Abstract
Mitochondrial dysfunction underlines a multitude of pathologies; however, studies are scarce that rescue the mitochondria for cellular resuscitation. Exploration into the protective role of mitochondrial Transcription factor A (TFAM) and its mitochondrial functions respective to cardiomyocyte death are in need of further investigation. TFAM is a gene regulator that acts to mitigate calcium mishandling and ROS production by wrapping around mitochondrial DNA (mtDNA) complexes. TFAM’s regulatory functions over serca2a, NFAT and Lon protease contribute to cardiomyocyte stability. Calcium and ROS dependent proteases, calpains and matrix metallo-proteinases (MMP’s) are abundantly found upregulated in the failing heart. TFAM’s regulatory role over ROS production and calcium mishandling leads to further investigation into the cardioprotective role of exogenous TFAM. In an effort to restabilize physiological and contractile activity of cardiomyocytes in HF models, we propose that TFAM-packed exosomes (TFAM-PE) will act therapeutically by mitigating mitochondrial dysfunction. Notably, this is the first mention of exosomal delivery of transcription factors in the literature. Here we elucidate the role of TFAM in mitochondrial rescue and focus on its therapeutic potential.
Keywords: mitochondria, TFAM, serca2a, Hsp60, calpain, MMPs
Mitochondrial Transcription Factor A (TFAM)
TFAM Roles and Regulation
Mitochondrial dysfunction is correlated with disease states at the core of oxidative phosphorylation and energy efficiency, essentially focused on mitochondrial DNA (mtDNA). MtDNA is a 16.5 Kb double stranded circular molecule encoding thirteen essential components of oxidative phosphorylation. Therefore, mtDNA is vital to the restoration of physiological conditions since efficient energy production is necessary for all basic human processes. As noted unanimously in the literature, the major regulator of mtDNA copy number in mammalian models is TFAM [1]. The mtDNA copy number is reflective of mtDNA transcription and ATP production. TFAM is a promoter specific enhancer of mtDNA and a member of the high mobility protein group [2]. This transcription factor is essential for mitochondrial DNA maintenance and coats mitochondrial DNA, playing a factor in mtDNA stability [3]. TFAM is a major, mitochondrial gene-regulator and as a transcription factor it modifies gene expression. TFAM is produced in the nucleus and is transported to the mitochondria (Fig 1). Nuclear regulatory factors including nuclear respiratory factors 1 and 2 (NRF 1, 2) seem to regulate TFAM; this is claimed to be a link between nuclear activity and energy production [4]. A finding through super-resolution microscopy reveals that mitochondrial nucleoids (mtDNA-protein complexes), contain a uniform size and TFAM is a main component [5]. TFAM acts as a packaging molecule by storing a single copy of mtDNA in a functional mitochondrial nucleoid [6]. Mitochondrial nucleoids contain essential enzymes of an integral antioxidant system including manganese superoxide dismutase (SOD2) and mitochondrial glutathione peroxidase (GPx1) [7]. These antioxidant enzymes mitigate oxidative damage and protect mtDNA. Components of the nucleoid create a safe environment for functional mtDNA processes such as transcription, transduction and repair [8]. The functionalities of TFAM act as a benefactor to cardiomyocyte cellular functions.
Fig 1. TFAM transport and LON Dependent TFAM Release.
TFAM is transcribed in the nucleus and transported to the mitochondria with the help of HSP60-HSP-70 complex. In view of increase in the ROS production in mitochondria, the Lon protease is up regulated, which then binds to the HSP60 of the HSP60-HSP-70 complex. This facilitates the release of TFAM from the HSP60-HSP-70 complex into the mitochondria. Subsequently, high levels in the mitochondria block ROS production.
TFAM transcriptional machinery is composed of two high mobility groups noted as HSP (heavy strand promoter) and LSP (light strand promoter). TFAM is necessary for mitochondrial-directed RNA-polymerase (POLRMT) to interact with the DNA promoter to initiate transcription producing the primary transcripts of mRNA, rRNA and tRNA [9]. As a major regulator of mtDNA transcriptional machinery TFAM has direct regulation over minor transcription factors, TFB1M and TFB2M. These transcription factors directly promote transcription, utilizing POLRMT [10]. TFBM1 is a dual function protein acting as a transcription factor and an RNA methyl-transferase enzyme [11]. Aside from indirect gene regulation through TFBM 1&2, TFAM also directly regulates promoter regions of DNA by binding to the promoter specific region, causing an unwinding reaction, initiating transcription [12]. It is thought that additional functions of the TFBM 1&2 include binding single stranded DNA to stabilize unwound promoter regions. A strong correlation between TFAM and TFBM 1&2 expression and initiation of mtDNA replication has been found. Additionally, overexpression of TFAM was clinically beneficial [13]. A significant component of mitochondrial transcription, the mitochondrial RNA polymerase, requires TFAM and TFBM2 for efficient initiation [14]. Mapping the binding sites of TFAM and TFBM2 resulted in novel functional aspects of TFBM2 binding. TFAM binding to an upstream promoter region initiates the preinitiation complex, which requires the binding of TFB2M for specific contact with promoter DNA. Loss of TFBM2 leads to nonspecific contact with DNA. Functionally, TFB2M separates the DNA strands and plays a role in initiating transcription. TFBM2 can reposition the specificity loop of mtRNAP to recognize specific promoters [14,15]. Mitochondrial protein formation is regulated by mitochondrial ribosomes, consisting of two subunits, the small (28S) mitochondrial ribosome subunit and the (55S) subunit. Post the regulatory functions of forming the initiation complex; TFB1M regulates the assembly of the 28S subunit by acting as a 12S ribosomal RNA methyltransferase. Observations have concluded that POLRMT’s activities to catalyze TFB1M’s methyltransferase function, results in protein formation [16]. POLRMT is essential for mitochondrial protein formation and gene regulation.
It is noted that both of these factors [TFB1M & TFB2M] are cooperative downstream effectors for mitochondrial biogenesis. More specifically, TFB2M functions to maintain mtDNA transcription. TFB1M’s 12S rRNA methyltransferase activity is essential for mitochondrial translation and metabolism. Hypermethylation caused by overexpression of TFB1M methyltransferase activity results in cell death and abnormal biogenesis [17]. To emphasize the importance of the methylation activity of TFB1M, studies show the effects of disrupting cardiac TFB1M. Complete loss of adenine di-methylation of rRNA decreased mitochondrial translation and reduced ribosomal function [18]. As noted above TFAM has direct regulation over TFB1M & TFB2M. Ablation of TFAM in neonatal mice led to sufficient decreases in the cardiac respiratory chain affecting mitochondrial biogenesis [19,2]. TFAM’s major regulatory activities of mtDNA led to direct regulation over mitochondrial gene expression affecting the 13 essential genes required for oxidative phosphorylation, effecting mitochondrial biogenesis and ATP production. Understanding the essence of mitochondrial biogenesis and oxidative phosphorylation is the key to cardiovascular pathologies. Mitochondrial dysfunction within cardiomyocytes leads to HF, but the core of mitochondrial defects is found within the stability of mtDNA. As stated previously, TFAM is a DNA-binding protein directly related to mitochondrial stability. Mutant uracil-DNA glycosylase 1 (mutUNG1) was found upregulated in transgenic mouse (Tet-on) models, which hampered mtDNA function and stability. Impairment of mtDNA via mutUNG1 upregulation resulted in rapid hypertrophic cardiomyopathy leading to HF and death. This culminated into pathological effects such as; reduced mtDNA replication & transcription, diminished mt respiration, impaired fission/fusion dynamics and majorly decreased cardiomyocyte contractility [20]. The impairment seen in mt dynamics raises the question of whether imbalance of mfn-2/Drp-1 leads to mtDNA decline or whether disturbance in the ratio is a compensatory factor. It would be interesting to observe the effects of exogenous TFAM administration into this mutant model.
TFAM Transport
TFAM is synthesized in the nucleus and transported to mitochondria. TFAM translocation to the mitochondria for initiation of transcription and translation requires the binding of chaperones creating a heat shock protein (HSP) complex. Deocaris and colleagues describe the intimate interaction between HSP60 and HSP70 [21]. Lon protease is an ATP dependent protease with chaperone properties and is shown to promote the association of heat shock protein complex (HSP60-HSP70). Kao and colleagues observed an increase in the stability of the HSP60-70 complex upon Lon binding the HSP60 subunit. Loss of Lon leads to HSP60 dislocation, increased protein degradation and apoptotic signaling activation upon HSP60 binding to P53 [22]. The HSP60-70 complex is Lon dependent and importantly TFAM binds to the HSP70 component. The interaction of TFAM bound to HSP-70 at the nucleus allows for transportation of TFAM into the mitochondria [23] (Fig 1). Lon can be observed as TFAM’s personal trainer keeping TFAM fit for mitochondrial biogenesis. Within this complex the literature states that Lon protease binds to the HSP-60 component while TFAM is bound to the HSP-70 portion of the complex. We speculate that it may be necessary for Lon to bind HSP60 resulting in a conformational change to HSP70 causing the release of TFAM within the mitochondria, acting to increase transcriptional activity and mitochondrial biogenesis. Lon protease has been observed as a cardioprotective molecule and this may be a route of its protective activity. Additionally, Ciesielski found that Mdj1 an HSP40 co-chaperone of HSP70 is necessary to maintain functional mtDNA. Overexpression of the J-domain reestablished mtDNA activity but a lack of Mdj1 leads to loss of functional mtDNA [24]. It is unexplored as to whether Mdj1 assists in the binding of TFAM to HSP70. The binding of Lon to HSP60 may cause a conformational change to the J subunit allowing for the release of TFAM, leading to increased mitochondrial stability.
An article by Naka and colleagues suggests that HSP70 transgenic mice are cardioprotective in a doxorubicin heart failure model, expressing that the transgenic model inhibited apoptotic signaling by mediating P53, Bax, caspase-3&9 and PARP-1 [25]. Upregulation of HSP-70 will increase mitochondrial biogenesis through increased TFAM transport. This acts to regulate calcium leading to decreased protease activity and apoptotic signaling. The presence of complex formation of HSP60-70 would down regulate the apoptotic (P53) pathway through increased complex reactivity. Santos and colleagues investigated the mechanisms of TFAM loss in diabetic models and TFAM’s association with HSP-70. This group discusses the posttranslational modification of TFAM by ubiquitination leading to decreased protein levels and activity, as found in high glucose conditions. Ubiquitination of TFAM prevents its transport into the mitochondria and ubiquitination inhibitor PYR-41 led to increased TFAM-transcriptional activity. Comparison of the overexpression of HSP-70 to the overexpression of TFAM resulted in HSP-70 failing to inhibit glucose actions to decrease TFAM transcriptional activity, while TFAM overexpression ameliorated decreased mitochondrial transcriptional activity and proteins [23]. Therefore we can conclude that exogenous TFAM may be beneficial to diabetic related pathologies. Santos describes TFAM binding to HSP60 but at a lower affinity when compared to HSP70 [23]. Lon’s proteolytic ability to cleave phosphorylated TFAM may be inhibiting complex formation in high ROS conditions thus preventing mitochondrial productivity.
HSP-70 regulation is important for proper functioning and Ko et al demonstrated the activity of the molecule Apoptzole (Az), an inhibitor of the HSP-70 ATPase activity. This group found that Az induced cancer cell death by inhibiting HSP-70 and activating caspase dependent apoptosis [26]. Functionally, HSP-70 transports TFAM a major stimulator of mitochondrial activity and is found upregulated in cancer cells. This is due to the cancer cell proliferation rate and need for increased mitochondrial function and production. Blocking the activity of HSP-70 would inhibit TFAM transport and result in cancer cell death. Interestingly, the cellular death is calcium dependent-calpain and apoptotic caspase activity. HSF-1 activates HSP70 and this activation is directly dependent on a sulfhydryl inducer, homocysteine could be a poential activator. Increased levels of HSF-1 inhibit the expression of NF-kB p65 resulting in decreased rates of cardiac hypertrophy [27]. Increased complex formation would inhibit HSP60 activation of the p65 subunit. Liu and colleagues found a novel regulatory mechanism involving NF-kB. They state that upregulation of NF-kB results in increased transcriptional regulation over hypertrophic and apoptotic signaling. The p65 subunit of NF-kB binds and synergistically promotes/upregulates NFAT’s transcriptional activity [28]. The malignant upregulation of NFAT and NF-kB are blocked/inhibited by activation of TFAM, this will be thoroughly discussed.
Overall, the up-regulated cytoplasmic calcium and ROS production observed in HF models would drive Lon to bind HSP-60. Our thoughts on this subject conclude that TFAM may be locked in the complex during increased ROS & calcium conditions. TFAM is phosphorylated and then broken down by the excess unbound Lon protease found within the cytoplasm. Loss of TFAM transport results in loss of mitochondrial efficiency and eventual apoptosis (Fig 1). This statement is made in regards to TFAM’s loss of activity in HF models.
Mitochondrial Dynamics (MFN-2: The Gate Keeper?)
Mitochondrial stability is pertinent to cardiomyocyte survival. Regulating heart failure is dependent upon the mitochondrial equilibrium of two proteins Mitofusin-2 (Mfn-2) and dynamin-related protein (Drp-1). Mfn-2 is noted for its role in mitochondrial fusion and Drp-1 for mitochondrial fission. Imbalance of these two proteins has been observed in cardiomyopathies. We have assessed the activity of MMP-9 an extracellular matrix protease in cardiac hypertrophy through treatment with mitochondrial division inhibitor, resulting in inhibition of abnormal cardiac mitophagy [29]. Analysis of the mitochondrial role of cardiomyocyte contractility via SiRNA silencing of mitochondrial fission revealed an increase in myocyte contractility. Inhibition of Mfn-2 decreased contractility and changed calcium and potassium ion concentrations. Interestingly, we observed that increasing fission caused a decrease in Serca-2a (sarco-endoplasmic reticulum calcium transporter) and an increase in cytochrome c leakage causing mitophagy [30]. In another study the cardiac tissue of Mfn-2 deficient mice resulted in cardiomyopathy. RNA analysis revealed a decrease in mtDNA content and observations of insertion-deletion mutations in Mfn-2 deficient cardiomyocytes [31]. Clustering of the mitochondrial nucleoid occurred in upregulated mitochondrial fission experiments utilizing a GTpase DRP-1 knockout vector during neonatal cardiomyocyte development [32]. This result suggests a regulatory role of Drp-1 over mitochondrial nucleoid development. Interesting observations that Mfn-2 deficient cardiomyocytes are protected from apoptotic pathways suggests that Mfn-2 has a signaling mechanism that triggers cell death in cardiomyocytes [33].
Mitofusin-2 mediates mitochondrial outer membrane fusion. Quite interestingly mitochondria are physically tethered to the SR by MFN-2 (Fig 3) resulting in direct regulation of calcium signaling [34,35]. Coupling/tethering of the mitochondria to the SR is crucial for calcium handling. MFN-2 may act as a gate-keeper for calcium transfer from the SR to the mitochondria. Therefore, loss of Mfn-2 would lead to loss of calcium signaling/storage in the SR, the mitochondria would store the calcium in the SR, at a decreased rate. Increased calcium concentration triggers depolarization of the mitochondrial membrane potential (MTPT) [36], leading to mitochondrial apoptosis and eventual cell death due to protease activity. Chen analyzed Mfn-2 activity in fruit flies, finding that Mfn-2 deficiency led to increased contraction. Mfn-2 deficiency was found to decrease contact length between the mitochondria and SR by 30% also reducing the presence of SR related proteins in the extra-mitochondrial matrix linking the two organelles [34]. Association of mitochondria and SR is a key component of cardiac calcium handling and contractility. Mitochondria are major regulators of cytoplasmic calcium, a transport system mediated by Mfn-2 to enhance the storage of calcium into the SR. Luongo expressed that the MCU calcium transporter is required in acute stress conditions [37]. In high stress conditions when cytoplasmic calcium levels are higher, the MCU will increase calcium transport into the mitochondria, perhaps activating the calcium storage pathways via Mfn-2. Decreased mitochondrial calcium levels inhibited the depolarization of the mitochondrial membrane and led to cardioprotection. Davidson and colleagues focused on calcium handling and secondary messenger NAADP, testing the hypothesis of cardioprotection using an NAADP antagonist Ned-K. Post ischemia-reperfusion they observed a cardiomyocyte protective mechanism by preventing the opening of the mitochondrial permeability transition pore [38].
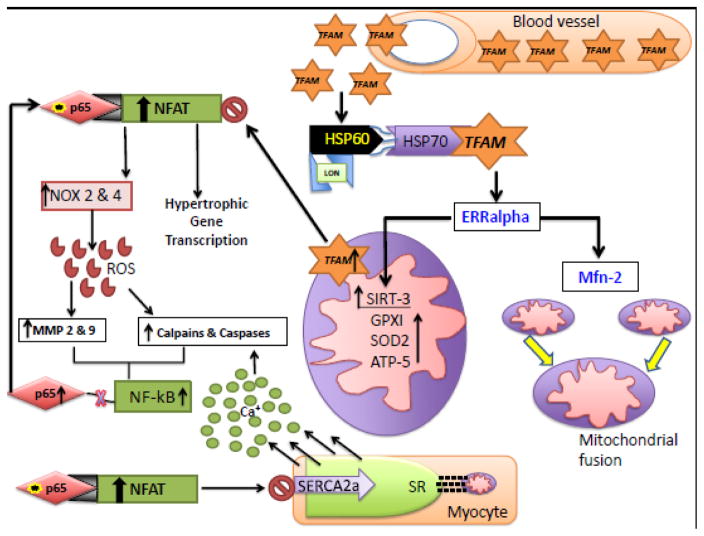
Fig 3. Proposed model.
TFAM-PE enter the blood stream and effectively fuse with the basolateral cellular membrane. Upon entering TFAM is transported to the mitochondria via the HSP60-70 complex. We speculate that TFAM acts as a secondary activator of ERRalpha inducing the up regulation of SIRT-3 and Mfn-2. Mfn-2 has regulatory function over calcium storage and SIRT-3 increases ROS mediators. TFAM’s major function of regulatory activity is to inhibit NFAT activation. Loss of TFAM would result in up regulation of NFAT causing hypertrophic gene expression and blockage of the SERCA2a ATPase pump and increase in cytoplasmic calcium levels inducing protease activity of calpains. Additionally, NFAT increases NOX 2 & 4 expression causing increased ROS generation, NFAT activation increases when calcium binds calcineurin. Increased ROS induces MMP activity. As an apoptotic trigger calcium induces the translocation of NF-kB resulting in release of the P65 subunit. The p65 subunit binds NFAT greatly stimulating NFAT activity. We speculate that exogenous TFAM will inhibit NFAT function and drive mitochondrial rescue.
Transcriptional regulation of Mfn-2 to maintain mt dynamic balance and calcium transport is dependent on PGC-1alpha signaling mechanisms. Chitra and Boopathy explain the importance of the PGC-1α/ERRα/Mfn2 axes, stating a positive correlation between the three variables. PGC-1α binds to ERRα and increases Mfn-2 expression [39]. Loss of ERRα leads to down-regulation of Mfn-2 expression [40]. TFAM is downstream of PGC-1alpha activation. The PGC-1alpha pathway encompasses another important mitochondrial biogenesis factor sirtiun-3 (SIRT-3). SIRT-3 is a mitochondrial deacetylase that acts to increase mitochondrial efficiency by enhancing ATP production and mitochondrial biogenesis (Fig 2). Mitochondrial DNA depletion and reduced replication were observed in heart failure, and found independent of PGC-1alpha [41]. This finding suggests other mechanisms act as secondary activators to increase mitochondrial biogenesis. Recent literature contains findings that transgenic TFAM mouse models with induced Volume-overload (VO) heart failure had cardio-protective effects when compared to wild type (WT). TFAM overexpression reduced MMP activity and mtROS production [42]. TFAM transgenic mouse (TG) model contains a higher survival rate post induced myocardial infarction (MI). Observations comparing MI induced WT and TFAM-TG resulted in decreased myocyte hypertrophy, oxidative stress, apoptosis and interstitial fibrosis in the TFAM-TG model. Many regulatory aspects of TFAM are essential for mitochondrial function.
Fig 2.
Primary activation of SIRT-3 deacetylase is via the PGC-1alpha/ERRalpha pathway. A potential secondary activator of SIRT-3 is TFAM, due to stimulated but decreased activation when eliminating the ERRalpha signal.
Epigenetics & miRNA
Epigenetic regulation of TFAM is a major subject for mitochondrial biogenesis. TFAM can be regulated by microRNAs and observations of post-transcriptional modifications of TFAM expression are performed by miRNA 155-5p. A significant negative correlation between miR-155-5p and TFAM expression was found in patient cardiac samples [43]. Yamamoto et al had an interesting finding regarding skeletal muscle and mitochondrial biogenesis. Their findings include that miRNA 494 has a negative regulation over mtDNA and TFAM [44]. Additionally, miRNA-214 is noted as a potential therapeutic target for cancer cell proliferation due to its regulation over TFAM. Regulatory miRNA’s are responsible for balancing TFAM expression in the control subject. TFAM can also be regulated epigenetically by methylation of the promoter region. Methylation of TFAM specific promoter regions would decrease expression of TFAM protein and would result in loss of mitochondrial function. Hydrogen sulfide was shown to regulate mt biogenesis via demethylation of TFAM. More specifically, exogenous H2S improved mtDNA copy number and increased TFAM expression. H2S functions to diminish DNA methlytransferase 3a (DNMT3a) resulting in TFAM demethylation, it also sulfhydrates interferon regulatory factor 1 (IRF-1) which increases the binding of IRF-1 to the DNMT3a promoter [45]. These treatment methods target TFAM’s regulation over mtDNA copy number [1]. An interesting side note is a cAMP regulatory pathway involving TFAM. Zhang observed cyclic nucleotide phosphodiesterase known as Prune, has stabilizing effects on mtDNA replication and TFAM by decreasing mitochondrial cAMP signaling [46]. TFAM’s role in mitochondrial stability is crucial to ATP and ROS production, therefore maintaining TFAM in various disease states may be essential to patients’ longevity and recovery. We have expressed that high levels of homocysteine activate N-methyl-D-aspartate receptor 1 (NMDAR) leading to abnormal DNA methylation. High homocysteine levels induce cardiac remodeling through epigenetic mechanisms such as methyltransferase activity via DNMT1 [47]. MiRNA and methylation are essential components of balancing TFAM expression.
Stated as the mitochondrial guardian, SIRT-3 acts to mediate ROS production via inducing ROS-detoxifying enzymes Glutathione Peroxidase 1 (GP1), Superoxide Dismutase 2 (SOD2), ATP synthase5c and cytochrome c. PGC-1alpha is a main regulator of SIRT-3 and is found upstream of the transcriptional activation of the SIRT-3 promoter region [48]. Stimulation occurs when PGC-1alpha-ERRalpha binds to the SIRT-3 promoter region, up-regulating SIRT-3 activity [49]. Although, the primary activation of SIRT-3 is PGC-1alpha/ERRalpha driven, it is unexplored whether TFAM could secondarily activate the SIRT-3 promoter region. Experimental observations of reduced mitochondrial DNA replication and mtDNA depletion in the failing heart are independent of reduced PGC-1alpha expression [41]. Synergistic actions may occur between TFAM and SIRT-3, due to their correlative functions in the mitochondria. Upon removal of ERR-alpha, SIRT-3 deacetylase activity is still present just at a decreased rate [48], supporting the claim that a secondary activator is present. In PGC-1alpha depletion, transcription factors NRF-1/2 and ERRalpha remain stagnant but interestingly increased TFAM levels were found [50]. Secondary activation of ERRalpha would increase activity of Mfn-2 and SIRT-3 (Fig 2). Hydrogen peroxide decreases SIRT-1 allowing for increased acetylation of p65 (NF-kB) subunit. Acetylation serves as the regulatory mechanism for the p65 subunit release from NF-kB, which is modulated by Sirtuin-1 deacetylase activity. Free P65 binds to the MMP-9 promoter increasing its transcription and driving apoptotic signaling [51]. The overview of epigenetic factors that regulate TFAM are provided in Fig 6.
Fig 6. Epigenetic regulation of TFAM.
Methylation of TFAM promoter region decreases the rate of TFAM release. Hypermethylation of TFAM coactivators PGC-1alpha and NRF-1 inhibit their transcriptional activity, hence they cannot act as co-activators for TFAM. Additionally, miRNA’s block TFAM mRNA production.
Role of TFAM in ischemic and non-ischemic cardiac dysfunction
The myocardial tissue death occurs from various forms of oxidative and nitrosative stress due to sepsis-induced myocardial dysfunction, ischemia/reperfusion after myocardial infarction, demand ischemia and ischemia [52–56]. Mitochondria, which occupy nearly 30% of the cardiomyocyte cytoplasmic volume, are the major site of damage during ischemic and non-ischemic cardiac dysfunction [57–59]. This damage disrupts the mitochondrial protein, DNA and lipid modifications leading to inhibition of energy production, contractile dysfunction, cell necrosis or apoptosis. To combat mitochondrial dysfunction, cardiomyocytes have evolved elegant systems of mitochondrial repair and anti-oxidant defense, which involves transcription of genes that regulate energy homeostasis and mitochondrial DNA repair. Mitochondrial resuscitation requires the coordination of both nuclear and mitochondrial genomes which involves synthesis of transcription factors in the nucleus and their transport to mitochondria e.g. NRF-1, NRF-2 and PGC-1α [60–63]. NRF-1 and its co-activator PGC-1α indirectly regulate TFAM (both mitochondrial transcription factor A and B), which enables transcription from the mitochondrial genome [64]. Oxidative stress resulting from ischemia/reperfusion injury or other cytotoxic insults in the myocardium leads to nuclear translocation of NRF-1 and upregulation of PGC-1α [65,66]. These two co-activators regulate the transcription of TFAM and contribute to mitochondrial biogenesis and repair mechanisms. These events suggest that during ischemia/reperfusion injury the transcription of TFAM is regulated which acts as a compensatory mechanism for mitochondrial biogenesis and repair. In an interesting study by Yue et al there is depletion of TFAM after ischemia/reperfusion injury, which is improved by lycopene. The authors describe that lycopene is cardioprotective by stabilizing TFAM [67]. This study suggests that presence of TFAM is required for mitochondrial repair and combating stress damage.
In failing hearts, initially TFAM levels go up as a compensatory mechanism but it progressively goes down as calcium mishandling and ROS production increase, as observed in later stages of heart failure. Restoring TFAM levels in HF models will increase mt biogenesis and stabilize cardiomyocyte function through increasing mitochondrial activity. In the presence of mitochondrial oxidative stress, TFAM is synthesized in the nucleus and transported to mitochondria however, when mitochondria become dysfunctional, there is loss of TFAM. Hence, it can be concluded that compensatory mechanisms that are cardioprotective during ischemia/reperfusion injury involve the presence of TFAM which otherwise get depleted in myocardial tissue death.
TFAM effects: matrix turnover
There is enhanced oxidative stress in myocardial tissue damage, which leads to remodeling of the extracellular matrix. Increase in ROS induces apoptosis, myocyte hypertrophy and interstitial fibrosis by activating matrix metalloproteases (MMPs) [68,69]. MMPs degrade a wide spectrum of extracellular proteins that lead to remodeling of the matrix [70]. The degradation of extracellular proteins provides an abnormal microenvironment of matrix in which the myocytes interact. The MMPs degrade collagen and elastin in the matrix. Collagen is replaced faster than elastin due to which there is change in the collagen/elastin ratio leading to interstitial fibrosis [71–73]. Collagen deposition decreases the tensile strength of walls leading to dilated cardiomyopathy. This compensatory mechanism accounts for cellular defense against these events implying an increase in anti-oxidant enzymes that involve increases mitochondrial transcription factor or TFAM. The lack of TFAM has been reported to cause dilated cardiomyopathy with reduced mtDNA [19]. In addition, the reduction of TFAM has been reported in many forms of heart failure [74–77]. The mice in which TFAM is overexpressed exhibit protection from LV remodeling, decreased oxidative stress, have preserved mtDNA copy number and mitochondrial function [78]. Moreover, TFAM overexpression attenuated histopathological changes such as interstitial fibrosis, apoptosis, myocyte hypertrophy and cardiac chamber dilation. In another study on TFAM transgenic mice, TFAM overexpression has been reported to increase modestly the mitochondrial number and reverse cardiomyopathy [79]. Hence TFAM plays an important role in myocardial protection against adverse cardiac remodeling and heart failure.
Protease Regulation
Calpains, MMP’s & Lon Protease
As we have reported earlier that mitochondrial dysfunction leads to protease activity and cardiomyocyte decline [80]. Proteases such as MMP’s, Calpains and Lon protease perform degradative roles in myocytes. MMPs are zinc dependent endopeptidases, capable of degrading the extracellular matrix (ECM) of cardiomyocytes causing cardiac remodeling. Our laboratory has assessed the activity of MMP-9 in cardiac hypertrophy through treatment with mitochondrial division inhibitor, resulting in inhibition of abnormal cardiac mitophagy [29]. Lon protease is a mitochondrial specific protease functioning to degrade oxidatively modified proteins. The literature states that a negative correlation exists between TFAM and Lon representing a reaction that affects mitochondrial bioenergetics. Phosphorylation of TFAM impairs TFAM binding to DNA, therefore decreasing transcription and protein production within the mitochondria. Lon protease degrades unbound TFAM, noting Lon as a regulator of TFAM and mtDNA abundance [81]. Increased Lon expression reduced TFAM levels and mtDNA copy number, Lon regulates mitochondrial transcription by degrading misfolded TFAM allowing for a proper balance of mtDNA and functional TFAM [82]. An additional function of the Lon protease is its ability to bind mtDNA at single stranded G rich regions [83]. Lu presents evidence of Lon protease having decreased binding to mtDNA under oxidative stress [84]. Lon protease upregulation at the (-623, +1) promoter region leads to its actions as a stress response protein [85]. Use of SiRNA confirms Lon proteases actions as a stress response protein, such as removal of oxidized proteins and diminished mitochondrial function [86].
TFAM activation decreases ROS production in two ways; via Lon protease and down regulation of nuclear factor of activated T-cells (NFAT). ROS driven apoptosis is managed by the mitochondrial Lon protease. ROS production has been reported as the major factor in hypoxia induced cardiomyocyte apoptosis. Under hypoxic conditions, hypoxia inducibility factor 1 alpha activates Lon protease binding to the promoter region [87]. Interestingly, c-myc interacts with HIF-1 alpha decreasing TFAM expression. C-myc binds TFAM gene and assists in mitochondrial biogenesis [88]. Down regulated TFAM causes mitochondrial oxidative phosphorylation dysfunction, resulting in increased ROS production [89]. This may be the merging interaction between Lon regulation of TFAM protein expression. Under hypoxic conditions the nuclear transcription factor HIF-1alpha regulates the transcription of cytochrome c oxidase cardiac isoform 7a1 (COX7a1) [90]. COX7a1 knockout mice have reduced cox activity leading to dilated cardiomyopathy [91]. HIF-1alpha initiates transcription of COX4-2 and Lon protease, Lon is responsible for the degradation of COX4-1 subunit in yeast [92]. Hypoxic conditions such as post MI, induce stress factors such as HIF1alpha and Lon to manage cardiomyocyte stability. Overall, up regulating TFAM reduces the negative effects of ROS driven apoptosis (Fig 3).
Nuclear Factor of Activated T-cells and ROS
Nuclear factor of activated T-cells (NFAT) is a nuclear transcription factor and activator of innate immunity. This calcium/calmodulin/calcineurin activated pathway plays an essential role in the activation of T cells and related cytokines [93]. Our immune system recognizes different pattern recognition receptors on immune cells like T cells, macrophages, and neutrophils. This activation initiates NF-kB and mitogen-associated protein kinases (MAPK). Toll-like receptors are essential for the activation of T cells. They are found spanning the membrane of immune cells and play a major role in inflammatory activation. A necessary component of Toll-like receptor (TLR) functional activity includes its regulation by NFAT [94]. Interestingly, prolonged stimulation of TLR-4 leads to translocation of NFAT into the mitochondria, which is noted to reduce ATP production [95]. Calcineurin a mechanistic component of NFAT upregulation is a calcium, calmodulin dependent serine/threonine phosphatase with a catalytic subunit calcineurin A (CnA) and a calcium binding subunit CnB. Calmodulin dephosphorylates NFAT at the N terminus causing the translocation of NFAT to the nucleus [93]. Studies regarding the role of TFAM in cardio-protection include the Fujino study stating that TFAM provides cardio-protection via inhibiting nuclear factor of activated T cells (NFAT). Interestingly, they observed that overexpression of TFAM in cultured cardiomyocytes increased mtDNA and blocked NFAT. [96] Based on the literature NFAT stimulates pathological hypertrophy. Coincidentally, the role of free HSP-60 affects NFAT activation of T cells at the binding domain of TLR-4. Exogenous HSP-60 acts to bind to TLR-4 and induce apoptotic signaling [97]. Kim et al state that this may be a mechanism of myocyte loss in HF. Li states that myocardial ischemia induces an apoptotic signaling cascade showing that HSP-60 increases the expression of caspases 3 & 8. In TLR-4 deficient mice I/R injury had reduced myocardial apoptosis and cytokine expression [98]. The interplay between HSP-60 and TLR-4 is essential in mediating apoptotic signaling within cardiovascular diseases. Contrastingly, the HSP60-70 complex plays a positive regulatory function within cardiomyocytes. Diabetic pathologies induce the over production of ROS and increases TLR-4 expression [99]. Interestingly this is NF-kB dependent, which is most likely due to NF-kB regulation over NFAT upregulation [100].
NFAT regulates the expression of mitochondrial ROS generating enzymes; NADPH Oxidases (NOX), specifically NOX 2&4. [101] Mitochondria and NOX enzymes are the leading source of ROS involved in cardiac pathology [102] TFAM up regulation and inhibition of NFAT activation would decrease ROS production by NOX 2&4 reducing MMP-2 & 9 proteolytic activities (Fig 3). Speculation regarding the activity of NOX enzymes led to the Sciaretta groups discovery that cardiac specific NOX4 is important in activation of cardiomyocyte autophagy [103]. Analysis of cardiomyocyte autophagy leads to the up regulation of the proteolytic factor calpain. Up regulation of ROS in cardiomyocytes induced the nuclear translocation of calpain2 and inhibition of NOX function resisted calpain 2 translocation [104]. Myocardial infarction models show that overexpression of human TFAM in mice mitigates cardiac dysfunction and increases mtDNA copy number [105]. TFAM up regulation is evident for cardiomyocyte stability, even more so in a diseased model. Intriguingly, NOX 5 is a calcium activated ROS generator and acts in pulsatile fashion. Banfi et al found that upon calcium activation increases NOX 5 activity, which highly induces large amounts of ROS production, it also acts as a proton channel [106]. NOX 5 activity is entirely dependent upon intracellular calcium levels and phosphorylation of NOX 5 sites Thr 494 and Ser 498 led to increased calcium sensitivity [107]. Calmodulin kinase could sensitize NOX 5 in the presence of calcium mishandling. Calpain 10 a calcium induced mitochondrial protease is necessary for functional mitochondrial activity. Smith observed that overexpression and loss of calpain 10 led to mitochondrial decline and degradation of mitochondrial calpain 10 is regulated by Lon protease. Lon protease cleaved calpain 10 at an increased rate during mitochondrial oxidation [108].
Mice with a triple KO of NOX 1, 2 and 4 had no effect on NADPH chemo-luminescent signaling in cardiac, renal and aortic tissue samples. They showed that overexpression of NOX 4 & 5 greatly increased ROS production [109]. TFAM is produced at the nucleus, therefore the decrease that we see in TFAM may be protease cutting TFAM or inhibiting the HSP-60/HSP-70 complex (Fig 1).
Serca2a & Calcium Mishandling
Calpains are cytosolic cysteine proteases activated by increased cytosolic calcium levels. Calpains are responsible for the proteolytic degradation of both contractile and cytosolic proteins. Mitigation of cardiomyocyte deterioration depends upon the regulation of calcium also known as calcium handling. A major factor in calcium handling within the cardiomyocyte is the sarco(endo)plasmic reticulum Ca2+ ATPase 2a (Serca2a) which is an ATP dependent calcium transporter. It is responsible for transporting calcium into the SR, while decreasing free calcium in the cytoplasm. When down regulated, excess calcium is found within the cytoplasm and activates calpains. The proteolytic activity of calpains leads to intracellular degradation of proteins such as Titin, an essential contractile protein [110]. Serca2a regulation of calcium is crucial to cardiomyocyte stability. Multiple laboratories have explored the therapeutic use of up regulating Serca2a in cardiomyopathies as discussed in a review by Sikkel et al. Serca2a is a key protein involved in the sequestration of Ca2+ into the SR and is reduced in the heart failure model. Within cardiomyocytes, G protein coupled stimulation of p21-activated kinase-1 (Pak-1) has been shown to transcriptionally regulate Serca2a [111]. Interestingly, sinoatrial node pacemaker cells utilize Pak-1 as a regulator of L-type calcium channels and delays potassium channels through direct activation of protein phosphatase 2a [112]. Regulatory activity of calcium channels within pacemaker cells allows for myocardial syncytium activity. According to Huang et al ERK 1, 2 suppress Serca2a transcription via NFkB related mechanism [113]. This activity was observed in a cardiac hypertrophic state. Regulation of Serca2a is essential to a functional myocardium. Increased cytosolic calcium activates the calcineurin/NFAT pathway, which acts to inhibit Serca2a gene transcription, resulting in Serca2a downregulation [114]. It is found in the literature that activation of the NFAT pathway directly stimulates hypertrophic genes.
Analysis of the critical function of Serca2a was observed in knockout (KO) models allowing for insight into the significance of its activity, heart failure induced death occurred after 7–10 weeks [115]. Li analyzed the calcium dynamics in these KO mice, stating that calcium maintenance is best regulated by increased activity of the sodium-calcium exchanger and L-type calcium currents [116]. Patch-clamp analysis in Serca2 KO mice revealed a decrease in the percent of calcium induced transient amplitudes [117]. This finding shows a variation of electrical activity found within KO mice. Serca2a activity has been noted to suppress cellular alternans found in ventricular arrhythmias [118]. A mouse study analyzing the effects of levosimendan (a calcium sensitizer) on KO models with HF, showed improved contractility, relaxation and reduced calcium transients [119]. Swift et al reveals the electrophysiological and compensatory remodeling of the myocardium post KO of serca2a. Observations included a great increase in T-Tubule density and the newly grown longitudinal T-tubules had Na+/Ca2+ exchanger proximal to SR ryanodine receptors, showing a major switch in calcium driven transients, now coming from cytoplasmic calcium as opposed to normal SR driven release [120]. Speaking of the importance of the Na+/Ca2+ exchanger (NCX) in calcium regulation in the absence of serca2a, brings us to the Lu study on HF showing an imbalance in these two major regulatory proteins. CaMKIIδB regulates NCX, it is stated that pharmacological inhibition of CaMKIIδB improved cardiac function [121]. Electrical activation of the calcium wave seems to be Serca2a dependent, it was observed that CAMKII inhibition decreased the Ca2+ spark, CAMKII shift of SR calcium potentiates the calcium wave [122].
Gene therapy involving Serca2a up regulation is a major topic of discussion in translational research/clinical medicine [123–125]. Serca2a gene therapy for HF models including acute ischemia/reperfusion, chronic pressure overload and chronic myocardial infarction resulted in a reduction in ventricular arrhythmias. Studies show that Serca2a gene delivery enhanced contractile function and restores electrical stability in the heart failure model both caused by SR Ca2+ leak [126]. Serca2a gene transfer reduced Ca2+ leak and ameliorated ventricular arrhythmias in HF models [127]. Use of adenoviral Serca2a vectors overexpress the Serca2a protein causing ameliorating affects in the HF model [127,128]. Phase 1 human clinical gene therapy trials support the in vitro and in vivo findings of other investigators resulting in positive cardiovascular events post intracoronary injection of Serca2a adenovirus vector [129]. In general, scientific literature supports the beneficial aspects of Serca2a gene therapy, as is readily reported [130].
Mitochondrial dysfunction affects both nuclear and mt expression of TFAM. Clinical HF trials utilize Serca2a gene therapy as a method of therapeutic repair. HF models have reduced mitochondrial function leading to reduced Serca2a expression due to loss of nuclear TFAM. Reduced mitochondrial biogenesis due to lack of TFAM expression reduces nuclear expression of Serca2a. Serca2a up regulation as discussed above is highly beneficial to the HF model. The regulation of Serca2a gene transcription by nuclear TFAM allows for gene and protein up regulation of Serca2a [131]. Watanabe et al observed a correlation between Serca2a mRNA levels and nuclear TFAM. TFAM’s regulation of the Serca2a promoter in cardiomyocytes leads to a potential therapeutic function. Therefore, up regulation of TFAM will result in up regulation of Serca2a, increased calcium handling into the SR and reduced protease activity. Due to calcium up regulation in the HF model, calpain activation is persistent. The literature suggests that continuous calpain up regulation is a major factor in the development of HF [132]. The use of calpain inhibitors improve cardiomyocyte electrical activity [133]. TFAM up regulation of Serca2a will decrease calcium and calpain activity in the cytoplasm ameliorating cardiomyopathies.
Cytoplasmic calcium regulation is the key to degradative activity within the myocardium. Calpains have been found within the mitochondrial matrix and contribute to mitochondrial apoptosis. The addition of calcium to isolated mitochondria inactivates the activity of the electron transport chain at Complex I by cleaving the ND6 subunit. It is shown that calpains I, II & X are expressed within the mitochondria. Post I/R injury Calpain I is upregulated and contributes to the opening of the mitochondrial permeability transition pore (mPTP) [134]. Recent reviews covering the importance of calcium regulation and apoptosis unveil the mechanistic application of proteolytic activity found within cell death [135–137]. To calibrate the importance of protease activity within the mitochondria, observations of mitochondrial calpain I activity were assessed. Calpain I is responsible for cleaving apoptosis inducibility factor (AIF) which is then translocated to the nucleus [138]. TFAM regulation over Serca2a would decrease cytoplasmic calcium levels and alleviate calpain activity within the cytoplasm. We have expressed that homocysteine induces the intramitochondrial translocation of calpain 1 resulting in an intramitochondrial oxidative burst propagating MMP9 activity [139]. Apoptotic inducing factor (AIF) is a pro-apoptotic factor located on the inner mitochondrial membrane and is activated by calpain cleavage; mitochondrial ROS increases the susceptibility of cleavage [135]. Additionally, pro-apoptotic factor Bid is cleaved by calpain and results in the release of cytochrome c as found in a myocardial ischemia/reperfusion model [140]. Overall, we can conclude that increased calcium leads to upregulated calpain protease activity, which results in activation of a pro-apoptotic pathway. As for mitochondrial calpain activity, SR regulation and proximity via fusion protein Mfn-2 (SR-‘mitochondrial coupling) shows significant importance for the physiological myocardium (Fig 3).
TFAM Directed Therapeutics
Cardioprotection & Diabetes
Multiple vehicle methods exist for the transportation of regulatory elements; such transport mechanisms include viruses, bacterial and viral vectors. The literature thoroughly describes the drawbacks of the various delivery methods with a particular focus on stimulation of immunogenic factors. Thomas and colleagues studied the effects of (recombinant) Rh-TFAM on aged mice. Findings in cardiac tissue of aged mice include loss of mtDNA gene copy numbers and reduced mitochondrial gene transcription. Intravenous injections of Rh-TFAM led to increased mt respiration in heart and muscle tissue. Age-related studies showed a 66% decrease in cardiac mtDNA gene levels. Post treatment, PGC-1 alpha was increased by 102% and NRF-2 increased by 30% within cardiac tissue. Additionally, SIRT3 (a major mitochondrial deacetylase) was increased post treatment. Increases in levels of mitochondrial electron transport chain subunits including complexes I & IV, ATP activity and reduced oxidative stress were found post treatment [141]. Increased delivery systems using TFAM may lead to enhanced functional components within mitochondria.
Exosomes range from 30–100nm in size and have the potential to be “therapeutic game changers” in the future of biomedical technologies. Infused within their membranous vessels are mRNA, miRNA, siRNA, proteins and nucleic acids that factor into changes in gene expression. Evidence of therapeutic advances utilizing an exosomal delivery system is found ubiquitously within the literature. We explored the outcomes of exercise on exosome content and found a positive correlation between highly up regulated mir29b & mir455 and exercise, resulting in decreased MMP-9 expression [142]. Our group has characterized the favorable results of decreasing MMP9 on increased differentiation of stem cells into cardiomyocytes [143].
In vitro cardiomyocyte contractility analysis via transfection of TIMP-4 (an MMP-9 inhibitor) led to an up regulation of serca2a and its regulator mir122a [144]. Up regulation of TFAM via an exosomal vehicle will not only regulate MMP activity but also calpain proteases (Fig 3). Wang introduces the cardioprotective functionality of pluripotent stem cell exosomes in their ability to deliver miRNA’s essential for inhibiting cardiomyocyte apoptosis post myocardial ischemia [145]. Our laboratory observed that Cur-Exosc reduced the effects of in vivo oxidative stress [146]. Cardiac progenitor derived exosomes are cardioprotective post myocardial ischemia/reperfusion [147]. Plasma exosomes also show cardioprotective aspects [148]. These cardioprotective observations are profoundly based on the content found within the exosomes leading to mitochondrial and nuclear DNA regulation. Many reviews have focused on the potential for exosomal microRNA content and its effects on cardiomyopathies [149]. Gray found that cardiac progenitor cell exosomes contained 11 miRNA’s that were upregulated in hypoxic conditions. Interestingly, hypoxic exosomes enhanced cardiac function and reduced fibrosis [150]. Recently we reported exercise effects on the miRNA content of cardiosomes. We speculate that specific miRNA’s such as mir455, mir29b, mir323-5p & mir466 are increased after exercise therefore being cardioprotective. These miRNA function by binding the 3′ end of the MMP9 promoter region inhibiting its degradative activity [151]. Exosomal delivery overexpressing HIF-1 alpha (known to induce HSP70) also showed miRNA-126 and 210 upregulation, which was found to improve cardiac progenitor cell survival rates [152].
TFAM expression by drugs or natural compounds
Exogenous routes of increasing TFAM levels will induce cytoprotective effects. Interestingly, natural components increase mitochondrial function and prevent disease. Compounds such as grape seed procyanidin B2 (GSPB2), daidzein, humanin and honokoil all increase mitochondrial copy number and biogenic function. GSPB2 inhibited glucose induced apoptosis and suppressed ROS production. Additionally, mRNA expressions of NRF-1, TFAM and mtDNA copy number were vastly increased. This group also showed activation of the PGC-1alpha pathway leading to SIRT-1 activation [153]. Yoshino et al describe observations of the dietary substance soy isoflavone daidzein on muscle mitochondria. This soy extract directly promotes TFAM expression increasing mitochondrial biogenesis through activation of PGC-1 alpha [154]. Interestingly, Qin et al investigated the effects of sulfur dioxide leading to cardiac dysfunction via mitochondrial impairment, overexpression of TFAM ameliorated mt decline [155]. Humanin is a natural mitochondrial peptide that greatly increased mtDNA copy number, upregulated TFAM, activated STAT3 and decreased caspase-3 activation [156]. Honokoil is a lignan isolated from magnolia tree bark and seed cones. In studying hypertrophy, honokoil a known anti-cancer, anti-oxidative and anti-inflammatory substance acts to reduce mitochondrial stress through activation of SIRT-3. Sirtuin-3 protects from cardiac hypertrophy via inhibition of NFAT and reduces ROS production. The PGC-1alpha Sirtuin & TFAM axis plays a major role in cardiomyocyte stability [157] Increasing the PGC-1alpha pathway via honokoil will increase TFAM levels leading to greater mitochondrial stability and reduced hypertrophy as observed. Therefore the cardioprotective nature of TFAM-packed exosomes is a novel and unexplored idea. Mei et al observed that cancer cell lines increased mtDNA copy number in the presence of chemotherapeutic treatments Cisplatin and Doxorubicin. This tumorigenic protective mechanism was diminished by transfection of shRNA-TFAM plasmids, which reduced mtDNA copy number and increased ROS production [158]. It is interesting that cell stability is dependent on mtDNA copy number.
Diabetic Mitochondrial Stability
Diabetic TFAM-transgenic mice showed significant prevention of downstream damage compared to mitochondrial DNA damage of WT mice [159]. Findings include that overexpression of TFAM inhibited rotenone-induced mt ROS production and nuclear factor-kappaB (NFkB) translocation. TFAM-TG models showed reduced mitochondrial damage with age [160]. Increased TFAM enhances mt stability and reduces oxidative stress. A patient based study analyzing single nucleotide polymorphisms within the TFAM gene (rs1937 and rs2306604) showed that mutations in the TFAM gene contribute to declining mitochondria biogenesis leading to disease [161]. The Aguirre-Reuda group found that overexpression of PGC-1alpha and TFAM protect against oxidative stress [162]. Overexpression of TFAM leads to a mitochondrial rescue, which impacts the survival rate of mitochondrial driven diseases. An interesting complimentary study by the Xu group expressed the beneficial effects of TFAM overexpression on mitigating ROS production [163].
Cardiac hypertrophic state is found among diabetes related cardiomyopathies. Adiponectin an adipocyte secreted hormone is stated to be cardioprotective. Functionally, adiponectin which is released from adipocytes increases insulin sensitivity, glucose uptake and fatty acid oxidation [164]. Diabetic mice with chikusetsu saponin IVa pretreatment after I/R injury showed ameliorating results by enhancing adiponectin levels. Adiponectin upregulation increased AMPK activity, thereby increasing GSK-3beta for increased glucose release for this adiponectin induced insulin sensitivity and enhanced glucose uptake [165]. Glycogen synthase kinase (GSK3b) phosphorylates glycogen stores initiating the release of glucose from glycogen storage. The cardioprotective aspects of adiponectin are found within its regulator activity over Serca2a. According to Yan et al, adiponectin increases CamKII phosphorylation of phospholamban, which directly increases Serca2a activity. It is also noted that this activity is stimulated by sphingosine-1-phosphate [166].
Extracellular signal-related kinase activation (ERK 1, 2) regulates mt function through direct phosphorylation of TFAM resulting in suppression of mt transcription [167]. TFAM phosphorylation impairs DNA binding and promotes degradation by Lon protease [81]. Adiponectin activity is inhibited in the pathological state of diabetes; it is interesting that diabetes related cardiomyopathies are greatly increased as compared to other synergistic pathologies.
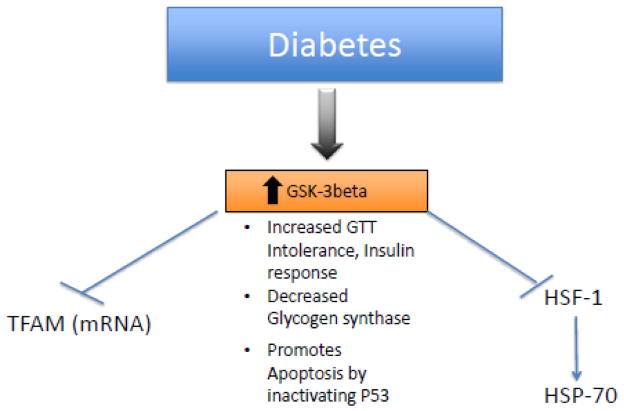
The Santos group expresses the effects of diabetes on mitochondrial biogenesis showing that hyperglycemia reduces mitochondrial biogenesis and the binding rate of HSP-70 and TFAM is 30% lower than normal [168]. The activator of heat shock proteins called heat shock transcription factor (HSF-1) is down-regulated by both GSK3beta and ERK, MAPK resulting in a loss of HSP-70, this action is performed by displacing HSF-1 from its active transcription site [169]. A pharmacological inhibitor of GSK3beta affected mitochondrial biogenesis resulting in loss of mRNA TFAM expression but had no effect on TFAM content. Also glycolysis inhibitor 2-Deoxyglucose resulted in an increase in mitochondrial DNA content and increased levels of PGC-1alpha, NRF-1 and TFAM, therefore decreasing glucose levels favors TFAM production [170]. Inactivation of GSK3beta assisted in recovery of the mitochondrial membrane potential by suppressing ROS and inhibiting the opening of the mitochondrial permeability transition pore [171]. Fig 4 represents events that connect diabetes with TFAM. We speculate that the mitochondrial membrane potential depolarization could propagate an ionic signal to other mitochondria attached to the same SR inducing depolarization of other interconnected mitochondrial membranes.
Fig 4.
In diabetes there is increase in the GSK-3 beta expression, which is known to inhibit TFAM and HSF-1 both necessary for mitochondrial biogenesis. There is Increased GTT Intolerance, decreased insulin response and decreased Glycogen synthase. It also promotes apoptosis by inactivating P53.
Final Statement
In conclusion, this review states that TFAM which is a mitochondrial transcription factor is synthesized in the nucleus and transported to mitochondria with the help of hsp60-70 complex and the binding of Lon protease to hsp60 plays a role in mitochondrial regulatory activity. TFAM upregulation within mitochondria reduces ROS production (Fig 5). The absence of TFAM in mitochondria of failing hearts plays a role mitochondrial dysfunction (Fig 5). TFAM is regulated by epigenetic factors like methylation of the mtDNA, TFAM promoter and microRNAs (Fig 6). As we have hypothesized in this review TFAM-PE may assist in mitochondrial related cardiomyopathies both dependent and independent of the negative synergistic effects of diabetes. TFAM regulates cardiomyocyte cytoplasmic calpain and calcium levels via increasing Serca2a mRNA and protein levels. Additionally, TFAM regulates ROS production via inhibiting NFAT activity, which is a hypertrophic stimulator and NOX regulator. Exogenous delivery of TFAM is beneficial to cardiac function.
Fig 5. Comparison of a healthy and failing heart shows the molecular transport of TFAM.
With respect to the increased levels of ROS in the failing heart which up-regulates lon protease activity. We speculate that upon Lon binding to the “HSP60-HSP70 complex, a locking mechanism occurs forcing TFAM to be trapped within the complex and not allow for TFAM transport to the mitochondria. Lack of TFAM transport leads to mitochondrial decline.
Acknowledgments
The study was supported by NIH grants HL-74185 and HL-108621, AHA grant 15POST23110021 to PC and NIH F31 grant 1F31HL132527-01 to GHK.
Footnotes
Conflicts of interest: The authors claim that there are no conflicts of interest
References
- 1.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13(9):935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 2.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature genetics. 1998;18(3):231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 3.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic acids research. 2003;31(6):1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A. 1994;91(4):1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kukat C, Wurm CA, Spåhr H, Falkenberg M, Larsson N-G, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(33):13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kukat C, Davies KM, Wurm CA, Spahr H, Bonekamp NA, Kuhl I, Joos F, Polosa PL, Park CB, Posse V, Falkenberg M, Jakobs S, Kuhlbrandt W, Larsson NG. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1512131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kienhofer J, Haussler DJ, Ruckelshausen F, Muessig E, Weber K, Pimentel D, Ullrich V, Burkle A, Bachschmid MM. Association of mitochondrial antioxidant enzymes with mitochondrial DNA as integral nucleoid constituents. Faseb j. 2009;23(7):2034–2044. doi: 10.1096/fj.08-113571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hensen F, Cansiz S, Gerhold JM, Spelbrink JN. To be or not to be a nucleoid protein: a comparison of mass-spectrometry based approaches in the identification of potential mtDNA-nucleoid associated proteins. Biochimie. 2014;100:219–226. doi: 10.1016/j.biochi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 10.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nature genetics. 2002;31(3):289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 11.Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nature genetics. 2003;33(1):23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 12.Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. The Journal of biological chemistry. 1992;267(5):3358–3367. [PubMed] [Google Scholar]
- 13.Correia RL, Oba-Shinjo SM, Uno M, Huang N, Marie SK. Mitochondrial DNA depletion and its correlation with TFAM, TFB1M, TFB2M and POLG in human diffusely infiltrating astrocytomas. Mitochondrion. 2011;11(1):48–53. doi: 10.1016/j.mito.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Morozov YI, Agaronyan K, Cheung AC, Anikin M, Cramer P, Temiakov D. A novel intermediate in transcription initiation by human mitochondrial RNA polymerase. Nucleic Acids Res. 2014;42(6):3884–3893. doi: 10.1093/nar/gkt1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morozov YI, Parshin AV, Agaronyan K, Cheung AC, Anikin M, Cramer P, Temiakov D. A model for transcription initiation in human mitochondria. Nucleic Acids Res. 2015;43(7):3726–3735. doi: 10.1093/nar/gkv235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Surovtseva YV, Shadel GS. Transcription-independent role for human mitochondrial RNA polymerase in mitochondrial ribosome biogenesis. Nucleic Acids Res. 2013;41(4):2479–2488. doi: 10.1093/nar/gks1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotney J, McKay SE, Shadel GS. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum Mol Genet. 2009;18(14):2670–2682. doi: 10.1093/hmg/ddp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9(4):386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, Thoren P, Larsson NG. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nature genetics. 1999;21(1):133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 20.Lauritzen KH, Kleppa L, Aronsen JM, Eide L, Carlsen H, Haugen OP, Sjaastad I, Klungland A, Rasmussen LJ, Attramadal H, Storm-Mathisen J, Bergersen LH. Impaired dynamics and function of mitochondria caused by mtDNA toxicity leads to heart failure. Am J Physiol Heart Circ Physiol. 2015 doi: 10.1152/ajpheart.00253.2014. ajpheart.00253.02014. [DOI] [PubMed] [Google Scholar]
- 21.Deocaris CC, Kaul SC, Wadhwa R. On the brotherhood of the mitochondrial chaperones mortalin and heat shock protein 60. Cell stress & chaperones. 2006;11(2):116–128. doi: 10.1379/CSC-144R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao TY, Chiu YC, Fang WC, Cheng CW, Kuo CY, Juan HF, Wu SH, Lee AY. Mitochondrial Lon regulates apoptosis through the association with Hsp60-mtHsp70 complex. Cell death & disease. 2015;6:e1642. doi: 10.1038/cddis.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos JM, Mishra M, Kowluru RA. Posttranslational modification of mitochondrial transcription factor A in impaired mitochondria biogenesis: implications in diabetic retinopathy and metabolic memory phenomenon. Exp Eye Res. 2014;121:168–177. doi: 10.1016/j.exer.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciesielski GL, Plotka M, Manicki M, Schilke BA, Dutkiewicz R, Sahi C, Marszalek J, Craig EA. Nucleoid localization of Hsp40 Mdj1 is important for its function in maintenance of mitochondrial DNA. Biochimica et biophysica acta. 2013;1833(10):2233–2243. doi: 10.1016/j.bbamcr.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naka KK, Vezyraki P, Kalaitzakis A, Zerikiotis S, Michalis L, Angelidis C. Hsp70 regulates the doxorubicin-mediated heart failure in Hsp70-transgenic mice. Cell Stress Chaperones. 2014;19(6):853–864. doi: 10.1007/s12192-014-0509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko SK, Kim J, Na DC, Park S, Park SH, Hyun JY, Baek KH, Kim ND, Kim NK, Park YN, Song K, Shin I. A small molecule inhibitor of ATPase activity of HSP70 induces apoptosis and has antitumor activities. Chem Biol. 2015;22(3):391–403. doi: 10.1016/j.chembiol.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Xu T, Zhang B, Yang F, Cai C, Wang G, Han Q, Zou L. HSF1 and NF-kappaB p65 participate in the process of exercise preconditioning attenuating pressure overload-induced pathological cardiac hypertrophy. Biochem Biophys Res Commun. 2015;460(3):622–627. doi: 10.1016/j.bbrc.2015.03.079. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, Chen Y, Auger-Messier M, Molkentin JD. Interaction between NFkappaB and NFAT coordinates cardiac hypertrophy and pathological remodeling. Circulation research. 2012;110(8):1077–1086. doi: 10.1161/circresaha.111.260729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Givvimani S, Munjal C, Tyagi N, Sen U, Metreveli N, Tyagi SC. Mitochondrial division/mitophagy inhibitor (Mdivi) ameliorates pressure overload induced heart failure. PloS one. 2012;7(3):e32388. doi: 10.1371/journal.pone.0032388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Givvimani S, Pushpakumar SB, Metreveli N, Veeranki S, Kundu S, Tyagi SC. Role of mitochondrial fission and fusion in cardiomyocyte contractility. International journal of cardiology. 2015;187:325–333. doi: 10.1016/j.ijcard.2015.03.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Sparks M, Bhandari P, Matkovich SJ, Dorn GW., 2nd Mitochondrial genome linearization is a causative factor for cardiomyopathy in mice and Drosophila. Antioxid Redox Signal. 2014;21(14):1949–1959. doi: 10.1089/ars.2013.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishihara T, Ban-Ishihara R, Maeda M, Matsunaga Y, Ichimura A, Kyogoku S, Aoki H, Katada S, Nakada K, Nomura M, Mizushima N, Mihara K, Ishihara N. Dynamics of mitochondrial DNA nucleoids regulated by mitochondrial fission is essential for maintenance of homogeneously active mitochondria during neonatal heart development. Molecular and cellular biology. 2015;35(1):211–223. doi: 10.1128/mcb.01054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papanicolaou KN, Khairallah RJ, Ngoh GA, Chikando A, Luptak I, O’Shea KM, Riley DD, Lugus JJ, Colucci WS, Lederer WJ, Stanley WC, Walsh K. Mitofusin-2 maintains mitochondrial structure and contributes to stress-induced permeability transition in cardiac myocytes. Molecular and cellular biology. 2011;31(6):1309–1328. doi: 10.1128/mcb.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Csordas G, Jowdy C, Schneider TG, Csordas N, Wang W, Liu Y, Kohlhaas M, Meiser M, Bergem S, Nerbonne JM, Dorn GW, 2nd, Maack C. Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic responses via interorganelle Ca(2+) crosstalk. Circulation research. 2012;111(7):863–875. doi: 10.1161/circresaha.112.266585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Li X, Guan Y, Guo X. Mitofusin-2-mediated tethering of mitochondria and endoplasmic reticulum promotes cell cycle arrest of vascular smooth muscle cells in G0/G1 phase. Acta biochimica et biophysica Sinica. 2015;47(6):441–450. doi: 10.1093/abbs/gmv035. [DOI] [PubMed] [Google Scholar]
- 36.Williams GS, Boyman L, Lederer WJ. Mitochondrial calcium and the regulation of metabolism in the heart. Journal of molecular and cellular cardiology. 2015;78:35–45. doi: 10.1016/j.yjmcc.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luongo TS, Lambert JP, Yuan A, Zhang X, Gross P, Song J, Shanmughapriya S, Gao E, Jain M, Houser SR, Koch WJ, Cheung JY, Madesh M, Elrod JW. The Mitochondrial Calcium Uniporter Matches Energetic Supply with Cardiac Workload during Stress and Modulates Permeability Transition. Cell reports. 2015;12(1):23–34. doi: 10.1016/j.celrep.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davidson SM, Foote K, Kunuthur S, Gosain R, Tan N, Tyser R, Zhao YJ, Graeff R, Ganesan A, Duchen MR, Patel S, Yellon DM. Inhibition of NAADP signalling on reperfusion protects the heart by preventing lethal calcium oscillations via two-pore channel 1 and opening of the mitochondrial permeability transition pore. Cardiovascular research. 2015 doi: 10.1093/cvr/cvv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chitra L, Boopathy R. Altered mitochondrial biogenesis and its fusion gene expression is involved in the high-altitude adaptation of rat lung. Respir Physiol Neurobiol. 2014;192:74–84. doi: 10.1016/j.resp.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Cartoni R, Leger B, Hock MB, Praz M, Crettenand A, Pich S, Ziltener JL, Luthi F, Deriaz O, Zorzano A, Gobelet C, Kralli A, Russell AP. Mitofusins 1/2 and ERRalpha expression are increased in human skeletal muscle after physical exercise. The Journal of physiology. 2005;567(Pt 1):349–358. doi: 10.1113/jphysiol.2005.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karamanlidis G, Nascimben L, Couper GS, Shekar PS, del Monte F, Tian R. Defective DNA replication impairs mitochondrial biogenesis in human failing hearts. Circulation research. 2010;106(9):1541–1548. doi: 10.1161/circresaha.109.212753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikeda M, Ide T, Fujino T, Arai S, Saku K, Kakino T, Tyynismaa H, Yamasaki T, Yamada K, Kang D, Suomalainen A, Sunagawa K. Overexpression of TFAM or twinkle increases mtDNA copy number and facilitates cardioprotection associated with limited mitochondrial oxidative stress. PLoS One. 2015;10(3):e0119687. doi: 10.1371/journal.pone.0119687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quinones-Lombrana A, Blanco JG. Chromosome 21-derived hsa-miR-155-5p regulates mitochondrial biogenesis by targeting Mitochondrial Transcription Factor A (TFAM) Biochimica et biophysica acta. 2015;1852(7):1420–1427. doi: 10.1016/j.bbadis.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto H, Morino K, Nishio Y, Ugi S, Yoshizaki T, Kashiwagi A, Maegawa H. MicroRNA-494 regulates mitochondrial biogenesis in skeletal muscle through mitochondrial transcription factor A and Forkhead box j3. American journal of physiology Endocrinology and metabolism. 2012;303(12):E1419–1427. doi: 10.1152/ajpendo.00097.2012. [DOI] [PubMed] [Google Scholar]
- 45.Li S, Yang G. Hydrogen Sulfide Maintains Mitochondrial DNA Replication via Demethylation of TFAM. Antioxid Redox Signal. 2015;23(7):630–642. doi: 10.1089/ars.2014.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang F, Qi Y, Zhou K, Zhang G, Linask K, Xu H. The cAMP phosphodiesterase Prune localizes to the mitochondrial matrix and promotes mtDNA replication by stabilizing TFAM. EMBO Rep. 2015;16(4):520–527. doi: 10.15252/embr.201439636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaturvedi P, Kalani A, Givvimani S, Kamat PK, Familtseva A, Tyagi SC. Differential regulation of DNA methylation versus histone acetylation in cardiomyocytes during HHcy in vitro and in vivo: an epigenetic mechanism. Physiological genomics. 2014;46(7):245–255. doi: 10.1152/physiolgenomics.00168.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PloS one. 2010;5(7):e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Ren X, Zhang Q, Li Z, Ma S, Bao J, Li Z, Bai X, Zheng L, Zhang Z, Shang S, Zhang C, Wang C, Cao L, Wang Q, Ji J. PGC-1alpha/ERRalpha-Sirt3 pathway regulates DAergic neuronal death by directly deacetylating SOD2 and ATP synthase beta. Antioxidants & redox signaling. 2015 doi: 10.1089/ars.2015.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uguccioni G, Hood DA. The importance of PGC-1alpha in contractile activity-induced mitochondrial adaptations. American journal of physiology Endocrinology and metabolism. 2011;300(2):E361–371. doi: 10.1152/ajpendo.00292.2010. [DOI] [PubMed] [Google Scholar]
- 51.Kowluru RA, Santos JM, Zhong Q. Sirt1, a negative regulator of matrix metalloproteinase-9 in diabetic retinopathy. Investigative ophthalmology & visual science. 2014;55(9):5653–5660. doi: 10.1167/iovs.14-14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrari R, Cargnoni A, Curello S, Boffa GM, Ceconi C. Effects of iloprost (ZK 36374) on glutathione status during ischaemia and reperfusion of rabbit isolated hearts. British journal of pharmacology. 1989;98(2):678–684. doi: 10.1111/j.1476-5381.1989.tb12643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loeper J, Goy J, Klein JM, Dufour M, Bedu O, Loeper S, Emerit J. The evolution of oxidative stress indicators in the course of myocardial ischemia. Free radical research communications. 1991;12–13(Pt 2):675–680. doi: 10.3109/10715769109145846. [DOI] [PubMed] [Google Scholar]
- 54.Loeper J, Goy J, Rozensztajn L, Bedu O, Moisson P. Lipid peroxidation and protective enzymes during myocardial infarction. Clinica chimica acta; international journal of clinical chemistry. 1991;196(2–3):119–125. doi: 10.1016/0009-8981(91)90064-j. [DOI] [PubMed] [Google Scholar]
- 55.Iqbal M, Cohen RI, Marzouk K, Liu SF. Time course of nitric oxide, peroxynitrite, and antioxidants in the endotoxemic heart. Critical care medicine. 2002;30(6):1291–1296. doi: 10.1097/00003246-200206000-00021. [DOI] [PubMed] [Google Scholar]
- 56.Supinski GS, Murphy MP, Callahan LA. MitoQ administration prevents endotoxin-induced cardiac dysfunction. American journal of physiology Regulatory, integrative and comparative physiology. 2009;297(4):R1095–1102. doi: 10.1152/ajpregu.90902.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laguens RP, Gomez-Dumm CL. Fine structure of myocardial mitochondria in rats after exercise for one-half to two hours. Circulation research. 1967;21(3):271–279. doi: 10.1161/01.res.21.3.271. [DOI] [PubMed] [Google Scholar]
- 58.Kane JJ, Murphy ML, Bissett JK, deSoyza N, Doherty JE, Straub KD. Mitochondrial function, oxygen extraction, epicardial S-T segment changes and tritiated digoxin distribution after reperfusion of ischemic myocardium. The American journal of cardiology. 1975;36(2):218–224. doi: 10.1016/0002-9149(75)90530-5. [DOI] [PubMed] [Google Scholar]
- 59.Jennings RB, Ganote CE. Mitochondrial structure and function in acute myocardial ischemic injury. Circulation research. 1976;38(5 Suppl 1):I80–91. [PubMed] [Google Scholar]
- 60.Evans MJ, Scarpulla RC. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes & development. 1990;4(6):1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- 61.Chau CM, Evans MJ, Scarpulla RC. Nuclear respiratory factor 1 activation sites in genes encoding the gamma-subunit of ATP synthase, eukaryotic initiation factor 2 alpha, and tyrosine aminotransferase. Specific interaction of purified NRF-1 with multiple target genes. The Journal of biological chemistry. 1992;267(10):6999–7006. [PubMed] [Google Scholar]
- 62.Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochimica et biophysica acta. 2002;1576(1–2):1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 63.Vercauteren K, Gleyzer N, Scarpulla RC. PGC-1-related coactivator complexes with HCF-1 and NRF-2beta in mediating NRF-2(GABP)-dependent respiratory gene expression. The Journal of biological chemistry. 2008;283(18):12102–12111. doi: 10.1074/jbc.M710150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Molecular and cellular biology. 2005;25(4):1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hickson-Bick DL, Jones C, Buja LM. Stimulation of mitochondrial biogenesis and autophagy by lipopolysaccharide in the neonatal rat cardiomyocyte protects against programmed cell death. Journal of molecular and cellular cardiology. 2008;44(2):411–418. doi: 10.1016/j.yjmcc.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 66.Suliman HB, Welty-Wolf KE, Carraway M, Tatro L, Piantadosi CA. Lipopolysaccharide induces oxidative cardiac mitochondrial damage and biogenesis. Cardiovascular research. 2004;64(2):279–288. doi: 10.1016/j.cardiores.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 67.Yue R, Xia X, Jiang J, Yang D, Han Y, Chen X, Cai Y, Li L, Wang WE, Zeng C. Mitochondrial DNA Oxidative Damage Contributes to Cardiomyocyte Ischemia/Reperfusion-Injury in Rats: Cardioprotective Role of Lycopene. J Cell Physiol. 2015 doi: 10.1002/jcp.24941. [DOI] [PubMed] [Google Scholar]
- 68.Spinale FG, Coker ML, Thomas CV, Walker JD, Mukherjee R, Hebbar L. Time-dependent changes in matrix metalloproteinase activity and expression during the progression of congestive heart failure: relation to ventricular and myocyte function. Circulation research. 1998;82(4):482–495. doi: 10.1161/01.res.82.4.482. [DOI] [PubMed] [Google Scholar]
- 69.Siwik DA, Tzortzis JD, Pimental DR, Chang DL, Pagano PJ, Singh K, Sawyer DB, Colucci WS. Inhibition of copper-zinc superoxide dismutase induces cell growth, hypertrophic phenotype, and apoptosis in neonatal rat cardiac myocytes in vitro. Circulation research. 1999;85(2):147–153. doi: 10.1161/01.res.85.2.147. [DOI] [PubMed] [Google Scholar]
- 70.Vu TH, Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes & development. 2000;14(17):2123–2133. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- 71.Tyagi SC. Dynamic role of extracellular matrix metalloproteinases in heart failure. Cardiovascular pathology : the official journal of the Society for Cardiovascular Pathology. 1998;7(3):153–159. doi: 10.1016/s1054-8807(97)00121-x. [DOI] [PubMed] [Google Scholar]
- 72.Weber KT, Sun Y, Tyagi SC, Cleutjens JP. Collagen network of the myocardium: function, structural remodeling and regulatory mechanisms. Journal of molecular and cellular cardiology. 1994;26(3):279–292. doi: 10.1006/jmcc.1994.1036. [DOI] [PubMed] [Google Scholar]
- 73.Mujumdar VS, Smiley LM, Tyagi SC. Activation of matrix metalloproteinase dilates and decreases cardiac tensile strength. International journal of cardiology. 2001;79(2–3):277–286. doi: 10.1016/s0167-5273(01)00449-1. [DOI] [PubMed] [Google Scholar]
- 74.Lebrecht D, Setzer B, Ketelsen UP, Haberstroh J, Walker UA. Time-dependent and tissue-specific accumulation of mtDNA and respiratory chain defects in chronic doxorubicin cardiomyopathy. Circulation. 2003;108(19):2423–2429. doi: 10.1161/01.CIR.0000093196.59829.DF. [DOI] [PubMed] [Google Scholar]
- 75.Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura K, Utsumi H, Hamasaki N, Takeshita A. Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circulation research. 2001;88(5):529–535. doi: 10.1161/01.res.88.5.529. [DOI] [PubMed] [Google Scholar]
- 76.Kanazawa A, Nishio Y, Kashiwagi A, Inagaki H, Kikkawa R, Horiike K. Reduced activity of mtTFA decreases the transcription in mitochondria isolated from diabetic rat heart. American journal of physiology Endocrinology and metabolism. 2002;282(4):E778–785. doi: 10.1152/ajpendo.00255.2001. [DOI] [PubMed] [Google Scholar]
- 77.Garnier A, Fortin D, Delomenie C, Momken I, Veksler V, Ventura-Clapier R. Depressed mitochondrial transcription factors and oxidative capacity in rat failing cardiac and skeletal muscles. The Journal of physiology. 2003;551(Pt 2):491–501. doi: 10.1113/jphysiol.2003.045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ikeuchi M, Matsusaka H, Kang D, Matsushima S, Ide T, Kubota T, Fujiwara T, Hamasaki N, Takeshita A, Sunagawa K, Tsutsui H. Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 2005;112(5):683–690. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- 79.Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, Medeiros DM, Valencik ML, McDonald JA, Kelly DP. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circulation research. 2004;94(4):525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- 80.Kunkel GH, Chaturvedi P, Tyagi SC. Resuscitation of a dead cardiomyocyte. Heart failure reviews. 2015;20(6):709–719. doi: 10.1007/s10741-015-9501-z. [DOI] [PubMed] [Google Scholar]
- 81.Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, Suzuki CK. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell. 2013;49(1):121–132. doi: 10.1016/j.molcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matsushima Y, Goto Y, Kaguni LS. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM) Proceedings of the National Academy of Sciences of the United States of America. 2010;107(43):18410–18415. doi: 10.1073/pnas.1008924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fu GK, Markovitz DM. The human LON protease binds to mitochondrial promoters in a single-stranded, site-specific, strand-specific manner. Biochemistry. 1998;37(7):1905–1909. doi: 10.1021/bi970928c. [DOI] [PubMed] [Google Scholar]
- 84.Lu B, Yadav S, Shah PG, Liu T, Tian B, Pukszta S, Villaluna N, Kutejova E, Newlon CS, Santos JH, Suzuki CK. Roles for the human ATP-dependent Lon protease in mitochondrial DNA maintenance. The Journal of biological chemistry. 2007;282(24):17363–17374. doi: 10.1074/jbc.M611540200. [DOI] [PubMed] [Google Scholar]
- 85.Pinti M, Gibellini L, De Biasi S, Nasi M, Roat E, O’Connor JE, Cossarizza A. Functional characterization of the promoter of the human Lon protease gene. Mitochondrion. 2011;11(1):200–206. doi: 10.1016/j.mito.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 86.Ngo JK, Davies KJ. Mitochondrial Lon protease is a human stress protein. Free Radic Biol Med. 2009;46(8):1042–1048. doi: 10.1016/j.freeradbiomed.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuo CY, Chiu YC, Lee AY, Hwang TL. Mitochondrial Lon protease controls ROS-dependent apoptosis in cardiomyocyte under hypoxia. Mitochondrion. 2015;23:7–16. doi: 10.1016/j.mito.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 88.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Molecular and cellular biology. 2005;25(14):6225–6234. doi: 10.1128/mcb.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mendelsohn AR, Larrick JW. Partial reversal of skeletal muscle aging by restoration of normal NAD(+) levels. Rejuvenation research. 2014;17(1):62–69. doi: 10.1089/rej.2014.1546. [DOI] [PubMed] [Google Scholar]
- 90.Hwang HJ, Lynn SG, Vengellur A, Saini Y, Grier EA, Ferguson-Miller SM, LaPres JJ. Hypoxia Inducible Factors Modulate Mitochondrial Oxygen Consumption and Transcriptional Regulation of Nuclear-Encoded Electron Transport Chain Genes. Biochemistry. 2015;54(24):3739–3748. doi: 10.1021/bi5012892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huttemann M, Klewer S, Lee I, Pecinova A, Pecina P, Liu J, Lee M, Doan JW, Larson D, Slack E, Maghsoodi B, Erickson RP, Grossman LI. Mice deleted for heart-type cytochrome c oxidase subunit 7a1 develop dilated cardiomyopathy. Mitochondrion. 2012;12(2):294–304. doi: 10.1016/j.mito.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129(1):111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 93.Vandewalle A, Tourneur E, Bens M, Chassin C, Werts C. Calcineurin/NFAT signaling and innate host defence: a role for NOD1-mediated phagocytic functions. Cell Commun Signal. 2014;12:8. doi: 10.1186/1478-811x-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Minematsu H, Shin MJ, Celil Aydemir AB, Kim KO, Nizami SA, Chung GJ, Lee FY. Nuclear presence of nuclear factor of activated T cells (NFAT) c3 and c4 is required for Toll-like receptor-activated innate inflammatory response of monocytes/macrophages. Cell Signal. 2011;23(11):1785–1793. doi: 10.1016/j.cellsig.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma B, Yu J, Xie C, Sun L, Lin S, Ding J, Luo J, Cai H. Toll-Like Receptors Promote Mitochondrial Translocation of Nuclear Transcription Factor Nuclear Factor of Activated T-Cells in Prolonged Microglial Activation. J Neurosci. 2015;35(30):10799–10814. doi: 10.1523/jneurosci.2455-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fujino T, Ide T, Yoshida M, Onitsuka K, Tanaka A, Hata Y, Nishida M, Takehara T, Kanemaru T, Kitajima N, Takazaki S, Kurose H, Kang D, Sunagawa K. Recombinant mitochondrial transcription factor A protein inhibits nuclear factor of activated T cells signaling and attenuates pathological hypertrophy of cardiac myocytes. Mitochondrion. 2012;12(4):449–458. doi: 10.1016/j.mito.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 97.Kim SC, Stice JP, Chen L, Jung JS, Gupta S, Wang Y, Baumgarten G, Trial J, Knowlton AA. Extracellular heat shock protein 60, cardiac myocytes, and apoptosis. Circulation research. 2009;105(12):1186–1195. doi: 10.1161/circresaha.109.209643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li Y, Si R, Feng Y, Chen HH, Zou L, Wang E, Zhang M, Warren HS, Sosnovik DE, Chao W. Myocardial ischemia activates an injurious innate immune signaling via cardiac heat shock protein 60 and Toll-like receptor 4. The Journal of biological chemistry. 2011;286(36):31308–31319. doi: 10.1074/jbc.M111.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dasu MR, Jialal I. Free fatty acids in the presence of high glucose amplify monocyte inflammation via Toll-like receptors. American journal of physiology Endocrinology and metabolism. 2011;300(1):E145–154. doi: 10.1152/ajpendo.00490.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wei M, Li Z, Xiao L, Yang Z. Effects of ROS-relative NF-kappaB signaling on high glucose-induced TLR4 and MCP-1 expression in podocyte injury. Mol Immunol. 2015 doi: 10.1016/j.molimm.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 101.Williams CR, Gooch JL. Calcineurin Abeta regulates NADPH oxidase (Nox) expression and activity via nuclear factor of activated T cells (NFAT) in response to high glucose. The Journal of biological chemistry. 2014;289(8):4896–4905. doi: 10.1074/jbc.M113.514869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sorescu D, Griendling KK. Reactive oxygen species, mitochondria, and NAD(P)H oxidases in the development and progression of heart failure. Congestive heart failure (Greenwich, Conn) 2002;8(3):132–140. doi: 10.1111/j.1527-5299.2002.00717.x. [DOI] [PubMed] [Google Scholar]
- 103.Sciarretta S, Yee D, Ammann P, Nagarajan N, Volpe M, Frati G, Sadoshima J. Role of NADPH oxidase in the regulation of autophagy in cardiomyocytes. Clinical science (London, England : 1979) 2015;128(7):387–403. doi: 10.1042/cs20140336. [DOI] [PubMed] [Google Scholar]
- 104.Chang H, Sheng JJ, Zhang L, Yue ZJ, Jiao B, Li JS, Yu ZB. ROS-Induced Nuclear Translocation of Calpain-2 Facilitates Cardiomyocyte Apoptosis in Tail-Suspended Rats. Journal of cellular biochemistry. 2015 doi: 10.1002/jcb.25176. [DOI] [PubMed] [Google Scholar]
- 105.Kang D, Kim SH, Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion. 2007;7(1–2):39–44. doi: 10.1016/j.mito.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 106.Banfi B, Molnar G, Maturana A, Steger K, Hegedus B, Demaurex N, Krause KH. A Ca(2+)-activated NADPH oxidase in testis, spleen, and lymph nodes. The Journal of biological chemistry. 2001;276(40):37594–37601. doi: 10.1074/jbc.M103034200. [DOI] [PubMed] [Google Scholar]
- 107.Jagnandan D, Church JE, Banfi B, Stuehr DJ, Marrero MB, Fulton DJ. Novel mechanism of activation of NADPH oxidase 5. calcium sensitization via phosphorylation. The Journal of biological chemistry. 2007;282(9):6494–6507. doi: 10.1074/jbc.M608966200. [DOI] [PubMed] [Google Scholar]
- 108.Smith MA, Schnellmann RG. Mitochondrial calpain 10 is degraded by Lon protease after oxidant injury. Arch Biochem Biophys. 2012;517(2):144–152. doi: 10.1016/j.abb.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rezende F, Lowe O, Helfinger V, Prior KK, Walter M, Zukunft S, Fleming I, Weissmann N, Brandes RP, Schroder K. Unchanged NADPH Oxidase Activity in Nox1-Nox2-Nox4 Triple Knockout Mice: What Do NADPH-Stimulated Chemiluminescence Assays Really Detect? Antioxid Redox Signal. 2015 doi: 10.1089/ars.2015.6314. [DOI] [PubMed] [Google Scholar]
- 110.Kunkel GH, Chaturvedi P, Tyagi SC. Resuscitation of a dead cardiomyocyte. Heart failure reviews. 2015 doi: 10.1007/s10741-015-9501-z. [DOI] [PubMed] [Google Scholar]
- 111.Wang Y, Tsui H, Ke Y, Shi Y, Li Y, Davies L, Cartwright EJ, Venetucci L, Zhang H, Terrar DA, Huang CL, Solaro RJ, Wang X, Lei M. Pak1 is required to maintain ventricular Ca(2)(+) homeostasis and electrophysiological stability through SERCA2a regulation in mice. Circ Arrhythm Electrophysiol. 2014;7(5):938–948. doi: 10.1161/circep.113.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ke Y, Lei M, Collins TP, Rakovic S, Mattick PA, Yamasaki M, Brodie MS, Terrar DA, Solaro RJ. Regulation of L-type calcium channel and delayed rectifier potassium channel activity by p21-activated kinase-1 in guinea pig sinoatrial node pacemaker cells. Circulation research. 2007;100(9):1317–1327. doi: 10.1161/01.RES.0000266742.51389.a4. [DOI] [PubMed] [Google Scholar]
- 113.Huang H, Joseph LC, Gurin MI, Thorp EB, Morrow JP. Extracellular signal-regulated kinase activation during cardiac hypertrophy reduces sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2) transcription. Journal of molecular and cellular cardiology. 2014;75:58–63. doi: 10.1016/j.yjmcc.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Prasad AM, Inesi G. Regulation and rate limiting mechanisms of Ca2+ ATPase (SERCA2) expression in cardiac myocytes. Mol Cell Biochem. 2012;361(1–2):85–96. doi: 10.1007/s11010-011-1092-y. [DOI] [PubMed] [Google Scholar]
- 115.Heinis FI, Andersson KB, Christensen G, Metzger JM. Prominent heart organ-level performance deficits in a genetic model of targeted severe and progressive SERCA2 deficiency. PLoS One. 2013;8(11):e79609. doi: 10.1371/journal.pone.0079609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li L, Louch WE, Niederer SA, Andersson KB, Christensen G, Sejersted OM, Smith NP. Calcium dynamics in the ventricular myocytes of SERCA2 knockout mice: A modeling study. Biophys J. 2011;100(2):322–331. doi: 10.1016/j.bpj.2010.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stokke MK, Hougen K, Sjaastad I, Louch WE, Briston SJ, Enger UH, Andersson KB, Christensen G, Eisner DA, Sejersted OM, Trafford AW. Reduced SERCA2 abundance decreases the propensity for Ca2+ wave development in ventricular myocytes. Cardiovascular research. 2010;86(1):63–71. doi: 10.1093/cvr/cvp401. [DOI] [PubMed] [Google Scholar]
- 118.Cutler MJ, Wan X, Laurita KR, Hajjar RJ, Rosenbaum DS. Targeted SERCA2a gene expression identifies molecular mechanism and therapeutic target for arrhythmogenic cardiac alternans. Circ Arrhythm Electrophysiol. 2009;2(6):686–694. doi: 10.1161/circep.109.863118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hillestad V, Kramer F, Golz S, Knorr A, Andersson KB, Christensen G. Long-term levosimendan treatment improves systolic function and myocardial relaxation in mice with cardiomyocyte-specific disruption of the Serca2 gene. J Appl Physiol (1985) 2013;115(10):1572–1580. doi: 10.1152/japplphysiol.01044.2012. [DOI] [PubMed] [Google Scholar]
- 120.Swift F, Franzini-Armstrong C, Oyehaug L, Enger UH, Andersson KB, Christensen G, Sejersted OM, Louch WE. Extreme sarcoplasmic reticulum volume loss and compensatory T-tubule remodeling after Serca2 knockout. Proc Natl Acad Sci U S A. 2012;109(10):3997–4001. doi: 10.1073/pnas.1120172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lu YM, Huang J, Shioda N, Fukunaga K, Shirasaki Y, Li XM, Han F. CaMKIIdeltaB mediates aberrant NCX1 expression and the imbalance of NCX1/SERCA in transverse aortic constriction-induced failing heart. PloS one. 2011;6(9):e24724. doi: 10.1371/journal.pone.0024724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stokke MK, Briston SJ, Jolle GF, Manzoor I, Louch WE, Oyehaug L, Christensen G, Eisner DA, Trafford AW, Sejersted OM, Sjaastad I. Ca(2+) wave probability is determined by the balance between SERCA2-dependent Ca(2+) reuptake and threshold SR Ca(2+) content. Cardiovascular research. 2011;90(3):503–512. doi: 10.1093/cvr/cvr013. [DOI] [PubMed] [Google Scholar]
- 123.Sikkel MB, Hayward C, MacLeod KT, Harding SE, Lyon AR. SERCA2a gene therapy in heart failure: an anti-arrhythmic positive inotrope. British journal of pharmacology. 2014;171(1):38–54. doi: 10.1111/bph.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lipskaia L, Chemaly ER, Hadri L, Lompre AM, Hajjar RJ. Sarcoplasmic reticulum Ca(2+) ATPase as a therapeutic target for heart failure. Expert Opin Biol Ther. 2010;10(1):29–41. doi: 10.1517/14712590903321462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Park WJ, Oh JG. SERCA2a: a prime target for modulation of cardiac contractility during heart failure. BMB Rep. 2013;46(5):237–243. doi: 10.5483/BMBRep.2013.46.5.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cutler MJ, Wan X, Plummer BN, Liu H, Deschenes I, Laurita KR, Hajjar RJ, Rosenbaum DS. Targeted sarcoplasmic reticulum Ca2+ ATPase 2a gene delivery to restore electrical stability in the failing heart. Circulation. 2012;126(17):2095–2104. doi: 10.1161/circulationaha.111.071480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lyon AR, Bannister ML, Collins T, Pearce E, Sepehripour AH, Dubb SS, Garcia E, O’Gara P, Liang L, Kohlbrenner E, Hajjar RJ, Peters NS, Poole-Wilson PA, Macleod KT, Harding SE. SERCA2a gene transfer decreases sarcoplasmic reticulum calcium leak and reduces ventricular arrhythmias in a model of chronic heart failure. Circulation Arrhythmia and electrophysiology. 2011;4(3):362–372. doi: 10.1161/circep.110.961615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xin W, Li X, Lu X, Niu K, Cai J. Improved cardiac function after sarcoplasmic reticulum Ca(2+)-ATPase gene transfer in a heart failure model induced by chronic myocardial ischaemia. Acta Cardiol. 2011;66(1):57–64. doi: 10.1080/ac.66.1.2064967. [DOI] [PubMed] [Google Scholar]
- 129.Zsebo K, Yaroshinsky A, Rudy JJ, Wagner K, Greenberg B, Jessup M, Hajjar RJ. Long-term effects of AAV1/SERCA2a gene transfer in patients with severe heart failure: analysis of recurrent cardiovascular events and mortality. Circulation research. 2014;114(1):101–108. doi: 10.1161/circresaha.113.302421. [DOI] [PubMed] [Google Scholar]
- 130.Hayward C, Banner NR, Morley-Smith A, Lyon AR, Harding SE. The Current and Future Landscape of SERCA Gene Therapy for Heart Failure: A Clinical Perspective. Hum Gene Ther. 2015;26(5):293–304. doi: 10.1089/hum.2015.018. [DOI] [PubMed] [Google Scholar]
- 131.Watanabe A, Arai M, Koitabashi N, Niwano K, Ohyama Y, Yamada Y, Kato N, Kurabayashi M. Mitochondrial transcription factors TFAM and TFB2M regulate Serca2 gene transcription. Cardiovascular research. 2011;90(1):57–67. doi: 10.1093/cvr/cvq374. [DOI] [PubMed] [Google Scholar]
- 132.Takahashi M, Tanonaka K, Yoshida H, Koshimizu M, Daicho T, Oikawa R, Takeo S. Possible involvement of calpain activation in pathogenesis of chronic heart failure after acute myocardial infarction. J Cardiovasc Pharmacol. 2006;47(3):413–421. doi: 10.1097/01.fjc.0000210074.56614.3b. [DOI] [PubMed] [Google Scholar]
- 133.Undrovinas A, Maltsev VA, Sabbah HN. Calpain inhibition reduces amplitude and accelerates decay of the late sodium current in ventricular myocytes from dogs with chronic heart failure. PloS one. 2013;8(4):e54436. doi: 10.1371/journal.pone.0054436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shintani-Ishida K, Yoshida K. Mitochondrial m-calpain opens the mitochondrial permeability transition pore in ischemia-reperfusion. International journal of cardiology. 2015;197:26–32. doi: 10.1016/j.ijcard.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 135.Orrenius S, Gogvadze V, Zhivotovsky B. Calcium and mitochondria in the regulation of cell death. Biochem Biophys Res Commun. 2015;460(1):72–81. doi: 10.1016/j.bbrc.2015.01.137. [DOI] [PubMed] [Google Scholar]
- 136.Bhosale G, Sharpe JA, Sundier SY, Duchen MR. Calcium signaling as a mediator of cell energy demand and a trigger to cell death. Ann N Y Acad Sci. 2015;1350(1):107–116. doi: 10.1111/nyas.12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Griffiths EJ, Balaska D, Cheng WH. The ups and downs of mitochondrial calcium signalling in the heart. Biochimica et biophysica acta. 2010;1797(6–7):856–864. doi: 10.1016/j.bbabio.2010.02.022. [DOI] [PubMed] [Google Scholar]
- 138.Cao G, Xing J, Xiao X, Liou AK, Gao Y, Yin XM, Clark RS, Graham SH, Chen J. Critical role of calpain I in mitochondrial release of apoptosis-inducing factor in ischemic neuronal injury. J Neurosci. 2007;27(35):9278–9293. doi: 10.1523/jneurosci.2826-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Moshal KS, Singh M, Sen U, Rosenberger DS, Henderson B, Tyagi N, Zhang H, Tyagi SC. Homocysteine-mediated activation and mitochondrial translocation of calpain regulates MMP-9 in MVEC. Am J Physiol Heart Circ Physiol. 2006;291(6):H2825–2835. doi: 10.1152/ajpheart.00377.2006. [DOI] [PubMed] [Google Scholar]
- 140.Chen M, He H, Zhan S, Krajewski S, Reed JC, Gottlieb RA. Bid is cleaved by calpain to an active fragment in vitro and during myocardial ischemia/reperfusion. The Journal of biological chemistry. 2001;276(33):30724–30728. doi: 10.1074/jbc.M103701200. [DOI] [PubMed] [Google Scholar]
- 141.Thomas RR, Khan SM, Smigrodzki RM, Onyango IG, Dennis J, Khan OM, Portelli FR, Bennett JP., Jr RhTFAM treatment stimulates mitochondrial oxidative metabolism and improves memory in aged mice. Aging (Albany NY) 2012;4(9):620–635. doi: 10.18632/aging.100488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chaturvedi P, Kalani A, Medina I, Familtseva A, Tyagi SC. Cardiosome mediated regulation of MMP9 in diabetic heart: Role of mir29b and mir455 in exercise. Journal of cellular and molecular medicine. 2015 doi: 10.1111/jcmm.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mishra PK, Chavali V, Metreveli N, Tyagi SC. Ablation of MMP9 induces survival and differentiation of cardiac stem cells into cardiomyocytes in the heart of diabetics: a role of extracellular matrix. Can J Physiol Pharmacol. 2012;90(3):353–360. doi: 10.1139/y11-131. [DOI] [PubMed] [Google Scholar]
- 144.Chaturvedi P, Kalani A, Familtseva A, Kamat PK, Metreveli N, Tyagi SC. Cardiac tissue inhibitor of matrix metalloprotease 4 dictates cardiomyocyte contractility and differentiation of embryonic stem cells into cardiomyocytes: Road to therapy. International journal of cardiology. 2015;184C:350–363. doi: 10.1016/j.ijcard.2015.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang Y, Zhang L, Li Y, Chen L, Wang X, Guo W, Zhang X, Qin G, He SH, Zimmerman A, Liu Y, Kim IM, Weintraub NL, Tang Y. Exosomes/microvesicles from induced pluripotent stem cells deliver cardioprotective miRNAs and prevent cardiomyocyte apoptosis in the ischemic myocardium. International journal of cardiology. 2015;192:61–69. doi: 10.1016/j.ijcard.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kalani A, Kamat PK, Chaturvedi P, Tyagi SC, Tyagi N. Curcumin-primed exosomes mitigate endothelial cell dysfunction during hyperhomocysteinemia. Life Sci. 2014;107(1–2):1–7. doi: 10.1016/j.lfs.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen L, Wang Y, Pan Y, Zhang L, Shen C, Qin G, Ashraf M, Weintraub N, Ma G, Tang Y. Cardiac progenitor-derived exosomes protect ischemic myocardium from acute ischemia/reperfusion injury. Biochem Biophys Res Commun. 2013;431(3):566–571. doi: 10.1016/j.bbrc.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vicencio JM, Yellon DM, Sivaraman V, Das D, Boi-Doku C, Arjun S, Zheng Y, Riquelme JA, Kearney J, Sharma V, Multhoff G, Hall AR, Davidson SM. Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J Am Coll Cardiol. 2015;65(15):1525–1536. doi: 10.1016/j.jacc.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 149.Das S, Halushka MK. Extracellular vesicle microRNA transfer in cardiovascular disease. Cardiovascular pathology : the official journal of the Society for Cardiovascular Pathology. 2015;24(4):199–206. doi: 10.1016/j.carpath.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 150.Gray WD, French KM, Ghosh-Choudhary S, Maxwell JT, Brown ME, Platt MO, Searles CD, Davis ME. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circulation research. 2015;116(2):255–263. doi: 10.1161/circresaha.116.304360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chaturvedi P, Kalani A, Medina I, Familtseva A, Tyagi SC. Cardiosome mediated regulation of MMP9 in diabetic heart: role of mir29b and mir455 in exercise. J Cell Mol Med. 2015;19(9):2153–2161. doi: 10.1111/jcmm.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ong SG, Lee WH, Huang M, Dey D, Kodo K, Sanchez-Freire V, Gold JD, Wu JC. Cross talk of combined gene and cell therapy in ischemic heart disease: role of exosomal microRNA transfer. Circulation. 2014;130(11 Suppl 1):S60–69. doi: 10.1161/circulationaha.113.007917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cai X, Bao L, Ren J, Li Y, Zhang Z. Grape seed procyanidin B2 protects podocytes from high glucose-induced mitochondrial dysfunction and apoptosis via the AMPK-SIRT1-PGC-1alpha axis in vitro. Food & function. 2016;7(2):805–815. doi: 10.1039/c5fo01062d. [DOI] [PubMed] [Google Scholar]
- 154.Yoshino M, Naka A, Sakamoto Y, Shibasaki A, Toh M, Tsukamoto S, Kondo K, Iida K. Dietary isoflavone daidzein promotes Tfam expression that increases mitochondrial biogenesis in C2C12 muscle cells. The Journal of nutritional biochemistry. 2015;26(11):1193–1199. doi: 10.1016/j.jnutbio.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 155.Qin G, Wu M, Wang J, Xu Z, Xia J, Sang N. Sulfur Dioxide Contributes to the Cardiac and Mitochondrial Dysfunction in Rats. Toxicological sciences : an official journal of the Society of Toxicology. 2016 doi: 10.1093/toxsci/kfw048. [DOI] [PubMed] [Google Scholar]
- 156.Sreekumar PG, Ishikawa K, Spee C, Mehta HH, Wan J, Yen K, Cohen P, Kannan R, Hinton DR. The Mitochondrial-Derived Peptide Humanin Protects RPE Cells From Oxidative Stress, Senescence, and Mitochondrial Dysfunction. Investigative ophthalmology & visual science. 2016;57(3):1238–1253. doi: 10.1167/iovs.15-17053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pillai VB, Samant S, Sundaresan NR, Raghuraman H, Kim G, Bonner MY, Arbiser JL, Walker DI, Jones DP, Gius D, Gupta MP. Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nature communications. 2015;6:6656. doi: 10.1038/ncomms7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mei H, Sun S, Bai Y, Chen Y, Chai R, Li H. Reduced mtDNA copy number increases the sensitivity of tumor cells to chemotherapeutic drugs. Cell death & disease. 2015;6:e1710. doi: 10.1038/cddis.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chandrasekaran K, Anjaneyulu M, Inoue T, Choi J, Sagi AR, Chen C, Ide T, Russell JW. Mitochondrial transcription factor A regulation of mitochondrial degeneration in experimental diabetic neuropathy. American journal of physiology Endocrinology and metabolism. 2015;309(2):E132–141. doi: 10.1152/ajpendo.00620.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hayashi Y, Yoshida M, Yamato M, Ide T, Wu Z, Ochi-Shindou M, Kanki T, Kang D, Sunagawa K, Tsutsui H, Nakanishi H. Reverse of age-dependent memory impairment and mitochondrial DNA damage in microglia by an overexpression of human mitochondrial transcription factor a in mice. J Neurosci. 2008;28(34):8624–8634. doi: 10.1523/jneurosci.1957-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang Q, Yu JT, Wang P, Chen W, Wu ZC, Jiang H, Tan L. Mitochondrial transcription factor A (TFAM) polymorphisms and risk of late-onset Alzheimer’s disease in Han Chinese. Brain Res. 2011;1368:355–360. doi: 10.1016/j.brainres.2010.10.074. [DOI] [PubMed] [Google Scholar]
- 162.Aguirre-Rueda D, Guerra-Ojeda S, Aldasoro M, Iradi A, Obrador E, Ortega A, Mauricio MD, Vila JM, Valles SL. Astrocytes protect neurons from Abeta1-42 peptide-induced neurotoxicity increasing TFAM and PGC-1 and decreasing PPAR-gamma and SIRT-1. Int J Med Sci. 2015;12(1):48–56. doi: 10.7150/ijms.10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xu S, Zhong M, Zhang L, Wang Y, Zhou Z, Hao Y, Zhang W, Yang X, Wei A, Pei L, Yu Z. Overexpression of Tfam protects mitochondria against beta-amyloid-induced oxidative damage in SH-SY5Y cells. Febs j. 2009;276(14):3800–3809. doi: 10.1111/j.1742-4658.2009.07094.x. [DOI] [PubMed] [Google Scholar]
- 164.Diez JJ, Iglesias P. The role of the novel adipocyte-derived hormone adiponectin in human disease. Eur J Endocrinol. 2003;148(3):293–300. doi: 10.1530/eje.0.1480293. [DOI] [PubMed] [Google Scholar]
- 165.Duan J, Yin Y, Cui J, Yan J, Zhu Y, Guan Y, Wei G, Weng Y, Wu X, Guo C, Wang Y, Xi M, Wen A. Chikusetsu Saponin IVa Ameliorates Cerebral Ischemia Reperfusion Injury in Diabetic Mice via Adiponectin-Mediated AMPK/GSK-3beta Pathway In Vivo and In Vitro. Mol Neurobiol. 2015 doi: 10.1007/s12035-014-9033-x. [DOI] [PubMed] [Google Scholar]
- 166.Yan W, Zhang F, Zhang R, Zhang X, Wang Y, Zhou F, Xia Y, Liu P, Gao C, Wang H, Zhang L, Zhou J, Gao F, Gao E, Koch WJ, Wang H, Cheng H, Qu Y, Tao L. Adiponectin regulates SR Ca(2+) cycling following ischemia/reperfusion via sphingosine 1-phosphate-CaMKII signaling in mice. Journal of molecular and cellular cardiology. 2014;74:183–192. doi: 10.1016/j.yjmcc.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 167.Wang KZ, Zhu J, Dagda RK, Uechi G, Cherra SJ, 3rd, Gusdon AM, Balasubramani M, Chu CT. ERK-mediated phosphorylation of TFAM downregulates mitochondrial transcription: implications for Parkinson’s disease. Mitochondrion. 2014;17:132–140. doi: 10.1016/j.mito.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Santos JM, Kowluru RA. Role of mitochondria biogenesis in the metabolic memory associated with the continued progression of diabetic retinopathy and its regulation by lipoic acid. Investigative ophthalmology & visual science. 2011;52(12):8791–8798. doi: 10.1167/iovs.11-8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.He B, Meng YH, Mivechi NF. Glycogen synthase kinase 3beta and extracellular signal-regulated kinase inactivate heat shock transcription factor 1 by facilitating the disappearance of transcriptionally active granules after heat shock. Molecular and cellular biology. 1998;18(11):6624–6633. doi: 10.1128/mcb.18.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ngamsiri P, Watcharasit P, Satayavivad J. Glycogen synthase kinase-3 (GSK3) controls deoxyglucose-induced mitochondrial biogenesis in human neuroblastoma SH-SY5Y cells. Mitochondrion. 2014;14(1):54–63. doi: 10.1016/j.mito.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 171.Sunaga D, Tanno M, Kuno A, Ishikawa S, Ogasawara M, Yano T, Miki T, Miura T. Accelerated recovery of mitochondrial membrane potential by GSK-3beta inactivation affords cardiomyocytes protection from oxidant-induced necrosis. PloS one. 2014;9(11):e112529. doi: 10.1371/journal.pone.0112529. [DOI] [PMC free article] [PubMed] [Google Scholar]