can provide information on peritumoral regions, which are typically not surgically removed and thus not analysed in the laboratory.1 Human tissues often exhibit a diversity of distinctive traits on radiographic images, many of which currently have no known clinical significance. Furthermore, routine clinical practice often includes follow-up imaging to monitor treatment response and disease progression.10 Advances in imaging technologies now provide better anatomic localization and allow for non-invasive measurements of functional and physiologic tissue- and lesion-specific properties.11 Potentially, one would benefit tremendously from radiogenomic biomarkers that measure gene expression at frequent intervals during therapy.
Oncologic diagnosis is quickly moving from the traditional histology-based approaches to molecular stratification.12 Therefore, the traditional radiology–pathology paradigm alone is no longer sufficient to radiologists. Radiogenomics represents the evolution of the radiology–pathology correlation from the histology level to the subcellular level.2 This systematic association between imaging traits and gene expression allows useful inference in both directions: imaging traits can be used to predict gene expressions in human cancers; conversely, image features can be predicted from gene signatures.2,3 The predictive capabilities of these signatures not only enable immediate translational potential, but also suggest potential molecular mechanisms that may give rise to imaging phenotypes.13
In order for personalized medicine to transpire, biomarkers must accurately reflect the underlying molecular cancerous machinery.14 Given the growing number of genomic, imaging and clinical biomarkers that were identified in patients with various types of cancers, there is a need to create integrative biomarkers to link multiple types of data and measurements.14 The objective of this study was to provide a comprehensive review of radiogenomic research in tumour characterization.
PUBLISHED STUDIES USING A RADIOGENOMIC APPROACH IN CANCER RESEARCH
We searched multiple electronic databases for original research studies that correlated imaging features by manual, semi-automatic or automatic assessment with the whole genome data. Our search terms included variations of1 different imaging modalities including “MR”, “scintigraphy” and “nuclear medicine”, “CT” or “PET”; and2 molecular signatures such as “genome”, “genomics”, “molecular profiling”, “mutation”, “sequence”, “gene”, “genetic” and “signature”. Studies that contained the word radiogenomic or imaging genomics were identified separately. We excluded studies that associated imaging features with patient response to radiation therapy, since this refers to a different field of research called radiation genomics. Radiomics involves extraction of many quantitative imaging features with computer algorithms. The extracted features can be related to genomics or proteomics. Only “radiomics approach to radiogenomics” is included in this review. For studies that associated imaging features with specific genes and expression of specific gene subsets (e.g. tumour molecular subtype), we grouped them under the category “semi-radiogenomic studies”. Studies that correlated imaging with markers measured by immunohistochemistry or fluorescent in situ hybridization [e.g. (R)-2hydroxyglutarate (2HG) metabolites from isocitrate dehydrogenase 1 mutation, p53 nuclear staining, anaplastic lymphoma kinase + status etc.] were not included under this category. Furthermore, we did not include studies that correlated BRCA1/2 gene mutations or other specific gene expressions/mutations with breast density on mammography.
Overall, 27 studies were included in the final analyses (Table 1).9,15–40 These studies were published between 2007 and 2015. 8 studies used data from The Cancer Genome Atlas (TCGA) and/or The Cancer Imaging Archive (TCIA);1,16,23,27,32,34,38,41 2 studies were multi-institutional;9, 19 and the remaining 17 studies used local institutional data.15,17,18,20–22,24–26,28,30,31,33,35–37,39,40 26 out of 27 studies were retrospective in design. The number of patients ranged from 10 to 104 patients, with a median of 38 patients. 8 (30%) studies used a validation data set to verify the association between imaging features and genomic data identified in the initial data set.20,22,23,27,28,34,37,40 Six types of cancers were studied: glioblastoma multiforme (GBM)/high-grade glioma (n = 14, 52%), non-small-cell lung cancer (NSCLC) (n = 3, 11%), hepatocellular carcinoma (HCC) (n = 3, 11%), breast cancer (n = 5, 19%), clear-cell renal cell carcinoma (CCRCC) (n = 1, 4%) and cervical cancer (n = 1, 4%). The imaging modalities used included fluorine 18 fludeoxyglucose positron emission tomography (PET) (n = 4, 15%), MRI [n = 18 (including two perfusion MR), 67%] and CT [n = 5 (including one perfusion CT), 19%].
Table 1.
Radiogenomic studies published in the literature
| Study | Year | Country | Data source | Number | Validation set | Cancer | Imaging modality | Number of features | Method of feature extraction | Radiologist involvement | Genomic data | Individual vs clusters | Pathway analysis | Wet-lab validation | Histology correlation | Outcome | Clinical outcomes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gevaert et al15 | 2012 | USA | Single institutional | 26 | No | NSCLC | PET | 180 | Both | Yes | Microarray | Clusters | Yes | No | No | Yes | RFS and OS |
| Gevaert et al16 | 2014 | USA | TCGA | 55 | No | GBM | MR | 79 | Manual | Yes | Microarray, DNA methylation, array CGH | Clusters | Yes | No | No | Yes | PFS and OS |
| Aerts et al9 | 2014 | Netherlands | Multi-institutional | 89 | No | NSCLC | CT | 440 | Semi-automated | No | Microarrays | Clusters | Yes | No | Yes | Yes | OS |
| Jamshidi et al17 | 2014 | USA | Single institutional | 23 | No | GBM | MR | 6 | Manual | Yes | Microarrays, array CGH | Clusters | Yes | No | No | No | NA |
| Kuo et al18 | 2007 | USA | Single institutional | 30 | No | HCC | CT | 6 | Manual | Yes | Microarrays | Clusters | No | No | Yes | No | NA |
| Nair et al19 | 2012 | USA | Multi-institutional | 25 | No | NSCLC | PET | 14 | Semi-automated | Yes | Microarrays | Both | Yes | No | No | Yes | OS |
| Segal et al20 | 2007 | USA | Single institutional | 28 | Yes | HCC | CT | 138 | Manual | Yes | Microarrays | Clusters | Yes | No | Yes | Yes | OS |
| Yamamoto et al21 | 2012 | USA | Single institutional | 10 | No | Breast cancer | MR | 26 | Manual | Yes | Microarrays | Both | Yes | No | No | No | NA |
| Yamamoto et al22 | 2015 | USA | Single institutional | 19 | Yes | Breast cancer | MR | 47 | Semi-automated | Yes | RNA sequencing | Individual | Yes | Yes | Yes | Yes | MFS |
| Zinn et al23 | 2011 | USA | TCGA | 26 | Yes | GBM | MR | 3 | Semi-automated | Yes | mRNA and micro-RNA | Individual | Yes | No | No | Yes | PFS and OS |
| Barajas et al24 | 2010 | USA | Single institutional | 12 | No | GBM | MR | 9 | Manual | Yes | Microarray | Both | Yes | No | Yes | No | NA |
| Diehn et al25 | 2008 | USA | Single institutional | 22 | No | GBM | MR | 10 | Manual | Yes | Microarray | Both | Yes | Yes | No | Yes | OS |
| Pope et al26 | 2008 | USA | Single institutional | 52 | No | GBM | MR | 1 | Manual | Yes | Microarray | Individual | No | Yes | Yes | Yes | OS |
| Zinn et al27 | 2012 | USA | TCGA | 78 | Yes | GBM | MR | 1 | Manual | NA | Microarray and micro-RNA | Individual | Yes | No | No | Yes | OS |
| Jamshidi et al28 | 2015 | USA | Single institutional | 70 | Yes | CCRCC | CT | 35 | Manual | Yes | Microarray | Both | No | No | No | Yes | OS |
| Colen et al29 | 2014 | USA | TCGA | 99 | No | GBM | MRI | 1 | Manual | Yes | Microarray and micro-RNA | Individual | Yes | No | Yes | Yes | OS |
| Carlson et al30 | 2007 | USA | Single institutional | 71 | No | HGG | MRI | 1 | Manual | NA | Microarray | Individual | No | No | No | Yes | OS |
| Colen et al31 | 2014 | USA | TCGA | 104 | No | GBM | MRI | 30 | Manual | Yes | Microarray | Both | Yes | No | No | Yes | OS |
| Jain et al32 | 2012 | USA | TCGA | 18 | No | GBM | CT | 2 | Manual | Yes | Microarray | Individual | Yes | No | No | No | NA |
| Naeini et al33 | 2013 | USA | Single institutional | 46 | No | GBM | MRI | 3 | Manual | NA | Microarray | Clusters | Yes | No | No | Yes | OS |
| Nicolasjilwan et al34 | 2015 | USA | TCGA | 68 | Yes | GBM | MRI | 30 | Manual | NA | Microarrays, array CGH | Clusters | Yes | No | No | No | NA |
| Pope et al35 | 2012 | USA | Single institutional | 38 | No | GBM | MRI | 1 | Manual | Yes | Microarray | Individual | Yes | No | Yes | Yes | OS |
| Osborne et al36 | 2010 | USA | Single institutional | 20 | No | Breast cancer | PET | 1 | Manual | NA | Microarray | Both | Yes | No | Yes | No | NA |
| Palaskas et al37 | 2011 | USA | Single institutional | 18 | Yes | Breast cancer | PET | 1 | Manual | NA | Microarray, array CGH | Clusters | Yes | Yes | Yes | No | NA |
| Zhu et al38 | 2015 | USA | TCGA and TCIA | 91 | No | Breast cancer | MRI | 38 | Semi-automated | NA | Microarray, array CGH, micro-RNA, somatic mutations | Clusters | Yes | No | No | No | NA |
| Miura et al39 | 2015 | Japan | Single institutional | 77 | No | HCC | MRI | 1 | Manual | Yes | Microarray | Individuals | Yes | No | Yes | Yes | PFS and OS |
| Halle et al40 | 2012 | Norway | Single institutional | 46 | Yes | Cervical cancer | MRI | 1 | Manual | Yes | Microarray | Clusters | Yes | Yes | Yes | Yes | PFS |
CCRCC, clear-cell renal cell carcinoma; CGH, comparative genomic hybridization; GBM, glioblastoma multiforme; HCC, hepatocellular carcinoma; HGG, high-grade glioma; MFS, metastatic-free survival; NA, not available; NSCLC, non-small-cell lung cancer; OS, overall survival; PET, positron emission tomography; PFS, progression-free survival; RFS, recurrence-free survival; TCGA, The Cancer Genome Atlas; TCIA, The Cancer Imaging Archive.
Imaging features and extraction
The number of imaging features extracted range from 1 to 440 with a median of 6. 5 (19%) studies used automatic or semi-automatic imaging feature extraction; 21 (78%) studies used manual feature extraction; and 1 (4%) study used a combination of automatic and manual imaging feature extractions. 19 (70%) studies involved board-certified radiologists in the process of imaging feature extraction. In one study, Aerts et al9 defined the region of interest in one study. 7 studies did not provide any information regarding reader qualification. 6 (22%) studies focused on building an association map between genomic data and imaging features, while the other 21 (78%) studies identified significant imaging features that correlated with genomic data.
We tabulated all the manually extracted and computationally derived imaging features from all radiogenomic studies (Table 2).9,15–40 There was a wide array of imaging features that were extracted by radiologists, depending on the imaging modality used and the type of cancer studied. The most common CT features that were extracted include tumour necrosis and tumour margin. For HCC, enhancement properties on different phases of CT were the commonly studied imaging features.18,20 Internal air bronchogram was a specific feature extracted for NSCLC.15 Most MRI studies focused on GBM and breast cancer. For GBM, three studies used Visually Accessible Rembrandt Images (VASARI), a comprehensive feature set consisting of 24 observations familiar to neuroradiologists to describe the morphology of brain tumours on routine contrast-enhanced MRI.42 The imaging features in VASARI that were most likely to have a significant relationship with genomic data included enhancement characteristics of the brain tumour and its extent of involvement.16,31,34 This relationship held for other studies of GBM which did not utilize VASARI. One study of breast cancer found the location, lymph node and stromal patterns to be significant imaging features with genomic data,21 while another study of HCC focused on the intensity of the tumour in the hepatobiliary-phase MR.39
Table 2.
Manually extracted imaging features from radiogenomic studies
| Study | Modality | Cancer | Significance definition | Manually extracted feature |
|---|---|---|---|---|
| Jamshidi et al28 | CT | CCRCC | Association with gene clusters | Pattern of tumour necrosis, tumour transition zone, tumour–parenchyma interaction, tumour–parenchyma interface |
| Kuo et al18 | CT | HCC | Association with mRNA and gene clusters | Internal arteries, texture heterogeneity, wash-in, washout, necrosis, tumour margin score |
| Segal et al20 | CT | HCC | Association with mRNA | Necrosis, internal septa, texture heterogeneity (arterial and venous phase), tumour margin score (minimum and maximum), enhancement pattern, internal arteries (density and necrosis edge), hypodense halo, washout, internal arteries (density), tumour–liver difference, corrected imaging area, necrosis density, capsule, wash-in, infiltration, tumour–liver difference, attenuation/heterogeneity score |
| Carlson et al24 | CT | HGG | Association with mRNA | Oedema |
| Gevaert et al15 | CT | NSCLC | Association with mRNA | Internal air bronchogram, complex shape, vascular convergence, lobulated margin, oval shape, irregular margin, pleural retraction, solid density, entering airway, right upper lobe apical location |
| Aerts et al9 | CT | NSCLC | Association with mRNA | None |
| Jain et al32 | CT | GBM | Association with mRNA | None |
| Osborne et al36 | PET | Breast cancer | Association with molecular subtypes | None |
| Palaskas et al37 | PET | Breast cancer | Association with Myc-overexpression | None |
| Nair et al19 | PET | NSCLC | Association with mRNA and gene clusters | None |
| Yamamoto et al21 | MRI | Breast cancer | Association with gene clusters | Enhancement pattern, size, shape, margin, location, T2 tumour signal interface between tissue and tumour, satellite lesions, multifocal disease, lymph node involvement, un-coordinated growth, stromal alterations |
| Yamamoto et al22 | MRI | Breast cancer | Association with IncRNA | None |
| Zhu et al38 | MRI | Breast cancer | Association with gene clusters | None |
| Halle et al40 | MRI | Cervical cancer | Association with gene clusters | None |
| Gevaert et al16 | MRI | GBM | Association with molecular subtypes | VASARI (deep white matter location, enhancement, enhancing margin characteristics, diffusion characteristics) |
| Colen et al29 | MRI | GBM | Association with mRNA | VASARI (enhancing tumour across midline/corpus callosum, deep white matter tract involvement, ependymal involvement) |
| Nicolasjilwan et al34 | MRI | GBM | Association with mRNA and CNV | VASARI (proportion of tumour contrast enhancement) |
| Jamshidi et al17 | MRI | GBM | Association with gene clusters | Contrast enhancement, necrosis, contrast-to-necrosis ratio, infiltrative vs oedematous T2abnormality, mass effect, subventricular zone involvement |
| Barajas et al24 | MRI | GBM | Association with mRNA and gene clusters | Lesion location, presence of contrast enhancement, central necrosis, degree of T2 oedema, mass effect |
| Diehn et al25 | MRI | GBM | Association with gene clusters | Contrast enhancement, necrosis, mass effect, pattern of T2oedema (infiltrative/oedematous), cortical involvement, SVZ involvement, C : N ratio, contrast/T2 ratio, degree of T2 oedema, T2 heterogeneity |
| Pope et al26 | MRI | GBM | Association with mRNA | Enhancement extent |
| Zinn et al23 | MRI | GBM | Association with mRNA and micro-RNA | None |
| Zinn et al27 | MRI | GBM | Association with mRNA and micro-RNA | None |
| Colen et al31 | MRI | GBM | Association with mRNA and micro-RNA | None |
| Naeini et al33 | MRI | GBM | Association with molecular subtypes | None |
| Pope et al35 | MRI | GBM | Association with mRNA | None |
| Miura et al39 | MRI | HCC | Association with mRNA | Intensity on hepatobiliary phase |
CCRCC, clear-cell renal cell carcinoma; C:N ratio, contrast:necrosis ratio; CNV, copy number variation; GBM, glioblastoma multiforme; HCC, hepatocellular carcinoma; HGG, high-grade glioma; mRNA, messenger RNA; NSCLC, non-small-cell lung cancer; PET, positron emission tomography; SVZ, subventricular zone; VASARI, Visually Accessible Rembrandt Images.
Bold type means significant relationship of imaging feature with genomic data.
There was relative uniformity for the computationally derived imaging features (Table 3).9,15–40 For CT, tumour intensity, texture and shape were the most commonly extracted features, especially for NSCLC. PET studies were most likely to focus on the standardized uptake value, regardless of tumour type. Cerebral blood volume is the most commonly derived feature on either perfusion CT or MRI.24,32 For MRI studies on GBM, the most common feature extracted to correlate with genomic data was the volume of the tumour.1,16,23,27,33 Several studies divided the tumour into regions with specific imaging characteristics such as enhancing, necrotic, oedema etc. and correlated the volume of each region with the patient's genomic data.1,16,23,33 For MRI studies on breast cancer, tumour volume was still a commonly extracted imaging feature. Otherwise, studies have focused on signal strength on specific sequences at different time points and contrast kinetic pattern.21,22,38
Table 3.
Computationally extracted imaging features from radiogenomics studies
| Study | Modality | Cancer | Computer-extracted features |
|---|---|---|---|
| Jain et al32 | CT | GBM | CBV, PS |
| Aerts et al9 | CT | NSCLC | Tumour intensity, shape, texture, wavelet features |
| Jamshidi et al28 | CT | CCRCC | None |
| Kuo et al18 | CT | HCC | None |
| Segal et al20 | CT | HCC | None |
| Carlson et al30 | CT | HGG | None |
| Osburne et al36 | PET | Breast cancer | SUV |
| Palaskas et al37 | PET | Breast cancer | SUV |
| Nair et al19 | PET | NSCLC | SUV intensity metrics, SUV distribution metrics, SUV spatial metrics |
| Gevaert et al15 | PET/CT | NSCLC | Histogram, texture, edge sharpness, edge shape, ROI size, SUV |
| Yamamoto et al21 | MRI | Breast cancer | T1 intrinsic signal,T2intrinsic signal strength, contrast kinetic pattern, median peak signal strength at different times, nadir signal strength at different times |
| Yamamoto et al22 | MRI | Breast cancer | Largest tumour volume, tumour roundness, entropy, skewness, kurtosis, GLCM contrast, GLCM homogeneity, GLCM energy, Hu’s seven moment invariants, average of wash-in slope, average of washout slope, plateau fraction, persistent fraction, heterogeneity of time intensity, ERF |
| Zhu et al38 | MRI | Breast cancer | Size phenotypes, shape phenotypes, morphological phenotypes, enhancement texture phenotypes, kinetic curve assessment, enhancement-variance kinetics |
| Barajas et al24 | MRI | GBM | CBV, PH, PSR, ADC |
| Gevaert et al16 | MRI | GBM | Necrotic, enhancing, oedema ROIs |
| Zinn et al23 | MRI | GBM | FLAIR volume |
| Zinn et al27 | MRI | GBM | Volume |
| Colen et al29 | MRI | GBM | Necrosis volume |
| Naeini et al33 | MRI | GBM | Contrast-enhancing volume, necrotic volume, contrast enhancement+necrotic volume, T2hyperintense volume, the ratio of oedema/(necrosis+contrast) |
| Pope et al35 | MRI | GBM | ADC |
| Jamshidi et al17 | MRI | GBM | None |
| Diehn et al25 | MRI | GBM | None |
| Pope et al26 | MRI | GBM | None |
| Colen et al31 | MRI | GBM | None |
| Nicolasjilwan et al34 | MRI | GBM | None |
| Halle et al40 | MRI | Cervical cancer | Abrix (enhancement-variance kinetics) |
| Miura et al39 | MRI | HCC | None |
ADC, apparent diffusion coefficient; C : N ratio, contrast:necrosis ratio; CBV, cerebral blood volume; CCRCC, clear-cell renal cell carcinoma; CNV, copy number variation; EFR, enhancing rim fraction; FLAIR, fluid-attenuated inversion recovery; GBM, glioblastoma multiforme; GLCM, gray-level concurrence matrix; HCC, hepatocellular carcinoma; HGG, high-grade glioma; NSCLC, non-small-cell lung cancer; PET, positron emission tomography; PH, peak height; PSR, percentage of signal intensity recovery; PS, permeability surface; ROI, region of interest; SUV, standardized uptake value; SVZ, subventricular zone.
Bold type means significant relationship of imaging feature with genomic data.
Genomic data
26 (96%) studies used data from RNA or complementary DNA microarray. Only one (4%) study used data from RNA sequencing.22 Among these 26 studies that used microarray data to correlate with imaging, 4 studies included micro-RNA,1,23,27,38 5 studies copy number variation,16,17,34,37,38 1 DNA methylation16 and 1 somatic mutation.38 11 (41%) studies grouped gene expression data into gene clusters or modules to associate with imaging features;9,15–18,20,33,34,37,38,40 9 (33%) studies directly associated individual elements with imaging features;1,22,23,26,27,30,32,35,39 and 7 (26%) studies used both approaches.19,21,24,25,28,31,36 23 (85%) studies performed pathway analysis, either in the initial clustering of genes to associate with imaging features (n = 10)9,15,20,21,24,33,34,37,38,40 or in the final analysis of significant genomic markers (n = 12).1,16,17,19,22,23,27,31,32,35,36,39 One study performed pathway analysis for both purposes.25
Outcome, histology and wet lab validation
18 (67%) studies included outcome data.1,9,15,16,19,20,22,23,25–28,30,31,33,35,39,40 12 studies focused on overall survival (OS);1,9,19,20,25–28,30,31,33,35 4 studies included both OS and progression-free survival;15,16,23,39 1 study focused on progression-free survival in cervical cancer;40 and 1 study used metastatic-free survival in breast cancer.22 Three studies stated overall follow-up time.22,26,30 12 (44%) studies correlated with histological data.1,9,18,20,22,24,26,35–37,39,40 The histological parameters that were evaluated ranged from tumour type and tumour stage to specific immunological expression of tumour markers such as oestrogen receptor (ER), progesterone receptor (PR) and HER2 in breast cancer.
Five (19%) studies attempted to verify significant associations that were identified through lab-based techniques.22,25,26,37,40 Two studies performed quantitative polymerase chain reaction (PCR) to verify the significant difference in gene expression among imaging phenotypes through association studies.22,26 Two studies performed gene expression analysis in corresponding cancer cell lines.37,40 One study performed immunological staining of epidermal growth factor receptor (EGFR) and found differentially expressed EGFR among different imaging phenotypes.25
Semi-radiogenomic studies
38 semi-radiogenomic studies were identified (Table 4).28,43–79 Studies were published between 2005 and 2015. All studies were retrospective in design. Five studies used data from TCGA/TCIA;45,46,48,49,57 one study used multi-institutional data;50 and one study combined both institutional data and data from TCGA/TCIA.44 The number of patients ranged from 25 to 539 with a median of 75. Only two studies had a validation data set. The type and distribution of cancers in these studies were similar to those for radiogenomic studies, except for two studies that focused on low-grade glioma47,58 and one study that focused on diffuse large B-cell lymphoma.59 The imaging modalities used included CT (n = 9), MRI (n = 21, including five perfusion) and PET (n = 6). Two studies used both CT and MR.46,53 The number of imaging features extracted ranged from 1 to 120 with a median of 5. 11 (29%) studies used semi-automatic image feature extraction.45,49,55,58,61–63,72,73,75,79 All studies (except eight studies which did not provide this information) had radiologist participation. 29 (76%) studies focused on individual genes;28,43,44,46, 47,48,50–53, 54,56,57,59,60,64,65, 66–71,74,76–78 7 (18%) studies used gene clusters or subsets derived from primary genomic data;28,45,50,57,59,61,63 and 2 (5%) studies used a combination of both.48,55 14 (37%) studies included outcome data.28,47,48,50,51,55,57,59,61,63,69–71,79 Only four studies correlated with histology.50,60,71,74 None of the study verified their results via wet-lab techniques.
Table 4.
Semi-radiogenomic studies published in the literature
| Study | Year | Country | Cancer | Imaging modality | Number of features | Method of extraction | Radiologist involvement | Individual genes vs gene clusters | Wet-lab validation | Histology | Outcome | Clinical outcomes | Data source | Number | Validation set |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Halpenny et al43 | 2014 | USA | Lung adenocarcinomas | CT | 14 | Manual | Yes | Individual genes | No | No | No | NA | Single | 30 | No |
| Karlo et al44 | 2014 | USA | CCRCC | CT | 10 | Manual | Yes | Individual genes | No | No | No | NA | Single and TCGA | 233 | No |
| Mazurowski et al45 | 2014 | USA | Breast cancer | MRI | 23 | Semi-automatic | Yes | Gene clusters | No | No | No | NA | TCGA | 48 | No |
| Shinagare et al46 | 2015 | USA | CCRCC | CT, MRI | 6 | Manual | Yes | Individual genes | No | No | No | NA | TCGA | 103 | No |
| Wang et al47 | 2015 | China | LGG | MRI | 1 | Manual | Yes | Individual genes | No | No | Yes | PFS and OS | Single | 146 | No |
| Gutman et al48 | 2013 | USA | GBM | MRI | 26 | Manual | Yes | Both | No | No | Yes | OS | TCGA | 75 | No |
| Gutman et al49 | 2015 | USA | GBM | MRI | 11 | Semi-automatic | NA | Individual genes | No | No | No | NA | TCGA | 75 | No |
| Banerjee et al50 | 2015 | USA | HCC | CT | 3 | Manual | Yes | Gene clusters | No | Yes | Yes | RFS and OS | Multi-institutional | 157 | No |
| Jamshidi et al28 | 2015 | USA | CCRCC | CT | 28 | Manual | Yes | Gene clusters | No | No | Yes | OS | Single | 70 | Yes |
| Carrillo et al51 | 2012 | USA | GBM | MRI | 9 | Manual | Yes | Individual genes | No | No | Yes | OS | Single | 202 | No |
| Drabycz et al52 | 2010 | Canada | GBM | MRI | 4 | Manual | Yes | Individual genes | No | No | No | NA | Single | 103 | No |
| Moon et al53 | 2012 | Korea | HGG | CT and MRI | 10 | Manual | Yes | Individual genes | No | No | No | NA | Single | 32 | No |
| Aghi et al54 | 2005 | USA | GBM | MRI | 4 | Manual | NA | Individual genes | No | No | No | NA | Single | 75 | No |
| Ellingson et al55 | 2013 | USA | GBM | MRI | 2 | Semi-automatic | NA | Both | No | No | Yes | PFS and OS | Single | 507 | No |
| Gupta et al56 | 2015 | USA | GBM | MRI (physiologic) | 3 | Manual | Yes | Individual genes | No | No | No | NA | Single | 106 | No |
| Jain et al57 | 2013 | USA | GBM | MRI (physiologic) | 3 | Manual | Yes | Gene clusters | No | No | Yes | OS | TCGA | 98 | No |
| Kickingereder et al58 | 2015 | Germany | LGG+anaplastic | MRI (physiologic) | 1 | Semi-automatic | Yes | Individual genes | No | No | No | NA | Single | 73 | No |
| Lanic et al59 | 2012 | France | DLBCL | PET | 1 | Manual | Yes | Gene clusters | No | No | Yes | PFS and OS | Single | 45 | No |
| Miles et al60 | 2014 | England | CRC | PET/CT | 3 | Manual | Yes | Individual genes | No | Yes | No | NA | Single | 33 | No |
| Ashraf et al61 | 2014 | USA | Breast cancer | MRI | 31 | Semi-automatic | Yes | Gene clusters | No | No | Yes | PFS | Single | 56 | No |
| Li et al62 | 2014 | USA | Breast cancer | MRI | 45 | Semi-automatic | NA | Individual genes | No | No | No | NA | Single | 103 | No |
| Macyszyn63 | 2015 | USA | GBM | MRI | 120 | Semi-automatic | No | Gene clusters | No | No | Yes | OS | Single | 105 | Yes |
| Rizzo et al64 | 2016 | Italy | NSCLC | CT | 19 | Manual | Yes | Individual genes | No | No | No | NA | Single | 285 | No |
| Izuishi et al65 | 2012 | Japan | CRC | PET | 1 | Manual | NA | Individual genes | No | No | No | NA | Single | 37 | No |
| Lee et al66 | 2016 | Korea | CRC | PET | 4 | Manual | Yes | Individual genes | No | No | No | NA | Single | 179 | No |
| Kawada et al67 | 2012 | Japan | CRC | PET | 2 | Manual | Yes | Individual genes | No | No | No | NA | Single | 51 | No |
| Tykocinski et al68 | 2012 | USA | GBS | MRI (physiologic) | 1 | Manual | NA | Individual genes | No | No | No | NA | Single | 132 | No |
| Kong et al69 | 2011 | Korea | GBS | MRI (physiologic) | 1 | Manual | Yes | Individual genes | No | No | Yes | PFS and OS | Single | 73 | No |
| Romano et al70 | 2013 | Italy | GBS | MRI | 1 | Manual | Yes | Individual genes | No | No | Yes | PFS and OS | Single | 47 | No |
| Sunwoo et al71 | 2013 | Korea | GBS | MRI | 1 | Manual | NA | Individual genes | No | Yes | Yes | PFS | Single | 65 | No |
| Ahn et al72 | 2014 | Korea | GBS | MRI | 9 | Semi-automatic | Yes | Individual genes | No | No | No | NA | Single | 43 | No |
| Sutton et al73 | 2015 | USA | Breast cancer | MRI | 14 | Semi-automatic | Yes | Individual genes | No | No | No | NA | Single | 95 | No |
| Kitao et al74 | 2010 | Japan | HCC | MRI | 1 | Manual | Yes | Individual genes | No | Yes | No | NA | Single | 38 | No |
| Lee et al75 | 2013 | Korea | NSCLC | CT | 9 | Semi-automatic | Yes | Individual genes | No | No | No | NA | Single | 153 | No |
| Glynn et al76 | 2010 | Korea | NSCLC | CT | 5 | Manual | Yes | Individual genes | No | No | No | NA | Single | 64 | No |
| Plodkowski et al77 | 2015 | USA | NSCLC | CT | 12 | Manual | Yes | Individual genes | No | No | No | NA | Single | 73 | No |
| Ozkan et al78 | 2015 | USA | NSCLC | CT | 5 | Manual | Yes | Individual genes | No | No | No | NA | Single | 25 | No |
| Yoon et al79 | 2015 | USA | NSCLC | CT and PET | 51 | Semi-automatic | NA | Individual genes | No | No | Yes | PFS and OS | Single | 539 | No |
CCRCC, clear-cell renal cell carcinoma; CGH, comparative genomic hybridization; GBM, glioblastoma multiforme; HCC, hepatocellular carcinoma; HGG, high-grade glioma; LGG, low-grade glioma; NA, not available; NSCLC, non-small-cell lung cancer; OS, overall survival; PET, positron emission tomography; RFS, recurrence-free survival; TCGA, The Cancer Genome Atlas; TCIA, The Cancer Imaging Archive.
LIMITATIONS AND PROMISES
Radiogenomics is an emerging field that links tumour genotype with imaging phenotypes. Since 2007, a number of studies have been published on radiogenomic characterization of certain cancers.9,15–40 These studies pioneered the feasibility of this approach and paved the way for future developments in the field. However, we noticed a number of issues from our analysis.
Study design
Only a handful of studies can be considered as “real” radiogenomics studies in the sense that they used whole genome data. The dimensionality of imaging, despite being rapidly increasing over time, is still orders of magnitude lower than that of whole-genome sequencing or molecular profiling.1 One of the limitations of the current radiogenomic research is the need to reduce the dimensionality of genomic data to match that of imaging. A common approach in analysing these data is to group individual genetic elements into gene modules before performing association analysis with imaging features. Given the tenuous imaging-to-genomics and genomics-to-outcome relationships, such an approach may further undermine the potential of imaging to predict patient outcomes, one of the primary goals of radiogenomic analyses.3
Standardization in imaging analysis
Traditionally, medical imaging has been a subjective or qualitative art. Recent advances in medical imaging acquisition and analysis allow the high-throughput extraction of specific imaging features to quantify the differences that oncologic tissues exhibit in medical imaging.80 Aerts et al9 evaluated a total of 440 CT features of the lung and head and neck cancers on the basis of four imaging characteristics:1 tumour intensity,2 shape,3 texture and4 wavelet features. These imaging features were extracted by an automated algorithm written in MATLAB® (MathWorks®, Natick, MA). Using a predefined vocabulary and analytical algorithm, Gevaert et al15 extracted 153 computational image features, 26 semantic image features and standardized uptake value from PET to characterize NSCLC in 26 patients. Grimm et al81 used computer vision algorithms to extract 56 imaging features from breast cancers including morphologic, texture and dynamic features. However, automatic extraction of quantitative imaging features, such as tumour morphology, texture and contrast kinetics, is limited to homogeneous tumours. For example, in the study of GBM, the most commonly computationally derived imaging feature was tumour volume. Other features were not routinely evaluated, likely because GBM commonly demonstrated significant intratumour heterogeneity.73,82 To overcome this limitation, Gevaert et al16 segmented the tumour into enhancing, oedematous and necrotic regions. Quantitative imaging features were then extracted from each region and correlated with genomic data.
Unfortunately, automatic imaging feature extraction was implemented in only a minority of studies. In the majority of studies, imaging features were manually assessed by radiologists. Manual analysis of images has certain disadvantages. In particular, manual extraction is subject to interobserver variability, random errors during manual contour tracing for mass volume etc. Furthermore, it is labour intensive. A future direction in the field of radiogenomics is the implementation of quantitative image analysis tools to allow comprehensive image feature extraction in a fast and reproducible manner. In addition, creating a lexicon and ontology of reproducible semantics and computed image features will permit images to be mineable in a manner similar to genomic data.
Segmentation conundrums
A variety of automated or semi-automated image segmentation methods are available. Some are based on the analysis of (often multiparametric) imaging signals in an unsupervised83,84 or supervised way.85 Oftentimes, anatomical statistical priors encode normal anatomy and hence find tumours as deviations from it.86 Segmentation methods that explicitly incorporate biophysical models of tumour growth, in a way to facilitate imaging-based segmentation, have also been proposed.87,88 Although validation of these methods is a very challenging and effort-demanding task, some international efforts for creating validation platforms have started to emerge. A prime example is the Brain Tumor Segmentation challenge organized annually, which uses TCIA and other public data sets, along with ground truth, to evaluate a variety of algorithms.41
Segmentation methods are usually a first step prior to extracting imaging features, which are used in conjunction to build biomarkers of gene expression. Commonly used features include volumetrics of enhancing and non-enhancing parts, and of surrounding oedema, textural properties of the tumour, which reflect the spatial heterogeneity of tumours, shape properties of tumour boundaries, which relate to infiltrative/aggressive tumour phenotypes, multiparametric histograms of various imaging measures, which relate to cell density, perfusion dynamics, gadolinium enhancement and water content, and various other properties. Such features have been found to jointly form good predictors of tumour molecular characteristics, especially when integrated via machine learning and other multiparametric analysis models.15,63,89
Functional imaging
Currently, automated extraction is limited to CT, which is the most widely used imaging modality in oncology with the ability to assess tissue density. Emerging functional and molecular imaging methods, such as PET/CT and dynamic contrast-enhanced (DCE) or diffusion-weighted MRI, have the potential to assess the in situ tumour's metabolic and proliferative activity with higher accuracy than traditional imaging methods.1 In the only prospective radiogenomic study published to date, Barajas et al correlated physiologic MR parameters with RNA expression patterns in enhancing vs peritumoral non-enhancing GBM biopsy samples.24 The authors found that T2* dynamic susceptibility-weighted contrast-enhanced perfusion-weighted and diffusion-weighted imaging measurements were significantly different between biopsy regions and correlated with GBM histopathological features of aggressiveness. In addition, the upregulated genes were associated with similar cellular malignant biologic processes that were observed to correlate with physiologic-based MRI measurements. In another study of 18 patients with GBM, Jain et al32 correlated CT perfusion parameters with genes that are related to angiogenesis regulation. Of the 92 angiogenesis-associated genes, 19 genes had significant correlation with the permeability surface area product and 9 genes had significant correlation with the cerebral blood volume. Unfortunately, both of these studies were hampered by the extremely small sample sizes. In the future, studies with a larger cohort size and variety of cancer types are needed to uncover the potential correlation between functional and molecular imaging parameters and genomic data.
Sample size
Studies included in our review are limited by a small sample size. In addition, these studies often lack complete characterization of the patients and suffer from poor integration of individual data sets. In fact, one of the greatest limitations that are often cited for these studies is the difficulty in obtaining original cohorts of patients with both appropriate imaging studies and adequate tissue samples for genomic analysis.3 However, it is important to keep in mind that routine imaging data are readily available in large quantities, many of which have corresponding archival tumour tissue available for various molecular analyses. These cases can be collected retrospectively and studied by investigators working at large clinical institutions. Furthermore, the cost of next-generation sequencing and other high-throughput molecular techniques has reduced to a fraction of what it was before.32 These assays can generate large amount of data that can potentially be harvested.
Molecular genetic analysis
As demonstrated by our review, most studies performed to date are limited to microarray data, since it is the earliest type of genomic analysis available to allow assessment of the differential changes in genome-wide gene expression levels. Three studies used micro-RNAs.1,23,27 Micro-RNAs are non-protein-coding small RNAs that serve as negative gene regulators by binding to a specific sequence in the 3′ UTR of a target gene.90 A single micro-RNA can potentially target hundreds of genes.90 Therefore, micro-RNAs were found to have important roles as tumour suppressors and oncogenes, as well as regulators of various cancer-specific cellular features, such as proliferation, invasion and metastasis.91,92 In one of the studies on radiogenomics of GBM, Zinn et al.23 incorporated micro-RNA data into the analysis of microarray association with imaging features. By correlating quantitative MRI data with microarray data, the authors found periostin (POSTN) as the top upregulated gene. Through additional micro-RNA analysis, they identified miR-219 as the top downregulated micro-RNA. miR-219 is known to have a potential binding site in the 3′ untranslated region (UTR) of the POSTN gene. This inverse correlation between POSTN and miR-219 suggests a potential role of miR-219 in downregulating POSTN in GBM mesenchymal transition and cellular invasion. More importantly, this signature can be non-invasively detected by routine MRI. In another study by Yamamoto et al,22 the authors used next-generation RNA sequencing to correlate the expression of long non-coding RNA with MRI phenotypes and the presence of early metastasis in breast cancer. Long non-coding RNAs represent an important class of regulatory RNAs that are longer than 200 nucleotides,93 exhibit exquisite cell and tissue specificity and are critical in maintaining tissue structure and organization.93,94 The above examples illustrate the importance of including multiple genomic data sets to derive maximum benefit from radiogenomic association maps. Using new genomic technologies such as next-generation DNA sequencing, single nucleotide polymorphism genotyping, chromatin immunoprecipitation and RNA sequencing into the fold has the potential to open up new frontiers for radiogenomic research.95 Moreover, understanding molecular pathways that result in these radiogenomically identified imaging features should be one of the primary goals of radiogenomic analyses, as it is a necessary path to demonstrate radiogenomics' clinical significance.
Study validation
Validation with prospectively collected independent cohorts is the most robust approach and gold standard for verifying an identified statistical association.96 However, in our revealed literature, validation data set was used in only eight studies (30%). The decision to not proceed with validation data set in other studies may have stemmed from the data availability issue, as previously discussed. Genomic data are the hardest to obtain because they may require fresh tissue specimens. Gevaert et al15 demonstrated a radiogenomic strategy to rapidly identify prognostically significant image biomarkers. By using specific genomic characteristics as intermediate, they linked imaging data in the first data set to survival in the second data set. Since long-term clinical follow-up may not be feasible in patients with both genomic and imaging data, the authors argued that their approach was able to leverage imperfect data sets to draw new conclusions. However, this approach requires the existence of large gene expression data sets where survival outcomes are available. TCGA is a publicly available resource that contains multidimensional genomic and clinical data set for multiple types of adult cancers.97 TCIA is another publicly available resource that contains imaging corresponding to these patients in TCGA.98 However, the usefulness of radiological data that are contained in the TCIA is limited by the lack of image sample registration (i.e. gene expression profiles cannot be matched to a specific location on imaging). Successful attempts have been made to account for these differences by rigid alignment and registration with proper segmentation.23 As the imaging acquisition protocols become increasingly standardized and outcome data become more mature, public databases such as TCGA and TCIA will not only serve as powerful validation tools, but more importantly, as the foundation for further radiogenomic discoveries.5
Histopathological correlation
Radiologic studies can be correlated with whole-genome mapping, histopathology and specific genes. Most of the studies in our review (15/27, 56%) did not perform histopathology association with imaging data. In one of the studies, Pope et al26 found that incomplete enhancing imaging phenotype was associated with increased levels of oligodendroglioma marker oligodendrocyte lineage transcription factor 2 and achaete-scute complex-like 1 than completely enhancing the imaging phenotype. The authors confirmed this finding with histopathology, which showed a higher percentage of substantial oligodenroglioma histologic component in the incomplete enhancing group vs the complete enhancing group. In another study by Colen et al,1 the authors found that patients with GBM with low volumes of necrosis had a high prevalence of X-linked genes, while those with high volumes of necrosis had a high prevalence of Y-linked genes. Subsequently, the authors showed that in contrast to male patients, female patients with low volumes of necrosis on MRI had a significant survival advantage. This result was confirmed by a separate validation data set of 368 patients, where the authors were able to demonstrate that in female patients, cell death on histology was associated with a survival advantage. In another study by Pope et al, the authors correlated differential gene expression in GBM with apparent diffusion coefficient (ADC) histogram. They found that 6 of the 13 genes with increased expression in ADC tumours were isoforms of collagen-binding proteins.35 In order to confirm this result, the authors performed immunohistochemistry in both high- and low-ADC tumours to compare the expression of decorin and collagen one, three and six isoforms. There was no significant correlation between ADC values and collagen immunoreactivity scores. However, multiple patterns of immunoreactivity, including perivascular, interstitial and cytoplastmic patterns, were associated with higher ADC.35
Result verification
A limited number of studies (5/27, 19%) used wet-lab techniques to verify significant findings from their radiogenomic analyses.22,25,26,37,40 For example, Diehn et al25 confirmed EGFR overexpression among imaging phenotypes of GBM with immunohistochemistry. Halle et al40 found that in cervical cancer, the most differentially expressed gene sets between tumours with high and low ABrix (ABrix is the amplitude, Kep the transfer rate from tissue to plasma) on DCE-MRI were hypoxia-related features. To verify this result, the authors subjected three cervical cancer cell lines to hypoxia and performed gene expression profiling between the normoxia- and hypoxia-treated cell lines. The authors found that HIF1α protein was upregulated in all three hypoxia-treated cell lines. On the other hand, only minor changes of HIF1α protein regulation were observed in the control. The protein expression of HIF1α was further evaluated by immunohistochemistry and correlated with DCE-MRI in additional 32 patients. These results demonstrated that tumours with low ABrix was significantly associated with higher HIF1α expression than those with high ABrix. Verifying histopathological and molecular correlations of radiogenomic data significantly improves the quality of the radiogenomic study. Most importantly, multidimensional evaluation of biologic data allows one to gain causative insight into the underlying significance of initially discovered imaging feature—genomic correlation.
Clinical translation
Given that radiogenomics is still at its infancy, the full potential of clinical translation is yet to be realized. Nevertheless, several studies have demonstrated early promise. One example is in the research of HCC. In HCC, microscopic venous invasion (MVI) is a well-established sign of poor prognosis. However, it is extremely difficult to predict MVI using conventional imaging methods such as MRI.99,100 Currently, MVI can only be reliably diagnosed by the histology of the explanted tissue when its clinical utility is marginal. In 2002, Chen et al101 identified a 91 gene expression signature via microarray analysis that had significant correlation with the presence of vascular invasion. In 2007, Segal et al20 found that these 91 genes in the “venous invasion signature” were associated with two predominant imaging traits on CT—the presence of “internal arteries” and absence of “hypodense halos”. In a study of 157 patients with HCC who underwent surgical resection or liver transplant, Banerjee et al again demonstrated that these two imaging biomarkers, along with “tumour–liver difference”, were able to predict histological MVI with high precision. In addition, this radiogenomic biomarker consisting of these three features was associated with early disease recurrence and poor OS.50 Therefore, this marker can be extremely useful in identifying patients who are less likely to benefit from surgical treatment or liver transplant. The example above illustrates the significant impact radiogenomic analysis can have on patient management.
Radiogenomics can have significant impact on routine radiology practice. Similar to histopathology, the goal of radiogenomics is to provide information on the tumour that can be used to guide treatment and predict survival. Ideally, all of this can be achieved non-invasively with routine imaging studies. For example, in patients with CCRCC, a prognostic multigene signature, termed radiogenomic risk score, was constructed and shown to predict disease-specific survival, independent of disease stage, disease grade and performance status.102 The radiogenomic risk score consists of four CT imaging features: the pattern of tumour necrosis, tumour transition zone, tumour–parenchyma interaction and tumour–parenchyma interface. If further validated, such a radiogenomic signature can potentially be used in a way that coronary calcium score is used to improve risk stratification for future cardiovascular events.103 If the radiogenomic risk score incorporates genomic data in addition to radiologic data, the radiologist can issue an addendum to the report once genomic data from pathology become available. This enhances the radiologist's role in patient care by providing the ordering physician important information beyond what is typically reported for CCRCC (e.g. lymph node involvement, renal vein invasion etc.). Furthermore, radiologists are likely to gain a crucial role in clinical trials that use such a radiogenomic signature to divide patients into different risk groups.
FUTURE DIRECTIONS
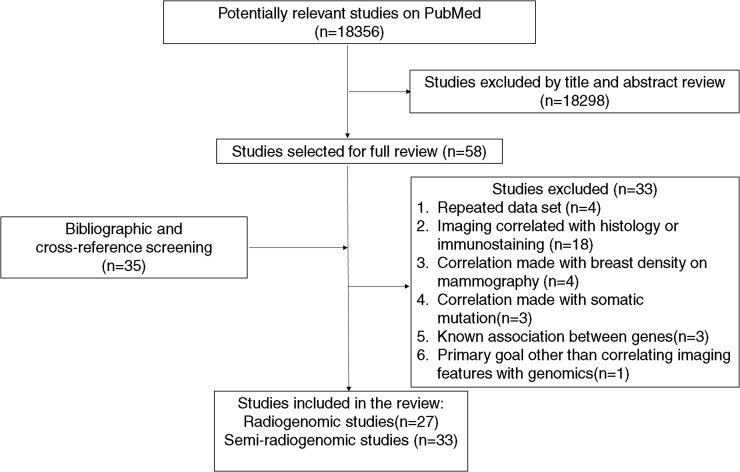
The emerging field of radiogenomics has shown the potential to provide additional insights into tumour biology based on imaging data. Current studies are limited to six types of common cancers: glioma, NSCLC, HCC, breast cancer and cervical cancer. Extension of existing research methods to other tumour types will likely uncover additional associations between molecular properties and imaging characteristics. An ideal design for a radiogenomic study is illustrated in Figure 1. Future studies should strive to incorporate as many elements shown as possible. Once a link between an imaging phenotype and a molecular signature is uncovered, imaging studies of previously treated patients (such as those on clinical trials) can be re-examined to assess the clinical significance of this new link. In the future, gene expression profiling by non-invasive imaging may supplement histologic examination for cancer diagnosis and prognosis (Figure 2).
Figure 1.
Literature search of published studies on radiogenomics.
Figure 2.
Ideal design for a radiogenomic study. CGH, comparative genomic hybridization; CHIP, chromatin immunoprecipitation; OS, overall survival; PET, positron emission tomography; PFS, progression-free survival; SNP, single nucleotide polymorphism.
Another opportunity in radiogenomics is in identifying imaging features that predict region-specific gene expression signatures within the tumour in the proper anatomic context of the patient. Intratumour heterogeneity, in addition to intertumour heterogeneity, has been increasingly recognized as the source of cancer's development of resistance to chemotherapy after initial response.104 Several studies have shown the existence of genomic differences between different regions of the same tumours and correlated them with imaging findings.24,105,106 Further development will require imaging modalities with high resolution for proper spatial registration.107 Targeted tissue specimens from a radiographically diverse region can be studied on a per tumour basis, per patient basis or on a population basis, to allow for additional levels of multiple hypothesis testing. In the near future, it may be possible to detect intertumoural differences in treatment response at the imaging level, thereby guiding personalized and tumour-specific treatment. While public repositories, such as those supported by TCGA and TCIA, continue to grow, it is important to procure, develop and evaluate additional data sets to ensure the depth and breadth of the sample population in each study.5
Given the non-invasive nature of medical imaging and its wide use in clinical practice, radiogenomics has the potential to impact on the treatment and prognosis of a wide range of human cancers. Identification of imaging phenotypes that are associated with distinct molecular phenotypes will help advance individualized patient care.
Contributor Information
Harrison X Bai, Email: hxbai@hotmail.com.
Ashley M Lee, Email: ashley.lee@hotmail.com.
Li Yang, Email: yangli762@gmail.com.
Paul Zhang, Email: Paul.Zhang2@uphs.upenn.edu.
Christos Davatzikos, Email: chris@usp.edu.
John M Maris, Email: JohnMaris357@sina.com.
Sharon J Diskin, Email: sharondiskin17@sina.com.
REFERENCES
- 1.Colen RR, Wang J, Singh SK, Gutman DA, Zinn PO. Glioblastoma: imaging genomic mapping reveals sex-specific oncogenic associations of cell death. Radiology 2015; 275: 215–27. doi: 10.1148/radiol.14141800 [DOI] [PubMed] [Google Scholar]
- 2.Kuo MD, Jamshidi N. Behind the numbers: decoding molecular phenotypes with radiogenomics–guiding principles and technical considerations. Radiology 2014; 270: 320–5. doi: 10.1148/radiol.13132195 [DOI] [PubMed] [Google Scholar]
- 3.Mazurowski MA. Radiogenomics: what it is and why it is important. J Am Coll Radiol 2015; 12: 862–6. doi: 10.1016/j.jacr.2015.04.019 [DOI] [PubMed] [Google Scholar]
- 4.Guo Z, Shu Y, Zhou H, Zhang W, Wang H. Radiogenomics helps to achieve personalized therapy by evaluating patient responses to radiation treatment. Carcinogenesis 2015; 36: 307–17. doi: 10.1093/carcin/bgv007 [DOI] [PubMed] [Google Scholar]
- 5.Jaffe CC. Imaging and genomics: is there a synergy? Radiology 2012; 264: 329–31. doi: 10.1148/radiol.12120871 [DOI] [PubMed] [Google Scholar]
- 6.Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol 2009; 70: 232–41. doi: 10.1016/j.ejrad.2009.01.050 [DOI] [PubMed] [Google Scholar]
- 7.Ding L, Wendl MC, Koboldt DC, Mardis ER. Analysis of next-generation genomic data in cancer: accomplishments and challenges. Hum Mol Genet 2010; 19: R188–96. doi: 10.1093/hmg/ddq391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomaszewski JJ, Uzzo RG, Smaldone MC. Heterogeneity and renal mass biopsy: a review of its role and reliability. Cancer Biol Med 2014; 11: 162–72. doi: 10.7497/j.issn.2095-3941.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014; 5: 4006. doi: 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang H, Lee HY, Lee KS, Kim JH. Imaging-based tumor treatment response evaluation: review of conventional, new, and emerging concepts. Korean J Radiol 2012; 13: 371–90. doi: 10.3348/kjr.2012.13.4.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frangioni JV. New technologies for human cancer imaging. J Clin Oncol 2008; 26: 4012–21. doi: 10.1200/JCO.2007.14.3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berman DM, Bosenberg MW, Orwant RL, Thurberg BL, Draetta GF, Fletcher CD, et al. Investigative pathology: leading the post-genomic revolution. Lab Invest 2012; 92: 4–8. doi: 10.1038/labinvest.2011.147 [DOI] [PubMed] [Google Scholar]
- 13.Bigos KL, Weinberger DR. Imaging genetics–days of future past. Neuroimage 2010; 53: 804–9. doi: 10.1016/j.neuroimage.2010.01.035 [DOI] [PubMed] [Google Scholar]
- 14.Mehta S, Shelling A, Muthukaruppan A, Lasham A, Blenkiron C, Laking G, et al. Predictive and prognostic molecular markers for cancer medicine. Ther Adv Med Oncol 2010; 2: 125–48. doi: 10.1177/1758834009360519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gevaert O, Xu J, Hoang CD, Leung AN, Xu Y, Quon A, et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data–methods and preliminary results. Radiology 2012; 264: 387–96. doi: 10.1148/radiol.12111607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gevaert O, Mitchell LA, Achrol AS, Xu J, Echegaray S, Steinberg GK, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 2014; 273: 168–74. doi: 10.1148/radiol.14131731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamshidi N, Diehn M, Bredel M, Kuo MD. Illuminating radiogenomic characteristics of glioblastoma multiforme through integration of MR imaging, messenger RNA expression, and DNA copy number variation. Radiology 2014; 270: 1–2. doi: 10.1148/radiol.13130078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol 2007; 18: 821–31. doi: 10.1016/j.jvir.2007.04.031 [DOI] [PubMed] [Google Scholar]
- 19.Nair VS, Gevaert O, Davidzon G, Napel S, Graves EE, Hoang CD, et al. Prognostic PET 18F-FDG uptake imaging features are associated with major oncogenomic alterations in patients with resected non-small cell lung cancer. Cancer Res 2012; 72: 3725–34. doi: 10.1158/0008-5472.CAN-11-3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 2007; 25: 675–80. doi: 10.1038/nbt1306 [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto S, Maki DD, Korn RL, Kuo MD. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. AJR Am J Roentgenol 2012; 199: 654–63. doi: 10.2214/AJR.11.7824 [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto S, Han W, Kim Y, Du L, Jamshidi N, Huang D, et al. Breast cancer: radiogenomic biomarker reveals associations among dynamic contrast-enhanced MR imaging, long noncoding RNA, and metastasis. Radiology 2015; 275: 384–92. doi: 10.1148/radiol.15142698 [DOI] [PubMed] [Google Scholar]
- 23.Zinn PO, Mahajan B, Sathyan P, Singh SK, Majumder S, Jolesz FA, et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One 2011; 6: e25451. doi: 10.1371/journal.pone.0025451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barajas RF, Jr, Hodgson JG, Chang JS, Vandenberg SR, Yeh RF, Parsa AT, et al. Glioblastoma multiforme regional genetic and cellular expression patterns: influence on anatomic and physiologic MR imaging. Radiology 2010; 254: 564–76. doi: 10.1148/radiol.09090663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A 2008; 105: 5213–18. doi: 10.1073/pnas.0801279105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pope WB, Chen JH, Dong J, Carlson MR, Perlina A, Cloughesy TF, et al. Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology 2008; 249: 268–77. doi: 10.1148/radiol.2491072000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zinn PO, Sathyan P, Mahajan B, Bruyere J, Hegi M, Majumder S, et al. A novel volume-age-KPS (VAK) glioblastoma classification identifies a prognostic cognate microRNA-gene signature. PLoS One 2012; 7: e41522. doi: 10.1371/journal.pone.0041522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jamshidi N, Jonasch E, Zapala M, Korn RL, Aganovic L, Zhao H, et al. The radiogenomic risk score: construction of a prognostic quantitative, noninvasive image-based molecular assay for renal cell carcinoma. Radiology 2015; 277: 114–23. doi: 10.1148/radiol.2015150800 [DOI] [PubMed] [Google Scholar]
- 29.Colen R, Foster I, Gatenby R, Giger ME, Gillies R, Gutman D, et al. NCI Workshop Report: clinical and computational requirements for correlating imaging phenotypes with genomics signatures. Transl Oncol 2014; 7: 556–69. doi: 10.1016/j.tranon.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlson MR, Pope WB, Horvath S, Braunstein JG, Nghiemphu P, Tso CL, et al. Relationship between survival and edema in malignant gliomas: role of vascular endothelial growth factor and neuronal pentraxin 2. Clin Cancer Res 2007; 13: 2592–8. doi: 10.1158/1078-0432.CCR-06-2772 [DOI] [PubMed] [Google Scholar]
- 31.Colen RR, Vangel M, Wang J, Gutman DA, Hwang SN, Wintermark M, et al. ; TCGA Glioma Phenotype Research Group. Imaging genomic mapping of an invasive MRI phenotype predicts patient outcome and metabolic dysfunction: a TCGA glioma phenotype research group project. BMC Med Genomics 2014; 7: 30. doi: 10.1186/1755-8794-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain R, Poisson L, Narang J, Scarpace L, Rosenblum ML, Rempel S, et al. Correlation of perfusion parameters with genes related to angiogenesis regulation in glioblastoma: a feasibility study. AJNR Am J Neuroradiol 2012; 33: 1343–8. doi: 10.3174/ajnr.A2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naeini KM, Pope WB, Cloughesy TF, Harris RJ, Lai A, Eskin A, et al. Identifying the mesenchymal molecular subtype of glioblastoma using quantitative volumetric analysis of anatomic magnetic resonance images. Neuro Oncol 2013; 15: 626–34. doi: 10.1093/neuonc/not008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicolasjilwan M, Hu Y, Yan C, Meerzaman D, Holder CA, Gutman D, et al. ; TCGA Glioma Phenotype Research Group. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. J Neuroradiol 2015; 42: 212–21. doi: 10.1016/j.neurad.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, Bittinger MA, et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol 2012; 107: 197–205. doi: 10.1007/s11060-011-0737-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osborne JR, Port E, Gonen M, Doane A, Yeung H, Gerald W, et al. 18F-FDG PET of locally invasive breast cancer and association of estrogen receptor status with standardized uptake value: microarray and immunohistochemical analysis. J Nucl Med 2010; 51: 543–50. doi: 10.2967/jnumed.108.060459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palaskas N, Larson SM, Schultz N, Komisopoulou E, Wong J, Rohle D, et al. 18F-fluorodeoxy-glucose positron emission tomography marks MYC-overexpressing human basal-like breast cancers. Cancer Res 2011; 71: 5164–74. doi: 10.1158/0008-5472.CAN-10-4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu J, Wang J, Zhen ZJ, Lu SY, Zhang F, Sun FF, et al. Brain metastasis in children with stage 4 neuroblastoma after multidisciplinary treatment. Chin J Cancer 2015; 34: 49. doi: 10.1186/s40880-015-0038-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura T, Ban D, Tanaka S, Mogushi K, Kudo A, Matsumura S, et al. Distinct clinicopathological phenotype of hepatocellular carcinoma with ethoxybenzyl-magnetic resonance imaging hyperintensity: association with gene expression signature. Am J Surg 2015; 210: 561–9. doi: 10.1016/j.amjsurg.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 40.Halle C, Andersen E, Lando M, Aarnes EK, Hasvold G, Holden M, et al. Hypoxia-induced gene expression in chemoradioresistant cervical cancer revealed by dynamic contrast-enhanced MRI. Cancer Res 2012; 72: 5285–95. doi: 10.1158/0008-5472.CAN-12-1085 [DOI] [PubMed] [Google Scholar]
- 41.braintumorsegmentation.org/ [homepage on the Internet]. Available from: http://www.cancer-pain.org/.
- 42. Available from: https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project. [homepage on the Internet].
- 43.Halpenny DF, Riely GJ, Hayes S, Yu H, Zheng J, Moskowitz CS, et al. Are there imaging characteristics associated with lung adenocarcinomas harboring ALK rearrangements? Lung Cancer 2014; 86: 190–4. doi: 10.1016/j.lungcan.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karlo CA, Di Paolo PL, Chaim J, Hakimi AA, Ostrovnaya I, Russo P, et al. Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology 2014; 270: 464–71. doi: 10.1148/radiol.13130663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI. Radiogenomic analysis of breast cancer: luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology 2014; 273: 365–72. doi: 10.1148/radiol.14132641 [DOI] [PubMed] [Google Scholar]
- 46.Shinagare AB, Vikram R, Jaffe C, Akin O, Kirby J, Huang E, et al. Radiogenomics of clear cell renal cell carcinoma: preliminary findings of The Cancer Genome Atlas-Renal Cell Carcinoma (TCGA-RCC) Imaging Research Group. Abdom Imaging 2015; 40: 1684–92. doi: 10.1007/s00261-015-0386-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Zhang T, Li S, Fan X, Ma J, Wang L, et al. Anatomical localization of isocitrate dehydrogenase 1 mutation: a voxel-based radiographic study of 146 low-grade gliomas. Eur J Neurol 2015; 22: 348–54. doi: 10.1111/ene.12578 [DOI] [PubMed] [Google Scholar]
- 48.Gutman DA, Cooper LA, Hwang SN, Holder CA, Gao J, Aurora TD, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 2013; 267: 560–9. doi: 10.1148/radiol.13120118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutman DA, Dunn WD, Jr, Grossmann P, Cooper LA, Holder CA, Ligon KL, et al. Somatic mutations associated with MRI-derived volumetric features in glioblastoma. Neuroradiology 2015; 57: 1227–37. doi: 10.1007/s00234-015-1576-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banerjee S, Wang DS, Kim HJ, Sirlin CB, Chan MG, Korn RL, et al. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology 2015; 62: 792–800. doi: 10.1002/hep.27877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carrillo JA, Lai A, Nghiemphu PL, Kim HJ, Phillips HS, Kharbanda S, et al. Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR Am J Neuroradiol 2012; 33: 1349–55. doi: 10.3174/ajnr.A2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drabycz S, Roldán G, de Robles P, Adler D, McIntyre JB, Magliocco AM, et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage 2010; 49: 1398–405. doi: 10.1016/j.neuroimage.2009.09.049 [DOI] [PubMed] [Google Scholar]
- 53.Moon WJ, Choi JW, Roh HG, Lim SD, Koh YC. Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology 2012; 54: 555–63. doi: 10.1007/s00234-011-0947-y [DOI] [PubMed] [Google Scholar]
- 54.Aghi M, Gaviani P, Henson JW, Batchelor TT, Louis DN, Barker FG, 2nd. Magnetic resonance imaging characteristics predict epidermal growth factor receptor amplification status in glioblastoma. Clin Cancer Res 2005; 11: 8600–5. doi: 10.1158/1078-0432.CCR-05-0713 [DOI] [PubMed] [Google Scholar]
- 55.Ellingson BM, Lai A, Harris RJ, Selfridge JM, Yong WH, Das K, et al. Probabilistic radiographic atlas of glioblastoma phenotypes. AJNR Am J Neuroradiol 2013; 34: 533–40. doi: 10.3174/ajnr.A3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta A, Young RJ, Shah AD, Schweitzer AD, Graber JJ, Shi W, et al. Pretreatment dynamic susceptibility contrast MRI perfusion in glioblastoma: prediction of EGFR gene amplification. Clin Neuroradiol 2015; 25: 143–50. doi: 10.1007/s00062-014-0289-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jain R, Poisson L, Narang J, Gutman D, Scarpace L, Hwang SN, et al. Genomic mapping and survival prediction in glioblastoma: molecular subclassification strengthened by hemodynamic imaging biomarkers. Radiology 2013; 267: 212–20. doi: 10.1148/radiol.12120846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kickingereder P, Sahm F, Radbruch A, Wick W, Heiland S, Deimling Av, et al. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep 2015; 5: 16238. doi: 10.1038/srep16238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lanic H, Mareschal S, Mechken F, Picquenot JM, Cornic M, Maingonnat C, et al. Interim positron emission tomography scan associated with international prognostic index and germinal center B cell-like signature as prognostic index in diffuse large B-cell lymphoma. Leuk Lymphoma 2012; 53: 34–42. doi: 10.3109/10428194.2011.600482 [DOI] [PubMed] [Google Scholar]
- 60.Miles KA, Ganeshan B, Rodriguez-Justo M, Goh VJ, Ziauddin Z, Engledow A, et al. Multifunctional imaging signature for V-KI-RAS2 Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations in colorectal cancer. J Nucl Med 2014; 55: 386–91. doi: 10.2967/jnumed.113.120485 [DOI] [PubMed] [Google Scholar]
- 61.Ashraf AB, Daye D, Gavenonis S, Mies C, Feldman M, Rosen M, et al. Identification of intrinsic imaging phenotypes for breast cancer tumors: preliminary associations with gene expression profiles. Radiology 2014; 272: 374–84. doi: 10.1148/radiol.14131375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Giger ML, Sun C, Ponsukcharoen U, Huo D, Lan L, et al. Pilot study demonstrating potential association between breast cancer image-based risk phenotypes and genomic biomarkers. Med Phys 2014; 41: 031917. doi: 10.1118/1.4865811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Macyszyn L, Akbari H, Pisapia JM, Da X, Attiah M, Pigrish V, et al. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol 2015. [Epub ahead of print]. doi: 10.1093/neuonc/nov127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rizzo S, Petrella F, Buscarino V, De Maria F, Raimondi S, Barberis M, et al. CT radiogenomic characterization of EGFR, K-RAS, and ALK mutations in non-small cell lung cancer. Eur Radiol 2016; 26: 32–42. doi: 10.1007/s00330-015-3814-0 [DOI] [PubMed] [Google Scholar]
- 65.Izuishi K, Yamamoto Y, Sano T, Takebayashi R, Nishiyama Y, Mori H, et al. Molecular mechanism underlying the detection of colorectal cancer by 18F-2-fluoro-2-deoxy-D-glucose positron emission tomography. J Gastrointest Surg 2012; 16: 394–400. doi: 10.1007/s11605-011-1727-z [DOI] [PubMed] [Google Scholar]
- 66.Lee JH, Kang J, Baik SH, Lee KY, Lim BJ, Jeon TJ, et al. Relationship between 18F-Fluorodeoxyglucose uptake and V-Ki-Ras2 kirsten rat sarcoma viral oncogene homolog mutation in colorectal cancer patients: variability depending on C-Reactive protein level. Medicine (Baltimore) 2016; 95: e2236. doi: 10.1097/MD.0000000000002236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kawada K, Nakamoto Y, Kawada M, Hida K, Matsumoto T, Murakami T, et al. Relationship between 18F-fluorodeoxyglucose accumulation and KRAS/BRAF mutations in colorectal cancer. Clin Cancer Res 2012; 18: 1696–703. doi: 10.1158/1078-0432.CCR-11-1909 [DOI] [PubMed] [Google Scholar]
- 68.Tykocinski ES, Grant RA, Kapoor GS, Krejza J, Bohman LE, Gocke TA, et al. Use of magnetic perfusion-weighted imaging to determine epidermal growth factor receptor variant III expression in glioblastoma. Neuro Oncol 2012; 14: 613–23. doi: 10.1093/neuonc/nos073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kong DS, Kim ST, Kim EH, Lim DH, Kim WS, Suh YL, et al. Diagnostic dilemma of pseudoprogression in the treatment of newly diagnosed glioblastomas: the role of assessing relative cerebral blood flow volume and oxygen-6-methylguanine-DNA methyltransferase promoter methylation status. AJNR Am J Neuroradiol 2011; 32: 382–7. doi: 10.3174/ajnr.A2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Romano A, Calabria LF, Tavanti F, Minniti G, Rossi-Espagnet MC, Coppola V, et al. Apparent diffusion coefficient obtained by magnetic resonance imaging as a prognostic marker in glioblastomas: correlation with MGMT promoter methylation status. Eur Radiol 2013; 23: 513–20. doi: 10.1007/s00330-012-2601-4 [DOI] [PubMed] [Google Scholar]
- 71.Sunwoo L, Choi SH, Park CK, Kim JW, Yi KS, Lee WJ, et al. Correlation of apparent diffusion coefficient values measured by diffusion MRI and MGMT promoter methylation semiquantitatively analyzed with MS-MLPA in patients with glioblastoma multiforme. J Magn Reson Imaging 2013; 37: 351–8. doi: 10.1002/jmri.23838 [DOI] [PubMed] [Google Scholar]
- 72.Ahn SS, Shin NY, Chang JH, Kim SH, Kim EH, Kim DW, et al. Prediction of methylguanine methyltransferase promoter methylation in glioblastoma using dynamic contrast-enhanced magnetic resonance and diffusion tensor imaging. J Neurosurg 2014; 121: 367–73. doi: 10.3171/2014.5.JNS132279 [DOI] [PubMed] [Google Scholar]
- 73.Sutton EJ, Oh JH, Dashevsky BZ, Veeraraghavan H, Apte AP, Thakur SB, et al. Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. J Magn Reson Imaging 2015; 42: 1398–406. doi: 10.1002/jmri.24890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kitao A, Zen Y, Matsui O, Gabata T, Kobayashi S, Koda W, et al. Hepatocellular carcinoma: signal intensity at gadoxetic acid-enhanced MR Imaging–correlation with molecular transporters and histopathologic features. Radiology 2010; 256: 817–26. doi: 10.1148/radiol.10092214 [DOI] [PubMed] [Google Scholar]
- 75.Lee HJ, Kim YT, Kang CH, Zhao B, Tan Y, Schwartz LH, et al. Epidermal growth factor receptor mutation in lung adenocarcinomas: relationship with CT characteristics and histologic subtypes. Radiology 2013; 268: 254–64. doi: 10.1148/radiol.13112553 [DOI] [PubMed] [Google Scholar]
- 76.Glynn C, Zakowski MF, Ginsberg MS. Are there imaging characteristics associated with epidermal growth factor receptor and KRAS mutations in patients with adenocarcinoma of the lung with bronchioloalveolar features? J Thorac Oncol 2010; 5: 344–8. doi: 10.1097/JTO.0b013e3181ce9a7a [DOI] [PubMed] [Google Scholar]
- 77.Plodkowski AJ, Drilon A, Halpenny DF, O'Driscoll D, Blair D, Litvak AM, et al. From genotype to phenotype: are there imaging characteristics associated with lung adenocarcinomas harboring RET and ROS1 rearrangements? Lung Cancer 2015; 90: 321–5. doi: 10.1016/j.lungcan.2015.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ozkan E, West A, Dedelow JA, Chu BF, Zhao W, Yildiz VO, et al. CT gray-level texture analysis as a quantitative imaging biomarker of epidermal growth factor receptor mutation status in adenocarcinoma of the lung. AJR Am J Roentgenol 2015; 205: 1016–25.doi: 10.2214/AJR.14.14147 [DOI] [PubMed] [Google Scholar]
- 79.Yoon HJ, Sohn I, Cho JH, Lee HY, Kim JH, Choi YL, et al. Decoding tumor phenotypes for ALK, ROS1, and RET fusions in lung adenocarcinoma using a radiomics approach. Medicine (Baltimore) 2015; 94: e1753. doi: 10.1097/MD.0000000000001753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: 441–6. doi: 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grimm LJ, Zhang J, Mazurowski MA. Computational approach to radiogenomics of breast cancer: luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms. J Magn Reson Imaging 2015; 42: 902–7. doi: 10.1002/jmri.24879 [DOI] [PubMed] [Google Scholar]
- 82.Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A 2013; 110: 4009–14. doi: 10.1073/pnas.1219747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ashraf AB, Gavenonis S, Daye D, Mies C, Feldman M, Rosen M, et al. A multichannel Markov random field approach for automated segmentation of breast cancer tumor in DCE-MRI data using kinetic observation model. Med Image Comput Comput Assist Interv 2011; 14: 546–53. [DOI] [PubMed] [Google Scholar]
- 84.Xu J, Greenspan H, Napel S, Rubin DL. Automated temporal tracking and segmentation of lymphoma on serial CT examinations. Med Phys 2011; 38: 5879–86. doi: 10.1118/1.3643027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zikic D, Glocker B, Konukoglu E, Criminisi A, Demiralp C, Shotton J, et al. Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel MR. Med Image Comput Comput Assist Interv 2012; 15: 369–76. [DOI] [PubMed] [Google Scholar]
- 86.Riklin-Raviv T, Van Leemput K, Menze BH, Wells WM, 3rd, Golland P. Segmentation of image ensembles via latent atlases. Med Image Anal 2010; 14: 654–65. doi: 10.1016/j.media.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gooya A, Pohl KM, Bilello M, Cirillo L, Biros G, Melhem ER, et al. GLISTR: glioma image segmentation and registration. IEEE Trans Med Imaging 2012; 31: 1941–54. doi: 10.1109/TMI.2012.2210558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Menze BH, Van Leemput K, Honkela A, Konukoglu E, Weber MA, Ayache N, et al. A generative approach for image-based modeling of tumor growth. Inf Process Med Imaging 2011; 22: 735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Akbari SB H, Rozycki M, Da X, Pisapia J, Bilello M, O’Rourke D, et al. Non-invasive Determination of Epidermal growth factor Receptor Variant III expression in glioblastoma through analysis of multi-parametric magnetic resonance imaging. Chicago, Illinois, USA: RSNA 101st Scientific Assembly and Annual Meeting; 2015. [Google Scholar]
- 90.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–69. doi: 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- 91.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med 2009; 60: 167–79. doi: 10.1146/annurev.med.59.053006.104707 [DOI] [PubMed] [Google Scholar]
- 92.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015; 15: 321–33. doi: 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013; 19: 651–69. doi: 10.1534/genetics.112.146704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10: 155–9. doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 95.Konig J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet 2011; 13: 77–83. [DOI] [PubMed] [Google Scholar]
- 96.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet 2013; 14: 507–15. doi: 10.1038/nrg3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.cancergenome.nih.gov. [homepage on the Internet]. National Cancer Institute National Human Genome Research Institute. The Cancer Genome Atlas (TCGA), 2014. Available from: http://cancergenome.nih.gov/. [Google Scholar]
- 98.cancerimagingarchive.net. [homepage on the Internet]. National Cancer Institute Cancer Imaging Program Frederick National Laboratory for Cancer Re-search, 2013. The Cancer Imaging Archive (TCIA). Available from: http://cancerimagingarchive.net/. [Google Scholar]
- 99.Chandarana H, Robinson E, Hajdu CH, Drozhinin L, Babb JS, Taouli B. Microvascular invasion in hepatocellular carcinoma: is it predictable with pretransplant MRI? AJR Am J Roentgenol 2011; 196: 1083–9. doi: 10.2214/AJR.10.4720 [DOI] [PubMed] [Google Scholar]
- 100.Griffin N, Addley H, Sala E, Shaw AS, Grant LA, Eldaly H, et al. Vascular invasion in hepatocellular carcinoma: is there a correlation with MRI? Br J Radiol 2012; 85: 736–44. doi: 10.1259/bjr/94924398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, et al. Gene expression patterns in human liver cancers. Mol Biol Cell 2002; 13: 1929–39. doi: 10.1091/mbc.02-02-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jamshidi N, Jonasch E, Zapala M, Korn RL, Brooks JD, Ljungberg B, et al. The radiogenomic risk score stratifies outcomes in a renal cell cancer phase 2 clinical trial. Eur Radiol 2015 Nov 11. [Epub ahead of print]. doi: 10.1007/s00330-015-4082-8 [DOI] [PubMed] [Google Scholar]
- 103.Polonsky TS, McClelland RL, Jorgensen NW, Bild DE, Burke GL, Guerci AD, et al. Coronary artery calcium score and risk classification for coronary heart disease prediction. JAMA 2010; 303: 1610–6. doi: 10.1001/jama.2010.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Longo DL. Tumor heterogeneity and personalized medicine. N Engl J Med 2012; 366: 956–7. doi: 10.1056/NEJMe1200656 [DOI] [PubMed] [Google Scholar]
- 105.Van Meter T, Dumur C, Hafez N, Garrett C, Fillmore H, Broaddus WC. Microarray analysis of MRI-defined tissue samples in glioblastoma reveals differences in regional expression of therapeutic targets. Diagn Mol Pathol 2006; 15: 195–205. doi: 10.1097/01.pdm.0000213464.06387.36 [DOI] [PubMed] [Google Scholar]
- 106.Zhang T, Wang Y, Fan X, Ma J, Li S, Jiang T, et al. Anatomical localization of p53 mutated tumors: a radiographic study of human glioblastomas. J Neurol Sci 2014; 346: 94–8. doi: 10.1016/j.jns.2014.07.066 [DOI] [PubMed] [Google Scholar]
- 107.Asselin MC, O'Connor JP, Boellaard R, Thacker NA, Jackson A. Quantifying heterogeneity in human tumours using MRI and PET. Eur J Cancer 2012; 48: 447–55. doi: 10.1016/j.ejca.2011.12.025 [DOI] [PubMed] [Google Scholar]