Abstract
The presence of invasion into the extra-hepatic portion of the portal vein or the development of distant metastases renders hepatocellular carcinoma (HCC) patients ineligible for the only potential curative options for this malignancy - tumor resection or organ transplantation. Gene expression profiling of murine HCC cell lines identified KLF6 as a potential regulator of HCC cell migration. KLF6 knockdown increases cell migration, consistent with the correlation between decreased KLF6 mRNA levels and the presence of vascular invasion in human HCC. Concordantly, single-copy deletion of Klf6 in a HCC mouse model results in increased tumor formation, increased metastasis to the lungs, and decreased survival, indicating that KLF6 suppresses both HCC development and metastasis. By combining gene expression profiling and chromatin immunoprecipitation coupled to deep sequencing, we identified novel transcriptional targets of KLF6 in HCC cells including VAV3, a known activator of the RAC1 small GTPase. Indeed, RAC1 activity is increased in KLF6 knockdown cells in a VAV3-dependent manner, and knockdown of either RAC1 or VAV3 impairs HCC cell migration. Together, our data demonstrate a novel function for KLF6 in constraining HCC dissemination through the regulation of a VAV3-RAC1 signaling axis.
Keywords: metastasis, KLF6, VAV3, mouse model, cell migration
Introduction
Hepatocellular carcinoma is a common malignancy worldwide that affects over 800,000 people per year, and is the 2nd-leading cause of cancer-related death1. Survival for patients with HCC is generally poor, with a 5-year survival rate of 16.1% in the United States. Importantly, patient prognosis is inversely correlated with tumor stage at the time of diagnosis, where the survival rate increases to 29% for patients with localized disease and eligibility for resection2. However, treatment options are limited for other patients, resulting in a 5-year survival rate of 10% and 3% for patients with regional spread or distant metastasis, respectively2. Given the prevalence of invasive and metastatic disease 3, and the absence of effective treatment strategies for these patients, studies defining the mechanisms underlying HCC metastasis may provide opportunities to improve the clinical outcome of a significant fraction of HCC patients.
Among the factors previously associated with HCC progression is inactivation of TP53 4, 5. Previous work from our laboratory demonstrated that while Trp53 gene deletion did not impact tumor development, yet promoted tumor progression and metastasis in a HCC mouse model, consistent with a role in HCC progression 6. Other studies from our laboratory demonstrated a role for insulin-like growth factor signaling in HCC cell migration and invasion 7. Recent expression profiling and genome sequencing approaches have identified expression changes associated with HCC development and progression8-13. While these studies identified several factors of potential prognostic and therapeutic significance, functional validation, particularly in vivo, remains lacking for many of these identified genes. Consequently, the mechanisms controlling HCC metastasis remain unclear. Therefore, we sought to identify factors that control the dissemination of HCC cells.
Here, we demonstrate that reduced expression of the zinc finger transcription factor KLF6 is associated with increased HCC cell migration and with the presence of vascular invasion in HCC patients. In agreement, liver-specific deletion of Klf6 promotes HCC dissemination to the lungs in mice. Moreover, shRNA-mediated knockdown of KLF6 in HCC cells results in an increased activity of the RAC1 small GTPase and enhances migration in a manner dependent on its activity. Combined gene expression profiling and chromatin immunoprecipitation experiments identified VAV3, a known activator of RAC1 function, as a novel KLF6 target gene that mediates its impact on HCC cell migration. Together, these findings identify a novel function of KLF6 in regulating Rho GTPase activity, and for the first time connect KLF6 and HCC dissemination.
Results
Identification of factors associated with HCC cell migration
BL185 is a murine HCC cell line, derived from a non-metastatic p53 null tumor, with an intrinsically low level of migration14. Isolated BL185 cells that migrated through the membranes of either a migration or invasion transwell insert were selected and expanded, generating subpopulations termed BL185-M1 and BL185-I1. These subpopulations display a higher absorbance by MTS assay over time, indicative of an increased proliferation rate (Supplemental Figure 1A). Additionally, the M1 and I1 subpopulations have increased soft agar colony formation relative to the parental cell line (Figure 1A). M1 and I1 also show a ten-fold higher rate of migration than the BL185 parent cell line (Figure 1B). Since migration assays serve as a surrogate for the initial steps of metastasis, these cell lines may serve as useful models for understanding HCC dissemination in vivo.
Figure 1.
(A) Soft agar colony formation by the parent BL185 cell line and the BL185-I1 and M1 subpopulations. Data are from a representative experiment performed in triplicate. (B) Transwell migration activity of the parent BL185 cell line and isolated subpopulations. Data are from a representative experiment performed in triplicate. (C) Validation of selected candidate genes identified by gene expression microarray by qRT-PCR. Fold changes values for the BL185-I1 and M1 subpopulations were calculated relative to the parent BL185 cell line. * p<0.05, ** p<0.01, *** p<0.001 by 2-sided student t-test.
We next identified gene expression changes in common between BL185-M1 and -I1, as compared to the parent BL185 line, using Affymetrix MG 430.2 microarrays. 313 genes were identified that displayed a greater than two-fold change in expression and had an adjusted p-value <0.01. Gene Ontology (GO) term analysis included categories related to cell cycle, angiogenesis, and migration, consistent with the phenotypes described above (Supplemental Table 1). These categories contain several genes that have well-characterized roles in tumor progression and metastasis, such as DLC1 15, 16 and Hif1α 17. We validated the differential expression of a subset the genes included within these GO terms by qRT-PCR (Figure 1C). Of note, direct evaluation of several factors associated with epithelial-to-mesenchymal transition (EMT) showed no significant changes (Supplemental Figure 1B)18.
KLF6 is decreased in human HCC and invasive HCC cells
One of the differentially expressed genes was Klf6. KLF6 is decreased at the mRNA level in human HCC19-21 and has tumor suppressor activity in this tumor type21. KLF6 mRNA levels are also decreased in several other human cancers, including those of the prostate, lung, and breast22-24. In prostate and breast cancers, increased expression of a dominant negative splice variant, KLF6-SV1, is associated with decreased survival and increased rates of metastasis25, 26. Finally, reported KLF6 target genes, including CDH1 (encoding E-Cadherin) and MMP9, are associated with metastasis in vivo27, 28. Thus, KLF6 was an attractive candidate for further analysis.
Quantitative RT-PCR confirmed a decrease in KLF6 mRNA levels in the M1 and I1 subpopulations relative to the BL185 parental cell line (Figure 1C). In agreement with KLF6 mRNA levels, all species of KLF6 protein are decreased in the M1 and I1 subpopulations (Figure 2A). Similarly, full-length KLF6 is decreased in human HCC cell lines when compared to an immortalized hepatocyte cell line, Thle2 (Figure 2B). KLF6 protein levels are also reduced in 3 out of 4 resected human HCC tumor samples compared to their matched normal liver samples (Supplemental Figure 2A). Consistent with previously published findings21, data from Oncomine demonstrate that KLF6 is reduced at the message level in HCC samples as compared to normal liver (Supplemental Figure 2B). Moreover, an expression profiling dataset in Oncomine revealed that HCC specimens with vascular invasion have lower KLF6 mRNA than those without vascular invasion (Figure 2C)29. These data are consistent with a reported connection between expression of the dominant-negative KLF6-SV1 splice variant and vascular invasion 30. Thus, reduced KLF6 levels correlate with enhanced HCC dissemination.
Figure 2.
(A) Immunoblot of KLF6 in BL185 cells and the BL185-I1 and M1 subpopulations. (B) Immunoblot of KLF6 in human HCC cell lines. The immortalized hepatocyte cell line Thle-2 is used as a control. (C) KLF6 mRNA levels in human HCC specimens with or without documented vascular invasion. Data are from the Wurmbach Liver dataset in Oncomine29. (D) Immunoblot detection of KLF6 knockdown in BL185 cells infected with lentiviruses encoding KLF6-targeting shRNAs. (E) Soft agar colony formation by BL185 cells following KLF6 knockdown. (F) Transwell migration activity of BL185 cells following KLF6 knockdown. (G) Immunoblot confirming ectopic expression of KLF6-SV1. Cells infected with the pBabe-puro vector are used as a control. β-actin serves as a loading control. (H) Transwell migration activity of BL185 cells with ectopic KLF6-SV1 expression. Migration activity for BL185 cells infected with pBabe-puro is set to 100%. Data are from a representative experiment performed in triplicate. Error bars represent standard deviation. * p<0.05, ** p<0.01 by 2-sided student t-test.
To determine if KLF6 could function as a potential prognostic marker for HCC, we performed immunostaining on a human tissue microarray containing 106 human HCC specimens. We found that HCC specimens displayed a range of nuclear, cytoplasmic, and negative staining, in roughly the same proportions as the tumor-associated normal tissue (where available) (Supplemental Figure 2C, D). Neither the absence of KLF6 staining nor KLF6 localization correlated with HCC grade or overall survival (data not shown), in contrast to previous studies that demonstrated an inverse correlation between KLF6 mRNA levels and tumor stage21.
KLF6 is a repressor of transformation-related phenotypes
To test whether reduced KLF6 levels in HCC cells stimulated phenotypes associated with tumor dissemination, KLF6 was stably knocked down using two independent shRNAs in the BL185 cell line (Figure 2D). KLF6 knockdown increased soft agar colony formation and enhanced cell migration (Figure 2E, F). Similarly, KLF6 knockdown increased cell migration in two additional HCC cell lines (Supplemental Figure 3A), confirming that this is not a cell line-specific effect.
The KLF6 SV1 splice variant is postulated to act as a dominant-negative protein and has been previously shown to enhance transformation and metastasis in other tumor types25, 26. Ectopic expression of KLF6-SV1 in BL185 cells increased cell migration by approximately 50% (Figure 2G, H), consistent with our findings following KLF6 knockdown.
Reduced expression of the KLF6 target CDH1 is associated with EMT18. Immunoblotting demonstrated that KLF6 knockdown cells have reduced E-cadherin levels (Supplemental Figure 3B). However, the levels of other EMT-associated markers are not significantly different between KLF6 knockdown cells and controls (Supplemental Figure 3B), suggesting that a classical EMT is not associated with KLF6-regulated cell migration in HCC cells.
Single-copy loss of Klf6 enhances HCC tumor formation and decreases survival
We next determined if decreased KLF6 levels promote HCC progression and metastasis in vivo using our previously described PyMT-driven RCAS-TVA HCC mouse model6, 14. In this model, hepatocytes and their progenitors are uniquely susceptible to RCAS virus infection6, 14. We previously demonstrated that delivery of RCAS-PyMT to Albumin-tva, Albumin-cre, Trp53flox/flox compound mice induces the development of metastatic HCC14. We crossed a conditional Klf6 allele into this model, such that half of the resulting progeny are Klf6 heterozygous in the liver while half are Klf6 wild type (WT) 31. (All progeny are also Albumin-tva, Albumin-cre, Trp53flox/flox.)
Mice were injected with RCAS-PyMT-producing DF1 chicken fibroblasts and a cohort of Klf6flox/WT and Klf6WT animals evaluated for tumor-free survival. We observed that Klf6flox/WT animals had significantly reduced survival relative to their Klf6WT counterparts (p=0.0398, Figure 3A). Necropsy demonstrated that these animals had HCC at the time of euthanasia, and proportionally more Klf6flox/WT mice developed HCC compared to Klf6WT mice (74% versus 38%) (Figure 3B). Analysis of RNA isolated from HCCs arising in either Klf6flox/WT or Klf6WT livers demonstrated that KLF6 was typically expressed at 50% lower levels in tumors arising in heterozygous livers, similar to the levels observed in non-tumor liver tissue (Supplemental Figure 4A, B).
Figure 3.
(A) Kaplan-Meier plot comparing tumor-free survival of mice of the indicated genotypes. p=0.0398 for difference between RCAS-PyMT injected Klf6WT and Klf6 Flox/WT mice (calculated by Log-rank test). (B) Tumor incidence in mice of the indicated genotypes. p=0.0258 by Fisher's exact test. (C) H&E stained tissue sections from HCCs arising in Klf6WT and Klf6 Flox/WT mice.
Despite increased tumor incidence and reduced survival in the Klf6flox/WT animals, tumor burden per mouse was not significantly different than that observed in Klf6WT mice (Supplemental Figure 4C). Moreover, Klf6 heterozygous HCCs do not display different histologies than Klf6WT tumors (Figure 3C). HCCs arising in Klf6 heterozygous and Klf6WT livers display similar frequencies of tumors with vacuolated cytoplasm as previously observed in the RCAS-TVA model (Figure 3C, top panels)6, 14. Tumors arising in both genotypes also commonly displayed more cellular, dense, and fibrotic areas with similar frequencies (Figure 3C, lower panels). Thus, reduction of Klf6 gene dosage does not impact HCC histology.
To determine if Klf6 deletion accelerated HCC development, we assessed an independent cohort of animals euthanized at 4.5 months of age, a time before most animals succumb to HCC. We observed that Klf6flox/WT mice had larger tumors than Klf6WT mice at this time point (Supplemental Figure 4D). Ki67 staining demonstrated no difference in cell proliferation between tumors arising in the two genotypes (Supplemental Figure 4E). Cleaved caspase 3 staining also revealed no differences (data not shown). Therefore, we infer that the difference in tumor size at 4.5 months reflects earlier initiation of tumor development in Klf6flox/WT livers, consistent with previous findings21.
Single-copy loss of Klf6 facilitates the growth of lung metastases
Despite no differences in tumor burden, proportionally more mice from the Klf6flox/WT survival cohort developed lung metastases than from the Klf6WT group (p=0.0453, Chi-Square test) (Figure 4A). A single metastatic focus was identified in the lungs of a Klf6WT mouse with HCC upon serial sectioning of the tissue and H&E staining (Figure 4C). In contrast, many of the tumor-bearing Klf6flox/WT mice had grossly visible lung metastases (Figure 4C).
Figure 4.
(A) Incidence of lung metastasis in HCC-bearing mice of the indicated genotypes. (B) Fold change in mRNA levels of genes associated with poor HCC prognosis in Klf6 Flox/WT HCCs as compared to Klf6WT HCCs. p<0.0001 by F-test. (C) H&E images (25X magnification) of lung metastases observed in Klf6WT and Klf6 Flox/WT mice.
HCCs isolated from the survival cohort of mice were assayed for expression of α-fetoprotein (AFP) and insulin-like growth factor 2 (IGF2), as these markers are associated with poor prognosis in HCC patients 32, 33. AFP and IGF2 mRNA levels are increased in several of the HCCs from the Klf6fl/wt group (Figure 4B) when compared to the average levels of the Klf6WT tumors. The variance in expression in tumors from the Klf6flox/WT group is significantly different from those in the wild-type group for both poor-prognosis genes (p<0.0001 by F-test).
Taken together with our in vitro data demonstrating that KLF6 knockdown increases HCC cell migration, our in vivo findings support a novel role for KLF6 in suppressing HCC dissemination in vivo.
Knockdown of KLF6 impacts activation of RAC1 and CDC42
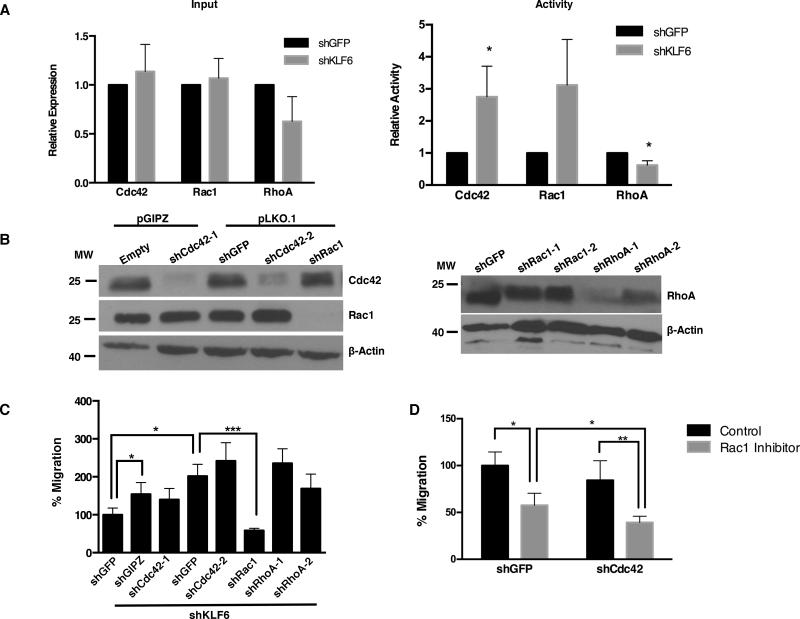
We next sought to characterize the mechanism of KLF6-mediated suppression of migration and dissemination in HCC. Rho family GTPases regulate cancer cell migration and metastasis, and have been implicated in HCC cell migration and metastasis34-36. We therefore assessed whether Rho GTPase activity is altered in HCC cells following KLF6 knockdown. We found that the levels of CDC42 and RAC1 are unchanged and the levels of RHOA are modestly decreased in KLF6 knockdown cells (Figure 5A, left). However, the activity of both CDC42 and RAC1 is increased in KLF6 knockdown cells, while RHOA activity is decreased relative to controls (Figure 5A, right). To determine the relative contributions of CDC42, RAC1, and RHOA to cell migration after KLF6 inhibition, BL185 cells with KLF6 knockdown were additionally infected with lentiviruses encoding shRNAs targeting these genes (Figure 5B). We observed that knockdown of RAC1, but not CDC42 or RHOA, reduced cell migration to baseline levels (Figure 5C). Moreover, treatment of KLF6 knockdown cells with a RAC1 inhibitor similarly reduced cell migration (Figure 5D). Combined knockdown of CDC42 and chemical inhibition of RAC1 further suppressed HCC cell migration, suggesting a context-dependent role for CDC42 in HCC cell migration (Figure 5D).
Figure 5.
(A) Quantification of relative RHO family protein levels in BL185 cells with KLF6 knockdown and non-silencing control (left panel). Quantification of the relative amounts of RHO family proteins in the active conformation (right panel). (B) Immunoblot of RAC1, CDC42, and RHOA in KLF6 knockdown cells with specific shRNAs targeting individual RHO family members. (C) Transwell migration activity of KLF6 depleted BL185 cells following subsequent knockdown of RAC1, CDC42, or RHOA. In columns 2 and 3, the indicated shRNAs are in the pGIPz vector. In all remaining lanes the indicated shRNAs are in the pLKO.1 vector. (D) Transwell migration activity of KLF6 depleted cells infected with a non-silencing control (shGFP) or shRNA targeting CDC42 and treated with the RAC1 inhibitor NSC2376 (gray bars), or water as a vehicle control (black bars). * p<0.05, ** p<0.01, *** p<0.001 by 2-sided student t-test.
The above findings suggest that KLF6 controls the expression of regulators of RAC1 and CDC42 activity. We therefore sought to identify KLF6 transcriptional targets that impact RHO activity, cell migration, and HCC metastasis.
KLF6 represses the expression of VAV3 and CDC42EP3
To determine transcriptional targets of KLF6 in HCC, we utilized two genome-wide approaches: gene expression profiling and ChIP-Seq. We performed gene expression profiling to identify genes differentially expressed upon KLF6 knockdown, utilizing three independently generated populations of KLF6 knockdown cells. Comparison with their corresponding non-silencing controls identified over 600 genes with a > 1.5-fold change in mRNA levels and an adjusted p-value < 0.01. To perform ChIP-Seq, we expressed a V5 epitope-tagged version of full-length KLF6 in BL185 cells and performed ChIP using an anti-V5 antibody. Libraries from two independent IP experiments were prepared for deep sequencing and compared to input chromatin as a control. ChIP peaks that were enriched in the KLF6 IP libraries were called and annotated to the nearest transcriptional start site (TSS). KLF6 binding was enriched around the TSS but was also found to occur at distant sites both upstream and downstream of TSSs (Supplemental Figure 5A). The location of KLF6 binding did not correlate with whether the target gene had increased or decreased expression in response to KLF6 knockdown (data not shown).
We then overlapped the gene lists from both experimental approaches to identify a collection of putative KLF6 transcriptional targets (Supplemental Table 2). Using ChIP coupled to PCR, we validated KLF6 binding to several of the putative target genes (Supplemental Figure 5B and data not shown). We also verified, using qRT-PCR, differential expression between cells with KLF6 knockdown and controls for a subset of the putative target genes (data not shown). Importantly, among the target genes identified was Cdh1, a known KLF6 target, providing validation of the robustness of the dataset 27.
Among the novel KLF6 target genes were genes encoding two modulators of RHO family GTPase function, VAV3 and CDC42EP3. KLF6 binds directly downstream of Cdc42ep3, and within the Vav3 gene, indicating that KLF6 is likely directly regulating their expression. VAV3 is a guanine exchange factor for several small GTPases, including RAC1 (and potentially CDC42), and is known to drive migration in several cancer cell lines37-39. CDC42EP3 is known to bind to active small GTPases and impact migration in fibroblasts40, 41.
In BL185 cells, VAV3 mRNA and protein levels are increased following KLF6 knockdown (Figure 6A, B), however the expression of the related Vav1 and Vav2 genes is not impacted (Supplemental Figure 6A). Like VAV3, CDC42EP3 levels are also increased following KLF6 knockdown (Supplemental Figure 6B). In addition, VAV3 levels are elevated in the highly migratory BL185-M1 and I1 cell lines, whereas CDC42EP3 levels were only modestly increased (Supplemental Figure 6C). Moreover, VAV3 mRNA levels are increased in tumors generated from Klf6flox/WT mice as compared to Klf6WT mice, although CDC42EP3 levels did not increase in this setting (Figure 6C, Supplemental Figure 6D). In human HCC cell lines, VAV3 and CDC42EP3 levels are increased as compared to an immortalized hepatocyte line Thle2, correlating with the reduced KLF6 levels in these HCC cell lines (Figure 6D, Supplemental Figure 6E). Together, these findings suggest that VAV3, and to a lesser extent CDC42EP3, may mediate, at least in part, the enhanced HCC cell migration induced following KLF6 inhibition.
Figure 6.
(A) mRNA levels of VAV3 in BL185 cells following KLF6 knockdown. (B) Immunoblot detection of VAV3 in BL185 cells following KLF6 knockdown. (C) qRT-PCR determination of VAV3 mRNA levels in tumors induced in Klf6WT and Klf6 Flox/WT mice. (D) Immunoblot detection of VAV3 in human HCC cell lines.
VAV3 and CDC42EP3 regulate HCC cell migration
To determine whether VAV3 functions downstream of KLF6 loss to stimulate HCC cell migration, we introduced lentiviral vectors encoding two independent VAV3 targeting shRNAs into BL185 cells with KLF6 knockdown and confirmed VAV3 knockdown by immunoblot (Figure 7A). Cell migration assays demonstrated that knockdown of VAV3 reduced cell migration to baseline levels (Figure 7B). Similarly, in 045-2a HCC cells with KLF6 knockdown, which also show increased VAV3 expression (Supplemental Figure 7A), concomitant VAV3 knockdown reduces cell migration (Supplemental Figure 7B). Likewise, introduction of two independent siRNAs targeting CDC42EP3 into BL185 cells reduced cell migration to baseline levels (Supplemental Figure 7C).
Figure 7.
(A) Immunoblot confirming VAV3 knockdown in KLF6 depleted BL185 cells. (B) Transwell migration activity of KLF6 depleted BL185 cells following subsequent knockdown of VAV3. (C) RAC1 activation, as detected by immunoprecipitation of RAC1-GTP and normalized to total RAC1, in BL185 cells with simultaneous knockdown of KLF6 and VAV3 and ectopic expression of wild type VAV3, VAV3 ISOIII (GEF mutant) or VAV3 W493L (PH mutant). The activity of cells with KLF6 knockdown is set to 100%. Data are the average of 3 independent experiments. (D) Transwell migration activity of the cell lines noted in (C). Migration activity for BL185 cells with KLF6 knockdown is set to 100%. Data are from a representative experiment performed in triplicate. Error bars represent standard deviation. * p<0.05, ** p<0.01, *** p<0.001 by 2-sided student t-test.
To ascertain whether VAV3-mediated activation of RAC1 activity potentially regulates HCC cell migration, we determined the impact of VAV3 expression on RAC1 activity. Concomitant VAV3 knockdown reduced RAC1 activity in BL185 cells with KLF6 knockdown (Figure 7C). Ectopic expression of wild type human VAV3 in these double knockdown cells significantly enhanced RAC1 activity, whereas GEF-inactive and pleckstrin homology domain mutant proteins failed to do so (Figure 7C) 42. In agreement, expression of wild type, but not mutant, VAV3 increased cell migration in double knockdown cells (Figure 7D).
Together, these data demonstrate that VAV3 is a critical factor that stimulates RAC1-mediated HCC cell migration downstream of KLF6 inhibition.
Discussion
The presence of invasive and metastatic disease renders HCC patients ineligible for any of the currently available curative options2, yet, the mechanisms that control the invasive and metastatic capabilities of HCC cells remain poorly understood.
Previous work demonstrated that heterozygous deletion of the gene encoding the transcription factor KLF6 in vivo increased hepatocyte proliferation and liver mass, and deletion of Klf6 in primary hepatocytes enhanced their proliferation in vivo21, 30. In mice with liver-specific deletion of Klf6, treatment with the carcinogen diethylnitrosamine resulted in increased hepatocarcinogenesis as compared to mice with intact KLF630. These data demonstrated a role for KLF6 as a tumor suppressor in HCC, yet data connecting KLF6 to HCC dissemination and metastasis were lacking. Here we demonstrate that KLF6 suppresses HCC cell migration, and that deletion of a single Klf6 allele promotes metastasis to the lungs, consistent with our observation that HCCs with vascular invasion have lower KLF6 mRNA levels than HCCs without vascular invasion. Given the frequency of invasive and metastatic disease in HCC patients, our work has important ramifications for understanding the mechanisms underlying HCC progression.
Our study is the first to demonstrate KLF6 regulation of metastasis in autochthonous mouse models, whereas prior studies that showed a role for KLF6 in regulating breast and prostate tumor metastasis utilized cancer cell lines implanted into immunocompromised mice25, 26, 43. Moreover, while these prior studies utilized the manipulation of the KLF6 splice variant KLF6-SV1, our study demonstrates for the first time that reduction in KLF6 levels is sufficient to promote metastasis. Thus, the effects previously observed following ectopic KLF6-SV1 expression likely reflect dominant-negative effects and not gain-of-function properties.
Our study differs from published studies that failed to demonstrate a role for KLF6 in constraining HCC dissemination21, 30. These differing results may reflect the different models employed (GEMM vs. carcinogen induced). However, they may also reflect differences in p53 gene status, as p53 is deleted in our model, but was intact in the previously published studies. Thus, the ability of KLF6 deficiency to promote HCC metastasis may be enhanced by inactivation of p53.
To elucidate the mechanisms underlying KLF6's ability to suppress HCC metastasis, we utilized gene expression profiling and ChIP-Seq to identify KLF6 regulated genes. Among the novel putative KLF6 target genes identified by our studies is VAV3, which has been shown to promote RAC1 and CDC42 activation 39. Indeed, our data demonstrate that RAC1 activity is increased following KLF6 knockdown in a VAV3-dependent manner. Consistent with an important role for VAV3 and RAC1 downstream of KLF6, HCC cell migration induced following KLF6 knockdown is dependent on these factors. Thus, we have identified a novel function for KLF6 in regulating the activity of RHO family GTPases, and link this function to a metastasis-associated phenotype. Confirmation that VAV3 and RAC1 regulate HCC metastasis downstream of KLF6 inhibition will require future in vivo studies.
Our studies described here highlight a novel role for a commonly inactivated tumor suppressor in restraining HCC metastasis. They also identify for the first time a transcriptional program regulated by KLF6 in tumor cells. Further characterization of the identified target genes and their potential roles in HCC development and dissemination will provide important insight into this deadly malignancy.
Materials and Methods
Mouse breeding, tumor analysis
All mouse strains used have been previously described6, 31, 44, 45. Klf6fl/fl mice were crossed with the RCAS-TVA HCC model6 to obtain Albumin-tv-a, p53fl/fl, Albumin-cre, Klf6fl/wt mice. These mice were then crossed with Albumin-tv-a, p53fl/fl, Albumin-cre, Klf6wt/wt mice to obtain littermates that were either heterozygous or wild type for Klf6 to be used for direct comparison. Mice were injected in the liver at 3 days old with 2×106 DF1 cells producing RCAS-GFP or RCAS-PyMT virus14 and assigned to either a survival study or designated for analysis at 4.5 months of age following genotyping. Mice were not randomly assigned to the studies. Group sizes were estimated based on investigators’ prior experience. In the survival study, mice were euthanized when illness became apparent. At 9 months of age, all remaining mice were euthanized and assessed for the presence of HCC. Kaplan-Meier statistics were performed using the Log-rank test to establish a p-value. Euthanized mice that did not have liver tumors were censored from the analysis. The pathologist was not blinded to mouse genotypes for tissue analysis.
Animals were housed in specific pathogen free facilities with abundant food and water. All studies were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee.
Tumor volume was calculated using the formula for the volume of an ellipsoid, and tumor burden was calculated by summing the tumor volumes per mouse.
To count and measure lung metastases, paraffin-embedded lungs from each mouse with a primary liver tumor were sectioned in 200μm steps. A section from each step was H&E stained and examined for the presence of metastases to lung. Using an eyepiece graticule, lung metastases were measured at their longest and shortest axes. Areas of the metastases were calculated using the formula for area of an ellipse.
Cell culture
All mouse HCC cell lines were cultured in high-glucose DMEM (Life Technologies) with 10% FBS and 1% Pen/Strep. All mammalian cell lines were grown at 37°C. Human HCC cell lines were obtained from ATCC. All other cell lines were from Lewis lab collections.
DF1 chicken fibroblast cells were cultured in growth medium at 39°C in high-glucose DMEM (Life Technologies) with 10% FBS and 1% Pen/Strep.
Stable knockdown cell lines were generated from lentiviral delivery of shRNAs directed to a mouse target mRNA. shRNA IDs are detailed in Supplemental Table 3.
The RAC1 inhibitor NSC23766 (Millipore, 553502) was added to cells as they were plated in a transwell migration chamber to a final concentration of 50μM.
In vitro migration, soft agar colony formation, and proliferation analysis
Transwell migration assays were performed as previously described 14. Soft agar colony formation was also performed as previously described 14, 46. Proliferation experiments were conducted via MTS assay in 96-well plates as previously described 14.
All experiments were performed using at least 3 biological replicates. Data presented are from a representative experiment performed in triplicate. Error bars represent the standard deviation of the dataset. Student's t-tests were used to calculate p-values.
Immunoblotting, Immunohistochemistry, and GTPase Activity Assays
Immunoblotting was performed as previously described 14. Antibodies for Western blotting were used as described in Supplemental Table 4.
Immunostaining was performed as previously described 6. KLF6 primary antibody (sc-7158) was added at a dilution of 1:250 in PBS with goat serum, and incubated overnight at 4°C.
Human HCC tissue microarrays were stained as described above. Samples were scored as either positive or negative for KLF6. Staining in KLF6 positive samples was classified as either primarily cytoplasmic or nuclear.
GTPase activity assays were performed as previously described 36.
KLF6 Expression in human HCC datasets
The Oncomine database was used for KLF6 expression data in Figure 2C and Supplemental Figure 2B. The data in Supplemental Figure 2B were derived from the Roessler Liver-2 dataset, and features 220 normal liver and 225 hepatocellular carcinoma samples 10. Figure 2C was derived from the Wurmbach Liver dataset and features 17 non-invasive and 18 invasive hepatocellular cancers 29.
Gene Expression Microarrays
Gene Expression profiling was conducted using Mouse Genome 430 2.0 arrays (Affymetrix, 900495). For profiling of the M1 and I1 populations, two independent RNA isolations were used as technical replicates. For expression profiling of KLF6 knockdown cells, three independent sets of control and shRNA infections were performed for analysis.
The RMA method in the Affymetrix package from Bioconductor 47 was used in R to summarize the probe level data and normalize the dataset to remove across-array-variation. Log transformed data were used in subsequent analyses. Moderated T statistics in the Limma package from Bioconductor 48 was used to determine whether a gene's expression level differs between treatments. Genes with an adjusted p-value <0.01 using the B-H method 49 and at least 1.5 fold difference in expression were considered significantly changed.
The raw data files in addition to normalized expression data have been deposited in the NCBI GEO Archive under accession numbers GSE54757 and GSE54762.
Gene Ontology terms were determined through the use of the Gene Ontology Enrichment Analysis Software Toolkit (GOEAST) program 50.
Quantitative RT-PCR
qRT-PCR was performed using SYBR Green (VWR, 95072) in an ABI 7300 machine using 50ng cDNA. Primers used for analysis are listed in Supplemental Table 5.
Ct values for each sample were averaged and normalized to β-actin Ct values. The Comparative Ct Method was used to calculate fold change, where fold change = 2−ΔΔCT.
ChIP-Sequencing
BL185 cells were transfected with pcDNA3.1 constructs containing KLF6-V5 or LacZ-V5. Chromatin was isolated from transfected cells according to the protocol specified in the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling). Either 10μL of V5 antibody (Abcam, 9116) or Rabbit IgG was added to pre-cleared chromatin for immunoprecipitation.
Immunoprecipitated DNA was prepared for deep sequencing using Illumina Genomic DNA Adapters and Primers. Libraries were sequenced on the Illumina platform using a single-end, long 76-basepair read. Sequence reads were mapped to the mm9 genome using the Illumina Genome Analyzer Pipeline. Reads that were uniquely mapped with two or fewer mismatches were retained for peak detection. Regions of KLF6 enrichment over input (referred to hereafter as “peaks”), were determined using the Model-based Analysis of ChIP-Seq algorithm (MACS, version 1.3.6 51) with the default settings and the following modified parameters: --gsize=2500000000 --tsize=76, --bw=150, --mfold=10. Peaks were assigned to nearest genes using the ChIPpeakAnno package 52 from Bioconductor. ChIP-sequencing raw files and annotated peaks are accessible in the GEO Archive under accession GSE54763.
Statistics
All error bars represent standard deviation, and data meets the assumptions of the tests unless otherwise noted. For all in vitro experiments, p-values were calculated using 2-sided student t-test.
Supplementary Material
Acknowledgements
The authors thank Genentech for sharing Klf6flox mice; Scott Friedman for providing the Klf6flox mice and sharing data prior to publication. The authors thank JiuFeng Cai and Victor Adelanwa for technical assistance; Dr. Kerry Burnstein for providing the VAV3 expression constructs; Karl Simin, Leslie Shaw, Scot Wolfe and members of the Lewis lab for helpful discussions and critical review of the manuscript. The assistance of the Memorial Sloan-Kettering Cancer Center Genomics Core and the University of Massachusetts Medical School Genomics and Deep Sequencing core facilities is gratefully acknowledged. Supported by NIH grant R01 CA121171 to BCL.
The funders played no role in the design, execution or interpretation of the study.
Footnotes
Conflict of Interest
The authors declare that they have no competing financial interests.
Supplementary Information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. European journal of cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Hayat MJ, Howlader N, Reichman ME, Edwards BK. Cancer statistics, trends, and multiple primary cancer analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. The oncologist. 2007;12:20–37. doi: 10.1634/theoncologist.12-1-20. [DOI] [PubMed] [Google Scholar]
- 3.Katyal S, Oliver JH, 3rd, Peterson MS, Ferris JV, Carr BS, Baron RL. Extrahepatic metastases of hepatocellular carcinoma. Radiology. 2000;216:698–703. doi: 10.1148/radiology.216.3.r00se24698. [DOI] [PubMed] [Google Scholar]
- 4.Buendia MA. Genetics of hepatocellular carcinoma. Seminars in cancer biology. 2000;10:185–200. doi: 10.1006/scbi.2000.0319. [DOI] [PubMed] [Google Scholar]
- 5.Villanueva A, Hoshida Y. Depicting the role of TP53 in hepatocellular carcinoma progression. Journal of hepatology. 2011;55:724–725. doi: 10.1016/j.jhep.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BC, Klimstra DS, Socci ND, Xu S, Koutcher JA, Varmus HE. The absence of p53 promotes metastasis in a novel somatic mouse model for hepatocellular carcinoma. Molecular and cellular biology. 2005;25:1228–1237. doi: 10.1128/MCB.25.4.1228-1237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen YW, Boyartchuk V, Lewis BC. Differential roles of insulin-like growth factor receptor- and insulin receptor-mediated signaling in the phenotypes of hepatocellular carcinoma cells. Neoplasia. 2009;11:835–845. doi: 10.1593/neo.09476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JS, Thorgeirsson SS. Genome-scale profiling of gene expression in hepatocellular carcinoma: classification, survival prediction, and identification of therapeutic targets. Gastroenterology. 2004;127:S51–55. doi: 10.1053/j.gastro.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Budhu A, Forgues M, Ye QH, Jia HL, He P, Zanetti KA, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 10.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer research. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome research. 2013;23:1422–1433. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Wholegenome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nature genetics. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 13.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. The Journal of clinical investigation. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YW, Klimstra DS, Mongeau ME, Tatem JL, Boyartchuk V, Lewis BC. Loss of p53 and Ink4a/Arf cooperate in a cell autonomous fashion to induce metastasis of hepatocellular carcinoma cells. Cancer research. 2007;67:7589–7596. doi: 10.1158/0008-5472.CAN-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue W, Krasnitz A, Lucito R, Sordella R, Vanaelst L, Cordon-Cardo C, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes & development. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X, Zimonjic DB, Park SW, Yang XY, Durkin ME, Popescu NC. DLC1 suppresses distant dissemination of human hepatocellular carcinoma cells in nude mice through reduction of RhoA GTPase activity, actin cytoskeletal disruption and downregulation of genes involved in metastasis. International journal of oncology. 2008;32:1285–1291. doi: 10.3892/ijo_32_6_1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X, Kang Y. Hypoxia and hypoxia-inducible factors: master regulators of metastasis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:5928–5935. doi: 10.1158/1078-0432.CCR-10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nature reviews Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 19.Kremer-Tal S, Narla G, Chen Y, Hod E, DiFeo A, Yea S, et al. Downregulation of KLF6 is an early event in hepatocarcinogenesis, and stimulates proliferation while reducing differentiation. Journal of hepatology. 2007;46:645–654. doi: 10.1016/j.jhep.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kremer-Tal S, Reeves HL, Narla G, Thung SN, Schwartz M, Difeo A, et al. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40:1047–1052. doi: 10.1002/hep.20460. [DOI] [PubMed] [Google Scholar]
- 21.Tarocchi M, Hannivoort R, Hoshida Y, Lee UE, Vetter D, Narla G, et al. Carcinogeninduced hepatic tumors in KLF6+/− mice recapitulate aggressive human hepatocellular carcinoma associated with p53 pathway deregulation. Hepatology. 2011;54:522–531. doi: 10.1002/hep.24413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gehrau RC, D'Astolfo DS, Dumur CI, Bocco JL, Koritschoner NP. Nuclear expression of KLF6 tumor suppressor factor is highly associated with overexpression of ERBB2 oncoprotein in ductal breast carcinomas. PloS one. 2010;5:e8929. doi: 10.1371/journal.pone.0008929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito G, Uchiyama M, Kondo M, Mori S, Usami N, Maeda O, et al. Kruppel-like factor 6 is frequently down-regulated and induces apoptosis in non-small cell lung cancer cells. Cancer research. 2004;64:3838–3843. doi: 10.1158/0008-5472.CAN-04-0185. [DOI] [PubMed] [Google Scholar]
- 24.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 25.Narla G, DiFeo A, Fernandez Y, Dhanasekaran S, Huang F, Sangodkar J, et al. KLF6- SV1 overexpression accelerates human and mouse prostate cancer progression and metastasis. The Journal of clinical investigation. 2008;118:2711–2721. doi: 10.1172/JCI34780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatami R, Sieuwerts AM, Izadmehr S, Yao Z, Qiao RF, Papa L, et al. KLF6-SV1 drives breast cancer metastasis and is associated with poor survival. Science translational medicine. 2013;5:169ra112. doi: 10.1126/scitranslmed.3004688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiFeo A, Narla G, Camacho-Vanegas O, Nishio H, Rose SL, Buller RE, et al. E-cadherin is a novel transcriptional target of the KLF6 tumor suppressor. Oncogene. 2006;25:6026–6031. doi: 10.1038/sj.onc.1209611. [DOI] [PubMed] [Google Scholar]
- 28.Das A, Fernandez-Zapico ME, Cao S, Yao J, Fiorucci S, Hebbel RP, et al. Disruption of an SP2/KLF6 repression complex by SHP is required for farnesoid X receptor induced endothelial cell migration. The Journal of biological chemistry. 2006;281:39105–39113. doi: 10.1074/jbc.M607720200. [DOI] [PubMed] [Google Scholar]
- 29.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, et al. Genomewide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 30.Vetter D, Cohen-Naftaly M, Villanueva A, Lee YA, Kocabayoglu P, Hannivoort R, et al. Enhanced hepatocarcinogenesis in mouse models and human hepatocellular carcinoma by coordinate KLF6 depletion and increased messenger RNA splicing. Hepatology. 2012;56:1361–1370. doi: 10.1002/hep.25810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leow CC, Wang BE, Ross J, Chan SM, Zha J, Carano RA, et al. Prostate-specific Klf6 inactivation impairs anterior prostate branching morphogenesis through increased activation of the Shh pathway. The Journal of biological chemistry. 2009;284:21057–21065. doi: 10.1074/jbc.M109.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iizuka N, Oka M, Tamesa T, Hamamoto Y, Yamada-Okabe H. Imbalance in expression levels of insulin-like growth factor 2 and H19 transcripts linked to progression of hepatocellular carcinoma. Anticancer research. 2004;24:4085–4089. [PubMed] [Google Scholar]
- 33.Izumi R, Shimizu K, Kiriyama M, Hashimoto T, Urade M, Yagi M, et al. Alphafetoprotein production by hepatocellular carcinoma is prognostic of poor patient survival. Journal of surgical oncology. 1992;49:151–155. doi: 10.1002/jso.2930490305. [DOI] [PubMed] [Google Scholar]
- 34.Chen L, Chan TH, Yuan YF, Hu L, Huang J, Ma S, et al. CHD1L promotes hepatocellular carcinoma progression and metastasis in mice and is associated with these processes in human patients. The Journal of clinical investigation. 2010;120:1178–1191. doi: 10.1172/JCI40665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding J, Huang S, Wu S, Zhao Y, Liang L, Yan M, et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nature cell biology. 2010;12:390–399. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 36.Chen YW, Chu HC, Ze-Shiang L, Shiah WJ, Chou CP, Klimstra DS, et al. p16 Stimulates CDC42-dependent migration of hepatocellular carcinoma cells. PloS one. 2013;8:e69389. doi: 10.1371/journal.pone.0069389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan B, Li Y, Zhao Q, Fan L, Wang D, Liu Y. Inhibition of gastric cancer cell growth and invasion through siRNA-mediated knockdown of guanine nucleotide exchange factor Vav3. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:1481–1488. doi: 10.1007/s13277-013-1204-2. [DOI] [PubMed] [Google Scholar]
- 38.Lin KT, Gong J, Li CF, Jang TH, Chen WL, Chen HJ, et al. Vav3-rac1 signaling regulates prostate cancer metastasis with elevated Vav3 expression correlating with prostate cancer progression and posttreatment recurrence. Cancer research. 2012;72:3000–3009. doi: 10.1158/0008-5472.CAN-11-2502. [DOI] [PubMed] [Google Scholar]
- 39.Zeng L, Sachdev P, Yan L, Chan JL, Trenkle T, McClelland M, et al. Vav3 mediates receptor protein tyrosine kinase signaling, regulates GTPase activity, modulates cell morphology, and induces cell transformation. Molecular and cellular biology. 2000;20:9212–9224. doi: 10.1128/mcb.20.24.9212-9224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joberty G, Perlungher RR, Macara IG. The Borgs, a new family of Cdc42 and TC10 GTPase-interacting proteins. Molecular and cellular biology. 1999;19:6585–6597. doi: 10.1128/mcb.19.10.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirsch DS, Pirone DM, Burbelo PD. A new family of Cdc42 effector proteins, CEPs, function in fibroblast and epithelial cell shape changes. The Journal of biological chemistry. 2001;276:875–883. doi: 10.1074/jbc.M007039200. [DOI] [PubMed] [Google Scholar]
- 42.Lyons LS, Burnstein KL. Vav3, a Rho GTPase guanine nucleotide exchange factor, increases during progression to androgen independence in prostate cancer cells and potentiates androgen receptor transcriptional activity. Mol Endocrinol. 2006;20:1061–1072. doi: 10.1210/me.2005-0346. [DOI] [PubMed] [Google Scholar]
- 43.Narla G, Difeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer research. 2005;65:1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- 44.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nature genetics. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 45.Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cellspecific gene knock-outs using Cre recombinase. The Journal of biological chemistry. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 46.Lewis BC, Shim H, Li Q, Wu CS, Lee LA, Maity A, et al. Identification of putative c-Mycresponsive genes: characterization of rcl, a novel growth-related gene. Molecular and cellular biology. 1997;17:4967–4978. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- 50.Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic acids research. 2008;36:W358–363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu LJ, Gazin C, Lawson ND, Pages H, Lin SM, Lapointe DS, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.