Abstract
Triple-negative breast cancer (TNBC) is a highly aggressive and metastatic form of breast cancer that lacks the estrogen, progesterone and HER2 receptors and is resistant to targeted and hormone therapies. TNBCs express high levels of the transmembrane glycoprotein, complement C1r/C1s, Uegf, Bmp1 (CUB)-domain containing protein 1 (CDCP1), which has been correlated with the aggressiveness and poor prognosis of multiple carcinomas. Full-length CDCP1 (flCDCP1) can be proteolytically cleaved, resulting in a cleaved membrane-bound isoform (cCDCP1). CDCP1 is phosphorylated by Src family kinases in its full-length and cleaved states, which is important for its pro-metastatic signaling. We observed that cCDCP1, compared with flCDCP1, induced a dramatic increase in phosphorylation of the migration-associated proteins: PKCδ, ERK1/2 and p38 mitogen-activated protein kinase in HEK 293T. In addition, only cCDCP1 induced migration of HEK 293T cells and rescued migration of the TNBC cell lines expressing short hairpin RNA against CDCP1. Importantly, we found that only cCDCP1 is capable of dimerization, which can be blocked by expression of the extracellular portion of cCDCP1 (ECC), indicating that dimerization occurs through CDCP1’s ectodomain. We found that ECC inhibited phosphorylation of PKCδ and migration of TNBC cells in two-dimensional culture. Furthermore, ECC decreased cell invasiveness, inhibited proliferation and stimulated apoptosis of TNBC cells in three-dimensional culture, indicating that the cCDCP1 dimer is an important contributor to TNBC aggressiveness. These studies have important implications for the development of a therapeutic to block CDCP1 activity and TNBC metastasis.
INTRODUCTION
Breast cancer is the second leading cause of death of women in the United States; most of these fatalities are caused by metastatic disease. Many therapies have been developed to target aggressive breast cancer, including receptor-targeted therapies (Herceptin) and anti-hormone therapies (aromatase inhibitors). However, triple-negative breast cancer (TNBC) patients are not candidates for many of these therapies, as they do not express the respective drug targets, HER2, estrogen and progesterone receptors. Moreover, TNBC is a highly aggressive form of breast cancer that is associated with advanced stage at diagnosis, early peak of recurrence and poorer outcome in comparison with non-TNBCs; in the advanced setting, responses to chemotherapy and radiation therapy lack durability.1 Finally, women who are diagnosed with TNBC tend to be younger than non-TNBC patients. Thus, there is an urgent need to develop a targeted therapy to inhibit the progression and metastasis of TNBC.
The transmembrane protein, complement C1r/C1s, Uegf, Bmp1 (CUB)-domain containing protein 1 (CDCP1), is upregulated in TNBC and is correlated with the cancer’s aggressiveness.2 Importantly, CDCP1 is involved in metastasis in multiple animal models of cancer, including lung adenocarcinoma, gastric scirrhous carcinoma, melanoma, and prostate and ovarian carcinomas.3–9 CDCP1 expression is induced by hypoxia in clear-cell renal cell carcinoma (ccRCC)10,11 and activated Ras in lung cancer,12 and its localization at the membrane is induced by epidermal growth factor receptor in ovarian cancer.13 The full-length, 135-kDa CDCP1 protein (flCDCP1) can be cleaved into a 70-kDa, membrane-bound cleaved CDCP1 (cCDCP1) and a 65-kDa portion that is shed on the extracellular side of the membrane. CDCP1 can be cleaved by plasmin, trypsin and matriptase,8,14,15 and evidence is growing to support the necessity of CDCP1 cleavage for its activity.2,3,8,16 In addition to cleavage, CDCP1 activity depends on phosphorylation; both flCDCP1 and cCDCP1 are phosphorylated by Src family kinases, Src, Fyn and Yes, first at Y734 and then at Y743, Y762, Y707 and Y806.4,14,17–23 CDCP1 can stimulate migration/invasion through a variety of signaling pathways including protein kinase C δ (PKCδ),10,24 protein kinase B (Akt),3,8,9 β1 integrin,3 matrix metalloproteinase secretion25 and others. Besides its well established role in cancer cell migration, CDCP1 was reported to degrade adherens junctions,2,18 protect against anoikis4 and inhibit autophagy,26 contributing to tumor progression.
CDCP1 contains three CUB domains located in its ectodomain, the specific role of which remains unclear. CUB domains are expressed in many developmentally regulated proteins and participate in interactions with CUB domains of other proteins.27 The first CUB domain is removed by CDCP1 cleavage, potentially stimulating important protein–protein interactions through CUB2 and CUB3, leading to the enhancement of CDCP1 signaling. Casar et al.3,8 recently reported a decrease in metastasis with inhibition of CDCP1 cleavage, hinting at the necessity of cleavage for CDCP1 activity. In support of this, Law et al.2 showed that inhibition of CDCP1 cleavage refined the borders of ex vivo tumors, proposing the inhibition of CDCP1 cleavage as a rational neoadjuvant therapy for TNBC. Although these data indicate that CDCP1 cleavage stimulates its activity, the mechanism of cCDCP1 activation has not been elucidated.
Here we demonstrate that CDCP1 expression is upregulated in TNBC and it is cleaved and phosphorylated in TNBC cell lines and primary TNBC specimens. We find that CDCP1 cleavage leads to cCDCP1 homodimerization, uncovering the mechanism of cCDCP1 activation. The homodimer acts as a docking station for Src to induce phosphorylation of a key CDCP1 downstream target, PKCδ, stimulating pro-migratory signaling. We provide evidence that dimerization is a key step in CDCP1 activation by breaking the dimer with the extracellular portion of cCDCP1 (ECC), which blocks CDCP1’s downstream signaling, leading to inhibition of migration, invasion, proliferation, and induction of apoptosis. Therefore, CDCP1 dimerization is a rational target for therapeutic intervention to inhibit metastasis of TNBC.
RESULTS
CDCP1 is overexpressed and cleaved in TNBC
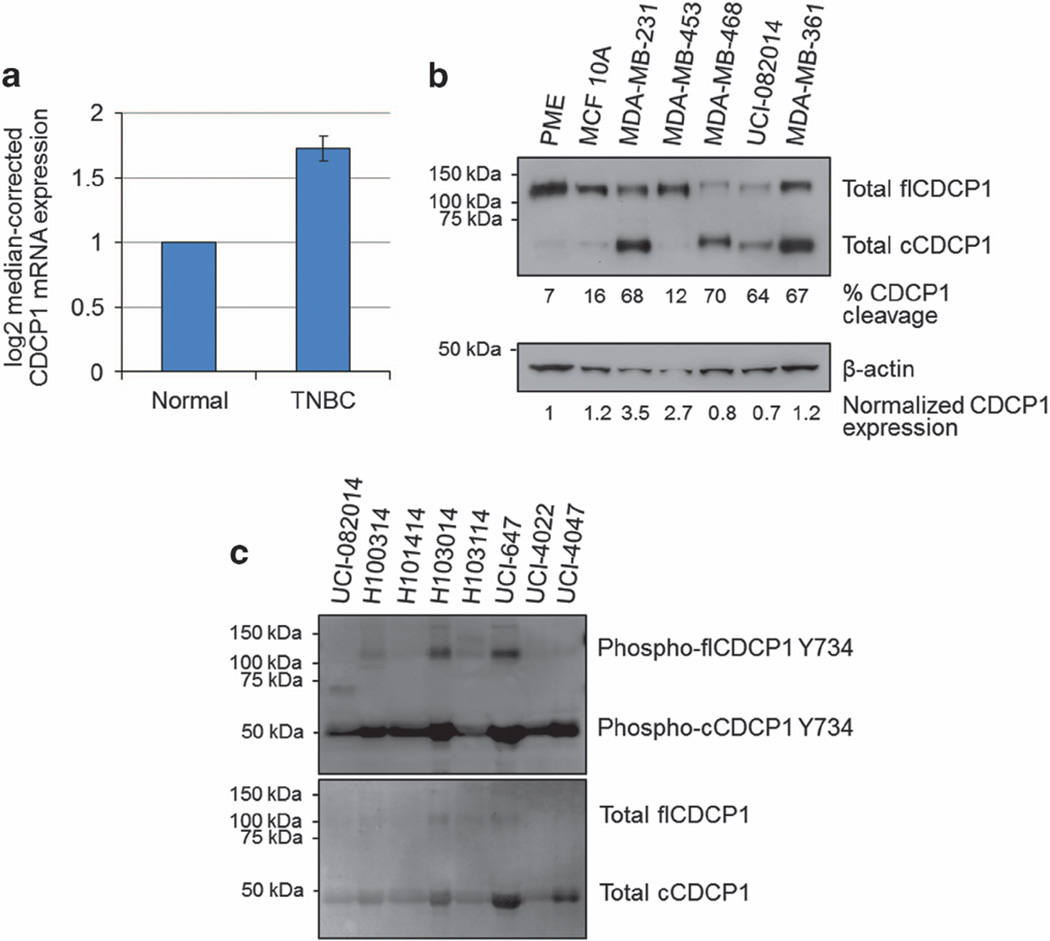
The data acquired from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) indicated that CDCP1 mRNA is upregulated in TNBC compared with normal breast tissue (Figure 1a), suggesting CDCP1’s role in breast cancer progression. Consistent with the data extracted from TCGA, we found that the average CDCP1 expression in MDA-MB-231, MDA-MB-453, MDA-MB-468 and UCI-082014 (Supplementary Table 1) TNBC cells was 1.8-fold higher than the average expression in primary mammary epithelium and immortalized non-tumorigenic MCF10A breast cells (Figure 1b).
Figure 1.
CDCP1 is overexpressed and cleaved in TNBC. (a) TCGA data showing normalized mRNA expression of CDCP1 in 57 TNBC and matched normal tissue samples, P = 0.002 calculated by one-way analysis of variance (ANOVA) with multiple comparison post-hoc t-test and error bars represent s.e.m. (b) WB analysis of CDCP1 expression in breast cancer cell lines. CDCP1 is mostly full-length in primary mammary epithelium (PME) cells and the immortalized non-tumorigenic breast cell line, MCF10A, and is mostly cleaved in TNBC cell lines and metastatic non-TNBC cells, MDA-MB-361. Quantified percent of CDCP1 cleavage was obtained by band densities in ImageJ (top row)=cCDCP1/(cCDCP1+flCDCP1)*100. Quantified relative CDCP1 expression was obtained from band densities in ImageJ (bottom row)=(flCDCP1+cCDCP1)/β-actin, CDCP1 expression in PME was normalized to 1 and all other lanes were normalized to that for comparison. (c) Primary human TNBC samples express cleaved and phosphorylated CDCP1. Equal protein amounts were loaded in each lane.
Recent reports indicated that cleavage stimulates CDCP1 protein activity.2,3,8,16 Figure 1b shows that CDCP1 is highly cleaved in all of the TNBC cell lines listed above (54% cleaved) compared with primary mammary epithelium and MCF10A cells, where CDCP1 exists primarily in the full-length form (12% cleaved). We also found that CDCP1 is cleaved in the non-TNBC cell line derived from a brain metastasis, MDA-MB-361 (Figure 1b). Furthermore, we found that eight out of eight TNBC patient samples analyzed by western blotting (WB) express CDCP1, which is mostly cleaved and phosphorylated (Figure 1c and Supplementary Figure 1).
Previously, we and others showed that CDCP1 expression was regulated by hypoxia-inducible factors in ccRCC.10,11 As breast tumors contain areas of low oxygen tension,28 we investigated whether CDCP1 is regulated by hypoxia in the TNBC cell line MDA-MB-231. We found that although the mRNA and protein levels of CDCP1 were not upregulated in hypoxia, the phosphorylation of cCDCP1 and its downstream target PKCδ were increased in hypoxia, indicating that CDCP1 activity increases (Supplementary Figure 2).
Both flCDCP1 and cCDCP1 interact with PKCδ and Src but only cCDCP1 stimulates downstream signaling
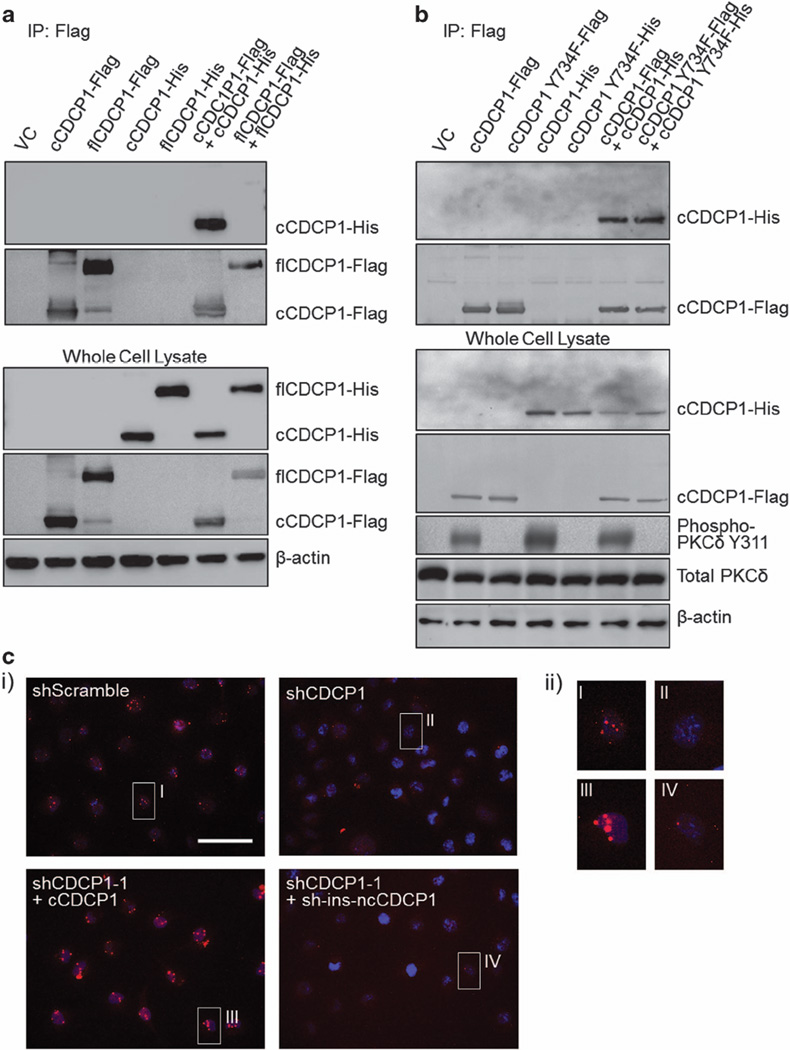
To test the functional difference between flCDCP1 and cCDCP1 we created Flag-tagged constructs of each CDCP1 isoform (Supplementary Figure 3). The signal peptide (SP) located at the N-terminus of flCDCP1 was added at the N-terminus of cCDCP1 to ensure proper localization. We overexpressed a vector control (VC), cCDCP1-Flag or flCDCP1-Flag in HEK 293T cells, which do not express endogenous CDCP1 and do not cleave exogenous flCDCP1. As expected, immunostaining confirmed that both CDCP1 isoforms localized on the cell membrane (Supplementary Figure 4). We found that although both isoforms of CDCP1 could be phosphorylated at Y734, only cCDCP1 could induce robust phosphorylation of its downstream target PKCδ, as well as ERK1/2 and p38 mitogen-activated protein kinase, other kinases known to be involved in migration29 (Figure 2a). Interestingly, previous findings indicate that CDCP1 expression is regulated by ERK1/2 signaling downstream of Ras in non-small cell lung carcinoma,12 suggesting a potential positive feedback loop between CDCP1 and ERK1/2. Based on previous reports that CDCP1 binds Src and PKCδ, stimulating PKCδ phosphorylation by Src at Y311,4,8,10,16,20,24 we compared the ability of both CDCP1 isoforms to associate with Src and PKCδ. We immunoprecipitated each CDCP1 isoform by Flag tag and probed the membrane with anti- Src and anti-PKCδ antibodies. We found that both flCDCP1 and cCDCP1 could associate with Src and PKCδ. However, only cCDCP1 co-precipitated phosphorylated PKCδ (Figure 2b). As Law et al.2 demonstrated the ability of dexamethasone to inhibit CDCP1 cleavage in TNBC cells, we treated MDA-MB-231 cells with a phosphate-buffered saline (PBS) vehicle control or 100 nm dexamethasone for 48 h. We found that inhibiting CDCP1 cleavage in TNBC cells inhibited PKCδ phosphorylation (Figure 2c). These data indicate the necessity of CDCP1 cleavage for signaling through PKCδ.
Figure 2.
WB analysis demonstrating that cCDCP1 activates PKCδ, ERK 1/2 and p38 mitogen-activated protein kinase (MAPK). (a) Cell lysates of HEK 293T transfected with VC, cCDCP1 or flCDCP1 and MDA-MB-231 cells were analyzed by WB. cCDCP1 stimulates phosphorylation of PKCδ, ERK 1/2 and p38 MAPK in HEK 293T cells, while flCDCP1 does not. TNBC cells, MDA-MB-231, express and phosphorylate high levels of cCDCP1 and its downstream targets, PKCδ, ERK1/2 and p38 MAPK. Cell lysates of HEK 293T and MDA-MB-231 were run on the same gel and equal amounts of protein were loaded. (b) HEK 293T cells were co-transfected with PKCδ and Src, along with cCDCP1-Flag or flCDCP1-Flag. Cell lysates were immunoprecipitated with anti-Flag antibody. Both cCDCP1 and flCDCP1 are able to bind to PKCδ and Src. However, only PKCδ bound to cCDCP1 is phosphorylated at Y311. (c) MDA-MB-231 cells overexpressing PKCδ were treated with a vehicle control (DMSO) or 100 nm dexamethasone (Dex) for 48 h. This treatment inhibited CDCP1 cleavage and caused a reduction of PKCδ phosphorylation at Y311 as analyzed by WB. Quantified phospho/total PKCδ ratio was obtained from band densities in ImageJ, the value for DMSO-treated cells was normalized to 1. Quantification shown is the average of three independent experiments.
cCDCP1 stimulates cell migration
Owing to the ability of cCDCP1 to induce robust phosphorylation of PKCδ, which we have previously linked to pro-migratory signaling,10 we aimed to analyze the effect of each CDCP1 isoform on cellular migration. To do this, we overexpressed VC, cCDCP1-Flag or flCDCP1-Flag in HEK 293T cells and analyzed migration through fibronectin-coated transwell inserts after 24 h. We found that cCDCP1 significantly induced migration of HEK 293T cells, whereas flCDCP1 did not (Figure 3a).
Figure 3.
CDCP1 cleavage stimulates cell migration. (a) Representative images of stained transwells (i) used for quantification of cell migration (ii). HEK 293T cells were transfected with VC, cCDCP1 or flCDCP1 as indicated. CDCP1 expression was verified by WB in Figure 2b. cCDCP1 induces migration of HEK 293T cells, while flCDCP1 does not. The cell number per field of HEK 293T cells transfected with VC was normalized to 1 and all other samples were normalized to that for comparison. (b) CDCP1 depletion by shRNA blocks migration of MDA-MB-231 cells, which can be restored by cCDCP1 overexpression, and not sh-ins-ncCDCP1 overexpression. The data represent average percent wound closure. (c) Representative images of stained transwells (i) used for quantification of cell migration (ii). CDCP1 depletion by shRNA blocks migration of MDA-MB-468 cells, which can be restored by cCDCP1 overexpression and not sh-ins-ncCDCP1 overexpression. All of the experiments in this figure were done in duplicate and repeated three times. **P ≤ 0.005, ***P ≤ 0.0005. P-values were analyzed by one way ANOVA with multiple comparison post-hoc t-test and error bars represent s.e.m.
Based on these data and the high level of CDCP1 cleavage in MDA-MB-231 cells (Figures 1b and 2a), we assessed the contribution of cCDCP1 in MDA-MB-231 cell migration in a scratch assay. We knocked down endogenous CDCP1 with two short hairpin RNAs (shRNAs) targeting two different sites on CDCP1’s mRNA. Migration of MDA-MB-231-shCDCP1-1 and MDA-MB-231-shCDCP1-7 cells was severely inhibited compared with MDA-MB-231-shScramble cells. Importantly, this migratory defect was rescued by overexpression of cCDCP1 and not shCDCP1-insensitive non-cleavable CDCP1 (sh-ins-ncCDCP1) (Figure 3b and Supplementary Figures 3 and 5). We validated these data in another TNBC cell line, MDA-MB-468, where CDCP1 knockdown inhibited migration through fibronectin-coated transwell inserts, which was rescued by cCDCP1 and not sh-ins-ncCDCP1 overexpression (Figure 3c).
cCDCP1 forms a dimer independently of its phosphorylation status
CUB domains have known roles in protein–protein interaction30–32 including the potential CUB domain–CUB domain interaction of CDCP1 with matriptase.14 flCDCP1 has three CUB domains (1, 2 and 3) and cCDCP1 has two CUB domains (2 and 3). Thus, we decided to test whether either isoform of CDCP1 could form a dimer. To do this, we created C-terminal His-tagged cCDCP1 and flCDCP1 constructs. We overexpressed Flag- and His-tagged CDCP1 constructs alone or in combination in HEK 293T cells (Figure 4a). The Flag tag was immunoprecipitated and dimerization was analyzed by immunoblotting for the His tag. Our results indicate that only cCDCP1 could form a dimer (Figure 4a).
Figure 4.
CDCP1 cleavage is necessary for dimer formation, which is independent of cCDCP1’s phosphorylation status. (a) HEK 293T cells were transfected with Flag- and His-tagged CDCP1 constructs alone or in combination (as indicated). After 48 h, CDCP1 was immunoprecipitated (IP) by the Flag tag. Co-immunoprecipitation (co-IP) samples were then analyzed by WB for the presence of flCDCP1-His tag or cCDCP1-His tag, indicating the formation of a dimer. Only cCDCP1-His tag was detected in the co-IP, indicating that cCDCP1 forms a dimer. (b) HEK 293T cells were transfected with either WT or Y734F mutant cCDCP1-Flag tag and cCDCP1-His tag constructs (as indicated). Samples were analyzed as in a. cCDCP1 Y734F-Flag is able to co-IP cCDCP1 Y734F-His but is unable to phosphorylate PKCδ. Thus, cCDCP1 dimerization is phosphorylation independent. (c) cCDCP1 forms a dimer endogenously in TNBC cells. (i) CDCP1 dimerization was monitored by the proximity ligation assay (PLA) using oligonucleotide-conjugated anti-CDCP1 antibody. MDA-MB-231-shCDCP1-1 and MDA-MB-231-shCDCP1-1+cCDCP1 were used as negative and positive controls, respectively. MDA-MB-231-shCDCP1-1+sh-ins-ncCDCP1 does not show signal, indicating the dimer is forming only through cCDCP1. Scale bar represents 50 µm. (ii) Larger pictures of the cells are shown in (i).
As interactions of CDCP1 with other proteins at the membrane are dependent on its phosphorylation,3,4,15,20,33 we analyzed the dependency of the dimer on CDCP1’s phosphorylation status. To do this, we created cCDCP1-Y734F-Flag and cCDCP1-Y734F-His constructs, which are phosphorylation incompetent (Supplementary Figure 6). We then repeated co-immunoprecipitation experiments in HEK 293T cells by expressing Flag- and His-tagged constructs of cCDCP1 and cCDCP1-Y734F alone or in combination and found that cCDCP1 was capable of forming a dimer independently of its phosphorylation status. However, overexpression of cCDCP1 Y734F-Flag did not stimulate phosphorylation of PKCδ at Y311 (Figure 4b).
To determine whether CDCP1 forms a dimer endogenously in TNBC cells, we used the Duolink proximity ligation assay. We found that CDCP1 formed a dimer in MDA-MB-231 cells visualized by the red punctate signal (Figure 4c). This signal was lost in MDA-MB-231-shCDCP1-1 cells and regained on rescue with cCDCP1 overexpression, but not with sh-ins-ncCDCP1 (Figure 4c and Supplementary Figure 7).
An extracellular fragment of cCDCP1 can block the cCDCP1 dimer formation
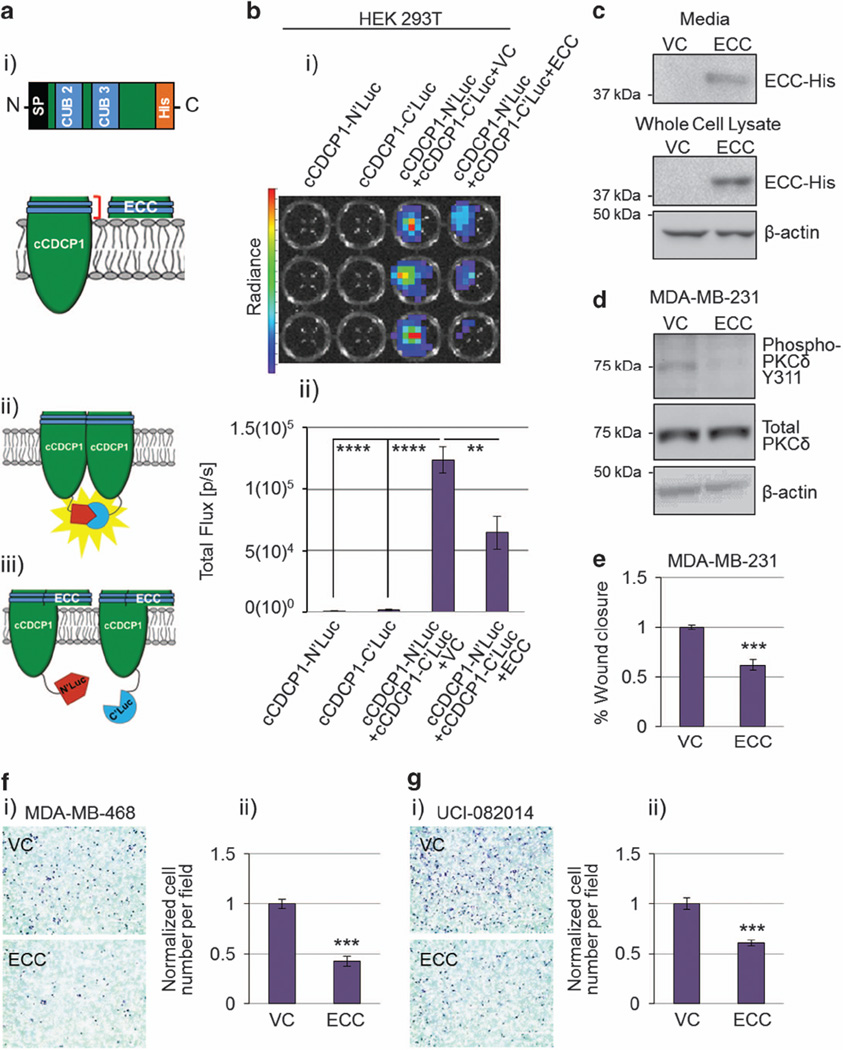
Based on the above data, we propose that cCDCP1 forms a dimer through its extracellular domain, specifically through one or both of its CUB domains. To test this hypothesis, we made a construct expressing the ECC with an N-terminal SP and a C-terminal His tag (Figure 5a (i)). To monitor CDCP1 dimerization, we used a firefly split-luciferase system34 in which the N-terminal domain or C-terminal domain of firefly luciferase was attached to the intracellular C-terminus of cCDCP1, resulting in cCDCP1-N’luc and cCDCP1-C’luc constructs, respectively (Figure 5a (ii and iii)). We also created flCDCP1-N’luc and flCDCP1-C’luc constructs. Using the split-luciferase system in HEK 293T cells we confirmed that only cCDCP1, not flCDCP1, forms a dimer and no heterodimer is formed between cCDCP1 and flCDCP1 (Figure 5b and Supplementary Figure 8). Next, we expressed the cCDCP1-N’luc and cCDCP1-C’luc constructs in combination with VC or ECC. Importantly, ECC was able to decrease the D-luciferin luminescence, indicating blockage of the cCDCP1 dimer formation (Figure 5b). The observed effect of ECC on cCDCP1 dimer blockage is specific, as ECC had no effect on cCDCP1 expression or luciferase enzyme activity (Supplementary Figure 9). As ECC has a SP at its N-terminus, which is necessary for secretion,14 we found secreted ECC in the media of transfected HEK 293T cells (Figure 5c).
Figure 5.
Co-expression of extracellular cleaved portion of CDCP1 (ECC) inhibits cCDCP1 dimerization and downstream signaling. (a) Schematics of the ECC construct (i), the cCDCP1-based split luciferase system for monitoring cCDCP1 dimerization (ii) and the tentative mechanism of how ECC breaks the dimer (iii). SP, signal peptide for proper translocation to the cell membrane/secretion; blue, CUB domains. (b) Split luciferase constructs were transfected into HEK 293T cells individually or together with VC or ECC, as indicated. After the addition of D-luciferin, the luminescence was imaged with an IVIS Lumina system. The representative image is shown in (i). The quantification shown in (ii) is the average of four independent experiments each done in triplicate. **P = 0.01, ****P < 1(10)− 5. P-values were analyzed by one-way ANOVA with multiple comparison post-hoc t-test. Error bars represent s.e.m. (c) HEK 293T secrete ECC-His into the media 48 h post transfection. (d and e) MDA-MB-231 cells overexpressing PKCδ were treated with VC or ECC conditioned media for 24 h. (d) ECC inhibits phosphorylation of PKCδ. (e) ECC inhibits cell migration in a scratch assay. (f and g) ECC inhibits migration of MDA-MB-468 (f) and UCI-082014 (g) TNBC cell lines through fibronectin-coated transwells when compared with VC. Representative images of stained transwells are shown in (i) and used for quantification of cell migration in (ii). Quantification is the average of three independent experiments each done in duplicate. ***P < 0.001. P-value was analyzed by paired t-test and error bars represent s.e.m.
To validate the importance of the cCDCP1 dimerization in pro-migratory signaling, we analyzed the effect of ECC on PKCδ phosphorylation and migration of MDA-MB-231 cells. We found that overexpressing ECC in MDA-MB-231 cells inhibited PKCδ phosphorylation (Figure 5d). We further found that transfer of ECC-conditioned media from ECC-transfected HEK 293T cells to MDA-MB-231 cells inhibited their migration (Figure 5e). We also validated the inhibitory effect of ECC on migration through transwell inserts using two additional TNBC cell lines, MDA-MB-468 and UCI-082014, expressing either VC or ECC (Figure 5f and g). Together, these data validate that cCDCP1 dimerization facilitates PKCδ phosphorylation and stimulates cell migration.
ECC inhibits TNBC cell invasiveness, decreases proliferation and induces apoptosis in 3D culture
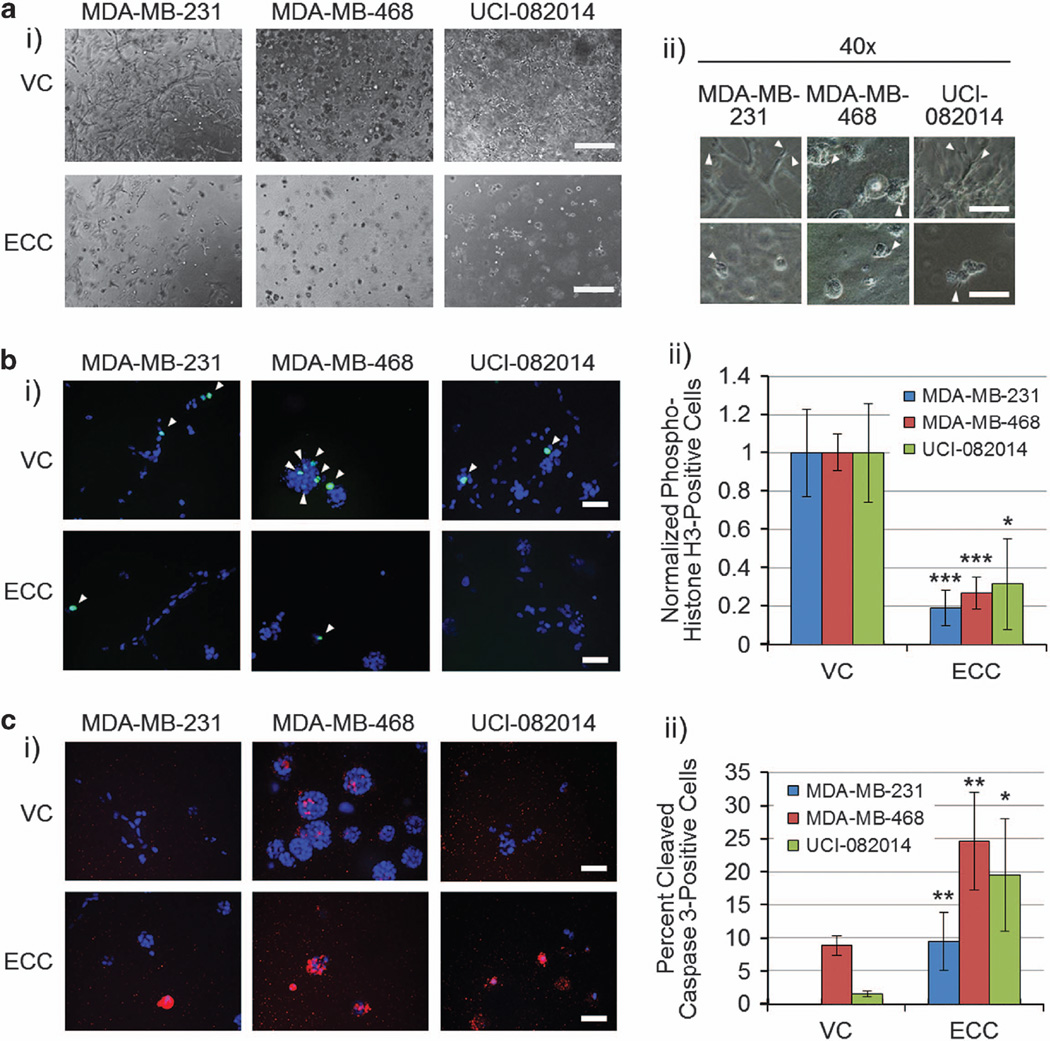
To analyze the effect of ECC on TNBC cells in three dimensions (3D),35 we cultured MDA-MB-231, MDA-MB-468 and UCI-082014 TNBC cells expressing VC or ECC in 1:1 collagen:Matrigel gels for 6 days. We found that TNBC cells expressing ECC had less and smaller stellate structures and invaded less into the matrix (Figure 6a), had decreased proliferation as analyzed by a mitotic marker, phospho-histone H3 staining (Figure 6b) and increased apoptosis as analyzed by cleaved caspase 3 staining (Figure 6c) when compared with TNBC cells expressing VC. The lack of any cleaved caspase 3 staining in the MDA-MB-231 VC cells (Figure 6c) is supported by reports showing increased resistance of MDA-MB-231 cells to apoptosis compared with MDA-MB-468.36–38 Staining of ECC using an anti-His-tag antibody revealed that after secretion, ECC concentration builds around the cells (Supplementary Figure 10), probably leading to saturation of cCDCP1 dimer inhibition at the membrane. Thus, the cCDCP1 inhibition by ECC leads to a more dramatic phenotype in 3D as opposed to two-dimensional culture, where ECC is secreted into the culture media.
Figure 6.
ECC inhibits TNBC cell invasiveness, decreases proliferation and induces apoptosis in 3D culture. (a, i) Phase-contrast images of the cell lines indicated grown in 3D. Scale bar, 400 µm. (ii) Phase-contrast images, arrows point to stellate structures invading the matrix. Scale bar, 200 µm. ECC inhibits TNBC cell invasiveness by blocking the stellate structure formation. (b, i) Phospho-histone H3 images of the cell lines indicated grown in 3D. Blue, 4',6-diamidino-2-phenylindole (DAPI); green, phospho-histone H3. Scale bar, 50 µm. White arrows point to DAPI and phospho-histone H3 co-stained nuclei. The results are quantified in (ii). The number of phospho-histone H3-positive nuclei in VC-transduced cells was normalized to 1. ECC inhibits TNBC cell proliferation in 3D as shown by the reduction of phospho-histone H3-positive nuclei in comparison with VC-expressing cells. (c, i) Cleaved caspase 3 images of the cell lines indicated grown in 3D. Blue, DAPI; red, cleaved caspase 3. Scale bar, 50 µm. The results are quantified in (ii). ECC stimulates TNBC cell apoptosis in 3D as shown by the increase of cleaved caspase 3 in comparison with VC-expressing cells. Quantification in b and c is the average of three independent experiments performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001. P-value was analyzed by one-way ANOVA with multiple comparison post-hoc t-test and error bars represent s.e.m.
DISCUSSION
Our studies have elucidated a new step in the CDCP1 activation cascade: cCDCP1 homodimerization, which is critical for phosphorylation of PKCδ and migration of TNBC cells. Figure 7 illustrates an updated model for CDCP1 activation, including its phosphorylation, cleavage and dimerization. We suggest that the cCDCP1 dimer acts as a docking station to facilitate PKCδ phosphorylation by Src: cCDCP1 dimer is important for binding of Src to one subunit and PKCδ to the other subunit, bringing Src and PKCδ into close proximity, allowing the phosphorylation of PKCδ by Src. This signaling cascade further leads to phospho-PKCδ-stimulated migration. Our model also explains the ability of flCDCP1 to bind to Src and PKCδ without stimulating PKCδ phosphorylation: Src and PKCδ are not bound to flCDCP1 at the same time. These data are in line with ex vivo data in TNBC2 and in vivo data in prostate cancer,3,8 showing that CDCP1 cleavage is required to induce cellular dissemination and multiple-organ metastasis. Although the positive and negative regulators of CDCP1 dimerization remain to be elucidated, we have shown that ECC represents a way to inhibit dimerization of cCDCP1, disrupting cCDCP1 pro-migratory signaling.
Figure 7.
Proposed model of CDCP1 activation and signal transduction. (1) CDCP1 is cleaved on the extracellular side of the membrane between CUB1 and CUB2. Src family kinases (SFKs) (Src/Fyn/Yes) can phosphorylate both flCDCP1 and cCDCP1. (2) SFKs and PKCδ bind to flCDCP1 or cCDCP1 protein monomers, presumably to overlapping/close binding sites, but signal transduction from SFKs to PKCδ does not occur. (3) cCDCP1 is capable of forming a dimer, which allows SFKs and PKCδ to bind to each individual subunit of a dimer, leading to PKCδ phosphorylation by SFKs. Activated PKCδ induces pro-migratory signaling. ECC is capable of inhibiting cCDCP1 dimerization, PKCδ phosphorylation and migration of TNBC cells. We propose that targeting CDCP1 dimerization will lead to blockade of SFK/PKCδ signaling and inhibit CDCP1-mediated migration and invasion in TNBC. Black arrows, phosphorylation by SFKs; gray arrows, binding; blue, CUB domains; red, binding site; P in red circle, phosphorylation.
Our data in 3D showing an increase in ECC-expressing TNBC cell apoptosis are in line with previous studies demonstrating that CDCP1 knockdown by si/shRNA induces apoptosis in lung and breast cancer cells cultured in anchorage-free conditions,4,39,40 a study showing that inhibition of CDCP1 cleavage in vivo induced apoptosis of prostate cancer cells8 and studies demonstrating that a function-blocking antibody against CDCP1 inhibited cell invasiveness in a 3D culture system and tumor growth in a xenograft model of breast cancer.41,42 The induction of apoptosis in the presence of extracellular matrix components, collagen I and Matrigel (collagen IV, laminin, enactin/nidogen-1 and proteoglycans), may indicate that ECC inhibits stellate structure formation, which is indicative of cell binding and anchoring to these substrates during invasion. This explanation is supported by a study showing preferential and direct binding of β1 integrin to cCDCP1 over flCDCP1.3 Our data suggest that cCDCP1 may promote β1 integrin activity, which is inhibited in the presence of ECC. It also remains to be investigated whether ECC is specific to the cCDCP1 homodimer or also inhibits cCDCP1 interactions with other proteins. The increased efficacy of ECC in 3D culture remains to be validated in an orthotopic mouse model of TNBC, where we expect the same accumulation of ECC extracellularly to inhibit cell–extracellular matrix adhesion, proliferation and induce apoptosis.
The ability of CDCP1 to form a dimer is supported by previous findings that treatment of NIH3T3 and MCF7 cells overexpressing exogenous CDCP1 with a bivalent anti-CDCP1 antibody stimulates CDCP1 activity, presumably by stimulating its clustering in lipid rafts.41 Kollmorgen et al.22 proposed that CDCP1 forms a dimer through the transmembrane or intracellular dimer interface but they did not investigate the dependence of dimer formation on CDCP1 cleavage. Although we do not exclude the possibility that CDCP1 forms a dimer through transmembrane or intracellular domains as well, our data support dimer formation through the extracellular CUB domains, with the secreted ECC portion capable of inhibiting dimerization of cCDCP1. Our data show that cCDCP1 forms a dimer, although we do not exclude the possibility that it is capable of forming a tetramer or higher molecular complex. Further research is needed to test this possibility.
It remains to be investigated how flCDCP1 stimulates signaling through PKCδ in ccRCC. For instance, 786-0 cells express almost exclusively flCDCP1 and we have shown previously that flCDCP1 stimulates migration of ccRCC cells through PKCδ.10 One striking difference between ccRCC and TNBC lies in abundance of lipid droplets, which is extremely high in ccRCC43 and rather low in TNBC.44,45 As CDCP1 is present in Triton X-100-insoluble lipid rafts,25,41 we propose that the high lipid content of ccRCC cells facilitates clustering of flCDCP1 at the membrane in lipid rafts, which leads to stimulation of PKCδ phosphorylation by Src. On the other hand, TNBC relies on CDCP1 cleavage to induce cCDPC1 dimerization to stimulate PKCδ phosphorylation by Src. In line with this, Casar et al.3,8 showed that PC3 and HEK 293T cells, which have low lipid content,46,47 are dependent on CDCP1 cleavage for cellular dissemination in vitro, in chick embryo and mouse models of metastasis.
Although we found that CDCP1 is not hypoxia inducible at the mRNA and protein levels in TNBC, the phosphorylation of cCDCP1 and PKCδ are hypoxia inducible. Importantly, we10 and others11 have shown that CDCP1 is hypoxia inducible through the hypoxia inducible factor pathway at the mRNA and protein levels in ccRCC. Cell-type-specific hypoxic induction of hypoxia inducible factor target genes has been described previously,48 which could explain the discrepancy between ccRCC and TNBC. Emerling et al.11 demonstrated that Src Y416 and CDCP1 Y734 phosphorylation are hypoxia inducible in ccRCC, which we replicated in TNBC. The mechanism of stimulation of these types of phosphorylation under hypoxia deserves further investigation.
The CDCP1 signaling cascade has not been investigated in depth in TNBC and other types of breast cancer. It was previously reported that cCDCP1 promotes TNBC migration by sequestering E-cadherin intracellularly2 and TNBC invasion by stimulating membrane type 1 matrix metalloproteinase activity.25 Our studies suggest that cCDCP1 promotes TNBC migration through dimerization, which is important for PKCδ phosphorylation and potentially ERK1/2, and p38 mitogen-activated protein kinase phosphorylation (requires further investigation). In addition, it was recently shown that CDCP1 directly interacts with HER2 and stimulates progression of HER2-positive tumors.49 On the other hand, a correlation between estrogen and progesterone receptor status and CDCP1 expression has not been found.50 Thus, the dependence of CDCP1 activity on cleavage and dimerization in hormone receptor-positive breast cancers remains to be investigated.
Our data validate the previously demonstrated therapeutic potential of targeting the cleaved isoform of CDCP1 to inhibit cancer metastasis.3 However, we provide novel insight into the CDCP1 activation mechanism and propose targeting a step downstream of CDCP1 cleavage: dimerization.
MATERIALS AND METHODS
Cell lines and media
HEK 293T, MDA-MB-231, MDA-MB-453, MDA-MB-468 and MDA-MB-361 cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM; Genesee, San Diego, CA, USA; #25-500) with 10% fetal bovine serum (FBS; Omega Scientific, Tarzana, CA, USA; #FB-12), 2 mm l-glutamine, 100 µg/ml penicillin and 100 µg/ml streptomycin. Cells were grown in mixed gas CO2 water-jacketed incubators (21% O2 and 5% CO2). MCF10A cells were grown in the same media as above supplemented with 20 µg/ml epidermal growth factor (PeproTech, Rock Hill, NJ, USA; #AF-100-15) and 10 µg/ml insulin (Roche, San Francisco, CA, USA; #11376497001). MDA-MB-231 and MDA-MB-468 cell lines, extensively used for in vitro experiments in this study, were validated using short tandem repeat profiling by the American Type Culture Collection (Manassas, VA, USA). All cells were regularly tested for mycoplasma contamination and confirmed to be negative.
Primary specimen acquisition and lysis
Fresh TNBC specimens were acquired from Hoag pathology and frozen on arrival. Frozen TNBC specimens were obtained from the UC Irvine Medical Center. For lysis, specimens were thawed on ice and then homogenized in cell lysis buffer (20 mm Tris-HCl pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm Na4P2O7, 1mm β-glycerophosphate, 1 mm Na3VO4) with protease inhibitors (Fisher Scientific, Pittsburgh, PA, USA; #P1-88266) and phosphatase inhibitors (Roche #04906845001).
Western blotting
Protocol was adapted from Razorenova et al.51 Briefly, cells were lysed using cold cell lysis buffer (see recipe above) with protease inhibitors (Fisher Scientific). Protein was measured using BCA Protein Assay kit (Pierce, Rockford, IL, USA; #PI-23225). Forty micrograms of protein was loaded/lane of 10% bis-acrylamide gels and transferred onto 0.2 µm nitrocellulose membranes (BioRad, Hercules, CA, USA). Membranes were incubated with primary antibodies overnight at 4 °C with gentle agitation, then incubated with secondary antibodies for 1 h at room temperature with gentle agitation and the signal was developed using Amersham ECL Prime WB substrate (GE Healthcare, Scottsdale, AZ, USA; #RPN2236) or SigmaFast BCIP/NBT substrate (Sigma Aldrich, St Louis, MO, USA; #B5655) for horseradish peroxidase or alkaline phosphatase conjugates, respectively. WB analysis of phospho-signaling downstream of CDCP1 was conducted 48 h post transfection: cells were serum starved for 6 h and then stimulated for 20 min with 10% FBS (Gibco, Carlsbad, CA, USA; #16000-044-1399) before lysing. All antibodies used for WB are listed in Supplementary Table 2.
Construction of shRNA and cDNA overexpression constructs for CDCP1
pLM-CMV-flCDCP1 was cloned by PCR using pDrive-flCDCP1 (Open-Biosystems, Huntsville, AL, USA; #IHS1382-8427202) as a template. As pDrive-flCDCP1 contained five mismatches when aligned to WT CDCP1 referenced in Cosmic database, all mismatches in pLM-CMV-flCDCP1 were reverted to WT sequence using the Lightning Site Directed Mutagenesis kit (Stratagene, Santa Clara, CA, USA; #210516).
pLM-CMV-flCDCP1-Flag and pLM-CMV-flCDCP1-His were cloned by PCR using pLM-CMV-flCDCP1 as a template.
pLM-CMV-cCDCP1-Flag and pLM-CMV-cCDCP1-His were cloned in two steps. First, cCDCP1 was amplified by PCR using pLM-CMV-flCDCP1 as a template. Second, the SP was cloned upstream of cCDCP1 by PCR using pLM-CMV-flCDCP1 as a template.
pLM-CMV-cCDCP1-Y734F-Flag and pLM-CMV-cCDCP1-Y734F-His were generated from pLM-CMV-cCDCP1-Flag and pLM-CMV-cCDCP1-His by site-directed mutagenesis using the Lightning Site Directed Mutagenesis kit (Stratagene).
pLM-CMV-ECC-His was cloned by PCR using pLM-CMV-flCDCP1 as a template.
pLM-CMV-cCDCP1-N’luc, pLM-CMV-cCDCP1-C’luc, pLM-CMV-flCDCP1-N’luc and pLM-CMV-flCDCP1-C’luc were cloned in two steps. First, N’luciferase and C’luciferase were PCR amplified from the constructs described by Choi et al.34 Second, cCDCP1 and flCDCP1 were cloned upstream of N’luc and C’luc. cCDCP1 was amplified from pLM-CMVc-CDCP1-Flag, which had already an SP. flCDCP1 was amplified from pLM-CMV-flCDCP1.
To make shRNA-insensitive flCDCP1 (pLM-CMV-sh-ins-flCDCP1), the third nucleotides of each codon in the regions of flCDCP1 recognized by the shRNAs were mutated to interfere with shRNA binding but carefully maintain WT flCDCP1 protein sequence. The 985-bp sh-ins-CDCP1 fragment with NheI and BamHI restriction sites at the 5′- and 3′-ends, respectively, was synthetized by GenScript (Piscataway, NJ, USA) and cloned to pUC57 by EcoRV. The fragment (+1 to +650 bp downstream of the start codon) was cloned to pLM-CMV-flCDCP1 by NheI and BamHI restriction sites.
To make shRNA-insensitive ncCDCP1 (pLM-CMV-sh-ins-ncCDCP1), the cleavage site in pLM-CMV-sh-ins-flCDCP1 was mutated: RK368-9QN. A gBlock containing CDCP1 fragment flanking the cleavage site (+977 to +1257 bp downstream of the start codon) with BamHI and AgeI was synthesized. This gBlock was cloned into pLM-CMV-sh-ins-flCDCP1 by BamHI and AgeI restriction sites.
Each construct originally had a puromycin resistance cassette. Puromycin was replaced with hygromycin and neomycin in pLM-CMV-cCDCP1-His and pLM-CMV-sh-ins-ncCDCP1, respectively. Hygromycin and neomycin resistance cassettes were obtained from pLM-CMV-H4-hygro and pLM-CMV-H4-neo plasmids using the SpeI and SalI restriction enzymes and cloned into SpeI- and SalI-digested pLM-CMV-cCDCP1-His and pLM-CMV-sh-ins-ncCDCP1, respectively.
pLKO.1shCDCP1-1 was purchased from OpenBiosystems (#RHS3979-98822176). pLKO.1shScrambled was purchased from Addgene (Cambidge, MA, USA; #1864). pLKO.1shCDCP1-7 was cloned using two annealed complimentary oligos and ligated to AgeI- and EcoRI-digested pLKO.1 empty vector (Addgene #8453).
All oligonucleotides and gBlocks in this section were synthesized by IDT (Coralville, IA, USA) and are all listed in the Supplementary Table 3.
Cell transfections with overexpression constructs
HEK 293T cells were transfected using Lipofectamine and Plus reagents (Life Tech, Carlsbad, CA, USA; #18324-012 and #11514-015) following the manufacturer’s protocol and used for experiments 2–3 days post transfection.
Virus production and infection of target cells
Detailed protocol is described in Razorenova et al.52 To produce virus, HEK 293T cells were transfected with each lentiviral plasmid along with packaging plasmids, pVSVG and ΔR8.2 using standard CaCl2 transfection protocol. Virus-containing media with 6 µg/ml polybrene filtered through a 0.45-µm PES filter was transferred to TNBC target cells, which were selected in antibiotic-containing media for minimum 1 week.
ECC detection in cell media
HEK 293T were transfected with pLM-CMV-ECC-His using Lipofectamine and Plus reagents following the manufacturer’s protocol (Life Tech) in a sixwell plate. Twenty-four hours after transfection, the media was changed to 1 ml fresh DMEM with 10% FBS. Forty-eight hours later, 50 µl of media was collected and denatured in 12.5 µl 5 × sample loading buffer (312 mm Tris-HCl pH 6.8, 10% SDS, 0.05% bromophenol blue, 50% glycerol, 10% β-mercaptoethanol). Samples were analyzed by WB.
Quantitative reverse transcriptase–PCR
Quantitative reverse transcriptase–PCR was performed as previously described10 using the ViiA 7 Real-Time PCR System (Life Tech). Human TATA-binding protein primers were chosen as an internal control. Primers are listed in Supplementary Table 3.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde for 10 min, then treated with 0.3% Triton X-100 in 1 × PBS for 5 min before blocking for 1 h in 5% bovine serum albumin in 1 × PBS. Cells were then incubated overnight in primary antibody at 4 °C, followed by incubation for 2 h in secondary antibody at room temperature in the dark. Antibodies used are listed in Supplementary Table 2. Both primary and secondary antibodies were diluted in 1% bovine serum albumin in 1 × PBS. Cell nuclei were counterstained with Hoechst 33342 at 1:500 in 1 × PBS for 1 min. Coverslips were mounted with Vectashield (Vector #H-1000) and imaged using a Nikon Eclipse Ti microscope with a 40 × or 63 × objective and NIS element AR3.10 software. Images for sh-ins-ncCDCP1-transfected HEK 293T cells were taken using a Zeiss Spinning Disc Confocal Microscope (Zeiss, Pleasanton, CA, USA) with a 63 × objective and AxioVision software (Zeiss).
Duolink proximity ligation assay
The assay was performed according to the manufacturer’s protocol (Sigma Aldrich; #DUO92008, #DUO92009 and #DUO92010). Primary antibody probes were made using goat anti-CDCP1 antibody (Abcam, Cambridge, MA, USA; #ab1377). Images were collected as for immunofluorescence staining analysis. Twelve-millimeter coverslips were used with 20 µl reaction volumes.
Co-immunoprecipitation
Four hundred micrograms of protein in 200 µl of cell lysis buffer (see recipe in ‘Primary specimen acquisition and lysis’) was incubated with 4 µl of anti- Flag antibody (Sigma Aldrich; #F1804) for 16 h at 4 °C with gentle agitation. Then, 20 µl Protein G Dynabeads (Life Tech; #10003D) were added, followed by incubation for 1 h at 4 °C. Immunoprecipitated proteins were collected using magnetic stand, washed three times for 5 min on ice with cell lysis buffer without protease or phosphatase inhibitors, boiled in 1 × sample loading buffer for 6 min and analyzed by WB.
Transwell migration
Transwell inserts (Corning Life Sciences Plastic, Corning, NY, USA; #3464) were coated in 50 mg/ml fibronectin (Sigma Aldrich; #F2006-2) in DMEM for 4 h at 37 °C. Cells were treated with 100 µm mitomycin C for 2.5 h before being detached with 2% EDTA in 1 × PBS. Sixty thousand HEK 293T, MDA-MB-468, and UCI-082014 cells in serum-free DMEM were added to the top chamber of the inserts immersed in 10% FBS-containing media in the bottom chamber. Cells were allowed to migrate for 24 h (HEK 293T), 16 h (MDA-MB-468), and 6 h (UCI-082014). Cells from the top of the transwell inserts were removed with a cotton swab and cells that migrated through the transwell inserts were fixed and stained using the Richard Allen 3 Step Staining Kit (Thermo Scientific, Grand Island, NY, USA; #3300), photographed and counted.
Scratch assay
MDA-MB-231 cells (2 00 000) were plated per well in a 6-well plate. Twenty-four hours later, cell monolayers were treated with mitomycin C for 2 h in serum-free DMEM and then scratched with a 200-µl pipette tip. The bottom of the plate was marked at three points on the scratch for triplicate measurements. Cells were washed once with 1 × PBS and incubated in DMEM with 2% FBS, 2 mm l-glutamine, 100 µg/ml penicillin and 100 µg/ml streptomycin. Pictures of the scratch were taken at the time of the scratch (T0) and at 6 h after the scratch (T6). Percent wound closure was calculated as (1 − (T6/T0))*100.
Split-luciferase assay
HEK 293T cells were plated at 10 000 cells per well in a black-walled 96-well plate. Sixteen hours later, they were transfected with CDCP1-N’luc, CDCP1-C’luc and either a VC or ECC using Lipofectamine and Plus reagents (Life Tech) following the manufacturer’s protocol. Forty-eight hours after transfection, dimerization was analyzed with D-luciferin (Promega, Madison, WI, USA; #E1605) on an IVIS Lumina Imager (PerkinElmer, Waltham, MA, USA).
3D assays
MDA-MB-231, MDA-MB-468 and UCI-082014 cell lines were plated at 3000 cells in 50 µl of 1:1 collagen (BD Biosciences, Franklin Lakes, NJ, USA; #354249):Matrigel (BD Biosciences; #356234) gel. Collagen was initially prepared at 6.4 mg/ml according to the manufacturer’s protocol before being mixed with Matrigel for a final concentration in the gels of 3.2 mg/ml collagen, similar to that in transformed breast tissue.53,54 Gels were plated onto 12-mm coverslips. Media on the cells was changed every 2 days and cells were allowed to proliferate/invade the gel for 6 days. Gels were fixed in 4% paraformaldehyde for 20 min, washed 4 × 15 min with 1 × PBS, permeabilized with 0.3% Triton X-100 in 1 × PBS for 15 min, washed 4 × 15 min with 1 × PBS, incubated in blocking solution (10% donkey serum, 5% bovine serum albumin, 0.1% Triton X-100 in 1 × PBS) overnight at 4 °C with gentle agitation. Antibodies used for fluorescent staining in 3D are provided in Supplementary Table 2. Phase-contrast images were taken using an EVOS × L Microscope (AMG, Bothell, WA, USA) with a 10 × objective. Fluorescent images were taken using a Zeiss Spinning Disc Confocal Microscope with a 20 × objective with numerical aperture 0.8 and AxioVision software. Quantification was the average of five images/gel for three biological replicates.
Supplementary Material
Acknowledgments
This work was supported by NIH Award F31CA196226 to HJW. We thank Drs Alice Police and Erin Lin for providing primary TNBC tissue, Dr Peter Kaiser for MDA-MB-231 and MDA-MB-468 cell lines, Dr Dan Mercola for MDA-MB-453 cell line, Dr Xing Dai for MCF10A cell line, Dr Albert Koong for split-luciferase vectors, Dr Inder Verma for pVSVG and ΔR8.2 lentivirus packaging plasmids, Dr Jae-Won Soh for pHACE-PKCδ construct, Dr Sanford Shattil for pRC-CMV-Src construct, Dr Peter Chumakov for pLM-CMV lentiviral vector, Miranda Paley from Dr Jennifer Prescher lab and Linan Liu from Dr Weian Zhao’s lab for help with IVIS Lumina Imager, and Dr David Fruman and Dr Hung Fan for critical reading of the manuscript.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
REFERENCES
- 1.Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012;30:1879–1887. doi: 10.1200/JCO.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- 2.Law ME, Corsino PE, Jahn SC, Davis BJ, Chen S, Patel B, et al. Glucocorticoids and histone deacetylase inhibitors cooperate to block the invasiveness of basal-like breast cancer cells through novel mechanisms. Oncogene. 2013;32:1316–1329. doi: 10.1038/onc.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casar B, Rimann I, Kato H, Shattil SJ, Quigley JP, Deryugina EI. In vivo cleaved CDCP1 promotes early tumor dissemination via complexing with activated β1 integrin and induction of FAK/PI3K/Akt motility signaling. Oncogene. 2014;33:255–268. doi: 10.1038/onc.2012.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uekita T, Jia L, Narisawa-Saito M, Yokota J, Kiyono T, Sakai R. CUB domain-containing protein 1 is a novel regulator of anoikis resistance in lung adenocarcinoma. Mol Cell Biol. 2007;27:7649–7660. doi: 10.1128/MCB.01246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uekita T, Tanaka M, Takigahira M, Miyazawa Y, Nakanishi Y, Kanai Y, et al. CUB-domain-containing protein 1 regulates peritoneal dissemination of gastric scirrhous carcinoma. Am J Pathol. 2008;172:1729–1739. doi: 10.2353/ajpath.2008.070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siva AC, Wild MA, Kirkland RE, Nolan MJ, Lin B, Maruyama T, et al. Targeting CUB domain-containing protein 1 with a monoclonal antibody inhibits metastasis in a prostate cancer model. Cancer Res. 2008;68:3759–3766. doi: 10.1158/0008-5472.CAN-07-1657. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Ong SE, Badu-Nkansah K, Schindler J, White FM, Hynes RO. CUB-domain-containing protein 1 (CDCP1) activates Src to promote melanoma metastasis. Proc Natl Acad Sci USA. 2011;108:1379–1384. doi: 10.1073/pnas.1017228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casar B, He Y, Iconomou M, Hooper JD, Quigley JP, Deryugina EI. Blocking of CDCP1 cleavage in vivo prevents Akt-dependent survival and inhibits metastatic colonization through PARP1-mediated apoptosis of cancer cells. Oncogene. 2012;31:3924–3938. doi: 10.1038/onc.2011.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Y, Wu AC, Harrington BS, Davies CM, Wallace SJ, Adams MN, et al. Elevated CDCP1 predicts poor patient outcome and mediates ovarian clear cell carcinoma by promoting tumor spheroid formation, cell migration and chemoresistance. Oncogene. 2015 doi: 10.1038/onc.2015.101. e-pub ahead of print 20 April 2015. [DOI] [PubMed] [Google Scholar]
- 10.Razorenova OV, Finger EC, Colavitti R, Chernikova SB, Boiko AD, Chan CK, et al. VHL loss in renal cell carcinoma leads to up-regulation of CUB domain-containing protein 1 to stimulate PKC{delta}-driven migration. Proc Natl Acad Sci USA. 2011;108:1931–1936. doi: 10.1073/pnas.1011777108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerling BM, Benes CH, Poulogiannis G, Bell EL, Courtney K, Liu H, et al. Identification of CDCP1 as a hypoxia-inducible factor 2α (HIF-2α) target gene that is associated with survival in clear cell renal cell carcinoma patients. Proc Natl Acad Sci USA. 2013;110:3483–3488. doi: 10.1073/pnas.1222435110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uekita T, Fujii S, Miyazawa Y, Iwakawa R, Narisawa-Saito M, Nakashima K, et al. Oncogenic Ras/ERK signaling activates CDCP1 to promote tumor invasion and metastasis. Mol Cancer Res. 2014;12:1449–1459. doi: 10.1158/1541-7786.MCR-13-0587. [DOI] [PubMed] [Google Scholar]
- 13.Adams MN, Harrington BS, He Y, Davies CM, Wallace SJ, Chetty NP, et al. EGF inhibits constitutive internalization and palmitoylation-dependent degradation of membrane-spanning procancer CDCP1 promoting its availability on the cell surface. Oncogene. 2014;34:1375–1383. doi: 10.1038/onc.2014.88. [DOI] [PubMed] [Google Scholar]
- 14.Bhatt AS, Erdjument-Bromage H, Tempst P, Craik CS, Moasser MM. Adhesion signaling by a novel mitotic substrate of src kinases. Oncogene. 2005;24:5333–5343. doi: 10.1038/sj.onc.1208582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown TA, Yang TM, Zaitsevskaia T, Xia Y, Dunn CA, Sigle RO, et al. Adhesion or plasmin regulates tyrosine phosphorylation of a novel membrane glycoprotein p80/gp140/CUB domain-containing protein 1 in epithelia. J Biol Chem. 2004;279:14772–14783. doi: 10.1074/jbc.M309678200. [DOI] [PubMed] [Google Scholar]
- 16.He Y, Wortmann A, Burke LJ, Reid JC, Adams MN, Abdul-Jabbar I, et al. Proteolysis-induced N-terminal ectodomain shedding of the integral membrane glycoprotein CUB domain-containing protein 1 (CDCP1) is accompanied by tyrosine phosphorylation of its C-terminal domain and recruitment of Src and PKCdelta. J Biol Chem. 2010;285:26162–26173. doi: 10.1074/jbc.M109.096453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wortmann A, He Y, Christensen ME, Linn M, Lumley JW, Pollock PM, et al. Cellular settings mediating Src Substrate switching between focal adhesion kinase tyrosine 861 and CUB-domain-containing protein 1 (CDCP1) tyrosine 734. J Biol Chem. 2011;286:42303–42315. doi: 10.1074/jbc.M111.227462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benes CH, Poulogiannis G, Cantley LC, Soltoff SP. The SRC-associated protein CUB domain-containing protein-1 regulates adhesion and motility. Oncogene. 2011;31:653–663. doi: 10.1038/onc.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leroy C, Shen Q, Strande V, Meyer R, McLaughlin ME, Lezan E, et al. CUB-domain-containing protein 1 overexpression in solid cancers promotes cancer cell growth by activating Src family kinases. Oncogene. 2015;34:5593–5598. doi: 10.1038/onc.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benes CH, Wu N, Elia AE, Dharia T, Cantley LC, Soltoff SP. The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell. 2005;121:271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 21.Spassov DS, Wong CH, Sergina N, Ahuja D, Fried M, Sheppard D, et al. Phosphorylation of Trask by Src kinases inhibits integrin clustering and functions in exclusion with focal adhesion signaling. Mol Cell Biol. 2011;31:766–782. doi: 10.1128/MCB.00841-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kollmorgen G, Bossenmaier B, Niederfellner G, Häring HU, Lammers R. Structural requirements for cub domain containing protein 1 (CDCP1) and Src dependent cell transformation. PLoS One. 2012;7:e53050. doi: 10.1371/journal.pone.0053050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hooper JD, Zijlstra A, Aimes RT, Liang H, Claassen GF, Tarin D, et al. Subtractive immunization using highly metastatic human tumor cells identifies SIMA135/CDCP1, a 135 kDa cell surface phosphorylated glycoprotein antigen. Oncogene. 2003;22:1783–1794. doi: 10.1038/sj.onc.1206220. [DOI] [PubMed] [Google Scholar]
- 24.Miyazawa Y, Uekita T, Hiraoka N, Fujii S, Kosuge T, Kanai Y, et al. CUB domain-containing protein 1, a prognostic factor for human pancreatic cancers, promotes cell migration and extracellular matrix degradation. Cancer Res. 2010;70:5136–5146. doi: 10.1158/0008-5472.CAN-10-0220. [DOI] [PubMed] [Google Scholar]
- 25.Miyazawa Y, Uekita T, Ito Y, Seiki M, Yamaguchi H, Sakai R. CDCP1 regulates the function of MT1-MMP and invadopodia-mediated invasion of cancer cells. Mol Cancer Res. 2013;11:628–637. doi: 10.1158/1541-7786.MCR-12-0544. [DOI] [PubMed] [Google Scholar]
- 26.Uekita T, Fujii S, Miyazawa Y, Hashiguchi A, Abe H, Sakamoto M, et al. Suppression of autophagy by CUB domain-containing protein 1 signaling is essential for anchorage-independent survival of lung cancer cells. Cancer Sci. 2013;104:865–870. doi: 10.1111/cas.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bork P, Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J Mol Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- 28.Chaudary N, Hill RP. Hypoxia and metastasis in breast cancer. Breast Dis. 2006;26:55–64. doi: 10.3233/bd-2007-26105. [DOI] [PubMed] [Google Scholar]
- 29.Huang C, Jacobson K, Schaller MD. MAP kinases and cell migration. J Cell Sci. 2004;117:4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- 30.Romero A, Varela PF, Sanz L, Töpfer-Petersen E, Calvete JJ. Crystallization and preliminary X-ray diffraction analysis of boar seminal plasma spermadhesin PSP-I/PSP-II, a heterodimer of two CUB domains. FEBS Lett. 1996;382:15–17. doi: 10.1016/0014-5793(96)00133-0. [DOI] [PubMed] [Google Scholar]
- 31.Ng D, Pitcher GM, Szilard RK, Sertié A, Kanisek M, Clapcote SJ, et al. Neto1 is a novel CUB-domain NMDA receptor-interacting protein required for synaptic plasticity and learning. PLoS Biol. 2009;7:e41. doi: 10.1371/journal.pbio.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HX, Mendes FA, Plouhinec JL, De Robertis EM. Enzymatic regulation of pattern: BMP4 binds CUB domains of Tolloids and inhibits proteinase activity. Genes Dev. 2009;23:2551–2562. doi: 10.1101/gad.1839309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gandji LY, Proust R, Larue L, Gesbert F. The tyrosine phosphatase SHP2 associates with CUB domain-containing protein-1 (CDCP1), regulating its expression at the cell surface in a phosphorylation-dependent manner. PLoS One. 2015;10:e0123472. doi: 10.1371/journal.pone.0123472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi CY, Chan DA, Paulmurugan R, Sutphin PD, Le QT, Koong AC, et al. Molecular imaging of hypoxia-inducible factor 1 alpha and von Hippel-Lindau interaction in mice. Mol Imaging. 2008;7:139–146. [PMC free article] [PubMed] [Google Scholar]
- 35.Cvetković D, Goertzen CG, Bhattacharya M. Quantification of breast cancer cell invasiveness using a three-dimensional (3D) model. J Vis Exp. 2014;(88) doi: 10.3791/51341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guiro K, Patel SA, Greco SJ, Rameshwar P, Arinzeh TL. Investigating breast cancer cell behavior using tissue engineering scaffolds. PLoS One. 2015;10:e0118724. doi: 10.1371/journal.pone.0118724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tu YF, Kaipparettu BA, Ma Y, Wong LJ. Mitochondria of highly metastatic breast cancer cell line MDA-MB-231 exhibits increased autophagic properties. Biochim Biophys Acta. 2011;1807:1125–1132. doi: 10.1016/j.bbabio.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 38.Arora R, Yates C, Gary BD, McClellan S, Tan M, Xi Y, et al. Panepoxydone targets NF-kB and FOXM1 to inhibit proliferation, induce apoptosis and reverse epithelial to mesenchymal transition in breast cancer. PLoS One. 2014;9:e98370. doi: 10.1371/journal.pone.0098370. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Lin CY, Chen HJ, Huang CC, Lai LC, Lu TP, Tseng GC, et al. ADAM9 promotes lung cancer metastases to brain by a plasminogen activator-based pathway. Cancer Res. 2014;74:5229–5243. doi: 10.1158/0008-5472.CAN-13-2995. [DOI] [PubMed] [Google Scholar]
- 40.Seidel J, Kunc K, Possinger K, Jehn C, Lüftner D. Effect of the tyrosine kinase inhibitor lapatinib on CUB-domain containing protein (CDCP1)-mediated breast cancer cell survival and migration. Biochem Biophys Res Commun. 2011;414:226–232. doi: 10.1016/j.bbrc.2011.09.062. [DOI] [PubMed] [Google Scholar]
- 41.Kollmorgen G, Niederfellner G, Lifke A, Spohn GJ, Rieder N, Harring SV, et al. Antibody mediated CDCP1 degradation as mode of action for cancer targeted therapy. Mol Oncol. 2013;7:1142–1151. doi: 10.1016/j.molonc.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandercock AM, Rust S, Guillard S, Sachsenmeier KF, Holoweckyj N, Hay C, et al. Identification of anti-tumour biologics using primary tumour models, 3-D phenotypic screening and image-based multi-parametric profiling. Mol Cancer. 2015;14:147. doi: 10.1186/s12943-015-0415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu B, Ackerman D, Sanchez DJ, Li B, Ochocki JD, Grazioli A, et al. HIF2α-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov. 2015;5:652–667. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nieva C, Marro M, Santana-Codina N, Rao S, Petrov D, Sierra A. The lipid phenotype of breast cancer cells characterized by Raman microspectroscopy: towards a stratification of malignancy. PLoS One. 2012;7:e46456. doi: 10.1371/journal.pone.0046456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S, Lee Y, Koo JS. Differential expression of lipid metabolism-related proteins in different breast cancer subtypes. PLoS One. 2015;10:e0119473. doi: 10.1371/journal.pone.0119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhau HE, He H, Wang CY, Zayzafoon M, Morrissey C, Vessella RL, et al. Human prostate cancer harbors the stem cell properties of bone marrow mesenchymal stem cells. Clin Cancer Res. 2011;17:2159–2169. doi: 10.1158/1078-0432.CCR-10-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim KH, Lee GY, Kim JI, Ham M, Won Lee J, Kim JB. Inhibitory effect of LXR activation on cell proliferation and cell cycle progression through lipogenic activity. J Lipid Res. 2010;51:3425–3433. doi: 10.1194/jlr.M007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sowter HM, Raval RR, Moore JW, Ratcliffe PJ, Harris AL. Predominant role of hypoxia-inducible transcription factor (Hif)-1alpha versus Hif-2alpha in regulation of the transcriptional response to hypoxia. Cancer Res. 2003;63:6130–6134. [PubMed] [Google Scholar]
- 49.Alajati A, Guccini I, Pinton S, Garcia-Escudero R, Bernasocchi T, Sarti M, et al. Interaction of CDCP1 with HER2 enhances HER2-driven tumorigenesis and promotes trastuzumab resistance in breast cancer. Cell Rep. 2015;11:564–576. doi: 10.1016/j.celrep.2015.03.044. [DOI] [PubMed] [Google Scholar]
- 50.Mamat S, Ikeda J, Enomoto T, Ueda Y, Rahadiani N, Tian T, et al. Prognostic significance of CUB domain containing protein expression in endometrioid adenocarcinoma. Oncol Rep. 2010;23:1221–1227. doi: 10.3892/or_00000753. [DOI] [PubMed] [Google Scholar]
- 51.Razorenova OV, Castellini L, Colavitti R, Edgington LE, Nicolau M, Huang X, et al. The apoptosis repressor with a CARD domain (ARC) gene is a direct hypoxia-inducible factor 1 target gene and promotes survival and proliferation of VHL-deficient renal cancer cells. Mol Cell Biol. 2014;34:739–751. doi: 10.1128/MCB.00644-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Razorenova OV, Ivanov AV, Budanov AV, Chumakov PM. Virus-based reporter systems for monitoring transcriptional activity of hypoxia-inducible factor 1. Gene. 2005;350:89–98. doi: 10.1016/j.gene.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miroshnikova YA, Jorgens DM, Spirio L, Auer M, Sarang-Sieminski AL, Weaver VM. Engineering strategies to recapitulate epithelial morphogenesis within synthetic three-dimensional extracellular matrix with tunable mechanical properties. Phys Biol. 2011;8:026013. doi: 10.1088/1478-3975/8/2/026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maller O, Hansen KC, Lyons TR, Acerbi I, Weaver VM, Prekeris R, et al. Collagen architecture in pregnancy-induced protection from breast cancer. J Cell Sci. 2013;126:4108–4110. doi: 10.1242/jcs.121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.