Abstract
Alterations in long non-coding RNAs (lncRNAs) are associated with human carcinogenesis. One group of lncRNAs, which are antisense in orientation to coding mRNAs (ASs), have been recently described in cancers but are poorly understood. We sought to identify ASs involved in human gastric cancer (GC) and to elucidate their mechanisms of action in carcinogenesis. We performed massively parallel RNA sequencing in GCs and matched normal tissues, as well as in GC-derived and normal gastric epithelial cell lines. One AS, designated KRT7-AS, was selected due to its marked upregulation and concordant expression with its cognate sense counterpart, KRT7, in GC tissues and cell lines. KRT7-AS formed an RNA-RNA hybrid with KRT7 and controlled KRT7 expression at both the mRNA and the post-transcriptional levels. Moreover, forced overexpression of the KRT7-overlapping region (OL) of KRT7-AS (but not its non-KRT7-overlapping portions) increased keratin 7 protein levels in cells. Finally, forced overexpression of full-length (FL) KRT7-AS or OL KRT7-AS (but not its non-KRT7-overlapping regions) promoted GC cell proliferation and migration. We conclude that lncRNA KRT7-AS promotes GC, at least in part, by increasing KRT7 expression.
Keywords: Antisense RNA, Long Noncoding RNA, Gastric Cancer, Tumor Development, Gene Regulation, Noncoding RNA, RNA-seq
INTRODUCTION
Gastric cancer (GC) is the 4th-most common cancer and the 2nd-highest cause of cancer death worldwide.1 As with other types of cancer, GCs are heterogeneous, and GC carcinogenesis is a multistep process involving diverse environmental and molecular genetic factors.2,3 Mechanisms underlying this process are still poorly understood. Recent studies have focused on long non-coding RNAs (lncRNAs) in human cancer pathogenesis, but little is known regarding their involvement in GC.
The term “noncoding RNAs” generally refers to RNAs that are transcribed into RNA but not translated into proteins. lncRNAs are defined as being greater than 200 nucleotides in length, which distinguishes them from small noncoding RNAs.4 However, as the structures and functions of noncoding transcripts become better characterized, it is becoming clear that classification based on this arbitrary definition is not very useful. Antisense (AS) transcripts (ASTs), defined as RNAs that are reverse complements of their endogenous sense counterparts, frequently do not encode proteins.5,6 Approximately 50-70% of lncRNAs are classified as ASTs, comprising a substantial proportion of the entire long noncoding transcriptome.5,7 For many years, their importance was overlooked due to their heterogeneity, low expression levels, and unknown functions. Recently, however, ASTs have garnered increased attention due to their highly locus-specific effects. One major emerging theme concerns the effects of ASTs on their neighboring protein-coding genes. As with the broader class of all lncRNAs, ASTs appear to function in a highly cell type-specific manner, exerting trans or cis effects on other genes that include suppression, activation, or homeostatic adjustment, and their mechanisms of action range from transcriptional regulation by gene promoter activation to post-transcriptional regulation by controlling mRNA stability and translatability.8-11 For example, aHIF, the antisense RNA transcript to HIF1-alpha, is overexpressed and acts to reduce HIF1-alpha protein levels in non-papillary clear-cell renal carcinoma12, breast cancer13 and paraganglioma14. ANRIL, an antisense noncoding RNA at the INK4b/ARF/INK4a locus, represses expression of its cognate sense mRNA cluster to promote cell growth in prostate cancer15, acute lymphoblastic leukemia16, glioma, and melanoma17. AChE-AS, another AS RNA, directly represses AChE expression via epigenetic modification of the AChE promoter region and exerts an anti-apoptotic effect in hepatocellular carcinoma cells.18 In contrast, bidirectional regulation between WDR83 and its natural AST DHPS shows concordant effects at both the transcriptional and translational levels in GC.19 Although divergent, these mechanisms suggest a potential mechanism by which transcriptional or post-transcriptional regulation of a sense gene is controlled through hybridization of an AST with its cognate sense mRNA to form a duplex in various diseases,20,21, 22 including cancer23. This mechanism has only recently been described, and such AST-sense RNA interactions may play pivotal roles in regulating inducible gene expression at the transcriptional or post-transcriptional levels in GC.
In the current study, we focused on the regulatory effects of one particular AST in GC. First, a next-generation sequencing analysis of human GC tissues and cell lines was performed, which identified marked and consistent upregulation of the AS lncRNA, RP3-416H24.1, relative to normal control tissues and cells. We further found that RP3-416H24.1 is a single antisense oligonucleotide RNA transcribed from the negative strand of Homo sapiens keratin 7, type II (KRT7), a protein-coding gene. The type II cytokeratins are basic or neutral proteins arranged in pairs of heterotypic keratin chains and co-expressed during differentiation of simple and stratified epithelial tissues.24 Although KRT7 overexpression has been observed in many neoplastic diseases, including cervical 25, esophageal squamous cell26, and colorectal cancers27 and is involved in several cancer-related pathways, including cytoskeletal signaling and the EGFR1 pathway,28-30 a knowledge gap remains. Most previous studies focused on correlations between KRT7 immunohistology and clinical pathology or prognosis, rather than on its molecular carcinogenic mechanism(s). We felt that by studying KRT7-AS in GC, we could gain insight into carcinogenic mechanisms of KRT7.
We first determined whether KRT7-AS was concordantly or discordantly upregulated with its cognate coding mRNA, KRT7, in GC-derived cell lines and tissues. We then established that KRT7-AS stabilizes KRT7 mRNA by forming an RNA-RNA duplex with it and increasing steady-state KRT7 expression at both the transcriptional and post-transcriptional levels. In addition, we demonstrated stimulatory effects of KRT7-AS on GC cell growth and cell cycle progression. The current findings suggest that the duplex formed by KRT7-AS and KRT7 mRNA increases steady-state KRT7 expression levels in GC. These data also justify studies of other ASTs that may promote carcinogenesis by stabilizing their cognate sense transcripts in other cancers.
RESULTS
Identification of an Overlapping AS Transcript at the KRT7 Gene Locus
First, in order to identify novel oncogenic lncRNAs in gastric cancer, we performed next-generation RNA sequencing (RNA-seq) on 9 different samples (2 matched normal tissue (NT)-GC tissue pairs; 1 normal gastric epithelial tissue; 3 unmatched GC tissues; 2 gastric cancer cell lines, NCI-N87 and MKN28; and 1 immortalized normal gastric epithelial cell line, HFE145). This analysis identified 22,740 lncRNAs that were expressed in GC tissues and cells, 12,882 of which were upregulated in GC tissues and cells relative to NTs and normal gastric cells. We prioritized for further study those lncRNAs showing the highest absolute expression levels in GC tissues and cells (normalized copy number >30; Online Supplementary Table S1). We excluded lncRNAs with normalized copy numbers <8 in any single tumor sample. This filtering process yielded 4 lncRNAs (H19, UCA1, RP11-297P16.4 and RP3-416H24.1). Notably, the lncRNA H19 is already known to be overexpressed in cancers of multiple organs31-34, including GC35, 36. Thus, the identification of H19 appears to underscore the validity of our filtering approach.
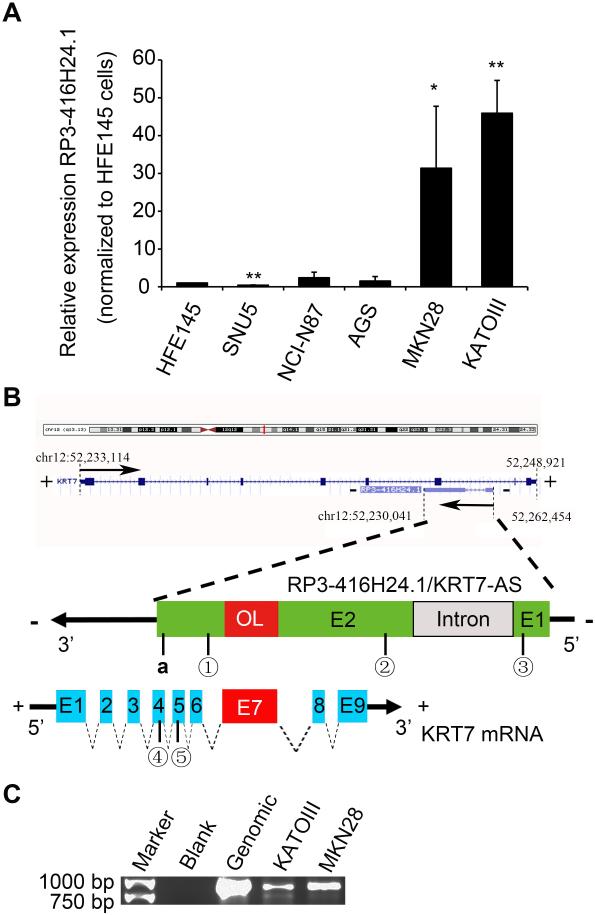
We then proceeded to validate differential expression of the three novel candidate lncRNAs: UCA1, RP11-297P16.4 and RP3-416H24.1. qRT-PCR analyses of these three lncRNAs in 5 GC-derived cell lines (NCI-N87, KATOIII, SNU5, MKN28, and AGS) vs. immortalized normal gastric epithelial cells (HFE145) revealed that RP3-416H24.1 was significantly overexpressed in two GC cell lines (MKN28 and KATOIII), by 31-fold and 45-fold, respectively (P< .01; Figure 1A, primers indicated in Figure 1B). RP3-416H24.1 was also overexpressed in NCI-N87 and AGS cells (2.4- and 1.5- fold, respectively; P>.05, Figure 1A), but significantly downregulated in SNU5 cells (0.4-fold; P<.01, Figure 1A). UCA1 exhibited diminished expression in KATOIII, SNU5, MKN28, and AGS gastric cancer cells but >1.8-fold increased expression in NCI-N87 gastric cancer cells (data not shown). RP11-297P16.4 exhibited decreased expression in NCI-N87 (0.0004-fold), SNU5 (0.0002-fold), and AGS (0.0005-fold) cells but increased expression in MKN28 and KATOIII cells (1.53- and 8.43-fold, respectively; data not shown). Since we were most interested in highly overexpressed lncRNAs, we chose RP3-416H24.1 and the GC cell lines MKN28 and KATOIII for further studies.
Figure 1. Identification of an overlapping antisense transcript at the KRT7 gene locus.
A. 5 gastric cancer-derived cell lines were tested: SNU5, NINC87, AGS MKN28 and KATOIII. KRT7-AS RNA expression was elevated in MKN28 and KATOIII. Expression level was normalized to HFE145 normal gastric epithelial cells.* P< .05; ** P< .01. B. Upper chart of this panel shows genome organization of the KRT7 gene at locus 12q13.13. The chart shows the start and end positions of these genes as indicated on the UCSC site (Genome Bioinformatics). Black arrows indicate transcription direction, and blue blocks are exons. The schema is in scale. “+”at both sides of strand represents the positive strand; “-“represents the negative strand. Lower chart of this panel is a schematic representation of RP3-416H24.1 (ensemble gene transcript ENST00000546686, on the minus DNA strand) and KRT7 mRNA (RefSeq gene NM_005556 on the positive DNA strand). The schema is not drawn to scale. Blocks with colors (green, blue and red) represent exons. Red block in KRT7-AS transcript represents overlapping region (OL). Red block in KRT7 mRNA represents the seventh exon and also OL region. “E” followed by number connotes exon and serial number. Block filed with grey color represents intronic portion of KRT7-AS. Black arrows represent direction of transcription. Dotted filed lines between colored blocks represent introns in KRT7. Primer sites were indicated as follows: a, KRT7-AS-specific RT-primer/KRT7-AS qPCR reverse primer; ①KRT7-AS qPCR forward primer; ②KRT7-AS PCR reverse primer; ③KRT7-AS PCR forward primer;④KRT7 mRNA qPCR forward primer; ⑤KRT7 mRNA qPCR reverse primer. “+”at both sides of strand represents the positive strand; “-“represents the negative strand. C. Antisense-specific RT-PCR screen for KRT7-AS. Antisense-specific RT-PCR detected expression of KRT7-AS RNA from the antisense strand in KATOIII and MKN28 cells. M, Gene ruler expression ladder (SM1551, Life technology); scale is illustrated at left. The cellular origin of the cDNA templates are: Lane 1, no template; Lane 2, genomic DNA PCR control (Applied Biosystems, Foster City, California, USA), Lane 3, KATOIII; Lane 4, MKN28;. cDNAs were reverse-transcribed using KRT7-AS-specific RT-primer. PCR products were amplified using KRT7-AS PCR forward/reverse primer pairs. Sites are indicated in Figure 1C as “a”,②,③.
From the UCSC genome browser, RP3-416H24.1 (ENST00000546686 in Ensemble database; Genbank HG504773.1) was identified as a single antisense RNA that is transcribed from the negative strand of the Homo sapiens keratin 7, type II (KRT7) locus, located at chromosomal band 12q13.13, and consists of two exons positioned 708 bp apart (Figure 1B). Supplementary Table S2 shows the sequence including 50 bp 5‘upstream to the transcription start site (TSS). There was no RNA polymerase binding consensus sequence (TATAAT and TTGACA) at positions -10 or -35 upstream of the TSS. It has been reported that AT-rich promoters exhibit lower expression and higher tissue specificity, properties known to define other lncRNA promoters; in addition, oligonucleotide repeats are frequently seen in lncRNA promoters.37,38 In our case, however, only 45% A/T content was found within 50bp upstream of the KRT7-AS 5’ transcription start site; nevertheless, mono-, di- and tri-nucleotide repeat-enriched patterns were indeed found. We heretofore refer to RP3-416H24.1 as KRT7 antisense (KRT7-AS) transcript. KRT7-AS shares a 213-nucleotide-long region with the seventh exon of KRT7 (from RefSeq NM005556) (Figure 1B); we heretofore refer to this at the overlapping region (OL).
Next, we used single strand-specific RT-PCR (SSPCR) to identify KRT7-AS RNA as originating from the reverse strand of the KRT7 locus in MKN28 and KATOIII GC cells. Antisense strand-specific cDNA was obtained via first-strand reverse transcription (RT) using an antisense-specific RT primer. The RT primer site is indicated in Figure 1B. Two primer sets were used to amplify KRT7-AS by PCR (primers are indicated in Figure 1B). Figure 1C shows that KRT7-AS was successfully amplified from antisense-specific RT cDNA, but that surprisingly, the “intron” separating exons 1 and 2 was not spliced out in either of the two GC cell lines studied (sequence-confirmed; data not shown). Unexpectedly, we were unable to amplify the complete sequence of KRT7-AS by 5’ and 3’ RACE PCR when attempting to validate the 5’ transcription start site (TSS). We then employed an indirect method to verify whether this was the correct TSS, or whether other alternate TSSs existed: we attempted to amplify PCR fragments initating -20 bp or -48 bp upstream of the 5’ TSS. (detailed in Supplementary Figure S1). Since DNase I treatment had been performed to exclude genomic DNA contamination during each RNA extraction (see Materials and Methods), these results proved the existence of a naturally-occurring AS RNA complementary to KRT7 in MKN28 and KATOIII cells, constituting a portion of the un-spliced primary transcript intron originating from the negative strand at the KRT7 gene locus, totaling 2 401 bp in length. A schematic representation is shown in Figure 1B.
Expression Patterns of KRT7-AS and KRT7 mRNA in Gastric Cancer
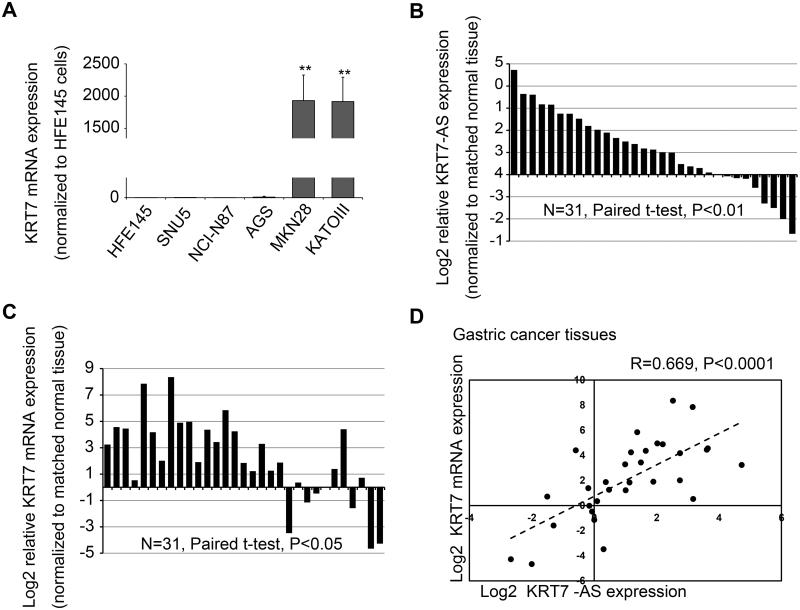
Many ASTs have been reported in eukaryotes, but their effects on their cognate coding genes remains unclear, including suppression or activation. In GC, modulatory effects of ASTs on neighboring genes has been reported only once before19. Therefore, we sought to elucidate the participation of KRT7-AS in KRT7 regulation. We first delineated KRT7-AS and KRT7 mRNA expression patterns in GC-derived cell lines and tissues. Among the 5 GC cell lines tested, KRT7 mRNA was significantly upregulated only in MKN28 and KATOIII cells (by 1 931- and 1 916-fold, respectively; P< .01, Figure 2A) among the 5 GC cell lines tested relative to HFE145 cells. Notably, KRT7 expression positively with KRT7-AS expression in all 5 GC cell lines tested (R=0.971, P< .01). In matched NC-GC tissue pairs obtained from 31 GC patients (23 men and 8 women, 63.77± 14.23 years of age), KRT7-AS expression was significantly elevated relative to corresponding normal tissue in the majority of GC tissues studied (22/31; average fold-change 4.10, paired t-test P-value < .01; Figure 2B). Similarly, KRT7 mRNA expression was significantly elevated compared with corresponding normal tissue in the majority of GC tissues studied (24/31; average fold-change 27.75, paired t-test P-value < .05; Figure 2C). Levels of KRT7-AS and KRT7 mRNA were positively correlated in GC tissues (R=0.699, P< .001; Figure 2D). Concordant co-regulation has been demonstrated for other sense-antisense RNA pairs.20,21 Interestingly, in the same samples, KRT7 mRNA levels were much higher than KRT7-AS levels. Specifically, In MKN28 and KATOIII cells, KRT7 mRNA was 61- and 41- fold higher than KRT7-AS, respectively (P< .01). Similarly, in 22 GC tissues, average expression of KRT7 mRNA was 7-fold higher than KRT7-AS (P< .01).Next, we sought to determine the subcellular distribution of KRT7-AS and KRT7 mRNAs in MKN28 and KATOIII cells. KRT7-AS was principally localized to the nucleus in both cell lines, suggesting that KRT7-AS may function mainly in the nucleus (Supplementary Figure S2). Moreover, in a panel of diverse human tissue-derived RNAs, we found a similar concordant expression pattern of KRT7-AS and its sense partner mRNA KRT7 (Supplementary Figure S3).
Figure 2. Expression pattern of KRT7-AS and KRT7 mRNAs in cell lines and tissues.
A. 5 GC-derived cell lines were measured: SNU5, NCI-N87, AGS, MKN28 and KATOIII. KRT7 mRNA was highly expressed in MKN28 and KATOIII human gastric cancer cells. Expression levels were normalized to HFE145 normal gastric epithelial cells. **, P< .01. B. Expression of KRT7-AS in 31 gastric cancer tissues. Expression levels are shown as log2-fold change vs. matching normal adjacent tissue. C. Expression of KRT7 mRNA in 31 gastric cancer tissues. Expression levels are shown as log2-fold change vs. matching normal adjacent tissue. D. Correlation between KRT7-AS and KRT7 mRNA expression in gastric cancer tissues.
KRT7-AS Controls KRT7 Expression at both the RNA and Protein Levels
To identify any direct link between the levels of KRT7-AS and KRT7 mRNAs, we first transiently downregulated KRT7-AS in MKN28 and KATOIII cells. siRNA target sites were designed to target only non-KRT7-overlapping regions of KRT7-AS (Supplementary Figure S4A). Downregulation of KRT7-AS by siRNAs in both cell lines ranged from 30-50% (P< .05, Supplementary Figure S4B). KRT7 mRNA levels were decreased by >30% (P< .01) after downregulation of KRT7-AS in MKN28 cells, whereas no significant change in KRT7 mRNA levels occurred in KATOIII cells (Supplementary Figure S4C). In addition, no differences in KRT7 protein expression were observed after KRT7-AS downregulation in either of the two GC cell lines studied (Supplementary Figure S4D).
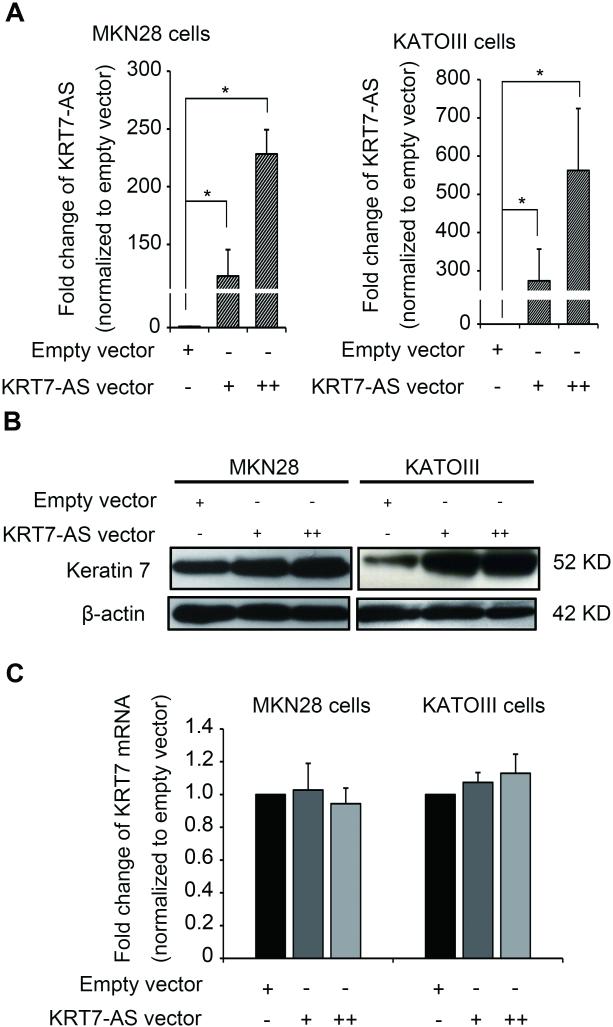
We then transiently overexpressed full-length KRT7-AS in MKN28 and KATOIII cells (Figure 3A): this resulted in significantly increased KRT7 protein levels in both cell lines (Figure 3B). In contrast, KRT7 mRNA levels did not change significantly in either cell line (Figure 3C). Thus, KRT7-AS appeared to augment KRT7 expression at the protein level, rather than at the level of mRNA accumulation.
Figure 3. Regulatory function of KRT7-AS overexpression to KRT7.
A. Overexpression of KRT7-AS in MKN28 and KATOIII cells after vector transfection. B. Western blot illustrates increased keratin 7 protein levels in both MKN28 and KATOIII cells after forced KRT7-AS overexpression. C. Unchanged KRT7 mRNA levels in MKN28 and KATOIII cells after forced KRT7-AS overexpression. Expression was normalized to empty vector-transfected cells.
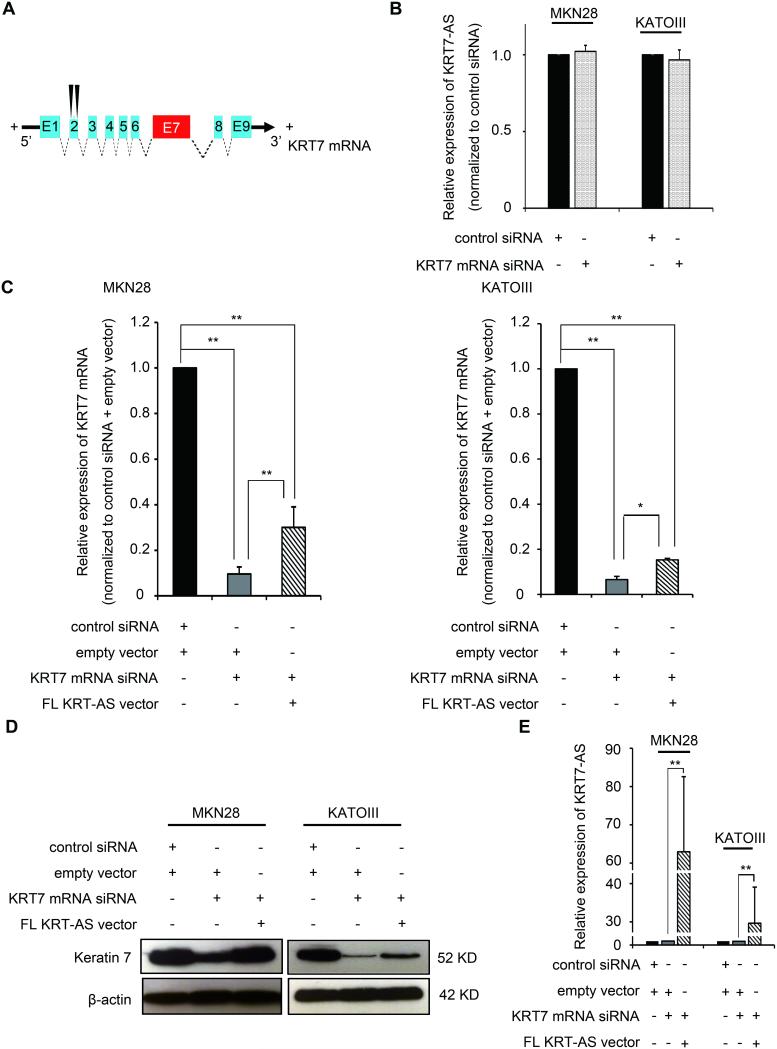
To further assess whether KRT7 mRNA exerted any effect on KRT7-AS expression, we measured KRT7-AS levels after transfecting cells with siRNAs directed exclusively against the non-overlapping portion of the KRT7 mRNA (Figure 4A). No change in KRT7-AS levels were detected after KRT7 mRNA knockdown (P> .05, Figure 4B). This observation suggested a lack of regulatory effect by KRT7 mRNA on its AS partner transcript.
Figure 4. Regulatory function of KRT7 mRNA downregulation to KRT7-AS and the rescue effect of KRT7-AS overexpression in KRT7 mRNA-downregulated cells.
A. Target sites of two siRNAs to KRT7 mRNA. Black triangles represent target sites of two different siRNAs specific to KRT7 mRNA which all avoid the region overlapping with KRT7-AS. B. No change of KRT7-AS transcript was detected after KRT7 mRNA downregulation. Expression levels were normalized to control-siRNA transfected cells. C. Left panel: Overexpression of KRT7-AS led to a three-fold upregulation of KRT7 mRNA in KRT7 mRNA-downregulation MKN28 cells. Right panel: Overexpression of KRT7-AS led to a two-fold upregulation of KRT7 mRNA in KRT7 mRNA-downregulated KATOIII cells. Expression levels were normalized to control-siRNA and empty vector transfected cells. *, P< .05; **, P< .01. D. Western blot illustrates increased keratin 7 protein levels in KRT7 mRNA-dowregulated MKN28 and KATOIII cells after forced KRT7-AS overexpression. E. Overexpression efficiency of KRT7-AS vector in the KRT7 mRNA-downregulated MKN28 and KATOIII cells. Expression levels were normalized to control-siRNA and empty vector transfected cells. **, P< .01.
Since forced KRT7-AS overexpression alone did not affect native KRT7 mRNA levels, we sought to determine whether it could increase KRT7 mRNA expression in cells pretreated with a KRT7-targeted siRNA. This experiment indeed showed that forced overexpression of KRT7-AS caused three-fold and two-fold increases of KRT7 mRNA levels in KRT7 siRNA-pretreated MKN28 and KATOIII cells, respectively (P< .05, Figure 4C). By Western blotting, forced overexpression of KRT7-AS also increased KRT7 protein levels in KRT7 siRNA-pretreated cells (Figure 4D). Figure 4E shows the efficiency of KRT7-AS overexpression in KRT7 siRNA-pretreated cells. We concluded that a substantial increase in KRT7 mRNA after forced KRT7-AS overexpression (but without KRT7 siRNA treatment) had not been previously detectable due to very high native KRT7 mRNA levels. In view of the above RT-PCR and Western blotting findings, we also inferred that regulation of KRT7 expression by KRT7-AS occurs at both the mRNA and protein levels.
KRT7-AS Increases KRT7 mRNA Stability by Forming an RNA Duplex
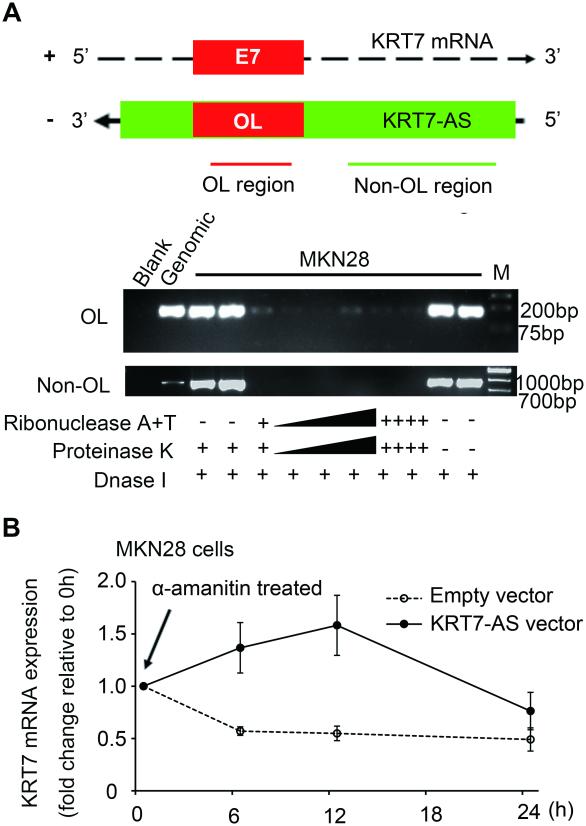
It has been previously demonstrated that AS RNAs can form duplexes with their cognate sense mRNAs, and that such complexes are protected from ribonuclease degradation.20,21 We therefore used an RNase protection assay (RPA) to verify the hypothesis that KRT7-AS and KRT7 mRNA form a protective duplex, specifically at their overlapping region (213 nucleotides). Total RNA from MKN28 cells was used to test this hypothesis. RT-PCR data revealed that this overlapping portion was at least partially protected from RNase degradation (Figure 5A).
Figure 5. KRT7-AS and KRT7 mRNA form a duplex RNA-RNA structure at their mutually overlapping region, which protects KRT7 mRNA from degradation.
A. Upper chart is a schematic representation of the PCR amplification region for overlapping (OL) and non-overlapping (non-OL) regions of KRT7-AS. Lower gel images illustrate OL and non-OL RT-PCR products in various samples. Total RNA was extracted and purified, single-stranded RNA was digested with increasing amounts of RNase A+T (indicated by the black wedge and multiple “++++”) and the remaining double-stranded RNA was subjected to RT-PCR to amplify the overlapping or non-overlapping regions of KRT7-AS. B. Stability of KRT7 mRNA over time was measured by qPCR relative to time 0 after blocking new RNA synthesis with α-amanitin (50 mM) (indicated with black arrow). MKN28 cells were transfected with KRT7-AS-containing vector or empty vector for 24 h, then further exposed to 50 mM α-amanitin for 6, 12 or 24 h. Cells were harvested, and the stability of the KRT7 mRNA was analyzed by qPCR. 18S RNA, a product of RNA polymerase I, was used as a control and was unchanged after α-amanitin treatment.
To test whether KRT7 mRNA stability was augmented by KRT7-AS, we used α-amanitin, an inhibitor of RNA polymerase II (Sigma Aldrich), to block new RNA synthesis in MKN28 cells over a 24-h period, then measured subsequent levels of KRT7 mRNA. 18s ribosomal RNA was used as an internal control (18s ribosomal RNA, a product of RNA polymerase I, is not affected by α-amanitin treatment).19,20 In MKN28 cells overexpressing KRT7-AS, we found increased stability of KRT7 mRNA vs. cells transfected with an empty vector (Figure 5B).
The Mutually Overlapping Region Plays a Dominant Role in KRT7-AS Function
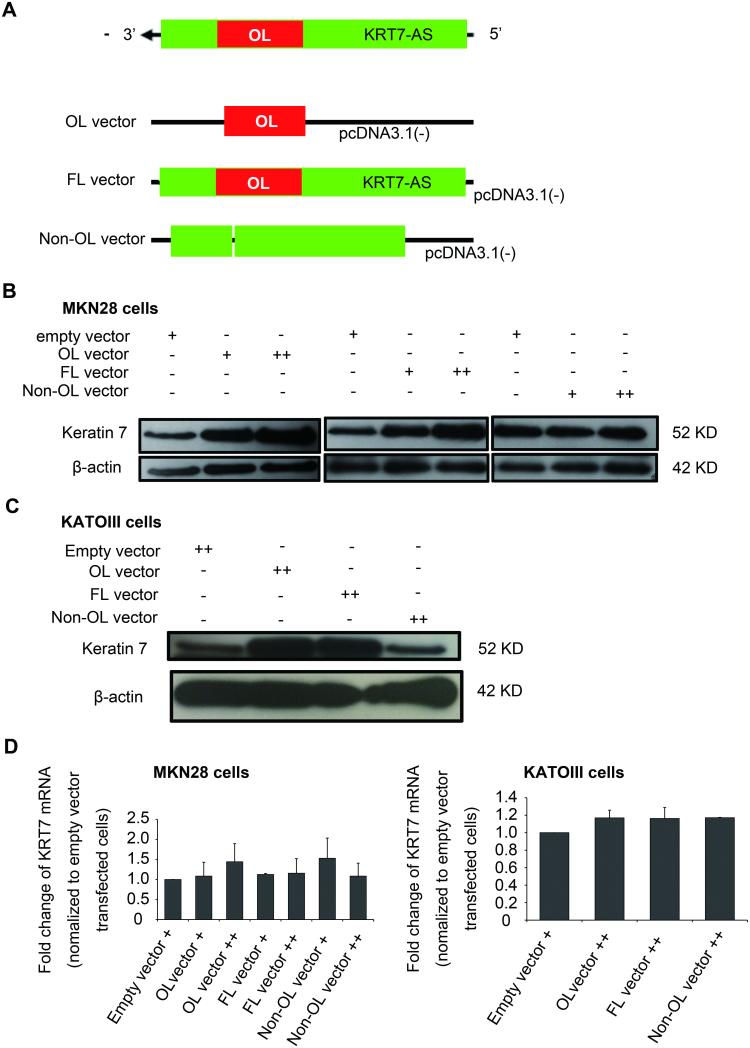
After verifying the existence of a KRT7-AS/KRT7 mRNA duplex, we hypothesized that the KRT7-overlapping region of KRT7-AS is more important than other portions of KRT7-AS. We constructed an overlapping-region (OL)-only vector, a non-overlapping region (non-OL) vector, and a full-length (FL) vector for transfection of MKN28 and KATOIII cells (Figures 6A). Overexpression efficiency of these three transfected vectors in MKN28 and KATOIII cells is shown in Supplementary Figure S5. The non-OL KRT7-AS vector failed to increase KRT7 protein levels in MKN28 and KATOIII cells, whereas both the OL and FL KRT7-AS vectors increased KRT7 protein levels, suggesting that this overlapping region is critical to the KRT7-buttressing function of KRT7-AS (Figure 6B, 6C). However, for all 3 vectors, no significant changes in KRT7 mRNA levels were detected (Figure 6D).
Figure 6. The OL region of KRT7-AS only increases KRT7 protein level, but has little effect on KRT7 mRNA level.
A. Schema of KRT7-AS full-length (FL) vector, overlapping (OL) vector, and non-overlapping (non-OL) vector construction. B. Keratin 7 protein level increased in MKN28 cells after OL- and FL-vector transfection, but no change occurred in non-OL vector-transfected MKN28 cells. C. Keratin7 protein level increased in KATOIII cells after OL- and FL-vector transfection, but no change occurred in non-OL vector-transfected KATOIII cells. D. KRT7 mRNA levels did not change substantially in KRT7-AS OL-, FL- or non-OL vector-transfected MKN28 cells and KATOIII cells. Expression was normalized to empty vector-transfected cells.
Overexpression of KRT7-AS in GC Cells Leads to Increased Proliferation and Migration
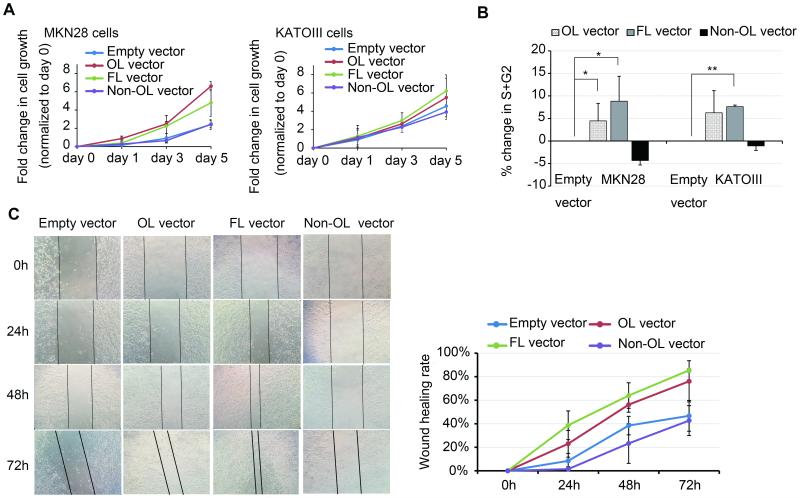
To determine potential functional consequences of activated KRT7-AS expression, in particular of its KRT7-overlapping portion, several in vitro assays were performed in MKN28 and KATOIII cells. Relative to cells transfected with empty vector, significantly increased growth of FL- and OL-KRT7-AS overexpressing cells was seen at day 5 in MKN28 cells (P< .05, Figure 7A). The growth rate of OL- and FL-vector-transfected cells was higher than that of empty vector- or non-OL vector-transfected cells at day 5 in KATOIII cells, although not significantly so (Figure 7A). Consistent with results of cell proliferation assays, KRT7-AS-OL and -FL overexpression caused significant accumulation of cells in the S+G1/M phases of the cell cycle (P< .05, flow cytometry results; Figure 7B). Finally, both KRT7-AS-OL and -FL overexpression caused substantial increases in cell migration. Specifically, scratch assays revealed that KRT7-AS-OL and -FL overexpression caused substantial increases in cell migration at 48h and 72h (Figure 7C), P< .05.
Figure 7. Overexpression KRT7-AS OL and FL promotes proliferation and migration of GC cells.
A. Cell proliferation assays in MKN28 and KATOIII cells. In MKN28 cells, KRT7-AS OL- and FL-overexpression induced significantly higher growth rates at day 5, P< .05. In KATOIII cells, no significant change was found. B. Cell cycle analyses in MKN28 and KATOIII cells. Relative to empty vector-transfected cells, KRT7-AS OL- and FL-overexpression induced a significant increase in cells at S+G1/M phase. *, P < .05; **, P< .01. C. Scratch assay results in MKN28 cells. Significantly more cells migrated in KRT7-AS OL- and FL- overexpressing cells than in empty vector-transfected cells or non-OL-overexpressing cells; P< .05.
Overexpression of KRT7-AS in GC Cells Partially Restores Decreased Proliferation Induced by KRT7 siRNA
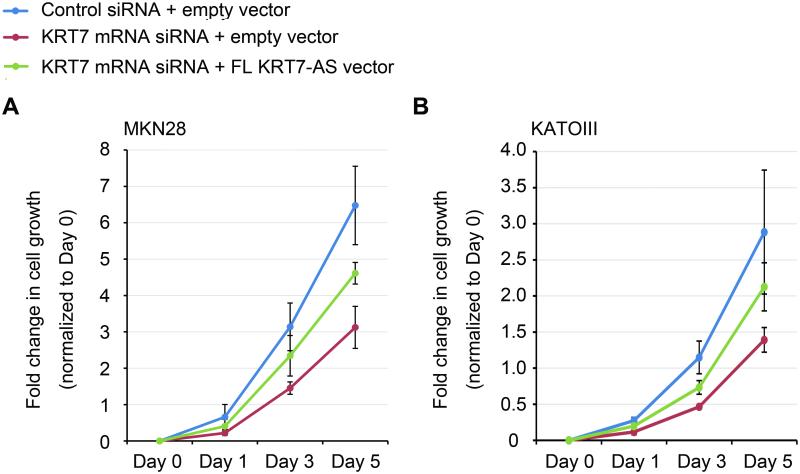
We observed a rescue effect of KRT7-AS FL-vector overexpression in MKN28 and KATOIII cells after decreased proliferation induced by KRT7 siRNAs (targeting the non-OL region of the KRT7 transcript). In MKN28 cells, the growth rate of KRT7 siRNA-treated cells was significantly slower than that of control siRNA-treated cells after Day 3 (P< .05, Figure 8A); a significantly higher growth rate of KRT7 siRNA + FL KRT7-AS vector- vs. KRT7 siRNA + empty vector-transfected cells was seen at Day 5 (P< .05, Figure 8A). In KATOIII cells, the growth rate of KRT7 siRNA-treated cells was significantly slower than that of control siRNA-treated cells after Day 1 (P< .05, Figure 8B); a significantly higher growth rate of KRT7 siRNA + FL KRT7-AS vector- vs. KRT7 siRNA + empty vector-transfected cells was seen after Day 3 (P< .05, Figure 8B).
Figure 8. Rescue effect on proliferation by FL KRT7-AS overexpression in KRT7 siRNA–treated cells.
Cells were treated with either control siRNA + empty vector, KRT7 siRNA + empty vector, or KRT7 siRNA + FL KRT7-AS vector in cell proliferation assays. A. Growth rate in MKN28 cells. B. Growth rate in KATOIII cells.
DISCUSSION
Mechanisms of action of the vast majority of antisense lncRNAs, if they exert any activity, remain largely unknown. Herein, we attempted to characterize an AST identified by RNA deep sequencing of GC tissues and cell lines. This study allowed us to make a number of observations. Firstly, we identified a previously unstudied antisense lncRNA, KRT7-AS (Genebank accession ID: HG504773.1). Unlike the protein-coding genes, there was no RNA polymerase binding consensus sequence in its promoter region; this finding is consistent with previous reports of lncRNA gene regulation.38 It has also been reported that relative to promoters of protein-coding genes, A/T-rich regions or mono-, di and tri-nucleotide repeat patterns occur in lncRNA promoters. These promoter patterns also correlate with lower expression levels but higher tissue specificity, properties known to define lncRNAs.37,39 Secondly, we found that KRT7-AS is concordantly upregulated with its coding sense partner, KRT7, in GC tissues and cell lines. Thirdly, we found that forced overexpression of KRT7-AS in GC cells resulted in increased KRT7 mRNA and protein accumulation in KRT7 siRNA-pretreated cells. Fourthly, we showed that KRT7-AS and KRT7 form an RNA-RNA duplex structure that protects KRT7 mRNA from RNase degradation. In addition, we found that the region of KRT7-AS that overlaps with KRT7 (designated “OL”) exerts the most dominant protective effect. Only forced overexpression of this OL region or the full-length KRT7-AS increased KRT7 protein levels, whereas the non-OL region did not. Finally, forced overexpression of OL- or FL-KRT7-AS resulted in increased GC cell growth, S-phase entry, and migration in MKN28 cells. A promotional effect on proliferation was even more obvious in KRT7 siRNA-pretreated cells. Taken together, these findings suggest that KRT7-AS stabilizes KRT7 mRNA by forming an RNA-RNA duplex, which in turn upregulates steady-state KRT7 expression at both the RNA and protein levels, thereby supporting GC development and/or progression.
Two types of regulation may occur between an AS and its cognate sense mRNA: discordant or concordant.5,6,40 Researchers have ascribed these patterns to the relative position of AS/sense pairs. According to the proximity between AS and sense partners in the genome, these pairs are classified as (1) nearby-to-head, when the 5' end of the sense gene is near the 5' end of the antisense transcript; (2) nearby-to-tail, when the 3' end of the sense gene is near the 3' end of the antisense gene; these two forms also referred to as “intergenic;” (3) head-to-head or divergent, when the 5' ends of both the sense and antisense genes align together; (4) tail-to-tail or convergent, when the 3' ends of both the sense and antisense genes align together; and (5) fully overlapping, when the antisense gene completely overlaps its sense counterpart.9 Nevertheless, to our knowledge, no functional relevance has been found for these categories thus far. Our data suggested that KRT7-AS and KRT7 position themselves “tail-to-tail” and are expressed in a concordant manner. Moreover, this finding raises an important question: why is the sense transcript expressed at a greater magnitude than its (potentially regulatory) AS partner, i.e., why does KRT7-AS exhibit lower expression than KRT7 mRNA? Although intense investigations have been conducted into mechanisms of concordant regulation,9-11,29 this phenomenon remains incompletely understood. In our case, we suggest that even without being expressed in equimolar amounts, ASTs can still exert biologically important functional effects on their sense partners. Alternatively, ASTs may have other regulatory effects on their sense partners, such as binding to or otherwise directly controlling their protein levels.
We explored the potential regulatory effect of KRT7-AS on KRT7 mRNA by siRNA assays. We transfected siRNAs targeting the non-overlapping region of KRT7-AS. Our siRNA-induced downregulation ranged from 30%-50%. Nevertheless, although KRT7 mRNA levels were decreased after siRNA-mediated KRT7-AS knockdown in MKN28 cells, no change was found in KATOIII cells; moreover, no effect on KRT7 protein levels was seen in either cell line. This lack of effect may have occurred because the KRT7-AS/KRT7 mRNA duplex exists in a steady state due to conformational changes in secondary or tertiary structure, resulting in low downregulation efficiency. To more clearly illustrate the regulatory effect of KRT7-AS on its sense partner, we studied KRT7 siRNA-pretreated cells. These experiments established that KRT7-AS controls KRT7 at both the mRNA and protein levels. Moreover, the OL portion of KRT7-AS alone exerted the same regulatory effect as did full-length (FL)-KRT7-AS. To our knowledge, this is the first data showing that the cognate sense gene-overlapping region of an AST can increase its protein levels. This finding diverges from previously reported concordant upregulation mechanisms. For example, Uchl1-AS controls Uchl1 only at the translational level, through 5’ overlapping sequence and an embedded SINEB2 repeat in a non-overlapping exon.22 Our findings, in conjunction with these previous studies, suggest that mechanisms of regulation between ASTs and their sense partners are more complex or subtle than previously believed.
Although endogenous RNA-RNA duplexes have been difficult to identify in human cells41, our RPA assays provided evidence that KRT7-AS and KRT7 mRNAs are capable of forming an RNA-RNA duplex at their complementary overlapping region. This duplex may act to alter the secondary or tertiary structure of KRT7 mRNA, thereby increasing its stability. Indeed, the stability of KRT7 mRNA was increased in KRT7-AS-overexpressing cells under α-amanitin treatment. This duplex forms at the exon adjacent to the poly(A) tail of KRT7 mRNA, where the deadenylation-dependent mRNA decay pathway is initiated, and the poly(A)-tail may be wrapped or folded inside a complicated secondary structure. This RNA duplex may therefore protect the 3’ end of KRT7 mRNA from being targeted by the ribonucleolytic RNA exosome.19,42 Another potential mechanism is associated with RNA duplex-mediated masking of microRNA binding sites within mRNAs. It has been reported that an AST can compete with microRNAs for binding sites, thereby obstructing microRNA-mediated regulation of target genes, which could increase mRNA stability and/or translation efficiency.19,43,44 Moreover, KRT7-AS may favor nuclear export of KRT7 mRNA or its interaction with the ribosome.22 However, these precise mechanisms require further study.
For some time now, KRT7 mRNA and protein levels have interested researchers because of their relationship to clinical prognosis in cancer patients. However, few studies have focused on potential KRT7 regulatory or carcinogenic mechanisms. Herein, we established KRT7-AS as a positive regulator of KRT7, which in turn exerts oncogenic effects. We evaluated the biological function of KRT7-AS in GC cells. We showed that KRT7-AS is involved in GC pathophysiology by increasing cell proliferation, promoting entry into S-phase, and stimulating migration.
In summary, we have shown that KRT7-AS expression levels are substantially increased in GC tissues vs. normal adjacent tissues, as well as in GC-derived cell lines. This elevated expression of KRT7-AS, its stimulative role in cell proliferation and the cell cycle, and its promotion of cell migration suggest that KRT7-AS upregulation is involved in the development and/or progression of GC, and that KRT7-AS (particularly its KRT7-overlapping portion) exerts these functions by forming an RNA-RNA duplex with its cognate sense transcript, thereby increasing KRT7 at both the mRNA and protein levels. Thus, KRT7-AS illustrates a novel type of long non-coding antisense RNA function in GC in particular, as well as in carcinogenesis in general.
MATERIALS AND METHODS
Cell culture
This study used 4 established human GC-derived cell lines (MKN28, KATOIII, AGS, NCI-N87, SNU5) as well as immortalized normal gastric epithelial cells (HFE145). See Supplementary Materials and Methods.
Tissue specimens
Primary tissue samples were obtained during surgical resections performed for clinical indications. All patients provided written informed consent under protocols approved by the institutional review board at the Johns Hopkins University School of Medicine. All tissue samples were pathologically confirmed as GC. Patient descriptions are listed in Supplementary Table S4). For detailed information, see Supplementary Materials and Methods.
RNA extraction, Nuclear and Cytoplasmic RNA separation, Next-generation RNA sequencing (RNA-seq), Small interfering RNAs, Plasmid construction and Cell transfection and treatment, Quantitative real-time polymerase chain reaction (PCR), PCR procedure, single strand-specific PCR (SS-PCR), Ribonuclease Protection Assay (RPA), Stability and α-amanitin Treatment, Western blotting, Cell Proliferation Assays, Cell Cycle Analysis and Scratch Assays
These procedures were performed as previously described.20,32,45 and in Supplementary Materials and Methods.
Statistical analyses
Results of experiments were displayed as mean ± standard deviation(SD). Statistical analyses were performed using SPSS software (SPSS, Chicago, Illinois, USA). Experimental results were evaluated using the two-tailed Student’s t-test or Pearson correlation test. Statistical significance was noted at P< .05, P< .01, or P< .001. Data were shown as mean ± standard deviation (SD). Error bars in figures indicate the SD value. Three independent triplicate experiments were performed for all cell biological assays, unless otherwise stated.
Supplementary Material
Supplementary Figure S4. Regulatory function of KRT7-AS downregulation to KRT7
A. Target sites of four siRNAs to KRT7-AS. Black triangles represent target sites of four different siRNAs specific to KRT7-AS which all avoid the region overlapping with KRT7 mRNA. B. Knockdown efficiency of KRT7-AS siRNAs in MKN28 and KATOIII gastric cancer cells. Expression levels were normalized to control-siRNA transfected cells. * P< .05, ** P< .01. C. KRT7 mRNA levels were lower in MKN28, but not in KATOIII cells, after KRT7-AS knockdown. Expression levels were normalized to control siRNA- transfected cells. ** P< .01. D. Western blot shows no change in KRT7 protein (keratin7) level after KRT7-AS knockdown.
Supplementary Figure S5 Overexpression efficiency of KRT7-AS FL-, OL- and non-OL vectors in MKN28 and KATOIII cells.
A. Schema representation of qPCR amplified region of KRT7-AS. Colored stick below the transcript with Part 1, 2 and 3 denotation represent designed detection regions of KRT7-AS. B. upper, middle and lower panels indicate qPCR result of three different part in KRT7-AS amplification by qPCR in MKN28 cells after transfected 3 vectors. C. upper, middle and lower panels indicate qPCR result of three different part in KRT7-AS amplification by qPCR in KATOIII cells after transfected 3 vectors. Expression levels were normalized to empty vector- transfected cells. * P< .05, ** P< .01.
Supplementary Figure S1. Verification of the start-point in KRT7-AS.
A. Indication of the sites of primers and possible PCR product. Small orange arrows show the forward primers. Small blue arrows show the reverse primer. “-“or “+” followed numbers show the first base position relative to the start-point. “-“means upstream of the start-point. “+“means downstream of the start-point. Numbers between the arrows indicate the possible PCR product. B. RT-PCR detected the expression of short fragment of KRT7-AS RNA from the antisense-specific strand reverse-transcribed cDNA. Cellular origin of the cDNA template is indicated above the lanes. M, Gene ruler expression ladder (SM1551, Life technology). Numbers above the lanes: 1, Fragment 1; 2, Fragment 2; 3, Fragment 3. Scale is illustrated at left.
Supplementary Figure S2. Subcellular distribution of KRT7-AS and KRT7 mRNA in MKN28 cells and KATOIII cells.
A. KRT7-AS distributes mainly in nucleus fractions of MKN28 and KATOIII cells. B. KRT7 mRNA distributes mainly in cytosolic fraction of MKN28, whereas distributes mainly in nucleus of KATOIII cells. RNAs were separated from cytosolic or nuclear fractions of both cells. Cyto, cytosolic; nuc, nucleus. **, P< .01.
Supplementary Figure S3 .Expression profiling of KRT7-AS and KRT7 mRNA in 20 different normal human organs.
A. Expression profiling of KRT7-AS in 20 different normal human organs. B. Expression profiling of KRT7 mRNA in 20 different normal human organs. Expression levels are shown as log2-fold change vs. skeletal muscle control cDNA.
ACKNOWLEDGEMENTS
S.J.M. is an American Cancer Society Clinical Research Professor and was supported by NIH grants CA190040, CA200483, CA85069, CA133012, CA173390 and Sidney Kimmel Comprehensive Cancer Center Core facilities. B.H. was supported by an Exchange Scholarship from the China Scholarship Council (CSC). B.H. is a recipient of a Joint Johns Hopkins University-Tongji University of China student scholarship.
Grant Support: B.H. was supported by an Exchange Scholarship from the China Scholarship Council (CSC). S.J.M. is an American Cancer Society Clinical Research Professor and was supported by NIH grants CA190040, CA200483, CA85069, CA133012, CA173390 and Sidney Kimmel Comprehensive Cancer Center Core facilities.
Footnotes
Binbin Huang and Jee Hoon Song share co-first authorship
Author Contributions: This study was conceived by Binbin H, JHS, Yulan C and SJM. All authors contributed to study design as well as to acquisition of samples or data. The data were analyzed and interpreted by Binbin H, JHS and SJM. This article was drafted by Binbin H and SJM, and was reviewed and approved by all authors in the current format. All who made significant contributions are listed as authors.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Contributor Information
Binbin Huang, Department of Gastroenterology, Tongji Hospital, Tongji University School of Medicine, Shanghai, China & Division of Gastroenterology, Departments of Medicine and Oncology and Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Jee Hoon Song, Division of Gastroenterology, Departments of Medicine and Oncology and Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Yulan Cheng, Division of Gastroenterology, Departments of Medicine and Oncology and Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
John M. Abraham, Division of Gastroenterology, Departments of Medicine and Oncology and Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, Maryland, USA
Sariat Ibrahim, Division of Gastroenterology, Departments of Medicine and Oncology and Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Zhenguo Sun, Division of Gastroenterology, Departments of Medicine and Oncology and Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Xiquan Ke, Division of Gastroenterology, Departments of Medicine and Oncology and Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
REFERENCES
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer Journal international du cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Houghton J, Wang TC. Helicobacter pylori and gastric cancer: a new paradigm for inflammation-associated epithelial cancers. Gastroenterology. 2005;128:1567–1578. doi: 10.1053/j.gastro.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi N, Kakizoe T. Synergistic interaction between Helicobacter pylori gastritis and diet in gastric cancer. Lancet Oncol. 2001;2:88–94. doi: 10.1016/S1470-2045(00)00225-4. [DOI] [PubMed] [Google Scholar]
- 4.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 6.Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Wight M, Werner A. The functions of natural antisense transcripts. Essays Biochem. 2013;54:91–101. doi: 10.1042/bse0540091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci. 2015;16:3251–3266. doi: 10.3390/ijms16023251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishizawa M, Ikeya Y, Okumura T, Kimura T. Post-transcriptional inducible gene regulation by natural antisense RNA. Front Biosci (Landmark Ed) 2015;20:1–36. doi: 10.2741/4297. [DOI] [PubMed] [Google Scholar]
- 11.Kimura T, Jiang S, Nishizawa M, Yoshigai E, Hashimoto I, Nishikawa M, et al. Stabilization of human interferon-alpha1 mRNA by its antisense RNA. Cell Mol Life Sci. 2013;70:1451–1467. doi: 10.1007/s00018-012-1216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J Natl Cancer Inst. 1999;91:143–151. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- 13.Cayre A, Rossignol F, Clottes E, Penault-Llorca F. aHIF but not HIF-1alpha transcript is a poor prognostic marker in human breast cancer. Breast Cancer Res. 2003;5:R223–230. doi: 10.1186/bcr652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Span PN, Rao JU, Oude Ophuis SB, Lenders JW, Sweep FC, Wesseling P, et al. Overexpression of the natural antisense hypoxia-inducible factor-1alpha transcript is associated with malignant pheochromocytoma/paraganglioma. Endocr Relat Cancer. 2011;18:323–331. doi: 10.1530/ERC-10-0184. [DOI] [PubMed] [Google Scholar]
- 15.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iacobucci I, Sazzini M, Garagnani P, Ferrari A, Boattini A, Lonetti A, et al. A polymorphism in the chromosome 9p21 ANRIL locus is associated to Philadelphia positive acute lymphoblastic leukemia. Leuk Res. 2011;35:1052–1059. doi: 10.1016/j.leukres.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression. PLoS Genet. 2010;6:e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xi Q, Gao N, Zhang X, Zhang B, Ye W, Wu J, et al. A natural antisense transcript regulates acetylcholinesterase gene expression via epigenetic modification in Hepatocellular Carcinoma. Int J Biochem Cell Biol. 2014;55:242–251. doi: 10.1016/j.biocel.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 19.Su WY, Li JT, Cui Y, Hong J, Du W, Wang YC, et al. Bidirectional regulation between WDR83 and its natural antisense transcript DHPS in gastric cancer. Cell Res. 2012;22:1374–1389. doi: 10.1038/cr.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michael DR, Phillips AO, Krupa A, Martin J, Redman JE, Altaher A, et al. The human hyaluronan synthase 2 (HAS2) gene and its natural antisense RNA exhibit coordinated expression in the renal proximal tubular epithelial cell. J Biol Chem. 2011;286:19523–19532. doi: 10.1074/jbc.M111.233916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 23.Salameh A, Lee AK, Cardo-Vila M, Nunes DN, Efstathiou E, Staquicini FI, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci U S A. 2015;112:8403–8408. doi: 10.1073/pnas.1507882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karantza V. Keratins in health and cancer: more than mere epithelial cell markers. Oncogene. 2011;30:127–138. doi: 10.1038/onc.2010.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambaudie E, Chereau E, Pouget N, Thomassin J, Minsat M, Charafe-Jauffret E, et al. Cytokeratin 7 as a predictive factor for response to concommitant radiochemotherapy for locally advanced cervical cancer: a preliminary study. Anticancer Res. 2014;34:177–181. [PubMed] [Google Scholar]
- 26.Oue N, Noguchi T, Anami K, Kitano S, Sakamoto N, Sentani K, et al. Cytokeratin 7 is a predictive marker for survival in patients with esophageal squamous cell carcinoma. Ann Surg Oncol. 2012;19:1902–1910. doi: 10.1245/s10434-011-2175-4. [DOI] [PubMed] [Google Scholar]
- 27.Harbaum L, Pollheimer MJ, Kornprat P, Lindtner RA, Schlemmer A, Rehak P, et al. Keratin 7 expression in colorectal cancer--freak of nature or significant finding? Histopathology. 2011;59:225–234. doi: 10.1111/j.1365-2559.2011.03694.x. [DOI] [PubMed] [Google Scholar]
- 28.Pujal J, Huch M, Jose A, Abasolo I, Rodolosse A, Duch A, et al. Keratin 7 promoter selectively targets transgene expression to normal and neoplastic pancreatic ductal cells in vitro and in vivo. FASEB J. 2009;23:1366–1375. doi: 10.1096/fj.08-115576. [DOI] [PubMed] [Google Scholar]
- 29.Kanduc D. Translational regulation of human papillomavirus type 16 E7 mRNA by the peptide SEQIKA, shared by rabbit alpha(1)-globin and human cytokeratin 7. J Virol. 2002;76:7040–7048. doi: 10.1128/JVI.76.14.7040-7048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandilands A, Smith FJ, Lunny DP, Campbell LE, Davidson KM, MacCallum SF, et al. Generation and characterisation of keratin 7 (K7) knockout mice. PLoS One. 2013;8:e64404. doi: 10.1371/journal.pone.0064404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byun HM, Wong HL, Birnstein EA, Wolff EM, Liang G, Yang AS. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 2007;67:10753–10758. doi: 10.1158/0008-5472.CAN-07-0329. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medrzycki M, Zhang Y, Zhang W, Cao K, Pan C, Lailler N, et al. Histone h1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014;74:6463–6473. doi: 10.1158/0008-5472.CAN-13-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, et al. The LncRNA H19 promotes epithelial to mesenchymal transition by functioning as MiRNA sponges in colorectal cancer. Oncotarget. 2015 doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alam T, Medvedeva YA, Jia H, Brown JB, Lipovich L, Bajic VB. Promoter analysis reveals globally differential regulation of human long non-coding RNA and protein-coding genes. PLoS One. 2014;9:e109443. doi: 10.1371/journal.pone.0109443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balbin OA, Malik R, Dhanasekaran SM, Prensner JR, Cao X, Wu YM, et al. The landscape of antisense gene expression in human cancers. Genome Res. 2015;25:1068–1079. doi: 10.1101/gr.180596.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munroe SH, Zhu J. Overlapping transcripts, double-stranded RNA and antisense regulation: a genomic perspective. Cell Mol Life Sci. 2006;63:2102–2118. doi: 10.1007/s00018-006-6070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid M, Olszewski P, Pelechano V, Gupta I, Steinmetz LM, Jensen TH. The Nuclear PolyA-Binding Protein Nab2p Is Essential for mRNA Production. Cell Rep. 2015;12:128–139. doi: 10.1016/j.celrep.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan JY, Sirey T, Honti F, Graham B, Piovesan A, Merkenschlager M, et al. Extensive microRNA-mediated crosstalk between lncRNAs and mRNAs in mouse embryonic stem cells. Genome Res. 2015;25:655–666. doi: 10.1101/gr.181974.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956–966. doi: 10.1053/j.gastro.2013.01.019. e954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S4. Regulatory function of KRT7-AS downregulation to KRT7
A. Target sites of four siRNAs to KRT7-AS. Black triangles represent target sites of four different siRNAs specific to KRT7-AS which all avoid the region overlapping with KRT7 mRNA. B. Knockdown efficiency of KRT7-AS siRNAs in MKN28 and KATOIII gastric cancer cells. Expression levels were normalized to control-siRNA transfected cells. * P< .05, ** P< .01. C. KRT7 mRNA levels were lower in MKN28, but not in KATOIII cells, after KRT7-AS knockdown. Expression levels were normalized to control siRNA- transfected cells. ** P< .01. D. Western blot shows no change in KRT7 protein (keratin7) level after KRT7-AS knockdown.
Supplementary Figure S5 Overexpression efficiency of KRT7-AS FL-, OL- and non-OL vectors in MKN28 and KATOIII cells.
A. Schema representation of qPCR amplified region of KRT7-AS. Colored stick below the transcript with Part 1, 2 and 3 denotation represent designed detection regions of KRT7-AS. B. upper, middle and lower panels indicate qPCR result of three different part in KRT7-AS amplification by qPCR in MKN28 cells after transfected 3 vectors. C. upper, middle and lower panels indicate qPCR result of three different part in KRT7-AS amplification by qPCR in KATOIII cells after transfected 3 vectors. Expression levels were normalized to empty vector- transfected cells. * P< .05, ** P< .01.
Supplementary Figure S1. Verification of the start-point in KRT7-AS.
A. Indication of the sites of primers and possible PCR product. Small orange arrows show the forward primers. Small blue arrows show the reverse primer. “-“or “+” followed numbers show the first base position relative to the start-point. “-“means upstream of the start-point. “+“means downstream of the start-point. Numbers between the arrows indicate the possible PCR product. B. RT-PCR detected the expression of short fragment of KRT7-AS RNA from the antisense-specific strand reverse-transcribed cDNA. Cellular origin of the cDNA template is indicated above the lanes. M, Gene ruler expression ladder (SM1551, Life technology). Numbers above the lanes: 1, Fragment 1; 2, Fragment 2; 3, Fragment 3. Scale is illustrated at left.
Supplementary Figure S2. Subcellular distribution of KRT7-AS and KRT7 mRNA in MKN28 cells and KATOIII cells.
A. KRT7-AS distributes mainly in nucleus fractions of MKN28 and KATOIII cells. B. KRT7 mRNA distributes mainly in cytosolic fraction of MKN28, whereas distributes mainly in nucleus of KATOIII cells. RNAs were separated from cytosolic or nuclear fractions of both cells. Cyto, cytosolic; nuc, nucleus. **, P< .01.
Supplementary Figure S3 .Expression profiling of KRT7-AS and KRT7 mRNA in 20 different normal human organs.
A. Expression profiling of KRT7-AS in 20 different normal human organs. B. Expression profiling of KRT7 mRNA in 20 different normal human organs. Expression levels are shown as log2-fold change vs. skeletal muscle control cDNA.