Abstract
Purpose
Hepatocyte growth factor (HGF)/Met signaling plays critical roles in pancreatic ductal adenocarcinoma (PDA) development and progression and is considered a potential therapeutic target for this disease. However, the mechanism of aberrant activation of HGF/Met signaling and resistance to Met inhibition in PDA remains unclear.
Experimental Design
The mechanistic role of cross-talk between Forkhead box M1 (FOXM1) and HGF/Met signaling in promotion of PDA growth and resistance to Met inhibition was examined using cell culture, molecular biology and mouse models; and the relevance of our experimental and mechanistic findings were validated using human PDA tissues.
Results
Met was markedly overexpressed in both PDA cell lines and pancreatic tumor specimens, and the expression of Met correlated directly with that of FOXM1 in human tumor specimens. Mechanistically, FOXM1 bound to the promoter region of the Met gene and transcriptionally increased the expression of Met. Increased expression of FOXM1 enhanced the activation of HGF/Met signaling and its downstream pathways, including RAS/extracellular signal-regulated kinase 1/2, phosphoinositide 3-kinase/AKT, and signal transducer and activator of transcription 3. Furthermore, activation of HGF/Met signaling increased the expression and transcriptional activity of FOXM1, and the cross-talk between FOXM1 and HGF/Met signaling promoted PDA growth and resistance to Met inhibition.
Conclusions
Collectively, our findings identified a positive feedback loop formed by FOXM1 and HGF/Met and revealed that this loop is a potentially effective therapeutic target for PDA.
Keywords: FOXM1, Met, resistance, growth, pancreatic cancer
Introduction
Pancreatic ductal adenocarcinoma (PDA), which is often just called pancreatic cancer, accounts for more than 90% of pancreatic cancer cases and is the seventh leading cause of cancer-related deaths worldwide, with an incidence that is increasing annually.1, 2 Although basic research of and surgery, radiation therapy, chemotherapy, and molecular targeted therapy for PDA have improved greatly, 75% of patients die within 12 months after diagnosis, and the 5-year overall survival rate is less than 5%.3 Thus further studies of the molecular drivers of and new therapeutic targets for PDA are urgently needed.
Met, a receptor tyrosine kinase, is predominantly expressed in epithelial and endothelial cells. Binding of Met with its ligand hepatocyte growth factor (HGF) leads to dimerization and phosphorylation at two tyrosine residues (Tyr1234 and Tyr1235) close to or within the catalytic pocket of Met. Furthermore, Met could associate and form dimerization with receptor originated from Nantes (RON), and RON ligand macrophage stimulating protein (MSP) induced a transphosphorylation of MetTyr1234/Tyr12354. These result in autophosphorylation of multiple cytoplasmic effectors; further activates a series of intracellular downstream signaling pathways, such as the RAS/extracellular signal-regulated kinase (ERK) cascade, the phosphoinositide 3-kinase (PI3K)/AKT axis, and signal transducer and activator of transcription proteins (STATs); and leads to the activation of a series of cellular processes, including proliferation, motility, morphologic changes, and invasion.5–7 Recently, researchers showed that Met is a marker for PDA stem cells.8 Met also plays critical roles in the interactions between PDA cells and fibroblasts and promotes cancer aggression and progression.9 Furthermore, PDA cells overexpressing Met are resistant to treatment with gemcitabine and radiation.8,10 Elevated expression of Met in PDA cells is frequently found and correlated with poor prognosis.11 Met can be mutated in other tumor types, but its mutation is rare in PDA cases according to the Cancer Genome Atlas database. Previous studies have demonstrated that hypoxia and expression of Neuromedin U and cytokines can lead to increased expression and activation of Met in PDA cells.12–14 However, the mechanism of elevated expression and activation of Met in PDA cases remains unclear.
Forkhead box M1 (FOXM1) is a transcription factor in the FOX superfamily that is characterized by a conserved winged helix DNA-binding domain.15 FOXM1 is a key regulator of cell-cycle progression, and a series of studies demonstrated that FOXM1 plays critical roles in epithelial-to-mesenchymal transition, invasion, metastasis, angiogenesis, and stem cell self-renewal and proliferation in PDA cases.16–18 Recently, we have reported that FOXM1 transactivates lactate dehydrogenase A and promotes the Warburg effect and PDA progression.19 However, the mechanism of elevated expression and the tumor-promoting role of FOXM1 in PDA cases remain to be elucidated.
In the present study, we have demonstrated that FOXM1 transcriptionally upregulates the expression of Met and plays critical roles in the activation of HGF/Met signaling. Meanwhile, activation of HGF/Met signaling leads to increased expression and transcriptional activity of FOXM1 via its downstream signaling of ERK1/2, AKT, and STAT3. HGF/Met and FOXM1 form a positive feedback loop which promotes PDA growth and resistance to Met inhibition
Results
Elevated expression of Met in PDA cells and its direct association with clinicopathologic characteristics
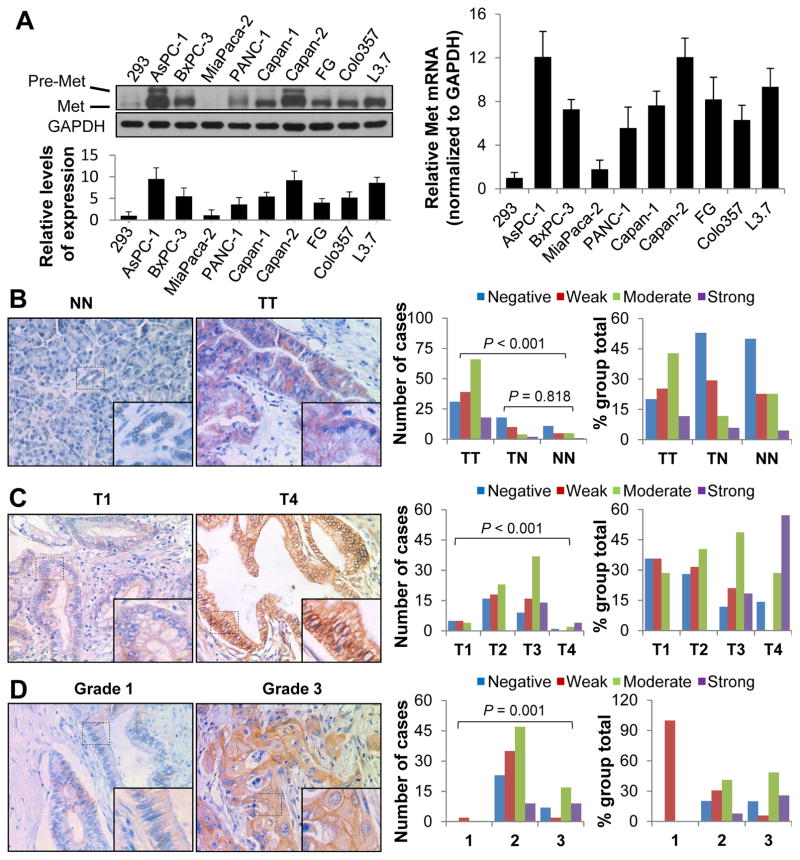
Authors have reported the tyrosine kinase Met to be overexpressed in multiple human cancers and that it drives tumor growth and metastasis. In the present study, we analyzed the expression of Met in PDA cell lines and pancreatic tumor specimens. As compared with its expression in normal human embryonic kidney 293 cells, Met mRNA and protein were markedly overexpressed in most of the PDA cell lines except for MiaPaca-2 (Fig. 1A), which was consistent with results of a previous study.4 We then investigated the expression of Met in a TMA as described previously.19 The clinicopathologic characteristics of the TMA were described in Supplementary Table S1. We found that the levels of Met expression in tumor specimens were markedly higher than those in the normal and tumor-adjacent tissue specimens (Fig. 1B). Moreover, we observed that stronger Met expression was correlated with higher disease pT classification (P<0.001) (Fig. 1C) and TNM stage (P<0.001) (Supplementary Table S2), and with increased grade of tumor differentiation (P=0.001) (Fig. 1D; Supplementary Table S2).
Figure 1.
Expression of Met and its association with clinicopathologic features in PDA patients. A, Shown was Western blot analysis of the Met protein expression levels in PDA cell lines (left upper panels) and relative protein levels of Met in PDA cell lines and standardization according to GAPDH (left lower panel). Total RNA were extracted and real-time PCR was used to analyze the mRNA levels of Met in those PDA cell lines (right panel). The experiments were performed three times. Data represent mean ± SEM of three independent experiments. B–D, A pancreatic tumor TMA was immunostained with a specific anti-Met antibody. Representative images of Met expression in normal pancreatic tissue (NN) vs. pancreatic tumor (TT) specimens (B, left panels), T1 stage vs. T4 stage tumor specimens (C, left panels), and grade 1 vs. grade 3 tumor specimens (D, left panels). Graphs showing that the levels of Met expression were significantly higher in pancreatic tumor (TT) than in tumor-adjacent normal tissue (TN) and normal pancreatic tissue (NN) specimens (P<0.001), but that the Met expression did not differ between TN and NN specimens (P=0.818) (B, middle and right panel). Met expression was positively associated with T stage (C, middle and right panel) and tumor differentiation (D, middle and right panel).
FOXM1 regulates the expression of Met in PDA cells
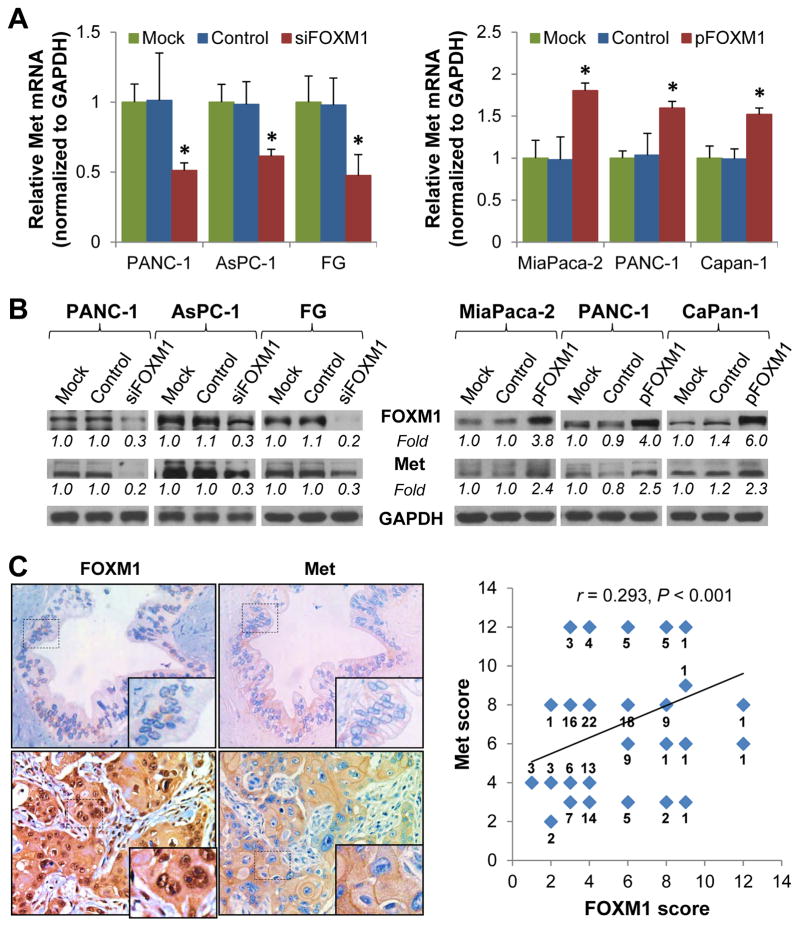
To identify the mechanisms underlying Met overexpression, we investigated the impact of altered FOXM1 expression on Met mRNA and protein expression in PDA cell lines using qPCR and Western blot analyses. Knockdown of FOXM1 expression in PANC-1, AsPC-1, and FG cells led to decreased Met mRNA and protein expression, whereas FOXM1 overexpression resulted in elevated Met expression (Fig. 2A, 2B and Supplementary Fig. S1A, S1B and S1C). To further confirm the positive correlation between FOXM1 and Met expression in PDA, we analyzed FOXM1 expression as for Met expression using the same TMA (Fig. 2C, left panels). Further statistical analysis revealed that the expression of Met was significantly correlated with that of FOXM1 in the TMA (r=0.293, P<0.001) (Fig. 2C, right panel), while a large variation between the expression of Met and FOXM1 also existed, suggesting a weak relationship between Met and FOXM1 expression and potentially complicated regulatory mechanisms of Met and FOXM1 expression. Nonetheless, our results indicated that Met may be a transcriptional target of FOXM1.
Figure 2.
Regulation of Met expression by FOXM1 in PDA cells. A and B, PANC-1, AsPC-1, and FG cells were transfected with siFOXM1 or control siRNA (left panels), and PANC-1, MiaPaca-2, and CaPan-1 cells were transfected with pcDNA3.1-FOXM1 or a control vector (right panels). The cells were cultured for 48 hr, and total RNA and protein were harvested. The Met mRNA expression levels in the cells were determined using qPCR (A). The FOXM1 and Met protein expression levels in the cells were determined using Western blot (B). C, Stains of the same cohorts of pancreatic tumor TMA sections for analysis of the related expression of FOXM1 and Met (left panels). The positive correlation of FOXM1 expression with Met expression was assessed using Pearson correlation coefficient analysis (n=154 pancreatic tumor TMA specimens, r=0.293, P<0.001) (right panel). Some of the dots on the graph represent more than one specimen. The numbers of overlapping scores were labeled. The levels of gene expression were quantitated and expressed as fold changes (italic numbers under each blot).
Met is a direct transcriptional target of FOXM1 in PDA cells
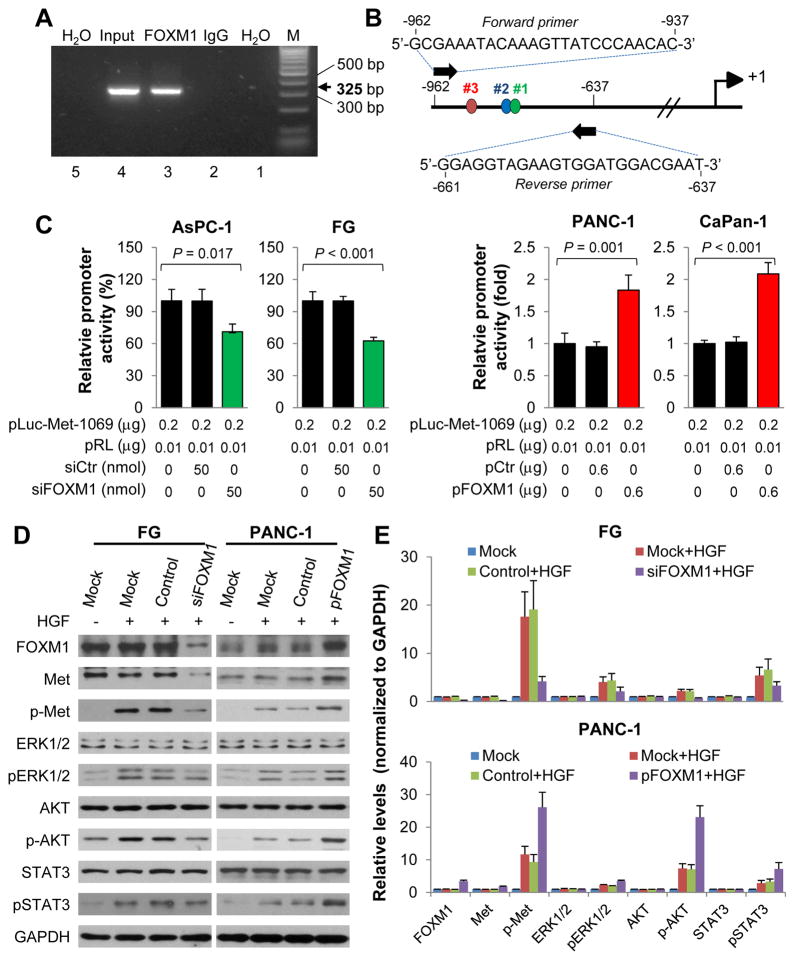
To determine whether Met is a transcriptional target of FOXM1, we analyzed the sequence of Met promoter for the potential FOXM1-binding elements 5’-TTT(G/A)AA(A/T)-3’, 5’-(C/T)TTCC(G/T)(G/T)-3’, 5’-(C/T)AAA(C/T)AA-3’, 5’-TAATCA-3’, and/or 5’-AGATTGAGTA-3’.31–34 We identified three putative FOXM1-binding elements in the Met promoter region (Fig. 3B and Supplementary Fig. S2A). We transfected PANC-1 cells with FOXM1-overexpressing plasmids and conducted a ChIP assay using these cells. An anti-FOXM1 antibody but not control IgG amplified a 325-bp DNA fragment of the Met promoter in the precipitates, suggesting that FOXM1 bound directly to the Met promoter (Fig. 3A and 3B). Conversely, knockdown of FOXM1 expression in PANC-1 and AsPC-1 cells resulted in decreased FOXM1 recruitment to Met promoter, while overexpression of FOXM1 in MiaPaca-2 and PANC-1 cells led to elevated FOXM1 recruitment to Met promoter (Supplementary Fig. S2B). These results demonstrated that FOXM1 bound directly to the Met promoter.
Figure 3.
Upregulation of HGF/Met signaling and activation of downstream pathways by FOXM1. A, A ChIP assay was performed using chromatins isolated from PANC-1 cells transfected with pcDNA3.1-FOXM1. Normal IgG was used as a control, and 1% of the total cell lysates were subjected to PCR before immunoprecipitation (input control). B, The primers of ChIP assay and FOXM1 putative binding sites in Met promoter reporter. C, The levels of Met promoter activity in PDA cells. AsPC-1 and FG cells were co-transfected with 0.2 μg of the Met promoter-luciferase construct pLuc-Met-1069 and 0 and 50 nmol/L siFOXM1 or control siRNA (siCtr) (left panels), and PANC-1 and CaPan-1 cells were co-transfected with 0.2 μg of pLuc-Met-1069 and 0 and 0.6 μg of pcDNA3.1-FOXM1 or a control vector (pCtr) (right panels). The promoter activity was measured using a dual luciferase assay kit. D, FG cells were transfected with siFOXM1 or control siRNA, and PANC-1 cells were transfected with pFOXM1 or a control vector. Forty-eight hr later, the cells were treated with 20 ng/mL HGF for 1 hr, and whole cell lysates were extracted. Phosphorylation of Met (Y1234/Y1235), AKT (S473), ERK1/2 (T202/Y204), and STAT3 (Y705) was analyzed using Western blotting with phosphorylation site-specific antibodies. The protein expression levels in each well were confirmed using anti-Met, anti-AKT, anti-ERK1/2, anti-STAT3, and anti-GAPDH antibodies (left panels) and quantitative results were determined by densitometry analysis (right panels). The experiments were performed three times. Data represent mean ± SEM of three independent experiments.
To investigate the Met transcription-regulatory role of FOXM1, we generated three Met promoter reporters, pLuc-Met-1259, pLuc-Met-1069 and pLuc-Met-580 (Supplementary Fig. S2A). We cotransfected Met promoter reporters with FOXM1 expression vectors or with siRNAs into AsPC-1, FG, PANC-1 and CaPan-1 cells. Overexpression of FOXM1 increased the MET promoter activity, whereas knockdown of FOXM1 decreased the MET promoter activity of both pLuc-Met-1259 and pLuc-Met-1069 reporters. But altered expression of FOXM1 had little effect on the promoter activity of pLuc-Met-580 (Fig. 3C and Supplementary Fig. S2C). Furthermore, the regulatory effect of FOXM1 on Met promoter activity of pLuc-Met-1259 and pLuc-Met-1069 were almost the same, suggesting that the major potential FOXM1 binding sites in Met promoter were #1 and #2. These results demonstrated that FOXM1 bound directly to the promoter region of Met and transcriptionally regulated the expression of Met.
FOXM1 upregulates HGF/Met signaling and activates its downstream pathways
Met is autophosphorylated at Tyr1234 and Tyr1235 when binds with HGF, leading to activation of a series of intracellular downstream signaling events, including RAS/ERK, PI3K/AKT, and STAT signaling. Therefore, we analyzed the effect of altered FOXM1 expression on HGF/Met downstream signaling. We transfected FG and PANC-1 cells with a FOXM1 expression vector, siFOXM1, or control vector and siRNA. Forty-eight hr later, we treated the cells with 20 ng/ml HGF for 1 hr and then determined the phosphorylation status of signaling proteins downstream from HGF/Met using Western blot. As shown in Fig. 3D and 3E, knockdown of FOXM1 expression in FG cells led to decreased HGF-dependent phosphorylation of Met at Y1234 and Y1235 and its downstream transducers, including ERK1/2 at T202 and Y204, AKT at S473, and STAT3 at Y705. In contrast, FOXM1 overexpression increased the phosphorylation of Met and its downstream signaling targets in PANC-1 cells.
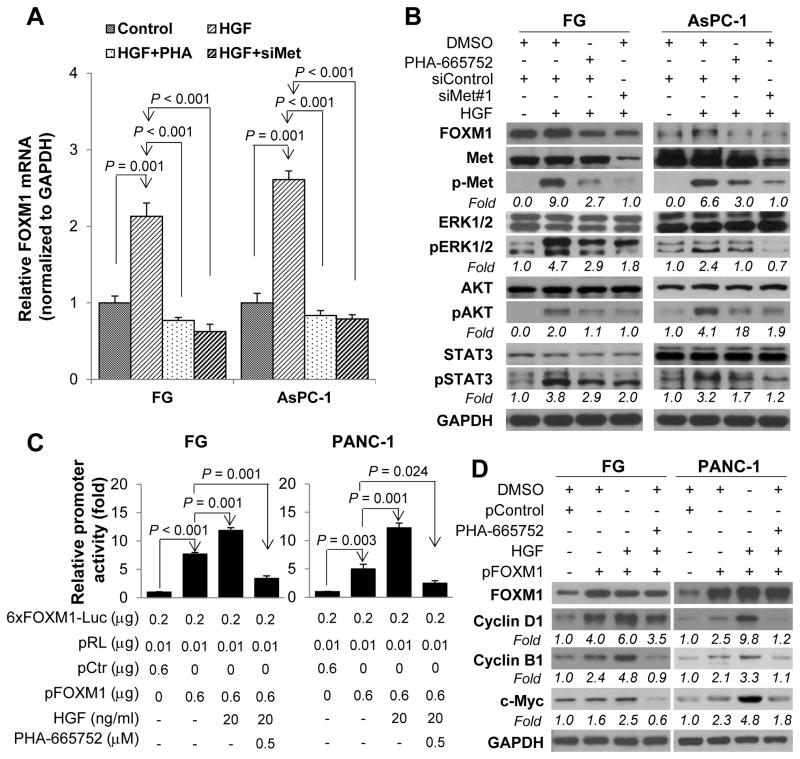
HGF/Met signaling stimulates the expression and transcriptional activity of FOXM1
Activation of RAS/ERK cascade, PI3K/AKT axis, and STAT3, which are the downstream signaling pathways for HGF/Met, increases PDA cell survival, proliferation, and motility via upregulation of FOXM1.35–37 Similarly, we found that activation of HGF/Met signaling in FG and AsPC-1 cells by HGF led to increased phosphorylation of ERK1/2, AKT, and STAT3 and upregulated FOXM1 mRNA and protein levels, whereas blockage of Met signaling by treatment with a selective Met inhibitor PHA-665752 or specific Met siRNAs abolished the upregulation of FOXM1 expression (Fig. 4A and 4B, and Supplementary Fig. S3 and S4). We then analyzed the effect of HGF/Met signaling on FOXM1 transcriptional activity by using a FOXM1-dependent luciferase reporter (6X-FOXM1-Luc) and FOXM1 typical downstream target gene Cyclin B1 promoter reporter. As shown in Fig. 4C and Supplementary Fig. S5, activation of HGF/Met signaling by HGF increased FOXM1 transcriptional activity, whereas blockage of HGF/Met signaling by PHA-665752 attenuated this effect of HGF. Furthermore, activation of HGF/Met signaling elevated the expression of FOXM1 downstream target genes, such as cyclin D1, cyclin B1, and c-Myc, whereas PHA-665752 suppressed the expression of FOXM1 downstream target genes (Fig. 4D and Supplementary Fig. S6).
Figure 4.
Stimulation of the expression and transcriptional activity of FOXM1 by HGF/Met signaling. FG and AsPC-1 cells were transfected with Met siRNA (siMet) or control siRNA for 24 hr and then treated with and without PHA-665752 (0.1 μM) and HGF (20 ng/mL) for 48 hr. A, Results of qPCR performed to analyze FOXM1 mRNA expression in the cells. B, Western blotting of the expression of FOXM1 protein and phosphorylation of Met (Y1234/Y1235), ERK1/2 (T202/Y204), AKT (S473), and STAT3 (Y705) in the cells. The protein expression levels in each well were confirmed using anti-Met, anti-AKT, anti-ERK1/2, anti-STAT3, and anti-GAPDH antibodies. C, Effect of HGF/Met signaling on FOXM1 transcriptional activity in FG and PANC-1 cells. The cells were transfected with a FOXM1-dependent luciferase (6xFOXM1-Luc) with and without pFOXM1 for 24 hr. Before the promoter’s activities in the cells was examined, cells were treated with or without PHA-665752 (0.5 μM) and HGF (20 ng/mL) for 3 hr. D, Regulation of the expression of FOXM1 downstream target genes by HGF/Met signaling. FG and PANC-1 cells were transfected with pFOXM1 or a control vector (pControl). Twenty-four hr later, the cells were treated with PHA-665752 (0.1 μM) and HGF (20 ng/mL) for 48 hr. Whole cell lysates were extracted, and the expression of FOXM1, cyclin D1, cyclin B1, and c-Myc was measured using Western blot. DMSO, dimethyl sulfoxide.
We next sought to determine which of the HGF/Met downstream signaling pathways played key roles in regulation of FOXM1 transcriptional activity by using ERK1/2 inhibitor FR180204, AKT inhibitor GSK690693, and STAT3 inhibitor S3I-201. We found that all three inhibitors blocked the stimulatory effect of HGF/Met signaling on FOXM1 transcriptional activity (Supplementary Fig. S7). These results demonstrated that FOXM1 upregulated HGF/Met signaling by transactivating Met and that activation of HGF/Met signaling stimulated the expression and transcriptional activity of FOXM1 via its downstream pathways, suggesting that HGF/Met and FOXM1 formed a positive feedback loop.
Cross-talk between FOXM1 and HGF/Met signaling promotes PDA growth
To assess the impact of cross-talk between FOXM1 and HGF/Met signaling on PDA growth, we analyzed their effects on PDA cell colony formation in vitro and subcutaneous pancreatic tumor growth in vivo. Specifically, activation of HGF/Met signaling by HGF markedly increased FG cell colony formation, whereas knockdown of FOXM1 expression and inhibition of Met signaling abolished the effect (Supplementary Fig. S8). For in vivo assay, we used PANC-1 cells with FOXM1 overexpression and FG cells with FOXM1 knockdown. The overexpression of FOXM1 markedly elevated Met expression in vivo and promoted tumor growth (Supplementary Fig. S9A). Conversely, FOXM1 knockdown markedly decreased Met expression and inhibited tumor growth (Supplementary Fig. S9B). These data suggested that this HGF/Met-FOXM1 positive feedback loop promoted PDA growth.
FOXM1 mediates PDA-cell resistance to Met inhibition
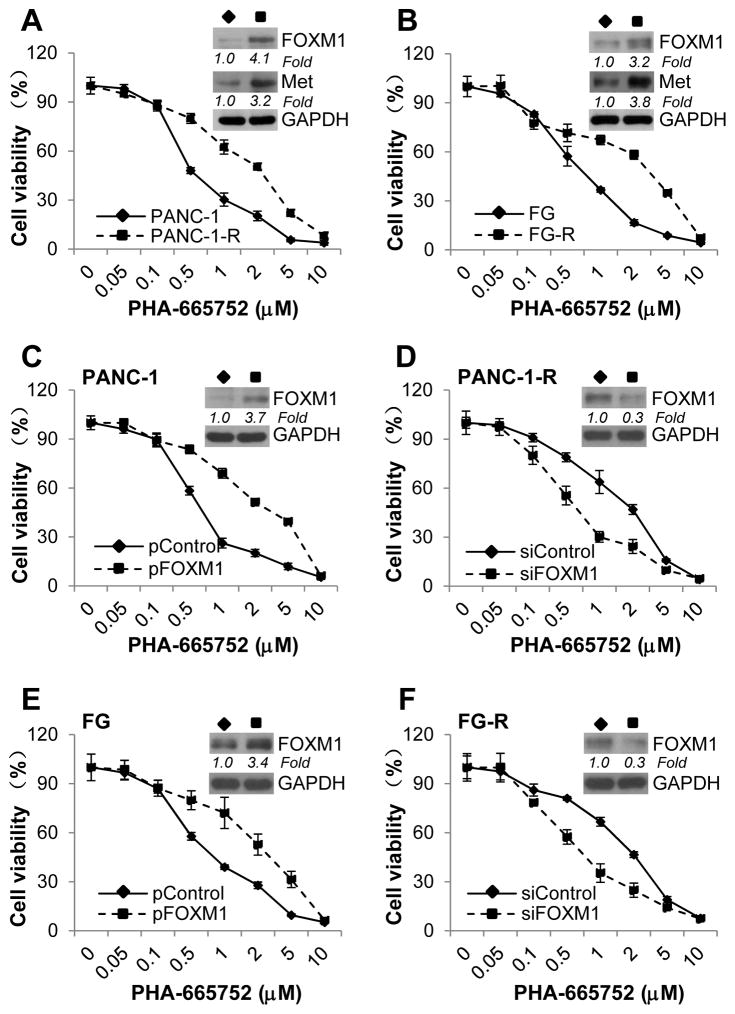
PHA-665752 is an ATP-competitive small-molecule inhibitor of Met that selectively inhibits the receptor kinase domain, disrupting the HGF/Met signaling pathway.29 Because FOXM1 forms a positive feedback loop with HGF/Met, we investigated whether FOXM1 played any roles in mediating PDA resistance to HGF/Met signaling inhibition. We generated PANC-1 and FG sublines resistant to PHA-665752 via continuous culture in increasing concentrations of the drug. The resulting sublines, PANC-1-R and FG-R, were more resistant to PHA-665752 than the parental cell lines (IC50, 0.53 μM versus 1.47 μM [P<0.001] and 0.58 μM versus 1.60μM [P<0.001], respectively) (Fig. 5A). Also, the PHA-665752–resistant cells had higher levels of FOXM1 and Met expression than the parental cells did (Fig. 5A; Supplementary Fig. S10A and S10B). The higher expression of FOXM1 in the PANC-1-R and FG-R cells implied that FOXM1 contributes to the resistance of PDA cells to Met inhibition. To further demonstrate the role of FOXM1 in Met resistance, we increased the expression of FOXM1 in parental PANC-1 and FG cells via gene transfection, and decreased the expression of FOXM1 in PHA-665752–resistant PANC-1-R and FG-R cells via knockdown with siRNA (Fig. 5C, 5D, 5E and 5F; and Supplementary Fig. S10C, S10D, S10E and S10F). As shown in Fig. 5C and 5E, overexpression of FOXM1 in the parental cells resulted in greater PHA-665752 resistance than in the control cells (IC50, 0.62 versus 1.96 [P=0.001] and 0.69 versus 1.82 [P=0.014], respectively). Conversely, knockdown of FOXM1 expression in PANC-1-R and FG-R cells led to less resistance to PHA-665752 than the control siRNA groups (IC50, 1.36 versus 0.56 [P<0.001] and 1.45 versus 0.56 [P=0.006], respectively) (Fig. 5D and 5F). These results demonstrated that elevated expression of FOXM1 was one of the mechanisms that mediated PDA cells’ resistance to Met inhibition.
Figure 5.
Mediation of PDA cell resistance to Met inhibition by FOXM1. A, Results of an MTT assay performed to determine the IC50 for PHA-665752 in PANC-1 and PANC-1-R cells (left panel) and in FG and FG-R cells (right panel). The insets show Western blots of the expression of FOXM1 and Met. B, PANC-1 cells were transfected with a control vector (pCtr) or pFOXM1 (left panel), and PANC-1-R cells were transfected with control siRNA (siCtr) or siFOXM1 (right panel). C, FG cells were transfected with a control vector (pCtr) or pFOXM1 (left panel), and FG-R cells were transfected with control siRNA (siCtr) or siFOXM1 (right panel). An MTT assay was performed to determine the IC50 for PHA-665752. The insets show Western blots of the expression of FOXM1. Relative protein levels of FOXM1 and Met in different groups were normalized to GAPDH. All the experiments were performed three times. Data represent mean ± SEM of three independent experiments with three triplicates in each experiment. The levels of gene expression were quantitated and expressed as fold changes (italic numbers under each blot).
FOXM1 inhibition increases PDA cells’ sensitivity to Met inhibition
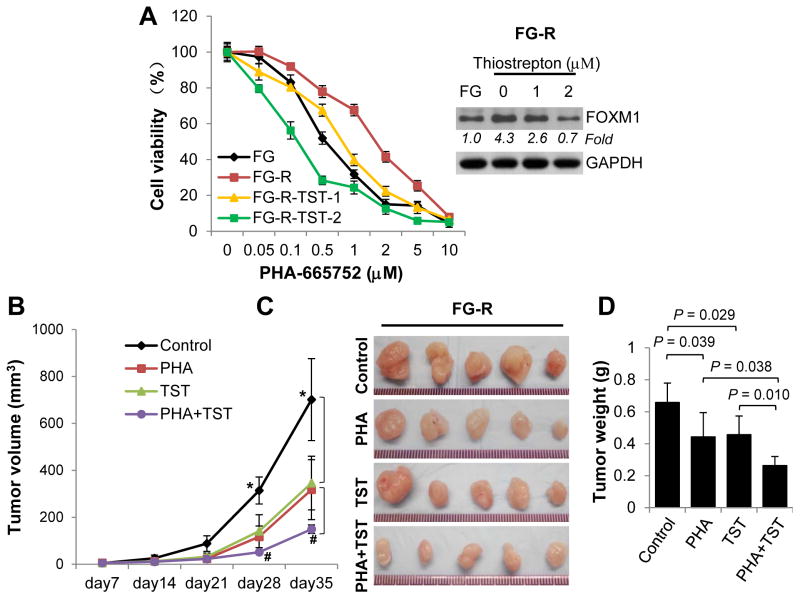
Our findings suggested that HGF/Met and FOXM1 form a positive feedback loop and that FOXM1 promotes PDA cells’ resistance to Met inhibition. We finally sought to determine whether inhibition of FOXM1 expression is an effective strategy of reversing resistance of PDA cells to Met inhibition. As shown in Fig. 6A, treatment of FG-R cells with the FOXM1 inhibitor TST decreased the expression of FOXM1 in a dose-dependent manner, and pretreatment of these cells with 2 μM TST significantly sensitized them to treatment with PHA-665752 (IC50: 0.55, 1.56, 0.63, and 0.19 μM in FG cells and in FG-R cells pretreated with 0, 1, and 2 μM TST, respectively; P<0.001).30 Furthermore, treatment with TST and PHA-665752 inhibited the growth of FG-R cells in a xenograft model as compared with control groups, and treatment with the combination of TST and PHA-665752 was more effective than that with either inhibitor alone (Fig. 6B, 6C and 6D).
Figure 6.
Increased PDA cell sensitivity to Met inhibition in vitro and in vivo resulting from treatment with TST. A, FG-R cells were treated with 0, 1, or 2 μM TST and various concentrations of PHA-665752 for 72 hr. The cells’ PHA-665752 resistance was determined via MTT assay. The inset shows a Western blotting analysis of FOXM1 expression. B–D, FG-R cells (1×106) were injected subcutaneously into the right scapular region in mice (n=5 for each group), which then received treatment with PHA-665752 (PHA) and/or TST. The resulting tumors in each group were measured once a week (B). Photographs of tumors isolated from the mice (C) and the weights of the tumors (D) are shown. * indicates significant difference between treated and control groups (P<0.05); # indicates significant difference between single (PHA or TST) and combined groups (PHA and TST) (P<0.05). The levels of gene expression were quantitated and expressed as fold changes (italic numbers under each blot).
Discussion
In the present study, we demonstrate for the first time that FOXM1 transcriptionally regulates the expression of Met; HGF/Met signaling stimulates the expression and transcriptional activity of FOXM1 via RAS/ERK1/2, PI3K/AKT, and STAT3 signaling pathways; and HGF/Met and FOXM1 form a positive feedback loop (Fig. 7). Our experimental and clinical evidence strongly suggest that this positive feedback loop promotes PDA growth and resistance to Met inhibition.
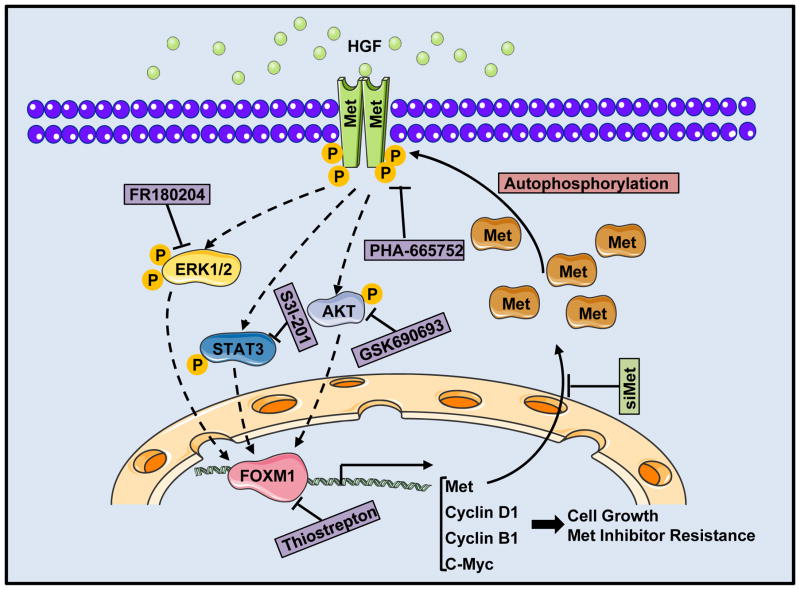
Figure 7.
A model of the HGF/Met-FOXM1 positive feedback loop. FOXM1 transcriptionally regulated the expression of Met, activated HGF/Met signaling, and stimulated the expression and transcriptional activity of FOXM1 via the RAS/ERK1/2, PI3K/AKT, and STAT3 signaling pathways, and the HGF/Met-FOXM1 positive feedback loop promoted PDA growth and resistance to Met inhibition. Treatment with HGF/Met signaling inhibitor, Met-specific siRNA (siMet), the FOXM1 inhibitor, or RAS/ERK1/2, PI3K/AKT, and STAT3 signaling pathway inhibitors abolished the positive feedback loop.
HGF/Met signaling is aberrantly induced or activated in different solid tumors and has been associated with poor prognosis.38 In PDAs, Met has been overexpressed and associated with clinicopathologic parameters and survival.11 Met also promotes the growth, invasion, metastasis, drug resistance, and stem cell maintenance of PDA.38 Expression of Neuromedin U and cytokines can elevate the expression and activation of Met in PDA cells, while mechanism underlying aberrant activation of HGF/Met signaling requires further study.12–14 In the present study, we systematically analyzed the Met expression patterns in PDA cell lines and tissue specimens. We found that expression of Met was markedly increased in most of PDA cell lines, and statistical analysis of the relationships of Met expression with clinicopathologic PDA parameters revealed that increased expression of Met in PDA specimens correlated with PDA progression and differentiation. Furthermore, we provided four lines of evidence supporting that FOXM1 transcriptionally upregulated the expression of Met and elevated the activation of HGF/Met signaling. First, we showed that FOXM1 and Met were concomitantly overexpressed in PDA specimens. Second, overexpression of FOXM1 increased the expression of Met at both the mRNA and protein level, whereas knockdown of FOXM1 expression did the opposite. Third, FOXM1 bound directly to the Met promoter region and regulated Met promoter activity. Fourth, stimulation by HGF caused overexpression of FOXM1, which elevated the phosphorylation of Met and activation of downstream signaling, including that of RAS/ERK1/2, PI3K/AKT, and STAT3. Conversely, both FOXM1 knockdown and blockade of Met signaling downregulated FOXM1 expression, and decreased the activation of Met and its downstream signaling. Therefore, FOXM1 is a critical regulator of aberrant activation of HGF/Met signaling in PDAs.
The oncogenic transcription factor FOXM1 is a key regulator of cell-cycle progression, and recent studies further established the roles of FOXM1 in promoting cancer growth, invasion, metastasis, angiogenesis, metabolism, and stem cell maintenance.16,19,39,40 FOXM1 is overexpressed in PDA cells and its overexpression is associated with PDA development and progression.16 However, the mechanism of elevated expression of FOXM1 and its promotive effect on PDA have remained unclear. In the present study, we found that FOXM1 transactivated Met and increased the activity of HGF/Met signaling and that activation of HGF/Met signaling stimulated the expression and transcriptional activity of FOXM1 in PDAs. Therefore, HGF/Met and FOXM1 form a positive feedback loop and promoted PDA growth in vitro and in vivo.
Given the critical roles of HGF and Met in cancer development and progression, a number of HGF/Met signaling inhibitors have been generated and have antitumor effects in many preclinical animal models. However, single-agent administration of Met inhibitors had only moderate clinical benefits in treatment of cancers with HGF/Met signaling abnormalities.7 Thus, studies of the mechanisms of resistance of PDA cells to Met inhibition are urgently needed. Previous studies demonstrated that overexpression of transforming growth factor-α led to activation of the epidermal growth factor receptor pathway and subsequent resistance to Met inhibition.41 In the present study, we showed that PDA cells resistant to the Met inhibitor PHA-665752 had higher levels of FOXM1 and Met expression than the control cells and that FOXM1 overexpression resulted in increased resistance to PHA-665752, whereas knockdown of FOXM1 expression led to reduced resistance to it. Furthermore, treatment with FOXM1 inhibitor TST and Met inhibitor PHA-665752 inhibited the growth of PHA-665752-resistant PDA cells in vitro and in vivo, and treatment with the combination of TST and PHA-665752 was more effective than that with either inhibitor alone. Recently, investigators have analyzed many approaches targeting HGF/Met signaling, including inhibition of HGF and of Met expression with antibodies or tyrosine kinase inhibitors, mostly in phase 1 clinical trials.38 Our present study has suggested an alternative approach to render tumor cells more sensitive to the existing inhibitors. In summary, our results demonstrate a positive feedback loop formed by HGF/Met and FOXM1 and its role in promoting PDA growth and resistance to Met inhibition. Therefore, we have not only identified a novel molecular mechanism underlying aberrant activation of HGF/Met and FOXM1 in PDA cells, but also have revealed that the combine use of both inhibitors for FOXM1 and HGF/Met signaling is a promising therapeutic strategy for PDA.
Materials and Methods
Cell culture and reagents
Human embryonic kidney 293 cells and the human PDA cell lines PANC-1, MiaPaCa-2, AsPC-1, BxPC-3, CaPan-1, and CaPan-2 were purchased from the American Type Culture Collection. The FG, COLO357, and L3.7 were described previously.17 All of these cell lines were maintained in plastic flasks as adherent monolayers in Eagle's minimal essential medium supplemented with 10% fetal bovine serum, sodium pyruvate, nonessential amino acids, L-glutamine, and a vitamin solution (Flow Laboratories). The cell lines were obtained directly from the American Type Culture Collection, which performs cell line characterization and authentication using short tandem repeat profiling, and were passaged in our laboratory for fewer than 6 months after receipt.
The Met tyrosine kinase inhibitor PHA-665752, ERK1/2 inhibitor FR180204, AKT kinase inhibitor GSK690693, STAT3 inhibitor S3I-201, Met ligand HGF, and FOXM1 inhibitor thiostrepton (TST) were obtained from Sigma-Aldrich.20–22
To generate PDA cell lines resistant to PHA-665752, parental PANC-1 and FG cells were treated with 0.5 μM PHA-665752. The concentration of PHA-665752 was progressively increased once every 2 weeks in 0.5-μM increments up to a final concentration of 2.5 μM. PHA-665752 was replenished every 3–5 days as needed. After 4 months, the two cell lines became sufficiently resistant to PHA-665752 and were referred to as PANC-1-R and FG-R, respectively. The half-maximal inhibitory concentrations (IC50) of the chemoresistant sublines were determined using MTT assays.
Human tissue specimens and immunohistochemical analysis
Tissue microarray (TMA) construction and immunohistochemical analysis were conducted as described previously.19 Briefly, a TMA containing primary tumor specimens from 154 patients with PDA was obtained from the PDA Tissue Bank at Shanghai Jiaotong University Affiliated First People’s Hospital (Shanghai, People’s Republic of China). In addition to 154 primary tumor specimens, the TMA contained 34 matched tumor-adjacent tissue specimens and 22 normal tissue specimens. The use of archived human PDA tissues was approved by MD Anderson Cancer Center institutional review board. The TMA was prepared and processed for immunostaining with anti-Met (#8198; Cell Signaling) and anti-FOXM1 (WH0002305M2; Sigma) antibodies. The staining results were scored by two investigators blinded to the clinical data as described previously.23
Plasmids and small interfering RNAs
The plasmid pcDNA3.1-FOXM1B (pFOXM1) and control vector plasmid pcDNA3.1 were described previously.24 Small interfering RNAs (siRNAs) targeting Met was synthesized by Dharmacon, and the sequence of siRNA#1 5'-GTGCAGTATCCTCTGACAG-3' and siRNA#2 5’-AAGTGCAGTATCCTCTGACAG-3’ were reported previously.25,26 SiRNA targeting FOXM1 (siFOXM1) was described previously.27
Transient transfection
Transfection of plasmids and siRNAs into PDA cells was performed using the transfection reagents Lipofectamine LTX and Lipofectamine 2000 CD (Invitrogen), respectively. For transient transfection, cells were transfected with plasmids or siRNAs at different doses as indicated for 48 hr before the performance of functional assays. PDA cells treated with a transfection reagent alone were included as mock controls.
Quantitative real-time polymerase chain reaction
Quantitative real-time reverse transcription-polymerase chain reaction (qPCR) analysis of expression of the Met and FOXM1 genes was performed using total RNA and SYBR Green reagent with an ABI Prism 7000HT sequence detection system.28 The sequences of the qPCR primers were as follows: Met, 5'-gtttgtccacagagacttggctg-3' (forward) and 5'-atccacttcactggcagctttg-3' (reverse); FOXM1, 5'-acgtccccaagccaggctc-3' (forward) and 5'-ctactgtagctcaggaataa-3' (reverse); and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5'-acagtccatgccatcactgcc-3' (forward) and 5'-gcctgcttcaccaccttcttg-3' (reverse).
Western blot analysis
Standard Western blot analysis was carried out using primary anti-FOXM1 (sc-502; Santa Cruz Biotechnology), anti-Met (sc-161; Santa Cruz Biotechnology), anti-phosphorylated Met (Tyr1234/Tyr1235 [#8218]; Cell Signaling Technology), anti-AKT (#9272; Cell Signaling Technology), anti-phosphorylated AKT (Ser472 [#9271]; Cell Signaling Technology), anti-p44/42 ERK1/2 (#9102; Cell Signaling Technology), anti-phosphorylated p44/42 ERK1/2 (Thr202/Tyr204 [#8201]; Cell Signaling Technology), anti-STAT3 (#9132; Cell Signaling Technology), and anti-phosphorylated STAT3 (Tyr705 [#9135]; Cell Signaling Technology) antibodies. Anti-mouse (Cell Signaling Technology) and anti-rabbit (Santa Cruz Biotechnology) antibodies were used as secondary antibodies. Equal protein-sample loading was monitored using an anti-GAPDH antibody (Santa Cruz Biotechnology). The bands were quantified using Quantity One analysis software, Version 4.6 (Bio-Rad) and the results of fold changes were expressed as numbers in italic font under individual blot.
Construction of the Met promoter reporter and dual luciferase assay
The promoter region of MET was delineated using the Gene2Promoter software program (Genomatix Software GmbH), and a fragment of 1259 bp containing MET 5' sequences from −1023 to +236 bp relative to the transcription initiation site was subcloned into the pGL3-basic vector (Promega). The final full-length reporter plasmid, which contained multiple FOXM1-binding sites, was designated pLuc-Met-1259. The deletion mutation reporter for this plasmids, pLuc-Met-1069 and pLuc-Met-580, were then generated. All the constructs were verified by sequencing the inserts and flanking regions of the plasmids. Cyclin B1 promoter reporter was a fragment of 968 bp containing CCNB1 5' sequences from −931 to +37 bp relative to the transcription initiation site, and was designated pLuc-cyclin B1. PDA cells were transfected with Met promoter reporters, siFOXM1, or an expression plasmid or transfected with pFOXM1, pLuc-cyclin B1 and a FOXM1-dependent luciferase reporter (6X-FOXM1-Luc), which was described previously,28 and treated with HGF or PHA-665752 and HGF with FR180204, GSK690693, or S3I-201. The transcriptional activity was normalized via co-transfection with a β-actin/Renilla luciferase reporter containing a full-length Renilla luciferase gene. The luciferase activity in the cells was quantified using a dual luciferase assay system (Promega) 24 hr after transfection.
Chromatin immunoprecipitation assay
Pancreatic tumor cells (2×106) were prepared for a chromatin immunoprecipitation (ChIP) assay with a ChIP assay kit (Millipore) according to the manufacturer’s protocol. The resulting precipitated DNA specimens were analyzed using PCR or qPCR to amplify a 325-bp region of the Met promoter. The PCR products were resolved electrophoretically on a 2% agarose gel and visualized using ethidium bromide staining.
Colony-formation assay
A colony-formation assay was performed to assess the proliferation of PDA cells. PANC-1 cells were transfected with siFOXM1 or control siRNA. Forty-eight hr later, 200 cells were plated in 24-well plates and treated with indicated groups. The media were changed every 2 days, and allowed to grow for 14 days. Cells were then fixed with 4% paraformaldehyde and stained with a 0.1% crystal violet solution for 10 minutes. PANC-1 Cell colonies (>20 cells each) were counted using an inverted microscope at 40× magnification. All experiments were performed in triplicate.
MTT assays
PDA cells were seeded at a density of 2×103 cells per well in 96-well plates. The cells were then treated with various does of PHA-665752 and TST. Seventy-two hr later, MTT (20 μL, 5 mg/L; Sigma) was added to each well, and the plates were continuously cultured for 4 hr. Dimethyl sulfoxide (200 μL) was then added to each well. The absorbance was measured at 570 nm.
Animal experiments
Pathogen-free female athymic nude mice were purchased from our institution. The animals were maintained in facilities approved by the Association for Assessment and Accreditation of Laboratory Animal Care International in accordance with the current regulations and standards of the U.S. Department of Agriculture and Department of Health and Human Services. All animal experiments were approved by the Institutional Animal Care and Use Committee. Both investigators on animal studies were blind to the group allocation and treatments in each group from start of the experiments and completion of collection of data related to the animal experimentation.
To study PDA cell growth in vivo, 1×106 PDA cells in 0.1 mL of Hank’s balanced salt solution were injected subcutaneously into the right scapular region in mice. The tumor-bearing mice were killed when they became moribund or on day 35 after injection and their tumors were removed and weighed. To determine the efficacy of PHA-665752 and TST on PHA-665752 resistance PDA cell growth, 1×106 cells were injected subcutaneously into the right scapular region in mice. PHA-665752 was dissolved in L-lactate (pH 4.8) and 10% polyethylene glycol, and TST was dissolved in N,N-dimethylacetamide, polyethylene glycol 400, and Tween 80 at a 2:7:1 v/v/v ratio. Seven days later, PHA-665752 and TST were intravenously injected into mice at 15 mg/kg per day and 50 mg/kg per day, respectively, for 21 days.29,30 The tumor-bearing mice were killed on day 35 after injection, and their tumors were removed and weighed.
Statistical analysis
The significance of the patient specimen data was determined using the Pearson correlation coefficient. A two-tailed χ2 test or the Fisher exact test was used to determine the significance of the differences among the covariates. All in vitro and in vivo experiments were repeated two to four times or as indicated, the data from independent experiments were presented as the mean ± standard error of the mean (SEM) or as indicated otherwise, and the significance of the data was determined using the Student t-test (two-tailed) or one-way analysis of variance (ANOVA). In all of the tests, P values less than 0.05 were considered statistically significant. The SPSS software program (version 13.0; IBM Corporation) was used for statistical analysis.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by grants R01-CA129956, R01-CA148954, R01CA152309, and R01CA172233 from the National Cancer Institute, National Institutes of Health (to K. Xie).
We thank Don Norwood for editorial assistance and Xuemei Wang, Associate Director of Quantitative Research at The University of Texas MD Anderson Cancer Center, for assistance with statistical analyses.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare no conflict of interest.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 4.Zhao S, Cao L, Freeman JW. Knockdown of RON receptor kinase delays but does not prevent tumor progression while enhancing HGF/MET signaling in pancreatic cancer cell lines. Oncogenesis. 2013;2:e76. doi: 10.1038/oncsis.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birchmeier C, Gherardi E. Developmental roles of HGF/SF and its receptor, the c-Met tyrosine kinase. Trends Cell Biol. 1998;8:404–410. doi: 10.1016/s0962-8924(98)01359-2. [DOI] [PubMed] [Google Scholar]
- 6.Zhang YW, Vande Woude GF. HGF/SF-met signaling in the control of branching morphogenesis and invasion. J Cell Biochem. 2003;88:408–417. doi: 10.1002/jcb.10358. [DOI] [PubMed] [Google Scholar]
- 7.Cui JJ. Targeting receptor tyrosine kinase MET in cancer: small molecule inhibitors and clinical progress. J Med Chem. 2014;57:4427–4453. doi: 10.1021/jm401427c. [DOI] [PubMed] [Google Scholar]
- 8.De Bacco F, Luraghi P, Medico E, Reato G, Girolami F, Perera T, et al. Induction of MET by ionizing radiation and its role in radioresistance and invasive growth of cancer. J Natl Cancer Inst. 2011;103:645–661. doi: 10.1093/jnci/djr093. [DOI] [PubMed] [Google Scholar]
- 9.Neesse A, Michl P, Frese KK, Feig C, Cook N, Jacobetz MA, et al. Stromal biology and therapy in pancreatic cancer. Gut. 2011;60:861–868. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 10.Logan-Collins J, Thomas RM, Yu P, Jaquish D, Mose E, French R, et al. Silencing of RON receptor signaling promotes apoptosis and gemcitabine sensitivity in pancreatic cancers. Cancer Res. 2010;70:1130–1140. doi: 10.1158/0008-5472.CAN-09-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu GH, Huang C, Qiu ZJ, Liu J, Zhang ZH, Zhao N, et al. Expression and prognostic significance of CD151, c-Met, and integrin alpha3/alpha6 in pancreatic ductal adenocarcinoma. Dig Dis Sci. 2011;56:1090–1098. doi: 10.1007/s10620-010-1416-x. [DOI] [PubMed] [Google Scholar]
- 12.Kitajima Y, Ide T, Ohtsuka T, Miyazaki K. Induction of hepatocyte growth factor activator gene expression under hypoxia activates the hepatocyte growth factor/c-Met system via hypoxia inducible factor-1 in pancreatic cancer. Cancer Sci. 2008;99:1341–1347. doi: 10.1111/j.1349-7006.2008.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ketterer K, Kong B, Frank D, Giese NA, Bauer A, Hoheisel J, et al. Neuromedin U is overexpressed in pancreatic cancer and increases invasiveness via the hepatocyte growth factor c-Met pathway. Cancer Lett. 2009;277:72–81. doi: 10.1016/j.canlet.2008.11.028. [DOI] [PubMed] [Google Scholar]
- 14.Otte JM, Kiehne K, Schmitz F, Folsch UR, Herzig KH. C-met protooncogene expression and its regulation by cytokines in the regenerating pancreas and in pancreatic cancer cells. Scand J Gastroenterol. 2000;35:90–95. doi: 10.1080/003655200750024597. [DOI] [PubMed] [Google Scholar]
- 15.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 16.Huang C, Du J, Xie K. FOXM1 and its oncogenic signaling in pancreatic cancer pathogenesis. Biochim Biophys Acta. 2014;1845:104–116. doi: 10.1016/j.bbcan.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C, Qiu Z, Wang L, Peng Z, Jia Z, Logsdon CD, et al. A novel FoxM1-caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res. 2012;72:655–665. doi: 10.1158/0008-5472.CAN-11-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao B, Wang Z, Ali S, Kong D, Banerjee S, Ahmad A, et al. Over-expression of FoxM1 leads to epithelial-mesenchymal transition and cancer stem cell phenotype in pancreatic cancer cells. J Cell Biochem. 2011;112:2296–2306. doi: 10.1002/jcb.23150. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Cui J, Shi M, Xie D, Wei D, Jia Z, Zheng S, et al. FOXM1 promotes the warburg effect and pancreatic cancer progression via transactivation of LDHA expression. Clin Cancer Res. 2014;20:2595–2606. doi: 10.1158/1078-0432.CCR-13-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sunaga N, Kaira K, Imai H, Shimizu K, Nakano T, Shames DS, et al. Oncogenic KRAS-induced epiregulin overexpression contributes to aggressive phenotype and is a promising therapeutic target in non-small-cell lung cancer. Oncogene. 2013;32:4034–4042. doi: 10.1038/onc.2012.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altomare DA, Zhang L, Deng J, Di Cristofano A, Klein-Szanto AJ, Kumar R, et al. GSK690693 delays tumor onset and progression in genetically defined mouse models expressing activated Akt. Clin Cancer Res. 2010;16:486–496. doi: 10.1158/1078-0432.CCR-09-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sen N, Che X, Rajamani J, Zerboni L, Sung P, Ptacek J, et al. Signal transducer and activator of transcription 3 (STAT3) and survivin induction by varicella-zoster virus promote replication and skin pathogenesis. Proc Natl Acad Sci U S A. 2012;109:600–605. doi: 10.1073/pnas.1114232109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Li Z, Kong X, Xie D, Jia Z, Jiang W, et al. Down-regulation of microRNA-494 via loss of SMAD4 increases FOXM1 and β-catenin signaling in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014;147:485–97. doi: 10.1053/j.gastro.2014.04.048. [DOI] [PubMed] [Google Scholar]
- 24.Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–3602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 25.Hu B, Guo P, Bar-Joseph I, Imanishi Y, Jarzynka MJ, Bogler O, et al. Neuropilin-1 promotes human glioma progression through potentiating the activity of the HGF/SF autocrine pathway. Oncogene. 2007;26:5577–5586. doi: 10.1038/sj.onc.1210348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh CY, Shin SM, Yeh HH, Wu TJ, Shin JW, Chang TY, et al. Transcriptional activation of the Axl and PDGFR-alpha by c-Met through a ras- and Src-independent mechanism in human bladder cancer. BMC Cancer. 2011;11:139. doi: 10.1186/1471-2407-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui J, Shi M, Xie D, Wei D, Jia Z, Zheng S, et al. FOXM1 Promotes the Warburg Effect and Pancreatic Cancer Progression via Transactivation of LDHA Expression. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-13-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu M, et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-beta-dependent cancer metastasis. J Clin Invest. 2014;124:564–579. doi: 10.1172/JCI71104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 2003;63:7345–7355. [PubMed] [Google Scholar]
- 30.Chiu WT, Huang YF, Tsai HY, Chen CC, Chang CH, Huang SC, et al. FOXM1 confers to epithelial-mesenchymal transition, stemness and chemoresistance in epithelial ovarian carcinoma cells. Oncotarget. 2015;6:2349–2365. doi: 10.18632/oncotarget.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanders DA, Gormally MV, Marsico G, Beraldi D, Tannahill D, Balasubramanian S. FOXM1 binds directly to non-consensus sequences in the human genome. Genome Biol. 2015;16:130. doi: 10.1186/s13059-015-0696-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Zhang N, Dai B, Liu M, Sawaya R, Xie K, et al. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res. 2008;68:8733–8742. doi: 10.1158/0008-5472.CAN-08-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wierstra I, Alves J. Despite its strong transactivation domain, transcription factor FOXM1c is kept almost inactive by two different inhibitory domains. Biol Chem. 2006;387:963–976. doi: 10.1515/BC.2006.120. [DOI] [PubMed] [Google Scholar]
- 34.Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26:6212–6219. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- 35.Lam AK, Ngan AW, Leung MH, Kwok DC, Liu VW, Chan DW, et al. FOXM1b, which is present at elevated levels in cancer cells, has a greater transforming potential than FOXM1c. Front Oncol. 2013;3:11. doi: 10.3389/fonc.2013.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Teh MT, Ji Y, Patel V, Firouzabadian S, Patel AA, et al. EPS8 upregulates FOXM1 expression, enhancing cell growth and motility. Carcinogenesis. 2010;31:1132–1141. doi: 10.1093/carcin/bgq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mencalha AL, Binato R, Ferreira GM, Du Rocher B, Abdelhay E. Forkhead box M1 (FoxM1) gene is a new STAT3 transcriptional factor target and is essential for proliferation, survival and DNA repair of K562 cell line. PLoS One. 2012;7:e48160. doi: 10.1371/journal.pone.0048160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avan A, Maftouh M, Funel N, Ghayour-Mobarhan M, Boggi U, Peters GJ, et al. MET as a potential target for the treatment of upper gastrointestinal cancers: characterization of novel c-Met inhibitors from bench to bedside. Curr Med Chem. 2014;21:975–989. doi: 10.2174/09298673113209990231. [DOI] [PubMed] [Google Scholar]
- 39.Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem. 2007;388:1257–1274. doi: 10.1515/BC.2007.159. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Ahmad A, Li Y, Banerjee S, Kong D, Sarkar FH. Forkhead box M1 transcription factor: a novel target for cancer therapy. Cancer Treat Rev. 2010;36:151–156. doi: 10.1016/j.ctrv.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi J, McTigue MA, Rogers A, Lifshits E, Christensen JG, Janne PA, et al. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer Res. 2011;71:1081–1091. doi: 10.1158/0008-5472.CAN-10-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.