Abstract
Introduction
Pancreatic adenocarcinoma remains one of the most clinically challenging cancers despite an in-depth characterization of the molecular underpinnings and biology of this disease. Recent whole-genome-wide studies have elucidated the diverse and complex genetic alterations which generate a unique oncogenic signature for an individual pancreatic cancer patient and which may explain diverse disease behavior in a clinical setting.
Areas covered
In this review article, we discuss the key oncogenic pathways of pancreatic cancer including RAS-MAPK, PI3KCA and TGF-β signaling, as well as the impact of these pathways on the disease behavior and their potential targetability. The role of tumor suppressors particularly BRCA1 and BRCA2 genes and their role in pancreatic cancer treatment are elaborated upon. We further review recent genomic studies and their impact on future pancreatic cancer treatment.
Expert opinion
Targeted therapies inhibiting pro-survival pathways have limited impact on pancreatic cancer outcomes. Activation of pro-apoptotic pathways along with suppression of cancer-stem-related pathways may reverse treatment resistance in pancreatic cancer. While targeted therapy or a ‘precision medicine’ approach in pancreatic adenocarcinoma remains an elusive challenge for the majority of patients, there is a real sense of optimism that the strides made in understanding the molecular underpinnings of this disease will translate into improved outcomes.
Keywords: cancer stem cells, expression signature, K-Ras pathway, molecular pathways, notch signaling, p53, pancreatic cancer, targeted treatment, Wnt signaling
1. Introduction
Pancreatic adenocarcinoma harbors one of the most aggressive tumor behaviors. The five-year survival rate in patients who undergo resection is 23.4% while this rate decreases to 6% for all stage patients [1,2]. A recent study reported that by 2020, pancreatic cancer-related mortality will catch up with the mortality rate of colon cancer, and by 2030 it will be the second most common cancer-related death after lung carcinoma although its incidence rate will remain out of the five most common cancers [3]. These observations demonstrate that advances in cancer research thus far haven’t had substantial impact on the natural course of pancreatic cancer. Therefore, a focus on the molecular biology of pancreas adenocarcinoma is herein emphasized to elucidate mechanisms for therapeutic opportunity and understand resistance to available therapies. We summarize the molecular pathways which play key roles in development and progression of pancreatic adenocarcinoma and discuss potential targetable molecular pathways and biomarkers.
1.1 Core pathways in pancreatic adenocarcinoma
1.1.1 Ras-MAPK pathway
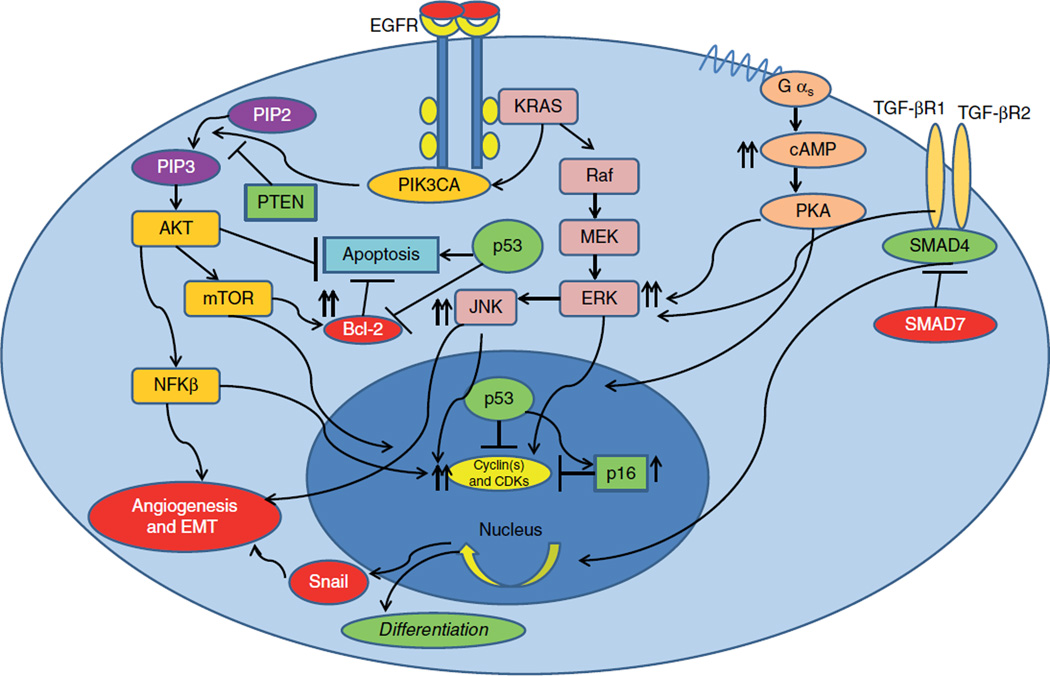
K-Ras (Kristen rat sarcoma viral oncogene homolog) mutation was one of the earliest discoveries in pancreatic cancer and has been reported in as high as 90% of pancreatic adenocarcinoma patients [4]. K-Ras is a small GTPase, and functions in one of the downstream signaling pathway receptor tyrosine kinases such as EGFR. Mutation in the K-Ras oncogene results in gain-of-function and constitutively activates extracellular signal regulated kinase pathway (MAPK; ERKs) [5]. Once ERK is activated, it translocates to the nucleus and promotes transcription activity for target genes that are involved in cell survival, growth and proliferation [6]. The Ras oncogene also interacts with other essential signaling molecules and functions as the maestro of survival pathways. In the early 1990s phosphatidylinositol-3-OH kinase (PIK3CA), an important signal transducing molecule for protein synthesis at the G1 phase of the cell cycle, was shown to be a target of the Ras oncogene [7]. Activation of PIK3CA by K-Ras and other growth stimuli receptors, such as EGFR, promotes cell tumor cell growth and prepares the cell for the next phase of cell cycle progression [8]. The K-Ras oncogene also interacts and activates other signaling molecules found to be related to stress response and cell growth such as c-Jun N-terminal kinase (JNK) and protein kinase C (PKC) [9] suggesting that the signaling network in pancreatic cancer cells is very complex and once a growth stimulus-related survival pathway is activated, simultaneously other cellular responses are also turned on to address cellular stress and altered microenvironment such as hypoxia and free radicals (Figure 1).
Figure 1. Core oncogenic pathways in pancreatic cancer cells.
Activation of diverse pro-survival pathways such as MAPK, PIK3CA-m-TOR and TGF-β in setting of mutant p53 and p16.
EMT: Epithelialmesenchymal transition; ERK: Extracellular signal regulated kinase pathway; JNK: Jun N-terminal kinase; PTEN: Phosphotase tensin homolog; PKA: Protein kinase A.
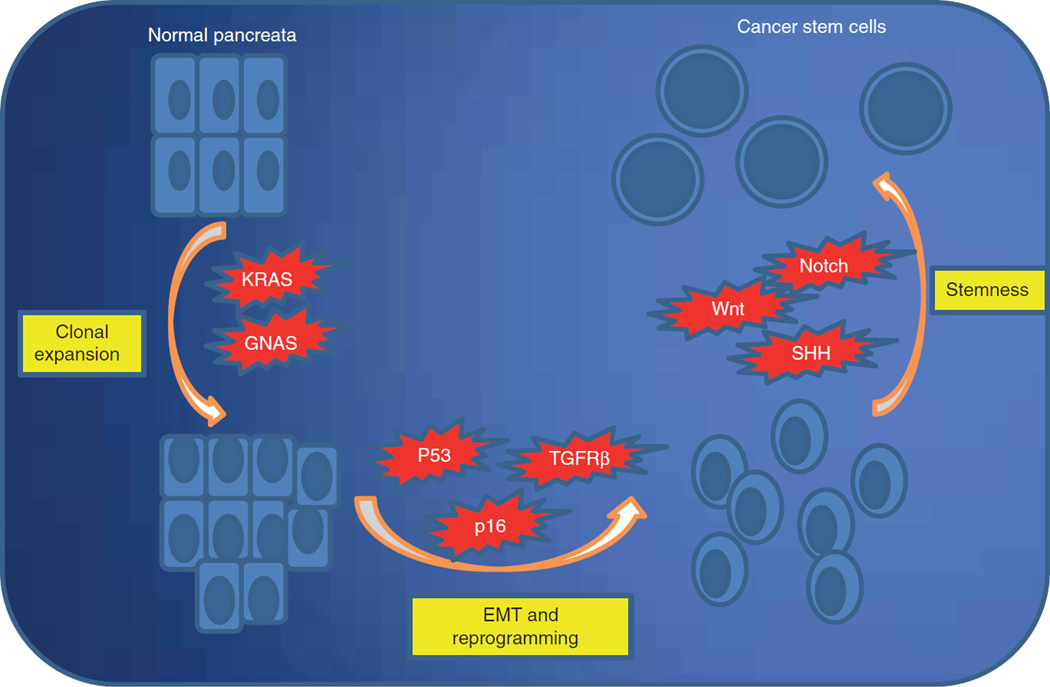
The occurrence of a K-Ras mutation has been shown to be evident at an early phase of pancreatic carcinogenesis [10]. K-Ras mutations can be detected in intraductal precancerous lesions (PanIN’s) suggesting that they confer a survival and growth advantage for mutant clones compared to wild-type ductal cells [11]. More strikingly, studies also suggest that K-Ras takes the initiative of cellular reprogramming processes to induce transition of the acinar cell into a malignant clone [12]. However, there is also strong evidence that in the presence of intact tumor suppressor response, it may induce cellular senescence [13]. For example, activation of oncogenic K-Ras also results in increased expression of p53 and p16, both of which promote premature cell senescence [14], indicating that accumulation of other mutations specifically in tumor suppressor genes are required to for acquisition of the features of a completely transformed cancer cell (Figure 2).
Figure 2. Model progression of pancreatic cancer.
Multistep activation of growth pathways from clonal expansion to gain of stemness properties; model progression of pancreatic cancer.
EMT: Epithelialmesenchymal transition.
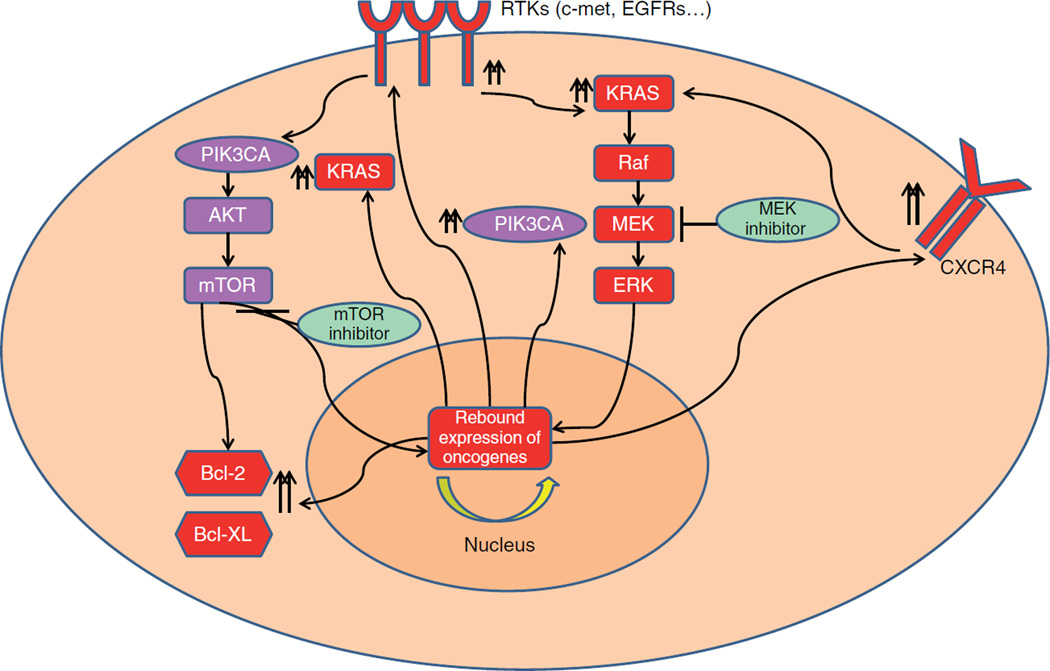
Given that K-Ras has a crucial role in pancreatic carcinogenesis, MEK, which is a downstream signal mediator of K-Ras signaling, was investigated as a therapeutic target. Initial preclinical studies with MEK inhibitors reported inhibition of tumor growth in xenograft models as a promising target for pancreatic cancer treatment [15]. However, clinical trials with MEK inhibitors have been conducted in multiple advanced stage tumor types and results have not been sufficiently promising to warrant further development as single agents [16,17]. Although K-Ras mutation is involved in pancreatic carcinogenesis inhibition of the K-Ras pathway has limited impact on pancreatic cancer cells. For example, a back-to-bench study observed a rebound activation of PIK3CA-mTOR pathway as a compensatory mechanism to MEK inhibition (Figure 3) [18]. Overall, the current evidence suggests that KRAS pathway suppression is overridden either by changing expression level of other prosurvival pathways or by activating other signal transducers that will be further discussed below.
Figure 3. Possible resistance mechanisms.
Rebound activation of PI3KCA and K-Ras pathways along with increased RTKs and anti-apoptotic proteins upon treatment with MEK and m-TOR inhibitors.
ERK: Extracellular signal regulated kinase pathway.
1.1.2 EGFR signaling in pancreatic cancer
The EGFR family (EGFR also known as HER) is a subgroup of receptor tyrosine kinases which have been shown to be active in many epithelial cancers such as colorectal, breast and lung cancers [19]. EGFRs are known to be activators of K-RAS, PIK3CA, Signal Transducer and Activator of Transcription (STATs), and phospholipase C (PLC) [20]. These pathways promote cell growth, proliferation and invasion via modifying gene expression profiles and creating a unique signature for each cancer cell.
Overexpression of EGFR has been identified in tissue studies of pancreatic adenocarcinoma [21]. Simultaneous upregulation of EGF along with EGFR overexpression [22] suggests that there might be a closed circuit in regulation of both receptor and ligand. Moreover clinical data has suggested that upregulated EGFR might be associated with more aggressive tumor behavior [23]. Increased EGFR activity has been also be related to higher rates of disease recurrence following surgery in pancreatic cancer [24]. Therefore, studies to investigate the role of EGFR signaling inhibitors in pancreatic cancers have been conducted.
In an early preclinical study, EGFR receptor blockage with cetuximab, a monoclonal antibody against EGFR, showed promising results and induced tumor regressions in orthotopic models of pancreatic cancer [25]. Additional work identified the potential benefit of the small molecule EGFR inhibitor, erlotinib, and suggested promise for combination with gemcitabine [26]. While initial clinical studies of cetuximab reported a modest benefit, a subsequent Phase III trial did not result in any survival advantage for the addition of cetuximab to gemcitabine (HR = 1.06) [27]. A study of the small tyrosine kinase inhibitor, erlotinib, along with gemcitabine, in 519 patients with locally advanced and metastatic pancreas adenocarcinoma, met statistical significance (HR of 0.82 [95% CI; 0.69 – 0.99]) but showed a borderline clinical benefit with less than one month increased overall survival in the experimental arm (Table 1) [28]. Considering the high rate of K-RAS mutations (> 90%) targeting EGFR alone does not appear to be a promising future strategy for pancreatic cancer treatment.
Table 1.
Clinical trials assessing the role of targeted agents in pancreatic cancer.
| Study | Study design | Targeting agent | Molecular mechanism |
Outcomes |
|---|---|---|---|---|
| Rinehart et al. [16] | Phase II clinical trial in multiple types of advanced cancers |
CI-1040 alone | Oral MEK inhibitor |
No significant clinical benefit (no objective responses) |
| Infante et al. [17] | Phase II randomized clinical trial in advance stage pancreatic cancer |
Trametinib (GSK1120212) combined with gemcitabine |
Oral MEK inhibitor |
No significant clinical benefit (95% CI, 0.67 – 1.44 p = 0.453) |
| Philip et al. [27] | Phase III randomized clinical trial in advanced stage pancreatic cancer |
Cetuximab combined with gemcitabine |
EGFR monoclonal antibody |
No significant clinical benefit (HR; = 1.06; 95% CI, 0.91 – 1.23; p = 0.23) |
| Moore et al. [28] | Phase III randomized clinical trial in advanced stage pancreatic cancer |
Erlotinib combined with gemcitabine |
EGFR small molecule tyrosine kinase inhibitor |
Superior to gemcitabine alone [HR of 0.77 (95% CI 0.64 – 0.92; p = 0.004)] |
| Wolpin et al. [41] | Phase II trial in gemcitabine- resistant pancreatic cancer |
Everolimus | Mammalian target of rapamycin (mTOR) inhibitor |
No complete or partial responses |
| Daud et al. [106] | Phase I trial in multiple advanced stage cancers |
MK-8776 ± gemcitabine |
Chk1 inhibitor | Two PR’s and 13 stable disease in N = 30 |
| Bramhall et al. [125] | Phase III double blind, placebo- controlled randomized trial in pancreatic cancer |
Marimastat combined with gemcitabine |
MMP inhibitor | No survival benefit in experimental arm vs gemcitabine alone (165.5 days vs 164 days p = 0.95) |
| Moore et al. [126] | Phase III randomized clinical trial in advanced stage pancreatic cancer |
Bay12 – 9566 | MMP inhibitor | Gemcitabine superior to Bay12 – 9566 (6.59 months vs 3.74 months p < 0.001) |
| Kim et al. [154] | A single arm pilot clinical trial in advanced stage pancreatic cancer |
Vismodegib combined with gemcitabine |
SHH inhibitor | Progression free survival and OS; 2.8 and 5.3 months respectively |
| Ross et al. (NCT01130142) |
Randomized clinical trial in advanced stage pancreatic cancer |
Saridegib combined with gemcitabine |
SHH inhibitor | Study suspended due to progressive disease in experimental arm |
1.1.3 GNAS and PIK3CA signaling pathways
GNAS is a very complex gene that encodes multiple proteins such as the G protein α-subunit (Gsα) which functions in signal transduction utilizing seven transmembrane receptors [29]. The activated Gsα induces an enzyme called adenyl cyclase which produces cyclic AMP (cAMP). Then, protein kinase A (PKA) which is an important signal transducer for cell growth and proliferation is activated by cAMP [30]. Gsα pathway also can interact and activate other signaling pathways such as Wnt and the Ras-MAPK pathways [31] and boost the growth of the malignant cell clones.
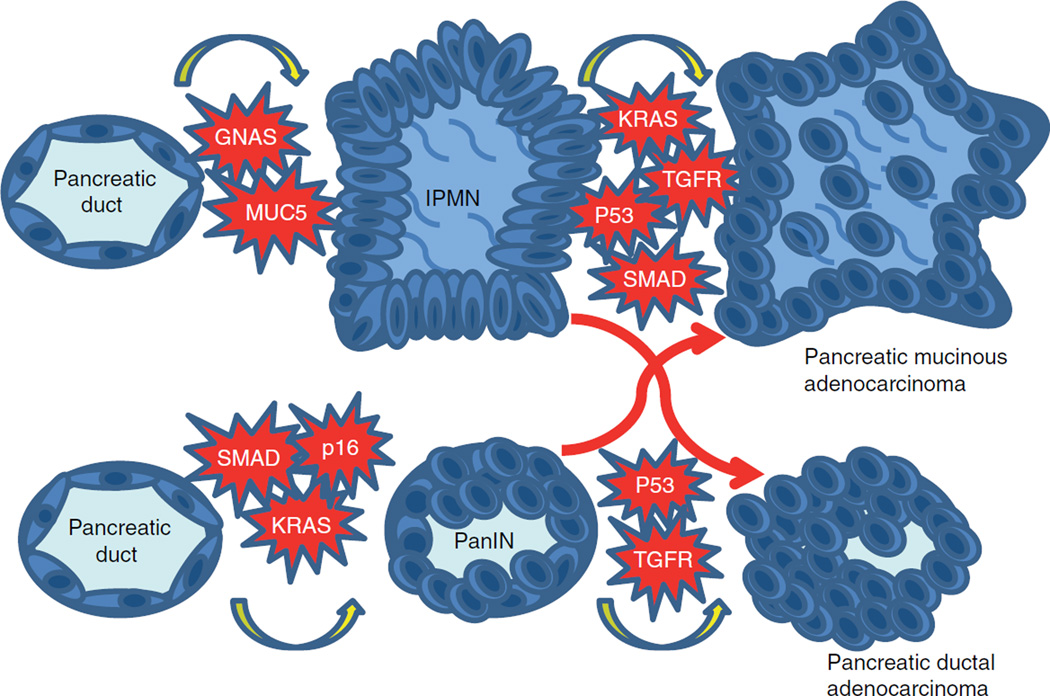
The presence of GNAS mutations in pancreatic cancer development appears to be more significant at an early stage of carcinogenesis and is more often seen in precursor lesions of pancreatic cancer termed intraductal papillary mucinous neoplasms (IPMNs) [32]. Activated GNAS signaling along with a K-RAS mutation induces IPMN, indicating a dual interaction in these two oncogenic pathway [32]. A study reported distinct disease behavior in pancreatic adenocarcinoma derived from IPMN based on their pathological features such as presence of concurrent IPMN indicating a distinct pathway for pancreatic adenocarcinogenesis in the setting of GNAS mutation [33]. Current evidence also indicates relatively frequent GNAS mutations in pancreatic carcinoma derived from IPMN while there was no GNAS mutation in de nova pancreatic adenocarcinoma suggesting a close relation between GNAS mutations and pancreatic carcinogenesis limited to the ones arising from premalignant mucinous lesions [32]. Together these findings suggest that GNAS is a key signaling molecule in pancreatic adenocarcinoma, and perhaps more specifically pertaining to the cancers that arise from IPMNs (Figure 4).
Figure 4. Models of premalignant lesion.
Model for development of IPMN and PanIN. Progression for both is not mutually exclusive and both lesions may progress into pancreatic mucinous adenocarcinoma and pancreatic ductal adenocarcinoma (red arrows).
IPMN: Intraductal papillary mucinous neoplasms; PanIN: Pancreatic intraepithelial lesions.
PIK3CA, another downstream signal mediator of receptor tyrosine kinases, initiates the Akt-mTOR pathway. PIK3CA-activated Akt-mTOR fuels cell growth and proliferation in cancer cells [34]. Mutant PIK3CA stimulates downstream signaling which turns on transcription activity and creates cancer cells that can survive in a nutrition-deprived microenvironment and invade the stroma [35]. PIK3CA-activated Akt-mTOR pathway also inhibits apoptosis by increasing the expression of anti-apoptotic Bcl-2 family proteins [36]. Moreover, overactive PIK3CA signaling abolishes K-Ras-induced senescence in the early phases of carcinogenesis [37] suggesting that PIK3CA-mTOR cascade boost Ras-MAPK mediated cell growth and abrogates activation of rebound growth control mechanisms by suppressing tumor suppressor gene activation.
Studies suggest that activation of PIK3CA occurs during the early phase of pancreatic carcinogenesis and is detectable in IPMN lesions [38]. Given the potential anti-apoptotic effect along with growth stimulus potential, the impact of PIK3CA on pancreatic carcinogenesis might be more significant [35]. For example, an in vitro study suggested successful apoptosis induction upon inhibition of PIK3CA upstream and downstream pathway [39]. A xenograft model of human pancreatic cancer also showed promising tumor growth suppression by mTOR inhibitors [40]. While preclinical studies provided promising results following inhibition of the PIK3CA/Akt/mTOR pathway, clinical trials have not shown a significant benefit with the oral mTOR inhibitors [41], as assessed in a Phase II clinical trial where no partial or complete responses were observed. Another Phase II clinical trial confirmed the lack of activity in that 15 of 16 (93.7%) patients enrolled in the study had progressive disease and one patient was reported as non evaluable [42]. After these disappointing clinical outcomes, studies have been conducted to investigate mechanisms explaining this resistance. A study on pancreatic cancer cells elucidated upregulation of Ras-MAPK pathway upon inhibition of PIK3CA-mTOR signaling (Figure 3) [43]. Another back-to-bench study reported activation of EGFR signaling as a rebound response to mTOR inhibition [44]. Furthermore, preclinical work identified stromal cell-derived factor-1 and the CXCR4 signaling loop as a resistance mechanism to mTOR pathway inhibition [45] suggesting that microenvironment factors may also mediate this resistance (Figure 3). Overall, the lack of response to mTOR inhibition in pancreatic cancer indicates a very dynamic signaling network operating in real time in pancreatic cancer cells that confers genetic plasticity and an ability to bypass or compensate for a defective growth signaling pathway.
1.1.4 Loss of PTEN function and dysregulation of PIK3CA pathway
Phosphotase tensin homolog (PTEN), a negative regulator of the PIK3CA pathway, has been shown to be involved in pancreatic carcinogenesis [46]. Loss of heterozygosity (LOH) in PTEN has been demonstrated in approximately 40% of pancreatic cancers [47]. In an animal model, loss of PTEN induced highly proliferative ductal cells [48] suggesting that PTEN loss may play an important role in early pancreatic carcinogenesis due to loss of a negative control mechanism on PIK3CA [49]. Furthermore, PTEN loss in the setting of a K-RAS mutation further fuels cell growth and enhances K-RAS-related carcinogenesis process [46]. An animal model of PTEN/KRAS mutant cell lines suggested that PTEN mutant tumors may respond to mTOR inhibitors, which was not observed in KRAS/p53 mutant cell lines [50]. However, the clinical utility of a PTEN mutation as a biomarker of PIK3CA pathway inhibitor treatment needs to be further studied.
1.1.5 TP53 mediated cell cycle control and tumor suppression
In a very early study from 1981, Jay et al. [51] discovered a phosphorylated 53 000 dalton protein, p53, in transformed cells while its expression was almost undetectable in resting cells and they hypothesized that p53 might be involved in cell cycle control which was later confirmed by other studies. p53 is capable of inducing expression of another small nuclear protein called p21 that shuts down the cell cycle via inhibition of cyclin dependent kinases (CDKs), more specifically CDK2 and arrests the cell cycle at the G1 phase [52] p53 is also involved in DNA damage response which is turned on by ataxia-telengectesia mutated (ATM) and Breast Cancer susceptibility gene 1 (BRCA1) [53]. The BRCA complex initiates the DNA repair process and if the repair process fails, downstream activation of p53 occurs via check point kinase 2 (CHK2 or CHEK2) which ultimately induces cell cycle arrest [54]. It is believed that cancer cells bearing mutant p53 or loss of both chromosomal copies, bypass this check point, gain mutations in DNA and become more tolerant and resistant to DNA damage-mediated cell cycle arrest [55]. The physiologic function of p53 is not limited to cell cycle control and DNA damage response. If cell cycle arrest occurs successfully, p53 also initiates an apoptotic process by activating redox pathway in mitochondria, generating reactive oxygen species and more importantly, inducing the expression of pro-apoptotic proteins such as Noxa [56]. Studies also suggest that p53-induced apoptosis mediates the cytotoxic effect of anticancer treatments in cancer cells [57]. Mutations that impair p53 signaling result in resistance to chemotherapeutic agents and treatment failure [58]. Furthermore, p53 also suppresses reprogramming processes, which are an important step for cancer stem cell generation via multiple mechanisms such as regulation of transcription factors and DNA damage response [59]. Overall, evidence from preclinical studies suggests that p53 is a very critical decision making protein not only in inhibition of carcinogenesis but also in self-regeneration of tumor cells.
p53 has been identified to be mutated in 40 – 75% of pancreatic adenocarcinoma cases [60]. Germline p53 mutations which result in Li-Fraumeni syndrome, are also associated with an increased risk of pancreatic cancer [61] indicating that inactivation of p53 has an essential role in pancreatic carcinogenesis. Somatic p53 mutations occur at a later stage of tumor development unlike early K-Ras oncogene activation [62]. Given the critical effect of p53-related pathways on pancreatic cancers, reactivation of p53 has been interrogated in preclinical studies. A study on cell lines suggested that the mutant p53 activator, PRIMA-1, accelerated apoptosis and sensitized the tumor cells to chemotherapy [63]. In another study with an animal model treated with nutlin (MDM2 inhibitor, a wild-type p53 activator) analogs also reported significant tumor inhibition in xenografts [64]. However, clinical studies are needed to elucidate the utility of p53 reactivation in pancreatic cancer patients. Collectively, these data point to a strong relationship between development of pancreatic cancer and p53 dysfunction. Moreover, whether resistance to conventional chemotherapy in pancreatic cancer is related to a disabled apoptotic process in the lack of this master tumor suppressor and can be reversed by targeting treatment warrants further investigation.
1.1.6 TGF-β/SMAD signaling pathway
TGF-β signaling is a very complex signaling network and the role of the TGF-β pathway in cellular homeostasis varies by genetic profile of the cancer cell. For example, TGF-β signaling can induce cell differentiation and acts as a tumor suppressor gene in non-malignant clones [65] while it can also promote angiogenesis and epithelial-mesenchymal transition (EMT) in cancer cells [66].
TGF-β functions as a tumor suppressor in multiple ways. For example, once signal transduction is initiated, Mother against decapentaplegic homologs (SMAD) proteins translocate to the nucleus to initiate transcription of target genes that are known to be involved in differentiation [67]. TGF-β signaling is not only involved in cell differentiation, but also has an operational function in cell cycle control as a tumor suppressor. One study demonstrated that TGF-β can inhibit CDK4 and stabilize retinoblastoma (Rb), which inhibits a cell cycle promoting transcription factor, E2K [68]. Furthermore, there are data suggesting that TGF-β can induce p53-independent expression of p21 which is another important cell cycle controlling protein [69]. TGF-β also suppresses expression of oncogenes. For example, myc which is a cell-cycle promoting transcription factor in proliferating cells is downregulated by the TGF-β signaling [70].
However, TGF-β receptor also cross-talks with oncogenic signaling pathways. For example, TGF-β can activate the Ras-MAPK pathway which also further increases the expression of TGF-β along with cell growth [71]. Moreover, both pathways synergistically induce EMT and the absence of any of these pathways can cause failure to gain mesenchymal plasticity in epithelial cells [72]. Snai1, a member of snail family, which is induced by the PIK3CA-Akt-mTOR pathway appears to function as an effector protein TGF-β-related EMT [73].
The data regarding the role of SMAD4 in pancreatic cancer are controversial. A study of surgically resected pancreatic cancers showed an improved overall survival in patients whose tumor expressed SMAD4 [74]. One postmortem study also suggests that loss of SMAD4 expression might be a marker of a more aggressive pancreatic cancer biology phenotype with higher potential for metastatic disease progression [75]. However, a clinical study in locally advanced pancreatic adenocarcinoma showed a correlation between local disease progression with retained SMAD4 expression rather than distant disease [76]. Another study indicated a correlation with inactivating mutations of SMAD4 and worse survival outcomes [77]. Although these studies suggest a biomarker role for SMAD4 for pancreatic cancer patients, the data are discordant. A recent study in surgically resected pancreatic tumors showed no association with disease recurrence and SMAD4 expression status [78]. Moreover, an early study reported an inverse relation between SMAD4 expression and survival outcomes [79]. An ongoing trial in locally advanced pancreas adenocarcinoma in North America, RTOG 1201, will evaluate the role of loss of SMAD4 as a biomarker for a more aggressive tumor biology and will correlate genotype with clinical outcome and the use of radiation therapies that may enhance loco-regional disease control (NCT01921751).
Given that preclinical observations suggest that TGF-β pathway also mediates EMT progress and disease progression, studies have been conducted with TGF-β pathway inhibitors. A study investigated a chimeric protein composed of extracellular domain of TGF-βR II as a decoy TGF receptor and IL2 in an animal model [80]. The authors reported inhibition of tumor generation in mice and via downregulation TGF signaling and activation of innate immune system. A clinical trial is currently investigating an antisense oligodeoxynucleotide of TGF-β2 called trabedersen and results are pending (NCT00844064). More translational and clinical studies are required to identify potential benefit of TGF-β signal inhibition in pancreatic cancer.
1.1.7 CDKN2A and its downstream pathway
CDKN2A gene, also known as p16INK4 and p14ARF, has an exceptional feature which allows encoding of two distinct proteins, p16INK4 and p14ARF, by alternative reading frames. p16INK4 is a major inhibitor of CDK4/CDK6 and Cyclin D1 complex, which promotes a signal for cell cycle progression via inhibiting Rb protein and unleashing the transcription factor E2F [81]. p16INK4 interferes with CDK4/CDK6 and CyclinD1 complex-mediated phosphorylation of Rb and ultimately induces cell cycle arrest [82]. The other product of CDKN2A gene, p14ARF, inhibits the Mdm-2 (Mouse double minute homolog) which is a negative regulator of p53 [83] indicating that the CDKN2A controls the cell cycle via two distinct mechanisms.
The role of CDKN2A has been widely studied in pancreatic cancer. CDKN2A has been shown to be mutated or deleted in more than 50% of pancreatic cancers and very often homozygous allelic loss occurs during late carcinogenesis [84]. There is also evidence suggesting that epigenetic silencing of CDKN2A is also involved in pancreatic carcinogenesis [85] denoting that multiple mechanisms silence CKDN2A during pancreatic cancer development. Moreover, germline mutations of CDKN2A, which cause the familial atypical multiple mole and melanoma syndrome (FAMMM), are related to an increased risk of pancreatic cancer [86]. Along with activation of oncogenes, loss of CDKN2A activity was found to be involved in gain of metastatic potential due to an unleashed EMT process [87].
Given the frequent mutations in check point tumor suppressor genes in pancreas adenocarcinoma, the role of check point regulating agents to reinstate CDKN2A activity have been investigated. A preclinical study showed suppression of pancreatic cancer cells growth in vitro and in vivo after being treated by a CDK4/6 inhibitor [88]. On the other hand, another preclinical study in cell lines reported increased invasiveness of pancreatic cancer cells via upregulation of TGF signaling pathway after programmed death (PD)-0332991 therapy, although it slowed growth of pancreatic cancer cells [89]. A Phase II study of this same targeting agent in breast cancer, Palbociclib, which has now been FDA approved in that disease, remains to have its impact on pancreatic cancer defined [90]. Currently, a clinical trial is recruiting patients to investigate the safety and efficacy of this novel agent in advanced stage solid tumors (NCT01522989).
1.1.8 ATM/BRCA2/Wee1 and DNA damage response pathway
ATM and BRCA2 are two well-known tumor suppressor genes that operate in synchronized fashion to facilitate DNA damage response in human cells. Mutations in the ATM gene cause a syndrome called ataxia telengectasia (name also used for nomenclature of gene) which is a multisystem disorder resulting from DNA damage. Mutations in ATM are also associated with certain cancers such as breast, adrenal and colorectal cancer. A recent genome wide study demonstrated that mutations in the ATM gene are associated with pancreatic adenocarcinoma [91]. Mutations in BRCA2/BRCA1 are also related to an increased risk of pancreatic adenocarcinoma [92,93].
ATM protein kinases are present as dimers in the resting state and upon irradiation autophosphorylation occurs and ATM molecules disassociate and are activated [94]. This activation may occur via direct DNA interaction or chromatin structure change [95]. Once ATM is activated it phosphorylates Chk2 [96] which subsequently inhibits Cdc25, a cell cycle promoter, and activates BRCA1 [97] which is another cell cycle regulator. ATM can also directly phosphorylate p53 [98]. In both pathways, the end point is to induce cell cycle arrest to prevent inheritance of mutated DNA in daughter cells. This defense mechanism prevents accumulations of deleterious mutations in next generation cells and induces apoptosis in selected unhealthy clones. As part of DNA damage response, BRCA2 operates downstream of DNA damage response and is activated by BRCA1/Chk2 signaling [99]. All these elements operate together in a very complex network and the fate of the cell is determined based on outcomes of DNA damage severity and reversibility [55].
Current evidence suggests that ATM and BRCA2 mutations occur at a relatively later stage of pancreatic cancer development [100]. Their exact role in pancreatic carcinogene-sis remains to be elucidated. In vitro studies suggest it might be related to resistance to chemotherapeutics via the lack of DNA damage response [101]. In contrast, tumors in BRCA1/BRCA2 mutations carriers lose the second copy of BRCA2, and cannot repair DNA damage which sensitizes cancer cells to apoptosis termed as synthetic lethality [102]. Growing evidence denotes that platinum-based chemotherapy approaches and poly-ADP ribose polymerase (PARP) inhibitors may change the course of disease in this subgroup of pancreatic adenocarcinoma patients [103].
Given that impaired DNA repair and check point control both sensitizes tumor cells and induce apoptosis in pancreatic cancer cells, the role of check point inhibitors has thus also been investigated. Wee-1, a check point kinase, phosphorylates Cdc2 and induces cell arrest [104]. Preclinical studies have investigated the role of inhibition of Wee-1 to induce apoptosis via generating damage in DNA during the mitotic phase. An animal model showed a fourfold increased tumor response to the combination of MK-1775, a Wee-1 inhibitor, with gemcitabine compared to gemcitabine alone [105], a concept which is now being tested in an early Phase IB clinical trial (NCT02037230). Similarly, radiation and chemotherapy sensitization in pancreatic cancer cells has been reported using other check point inhibitors [106], and this promising preclinical evidence warrants further clinical investigation (Figure 2).
1.1.9 SWI/SNF and MLL gene complex-mediated chromatin modification
Growing evidence implicates that the genetic expression profile of cancer cells is not only determined by signal activated transcription factors but also by modification in chromatin and DNA structural changes [107]. Hypermethylation in tumor suppressor gene promoter regions, which suppress transcription and deacetylation of histone structure which loosens the DNA chromatin structure and increases gene expression in protooncogenes, are key examples of epigenetic modification of gene expression.
Recent global genomic analyses have uncovered the impact of epigenetic modification changes and related genes in pancreatic cancer [108]. In one study, Jones et al. reported mutations in ARID1A, which is a member of SWI/SNF family, in pancreatic cancer [109]. More recently, another genomic analysis study revealed mutations in another member of this family, ARID2 [91]. This family functions in chromatin modification via helicase and ATPase activity and change the expression of target genes [110]. A recent study suggests that ARID1A might be a new tumor suppressor gene, as inactivating mutations lead to an increase in carcinogenesis and re-expresssion in cancer cells harboring the mutant gene and inhibit tumor growth [111]. In the same study, the authors reported that mutation in this gene results in decreased expression of tumor suppressor genes such as p21 and SMADs. A recent study identified that ARID1B, A homolog of ARID1A, is required for the survival of cancer cells bearing mutant ARID1A suggesting targetability of SWI/SNF pathway [112]. On the other hand, a syndrome resulting from SWI/SNF dysfunction called Coffin-Siris Syndrome [113] is not associated with an increased risk of any cancer suggesting that it is by itself not sufficient to activate a carcinogenesis process; however, it might be a secondary compensatory process in cancer cells permitting adjustment in their genetic signature by modulating their chromatin structures.
Similarly, mutations in another chromatin-modifying transcription coactivator, MLL3, have been identified in pancreatic cancer [91]. MLL3 is a member of the MLL (mixed linage leukemia) gene families, which are known tumor suppressors and function in histone methylation [114]. Histone methylation can modify amino acids in the structure of histone bodies, and based on the methylated amino acid and location, it can activate or repress expression of target genes [115]. Studies suggest that MLL is a co-activator of p53 and enhances the expression of p53 target genes [116] which may explain its potential tumor suppressor effect. The exact role of MLL3 in pancreatic cancer development remains to be elucidated. Given high gene expression in cancer cells, one plausible explanation is that epigenetic changes aforementioned in cancer cells advance their genetic plasticity to adapt to the dynamic microenvironment changes such as metabolic distress and metastasis. The impact of targeted inhibition of epigenetic modification on pancreatic adenocarcinoma remains to be investigated.
1.1.10 ADAMs (a disintegrin and metalloproteinase) and MMPs (matrix metalloproteinase)
ADAM is a family of metalloproteinases which is composed of 21 genes. Although 13 of the 21 members function as proteases they also have important roles in cell adhesion and migration [117]. During inflammation their expression is up-regulated and they mediate recruitment of activated inflammatory cells [118]. Studies suggest that ADAM8 and ADAM9 are overexpressed in pancreatic cancer and may facilitate invasion and dissemination of pancreatic cancer cells [119]. Up-regulation of ADAM9 was found to be related to poor tumor differentiation and worse survival outcomes [120]. A preclinical study reported anti-inflammatory macrophage-mediated ADAM8 expression [121] suggesting a close interaction between cancer cells and the tumor environment during the invasion of tumor cells. A recent study in an animal model demonstrated decreased tumor burden and metastatic lesions in mice which were treated with an ADAM8 inhibitor [122] and suggested that ADAM8 could be a druggable target.
MMPs are also important regarding tumor inflammation and are involved in matrix degradation. Given that invasion of tumors requires proteolysis in connective tissue, their role in cancer progression has been also studied extensively. Consistent with their physiologic role, their expression was found to be related to increased invasion of cancer cells in tumor stroma [123]. Therefore, in the late 1990s studies were initiated using MMP inhibitors as therapeutic agents. Given initial promising results in preclinical studies [124], an orally available broad spectrum MMP inhibitor, Marimastat, was evaluated in a randomized clinical trial; however, no benefit was observed when compared to and combined with conventional chemotherapy (p = 0.95) [125]. Another MMP inhibitor, BAY 12 – 9566, was also evaluated in pancreatic cancer in a randomized trial and gemcitabine alone was found to be superior to the MMP inhibitor alone arm with regard to overall survival (p < 0.001) [126]. Collectively, these data indicate that MMPs have limited impact on pancreatic cancer therapeutically. The lack of efficacy of these drugs could be due to multiple pathways involved in matrix degradation such as ADAMs as aforementioned.
1.2 Cancer immune surveillance and immune editing: immune-modulating agents and other novel approaches
The ability of cancer cells to evade immune surveillance has become a major focus of attention in the field of cancer research. Mutations which generate antigenic peptides on cancer cells result in activation of immune system. However, in many cancers, this immune response fails to eradicate the cancer cells and tumor continues to grow in part via immune editing processes. Recent research identified targetable peptides that are responsible for the failure of an immune response such as PD-1 receptor and ligand (PD-L1). PD-L1 antibody has been assessed in early safety studies with notable activity in solid tumors [127] and is currently being investigated in pancreatic cancer in Phase I study (NCT01693562). Other immune modulating agents targeting PD-1, CTLA-4, CXCR4 and indoleamine 2,3-dioxygenase (IDO) inhibitors are also currently under investigation in pancreatic adenocarcinoma for safety and clinical activity in Phase I and Phase II studies (Table 2). A novel approach to augment tumor suppressor gene activity in nucleus via nuclear transport inhibitors has been also investigated. A study of an orthotopic mice model suggested improved antitumor effect of gemcitabine when combined with KPT-330 (selinexor) in pancreatic cancer [128]. Currently, the safety and efficacy profile of this agent are also being examined in Phase I/II studies. The potential role of immune modulating agents in pancreatic cancer treatment clearly warrants further translational and clinical investigation.
Table 2.
Targeted agents currently being investigated.
| Author/NCT # | Study Design |
Agent | Target | Observations |
|---|---|---|---|---|
| Azmi et al. [64] | Animal model |
Nutlin analogs | MDM2 inhibitor to enhance p53 activity |
Decrease tumor burden in xenografts |
| Penafuerte et al. [80] | Animal model |
FIST | Decoy TGF-β receptor and IL-2 activator |
Abrogated tumor growth and inhibited angiogenesis in mice |
| NCT00844064 | Phase I study | Trabedersen | Antisense oligodeoxynucleotide of TGF-β2 |
Final results pending |
| Heilmann et al. [88] | Animal model |
PD-0332991 | CDK4/6 inhibitor | Inhibition of tumor growth |
| NCT01522989 | Phase I study | PD-0332991 | CDK4/6 inhibitor | Final results are pending |
| Rajeshkumar et al. [105] |
Animal model |
MK-1775 | Wee-1 inhibitor | Tumor sensitization to gemcitabine in p53 mutant xenograft model |
| NCT02037230 | Phase I study | MK-1775 | Wee-1 inhibitor | Final results are pending |
| NCT00413686 | Phase I study | AZD7762 | Chk inhibitor | Final results are pending |
| Gurney et al. [142] | Animal model |
OMP-18R5 | Wnt receptor inhibitor | Anti-cancer stem cell effect and inhibition of tumor growth |
| NCT02005315 | Phase I study | OMP-18R5 | Wnt receptor inhibitor | Final results pending |
| NCT01621243 | Phase I/II study |
OMP-59R5 | Notch 2/3 | Final results pending |
| NCT02050178 | Phase I study | OMP-54F28 | Wnt receptor and ligand inhibitor | Final results pending |
| NCT02179970 | Phase I study | Plerixafor | CXCR4 inhibitor | Final results pending |
| NCT02077881 | Phase I/II study |
Indoximod | IDO inhibitor | Final results pending |
| NCT01693562 | Phase I/II study |
MEDI4736 | PD-L1 inhibitor | Final results pending |
| NCT02054806 | Phase I study | Pembrolizumab | PD-1 inhibitor | Final results pending |
| NCT01473940 | Phase I study | Ipilimumab | CTLA-4 inhibitor | Final results pending |
| NCT02301130 | Phase I study | Mogamulizumab | CCR4 antibody | Final results pending |
| Kazim et al. [128] | Animal model |
KPT-330 (selinexor) | Nuclear export inhibitor | Promote nuclear accumulation of tumor suppressors |
| NCT02178436 | Phase I/II | KPT-330 (selinexor) | Nuclear export inhibitor | Final results pending |
IDO: Indoleamine 2,3-dioxygenase; PD: Programmed death.
1.3 Pancreatic cancer stem cells and associated signaling pathways
Tumor heterogeneity in cancer in general has been reported in many preclinical and clinical studies and has led to extensive investigation of this entity. Cancer stem cells are a subgroup of the cancer cell population which have a high potency to generate tumors, resist therapies, and are capable of self-renewal and dissemination to distant organs. A study identified a subclone of pancreatic cancer cells characterized with biomarkers including CD44, CD133, and epithelial-specific antigen (ESA) that exhibit cancer stem cell behaviors [129]. Several studies also evaluated these subgroups of cancer cells in pancreatic cancers and reported many active signaling pathways, more significantly active compared to non-stem cancer cells (Figure 5). Given the aggressive behavior observed in pancreatic cancer, studies have also investigated targeting stem cells in pancreatic adenocarcinoma; the data are summarized below.
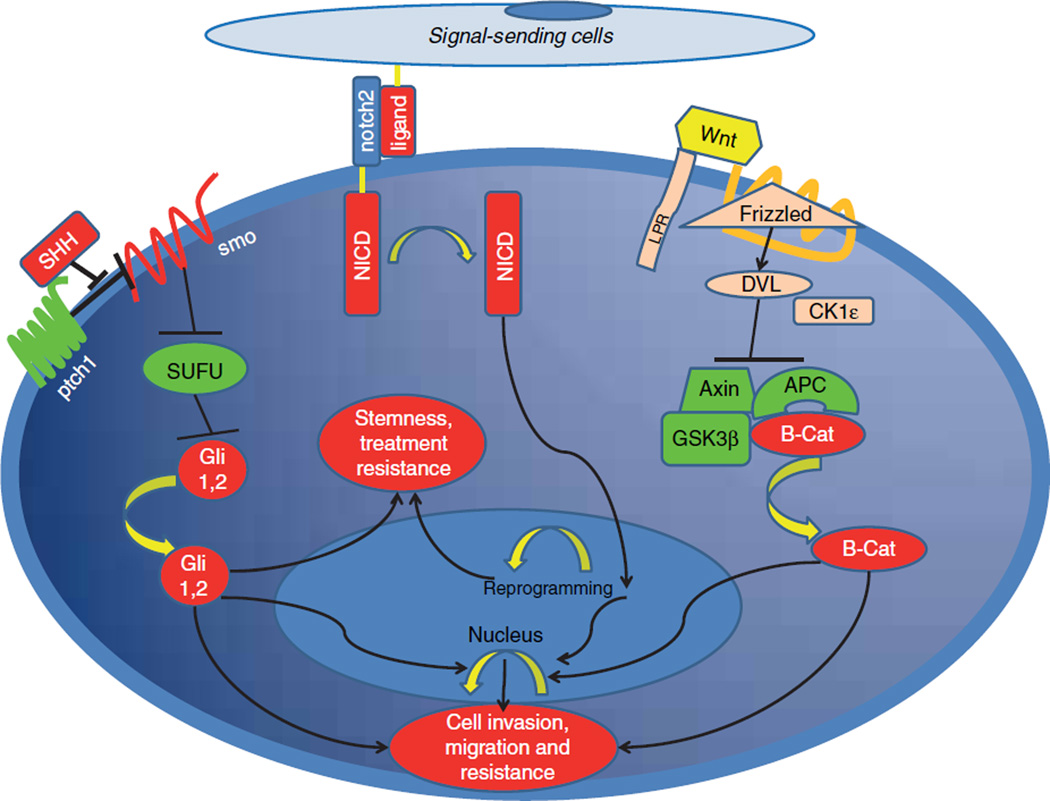
Figure 5. Oncogenic pathways in cancer stem cells.
Activation of Sonic Hedgehog, Wnt and Notch pathways confers stemness properties and induce treatment resistance and metastasis.
1.3.1 Wnt signaling pathway
Aberrant activation of the Wnt pathway has been documented in up to 65% of pancreatic cancer patients [130]. The underlying reason behind the activation of Wnt signaling in pancreatic cancer remains to be elucidated. However, given that the severity of dysplasia in IPMN correlates with up-regulated Wnt signaling, it could be a later event in pancreatic carcinogenesis [131]. Consistent with that, the activity of the Wnt pathway may be more significant in pancreatic cancer stem cells [132] suggesting that Wnt signaling has an accelerating-multi-phase role in exocrine pancreatic cancer development (Figure 2).
Wnt genes have been investigated for more than two decades and their role in embryogenesis and carcinogenesis has been well-documented [133]. Once activation occurs in Wnt signaling, CK1ε and DVL (Dishevelled Segment Polarity protein) dissolve the β-catenin (β-catenin) degradation complex which results in removal of the inhibitory signal on β-catenin. In normal cellular homeostasis, β-catenin is inactivated by an inhibitory complex composed of glycogen storage kinase 3 (GSK3). Adenomatous polyposis coli (APC) and Axin proteins enable degradation of β-catenin via the ubiquitin-protesome pathway [134]. If the degradation process is inhibited by upstream Wnt signaling, β-catenin is stabilized in the cytoplasm and subsequently moves to the nucleus to initiate transcription of certain genes such as c-myc, cyclin D1 and MMP-7 [135]. Multiple mechanisms have been implicated for increased Wnt signaling in pancreatic cancer. For example, one study demonstrated increased hypermethylation in Wnt signal inhibitors genetic locus [136] which results in lack of negative regulation of β-catenin. Another study reported increased expression of Wnt-1 and frizzled receptors along with up-regulated β-catenin in pancreatic adenocarcinoma cells [130]. In the same study, Zeng et al. reported on somatic mutations in β-catenin impairing its interaction with inhibitor protein kinase, GSK3.
The consequences of overactive Wnt signaling pathway have been extensively studied in pancreatic cancer. As stated above, activation of β-catenin results in promotion of cell cycle progress [135] via upregulated expression of target genes such as cyclin D1 and c-myc. An early study reported increased β-catenin activity and nuclear accumulation in solid and pseudopapillary neoplasms suggesting involvement of Wnt pathway in development of diverse pancreatic tumors [137]. A recent study investigated circulating pancreatic cancer cells and concluded that enrichment in Wnt signaling resulted in increased metastasis [138]. Increased β-catenin activity has been also shown to be related to reprogramming processes which suggests a potential role for cancer stem development process [139]. Furthermore, there is strong evidence suggesting that the cell renewal capacity of cancer stem cells might be related to Wnt signaling along with activated notch signaling [140]. Treatment resistance in cancer stem cells has been also attributed to activation of Wnt signaling pathway [141]. However, given that many other signaling pathways which are not observed in non-stem cancer cells operate in cancer stem cells, more evidence is required to establish etiology behind treatment resistance in cancer stem cells.
A monoclonal antibody for the frizzled receptor, OMP-18R5, is under investigation and has shown significant activity against pancreatic cancer cells in animal models along with conventional chemotherapy [142]. Currently, a Phase I study is being conducted for the safety profile and preliminary efficacy evaluation of this agent combined with nab-paclitaxel and gemcitabine (NCT02005315). A fusion protein against both frizzled receptor and Wnt ligand, OMP-54F28, is also in Phase I (NCT02050178).
1.3.2 Sonic hedgehog signaling pathway
Sonic Hedgehog (SHH) was first discovered in the 1980s as a segment polarization gene which plays a very critical role in Drosophila embryogenesis and in which mutation of SHH results in embryonic lethality [143]. The function of this important gene was later investigated in vertebrates and a similar critical functioning of SHH was observed in human organogenesis during embryonic development [144]. Given the crucial role of SHH in embryogenesis mediated by pluripotent stem cells, studies have investigated SHH signaling in cancer stem cells. Two transmembrane receptors have been established for hedgehog signaling: patched and smoothened. In the absence of a polypeptide ligand also called sonic hedgehog, patched receptor activation inhibits smoothened, which is a main signal transducing receptor [145]. However, once the ligand binds to patched receptor, the smoothened receptor is released and activates a transcription factor called Gli1 [144]. Thereafter, Gli1 is stabilized in the cytoplasm, and translocates to the nucleus where it induces transcription of target genes such as Wnt, JAG2, Snail and stem cell markers such as CD44 and CD133 and other genes involved in cell growth and the EMT process [146].
Similar to Wnt signaling, the SHH pathway is also functional at different stages of pancreatic carcinogenesis. One study demonstrated that SHH expression is increased both in early PanINs and pancreatic adenocarcinoma and maintains its activity even at later stages of disease [147]. Compatible with these observations, pancreatic cancer stem cells have been shown to express a 46-fold increased hedgehog pathway genes, in contrast to only fourfold increase in non-stem cancer cells [132] suggesting that there is accelerated activation throughout the carcinogenesis. Current thinking suggests that SHH pathway activations also enhance cancer stem cell maintenance and self renewal [148]. Beyond its effect on cancer cells, SSH has been implicated in tumor and stroma in pancreatic cancer. One study demonstrated up-regulated SHH-mediated desmoplasia and suggested that this might be associated with chemotherapy resistance [149]. Consistently, animal models demonstrate enhanced drug delivery with inhibition of the SHH pathway [150]. A case report of medulloblastoma, a disease with frequent SHH mutations, suggested very promising tumor response to an SHH inhibitor and stimulated clinical development [151]. A Phase I study also indicated potential utility of SHH inhibitors in solid tumors such as basal cell carcinomas [152] and showed substantial activity. However, clinical trials investigating the impact of SHH inhibition in pancreatic cancer have observed quite disparate results compared to the preclinical promise. One clinical trial comparing the efficacy of a SHH pathway inhibitor in combination with chemotherapy was terminated early due to increased mortality in the SHH treatment arm [153]. Another clinical trial studied an SHH inhibitor in combination with gemcitabine showed no significant improvement in the experimental arm was observed compared to gemcitabine alone [154]. Moreover, in the same study, the authors reported no change in pancreatic cancer stem cell populations either although significant changes in desmoplasia were observed. Following these disappointing outcomes, a back-to-bench study in animal models elucidated that stromal desmoplasia was functioning as a restraint rather than as a tumor support [155]. Although currently these agents are being investigated with other treatment modalities (NCT01088815, NCT01130142), this experience form bench to clinic and back to bench suggest that SHH’s role is very complex and that stromal response might as much act a defense mechanism of the organ to prevent dissemination of local disease (similar to Ghon’s complex in tuberculosis). Moreover, pancreatic cancer stem cells may not be addicted to SHH signaling. Although current evidence does not support druggability of the SHH pathway for pancreatic adenocarcinoma, further preclinical and clinical studies will elucidate the role of SHH inhibitors on pancreatic cancer treatment.
1.3.3 SLIT/ROBO and axon guidance pathway
Recent pancreatic cancer whole exome sequencing has highlighted novel genetic alterations that have not been reported previously in pancreatic cancer development: Axon guidance pathway [91]. SLIT/ROBO-mediated axonal guidance pathway alterations have been found to be involved in an important step of development of the CNS that guides axons to cross the midline and which generate a symmetric mirror image and crosstalk between the two brain hemispheres [156]. Following these discoveries, early evidence suggests a potential influence of axonal guidance pathway induction in tumor angiogenesis [157]. However, emerging evidence also implicates the Slit gene, which is a ligand of ROBO receptors, and is frequently silenced in varied cancers and is associated with worse outcomes with increased cancer stem cell activity [158]. In a recent pancreatic genome analysis study [91], higher expression of ROBO3, a negative regulator of ROBO1/2, is associated with worse outcomes, whereas high expression of ROBO2 expression resulted in improved survival in pancreatic cancers indicating a tumor suppressor role for the SLIT/ROBO pathway in pancreatic cancer. The totality of these observations implicate a dual role for axonal guidance pathways in pancreatic carcinogenesis, in that while it augments the angiogenic effect on the tumor microenvironment and endothelial cells, it also suppresses tumor cell growth via interacting/regulating various growth signaling pathways [159]. To enlighten the precise effect of this novel pathway on cancer stem cells and disease progression along with its targetability, further mechanistic studies are required.
1.3.4 Notch signaling
Notch signaling has essential functions in the development and differentiation of cells and tissues in many organs and is activated by direct cellular surface interaction. The role of Notch signaling in cancer development was initially discovered in T cell lymphoblastic leukemia [160]. Further studies have demonstrated that the Notch pathway has a crucial role in many human malignancies such as lung, breast cancer as well as pancreatic cancer [161]. One study suggested that Notch mediates the EMT process in the setting of TGF-a stimulation in pancreata and advances metastases to distant organs [162]. Although current evidence indicates late stage activation of Notch signaling along with other signaling pathways which facilitate metastasis in pancreatic cancer progression [163], there is also evidence denoting a potential role in a reprogramming stage of normal acinar cells into ductal adenocarcinoma in the setting of K-Ras mutations [12], supporting the hypothesis that constitutively activated growth signaling might be a key step for malignant transformation particularly in the lack of tumor suppressor activity. More strikingly, increased notch activity fuels EMT processes that give rise to CD44-positive pancreatic cancer stem cells [164] which bear significant resistance to conventional chemotherapy and are associated with an aggressive tumor behavior [165]. An animal model investigating druggability of the notch pathway demonstrated attenuated invasiveness of pancreatic cancer cells upon suppression of Notch activity via abolishing the nuclear factor-κB signaling [166]. There is also evidence indicating that chemotherapy resistance might be reversed by targeted inhibition of notch pathway [167]. Collectively, accumulating evidence suggests that targeting the notch signaling is a viable therapeutic strategy and clinical trials have been initiated to assess the value of notch and stem cell inhibitors in the clinic [168].
2. Conclusion
Collectively, the molecular pathways aforementioned denote a complex network functioning behind the scene in pancreatic cancer. Addiction of pancreatic cancer cells to oncogenic pathways such Kras-MAPK signaling appears to be limited as multiple prosurvival pathways compensate a specific inhibited pathway. Enhancing the activity of pro-apoptotic pathways via inducing mitosis by targeting agents such as wee-1 or check point inhibitors may abrogate resistance to conventional chemotherapy particularly platinum-based agents given that recent evidence in BRCA mutant patients denotes better treatment outcomes with platinum-based agents. Moreover, down regulation of cancer stem cell associated signaling networks such as Wnt and Notch pathways which have been related to lack of response to standard chemotherapeutic agents, may overcome treatment resistance in pancreatic cancer and change the course of disease. Further genomic and mechanistic studies may advance our understanding of the relation between aggressive disease behavior and the genetic fingerprint of pancreatic cancer and may elucidate further targetable pathways.
3. Expert opinion
Current state-of-the-science outlined here-to-fore indicates a strong relation between genetic alteration and clinical outcome in pancreatic cancer. Genome wide analyses of pancreatic cancers have revealed diverse genetic alterations that modify the gene expression profile of cancer cells and create a unique signature for cancer cell subclones that may determine the fate of the disease. These diverse genetic modifications yield varied subclones with different behaviors that incur one of the biggest challenges in modern cancer therapies, the issue of resistance particularly pertinent in the current precision medicine treatment era. New generation cancer treatments, for example, small molecule tyrosine kinase inhibitors, and monoclonal antibodies targeting certain pathways, suppress the clones that are addicted/dependent on the pathway. However, this initial promising response is generally followed by disease progression due to other clones that can bypass such signaling pathways via their genetic plasticity. These resistant clones grow and ultimately give rise to progressive disease, after an initial honeymoon period with decreased tumor bulk due to impact of targeting agent on sensitive tumor clones.
Another obstacle in cancer therapy relates to the role of cancer stem cells, which are more resistant to therapy due to their genetic plasticity, which advances their capability to generate new clones and develop distant organ metastasis. Studies aforementioned denote a potential relation between cancer stem cells and an aggressive disease behavior. Pathways more active in cancer stem cells such as hedgehog signaling have been studied in clinical trials however the results thus far in pancreatic cancer have been disappointing [154]. Therefore, further translational studies are warranted to reveal the exact role of cancer stem cell related signaling pathways on aggressive nature of pancreatic cancer and reversibility of this challenging situation via modulation of these pathways. Currently, trials are investigating the role of other signal pathway inhibitors functioning in cancer stem cells such as the Notch pathway and results are pending at this time.
A recent genome wide study identified a subgroup of disease, which is more sensitive to chemotherapy due to their genetic signature and instability, for example, homologous repair deficient pancreatic cancers with mutations in BRCA 1 or BRCA2 [169]. These observations show the impact of genetic expression profile of the cancer cells on disease behavior and provide a future insight to test utility of targeting BRCAs via small molecule inhibitors or small interfering RNAs (siRNA) with or without wee-1 inhibitors to advance treatment effect on pancreatic cancer. Therefore, further translational and clinical studies are warranted to evaluate this approach in pancreatic cancer era.
The clinical utility of targeted immunotherapeutic agents are currently being actively investigated in pancreatic cancer. Although initial preclinical and early clinical studies indicate a potential antitumor effect, ongoing trials will clarify the key settings which immune therapy strategies and in what combinations have efficacy in this disease.
Highlights.
Pancreas adenocarcinoma is a disease with a rising incidence and is a disease characterized by a multiplicity of challenges including late diagnosis, relative treatment resistance and genomic complexities.
The article herein evaluates the core genomic pathways that are implicated in both pathogenesis and therapeutic application in this disease.
Particular areas of focus include the RAS pathway, EGFR signaling, homologous repair aberrations, stem cell targeted approaches and the emerging era of immune therapeutics in this disease.
The data regarding the importance of the relevant pathways are summarized and the clinical/translational aspects are reviewed with regard to completed and ongoing clinical trials.
While targeted therapy or a ‘precision medicine’ approach for a given patient with pancreas adenocarcinoma remains an elusive challenge for the majority, there is a real sense of optimism that the strides made in understanding the molecular underpinnings of this disease will translate into improved outcomes.
Footnotes
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Sener SF, Fremgen A, Menck HR, Winchester DP. Pancreatic cancer: a report of treatment and survival trends for 100,313 patients diagnosed from 1985–1995, using the National cancer database. J Am Coll Surg. 1999;189(1):1–7. doi: 10.1016/s1072-7515(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 4.Almoguera C, Shibata D, Forrester K, et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53(4):549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 5.Kyriakis JM, App H, Zhang XF, et al. Raf-1 activates MAP kinase-kinase. Nature. 1992;358(6385):417–421. doi: 10.1038/358417a0. [DOI] [PubMed] [Google Scholar]
- 6.Bonni A, Brunet A, West AE, et al. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and-independent mechanisms. Science. 1999;286(5443):1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Viciana P, Warne PH, Dhand R, et al. Phosphatidylinositol-3-OH kinase direct target of Ras. Nature. 1994;370(6490):527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 8.Shaw RJ, Cantley LC. Ras, PI (3) K and mTOR signalling controls tumour cell growth. Nature. 2006;441(7092):424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 9.Minden A, Lin A, McMahon M, et al. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266(5191):1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 10.Klöppel G. Ki-ras oncogene activation in preinvasive pancreatic cancer. Gastroenterology. 1992;102:230–236. doi: 10.1016/0016-5085(92)91805-e. [DOI] [PubMed] [Google Scholar]
- 11.Tada M, Ohashi M, Shiratori Y, et al. Analysis of K-ras gene mutation in hyperplastic duct cells of the pancreas without pancreatic disease. Gastroenterology. 1996;110(1):227–231. doi: 10.1053/gast.1996.v110.pm8536861. [DOI] [PubMed] [Google Scholar]
- 12.De La OJP, Emerson LL, Goodman JL, et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2008;105(48):18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartkova J, Rezaei N, Liontos M, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 14.Serrano M, Lin AW, McCurrach ME, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 15.Matsui Y, Goto M, Iwakawa M, et al. Modified radiosensitivity of pancreatic cancer xenografts by farnesyl protein transferase inhibitor and MEK inhibitor. Oncol Rep. 2003;10(5):1525–1528. [PubMed] [Google Scholar]
- 16.Rinehart J, Adjei AA, LoRusso PM, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22(22):4456–4462. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 17.Infante JR, Somer BG, Park JO, et al. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer. 2014;50(12):2072–2081. doi: 10.1016/j.ejca.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 18.Pettazzoni P, Viale A, Shah P, et al. Genetic events that limit the efficacy of MEK and RTK inhibitor therapies in a mouse model of KRAS-driven pancreatic cancer. Cancer Res. 2015;75(6):1091–1101. doi: 10.1158/0008-5472.CAN-14-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hynes NE, Stern DF. The biology of erbB-2/nue/HER-2 and its role in cancer. Biochim Biophys Acta. 1994;1198(2):165–184. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 20.Hackel PO, Zwick E, Prenzel N, Ullrich A. Epidermal growth factor receptors: critical mediators of multiple receptor pathways. Curr Opin Cell Biol. 1999;11(2):184–1849. doi: 10.1016/s0955-0674(99)80024-6. [DOI] [PubMed] [Google Scholar]
- 21.Lemoine NR, Hughes CM, Barton CM, et al. The epidermal growth factor receptor in human pancreatic cancer. J Pathol. 1992;166(1):7–12. doi: 10.1002/path.1711660103. [DOI] [PubMed] [Google Scholar]
- 22.Korc MA, Chandrasekar B, Yamanaka Y, et al. Overexpression of the epidermal growth factor receptor in human pancreatic cancer is associated with concomitant increases in the levels of epidermal growth factor and transforming growth factor alpha. J Clin Invest. 1992;90(4):1352. doi: 10.1172/JCI116001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamanaka Y, Friess H, Kobrin M, et al. Coexpression of epidermal growth factor receptor and ligands in human pancreatic cancer is associated with enhanced tumor aggressiveness. Anticancer Res. 1992;13(3):565–569. [PubMed] [Google Scholar]
- 24.Tobita K, Kijima H, Dowaki S, et al. Epidermal growth factor receptor expression in human pancreatic cancer: significance for liver metastasis. Int J Mol Med. 2003;11(3):305–309. [PubMed] [Google Scholar]
- 25.Bruns CJ, Harbison MT, Davis DW, et al. Epidermal growth factor receptor blockade with C225 plus gemcitabine results in regression of human pancreatic carcinoma growing orthotopically in nude mice by antiangiogenic mechanisms. Clin Cancer Res. 2000;6(5):1936–1948. [PubMed] [Google Scholar]
- 26.Ng SS, Tsao MS, Nicklee T, Hedley DW. Effects of the epidermal growth factor receptor inhibitor OSI-774, tarceva, on downstream signaling pathways and apoptosis in human pancreatic adenocarcinoma 1 supported by the National cancer institute of Canada and the pat myhal fund for pancreatic cancer research. Mol Cancer Ther. 2002;1(10):777–783. [PubMed] [Google Scholar]
- 27.Philip PA, Benedetti J, Corless CL, et al. Phase III study comparing gemcitabine plus cetuximab versus gemcitabine in patients with advanced pancreatic adenocarcinoma: Southwest oncology group–directed intergroup trial S0205. J Clin Oncol. 2010;28(22):3605–3610. doi: 10.1200/JCO.2009.25.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National cancer institute of Canada clinical trials group. J Clin Oncol. 2007;25(15):1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 29.Weinstein LS, Liu J, Sakamoto A, et al. Minireview: GNAS: normal and abnormal functions. Endocrinology. 2004;145(12):5459–5464. doi: 10.1210/en.2004-0865. [DOI] [PubMed] [Google Scholar]
- 30.Grieco D, Porcellini A, Avvedimento EV, Gottesman ME. Requirement for cAMP-PKA pathway activation by M phase-promoting factor in the transition from mitosis to interphase. Science. 1996;271(5256):1718–1723. doi: 10.1126/science.271.5256.1718. [DOI] [PubMed] [Google Scholar]
- 31.Wilson C, McIntyre R, Arends M, Adams D. The activating mutation R201C in GNAS promotes intestinal tumourigenesis in ApcMin/+ mice through activation of Wnt and ERK1/2 MAPK pathways. Oncogene. 2010;29(32):4567–4575. doi: 10.1038/onc.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3(92):92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi K, Kanemitsu S, Hatori T, et al. Pancreatic ductal adenocarcinoma derived from IPMN and pancreatic ductal adenocarcinoma concomitant with IPMN. Pancreas. 2011;40(4):571–580. doi: 10.1097/MPA.0b013e318215010c. [DOI] [PubMed] [Google Scholar]
- 34.Osaki M, Oshimura MA, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9(6):667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 35.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7(6):561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nature Rev Cancer. 2002;2(4):277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 37.Kennedy AL, Morton JP, Manoharan I, et al. Activation of the PIK3CA/AKT pathway suppresses senescence induced by an activated RAS oncogene to promote tumorigenesis. Mol Cell. 2011;42(1):36–49. doi: 10.1016/j.molcel.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schönleben F, Qiu W, Ciau NT, et al. PIK3CA mutations in intraductal papillary mucinous neoplasm/carcinoma of the pancreas. Clin Cancer Res. 2006;12(12):3851–3855. doi: 10.1158/1078-0432.CCR-06-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bondar VM, Sweeney-Gotsch B, Andreeff M, et al. Inhibition of the phosphatidylinositol 3’-kinase-AKT pathway induces apoptosis in pancreatic carcinoma cells in vitro and in vivo. Mol Cancer Ther. 2002;1(12):989–997. [PubMed] [Google Scholar]
- 40.Ito D, Fujimoto K, Mori T, et al. In vivo antitumor effect of the mTOR inhibitor CCI-779 and gemcitabine in xenograft models of human pancreatic cancer. Int J Cancer. 2006;118(9):2337–2343. doi: 10.1002/ijc.21532. [DOI] [PubMed] [Google Scholar]
- 41.Wolpin BM, Hezel AF, Abrams T, et al. Oral mTOR inhibitor everolimus in patients with gemcitabine-refractory metastatic pancreatic cancer. J Clin Oncol. 2009;27(2):193–198. doi: 10.1200/JCO.2008.18.9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Javle MM, Shroff RT, Xiong H, et al. Inhibition of the mammalian target of rapamycin (mTOR) in advanced pancreatic cancer: results of two phase II studies. BMC Cancer. 2010;10(1):368. doi: 10.1186/1471-2407-10-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soares HP, Ming M, Mellon M, et al. Dual PI3K/mTOR inhibitors induce rapid over-activation of the MEK/ERK pathway in human pancreatic cancer cells through suppression of mTORC2. Mol Cancer Ther. 2015;14(4):1014–1023. doi: 10.1158/1535-7163.MCT-14-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei F, Zhang Y, Geng L, et al. mTOR inhibition induces EGFR feedback activation in association with its resistance to human pancreatic cancer. Int J Mol Sci. 2015;16(2):3267–3282. doi: 10.3390/ijms16023267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weekes CD, Song D, Arcaroli J, et al. Stromal cell-derived factor 1alpha mediates resistance to mTOR-directed therapy in pancreatic cancer. Neoplasia. 2012;14(8) doi: 10.1593/neo.111810. 690-IN6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill R, Calvopina JH, Kim C, et al. PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer Res. 2010;70(18):7114–7124. doi: 10.1158/0008-5472.CAN-10-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okami K, Wu L, Riggins G, et al. Analysis of PTEN/MMAC1 alterations in aerodigestive tract tumors. Cancer Res. 1998;58(3):509–511. [PubMed] [Google Scholar]
- 48.Stanger BZ, Stiles B, Lauwers GY, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. 2005;8(3):185–195. doi: 10.1016/j.ccr.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 49.Asano T, Yao Y, Zhu J, et al. The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene. 2004;23(53):8571–8580. doi: 10.1038/sj.onc.1207902. [DOI] [PubMed] [Google Scholar]
- 50.Morran DC, Wu J, Jamieson NB, et al. Targeting mTOR dependency in pancreatic cancer. Gut. 2014;63(9):1481–1489. doi: 10.1136/gutjnl-2013-306202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jay G, Khoury G, DeLeo AB, et al. p53 transformation-related protein: detection of an associated phosphotransferase activity. Proc Natl Acad Sci USA. 1981;78(5):2932–2936. doi: 10.1073/pnas.78.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55(22):5187–5190. [PubMed] [Google Scholar]
- 53.Bartek J, Lukas J. Mammalian G1-and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13(6):738–747. doi: 10.1016/s0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 54.Hirao A, Kong YY, Matsuoka S, et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287(5459):1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 55.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432(7015):316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 56.Polyak K, Xia Y, Zweier JL, et al. A model for p53-induced apoptosis. Nature. 1997;389(6648):300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 57.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74(6):957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 58.Lowe SW, Bodis S, McClatchey A, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994:807–807. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 59.Hong H, Takahashi K, Ichisaka T, et al. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature. 2009;460(7259):1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruggeri B, Zhang S, Caamano J, et al. Human pancreatic carcinomas and cell lines reveal frequent and multiple alterations in the p53 and Rb-1 tumor-suppressor genes. Oncogene. 1992;7(8):1503–1511. [PubMed] [Google Scholar]
- 61.Birch JM, Alston RD, McNally R, et al. Relative frequency and morphology of cancers in carriers of germline TP53 mutations. Oncogene. 2001;20(34):4621–4628. doi: 10.1038/sj.onc.1204621. [DOI] [PubMed] [Google Scholar]
- 62.Casey G, Yamanaka Y, Friess H, et al. p53 mutations are common in pancreatic cancer and are absent in chronic pancreatitis. Cancer Lett. 1993;69(3):151–160. doi: 10.1016/0304-3835(93)90168-9. [DOI] [PubMed] [Google Scholar]
- 63.Izetti P, Hautefeuille A, Abujamra AL, et al. PRIMA-1, a mutant p53 reactivator, induces apoptosis and enhances chemotherapeutic cytotoxicity in pancreatic cancer cell lines. Invest New Drugs. 2014;32(5):783–794. doi: 10.1007/s10637-014-0090-9. [DOI] [PubMed] [Google Scholar]
- 64.Azmi AS, Philip PA, Wang Z, et al. Reactivation of p53 by novel MDM2 inhibitors: implications for pancreatic cancer therapy. Curr Cancer Drug Targets. 2010;10(3):319. doi: 10.2174/156800910791190229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Massague J. The TGF-beta family of growth and differentiation factors. Cell. 1987;49(4):437–438. doi: 10.1016/0092-8674(87)90443-0. [DOI] [PubMed] [Google Scholar]
- 66.Ellenrieder V, Hendler SF, Boeck W, et al. Transforming growth factor beta1 treatment leads to an epithelial–mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 2001;61(10):4222–4228. [PubMed] [Google Scholar]
- 67.Nakao A, Imamura T, Souchelnytskyi S, et al. TGF-βeta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16(17):5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ewen ME, Sluss HK, Whitehouse LL, Livingston DM. TGF beta inhibition of Cdk4 synthesis is linked to cell cycle arrest. Cell. 1993;74(6):1009–1020. doi: 10.1016/0092-8674(93)90723-4. [DOI] [PubMed] [Google Scholar]
- 69.Datto MB, Li Y, Panus JF, et al. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA. 1995;92(12):5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Warner BJ, Blain SW, Seoane J, Massague J. Myc downregulation by transforming growth factor beta required for activation of the p15Ink4b G1 arrest pathway. Mol Cell Biol. 1999;19(9):5913–5922. doi: 10.1128/mcb.19.9.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mulder KM. Role of Ras and Mapks in TGFbeta signaling. Cytokine Growth Factor Rev. 2000;11(1):23–35. doi: 10.1016/s1359-6101(99)00026-x. [DOI] [PubMed] [Google Scholar]
- 72.Janda E, Lehmann K, Killisch I, et al. Ras and TGF beta cooperatively regulate epithelial cell plasticity and metastasis dissection of Ras signaling pathways. J Cell Biol. 2002;156(2):299–314. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cho HJ, Baek KE, Saika S, et al. Snail is required for transforming growth factor-beta-induced epithelial–mesenchymal transition by activating PI3 kinase/Akt signal pathway. Biochem Biophys Res Commun. 2007;353(2):337–343. doi: 10.1016/j.bbrc.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 74.Tascilar M, Skinner HG, Rosty C, et al. The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clin Cancer Res. 2001;7(12):4115–4121. [PubMed] [Google Scholar]
- 75.Iacobuzio-Donahue CA, Fu B, Yachida S, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27(11):1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crane CH, Varadhachary GR, Yordy JS, et al. Phase II trial of cetuximab, gemcitabine, and oxaliplatin followed by chemoradiation with cetuximab for locally advanced (T4) pancreatic adenocarcinoma: correlation of Smad4 (Dpc4) immunostaining with pattern of disease progression. J Clin Oncol. 2011;29(22):3037–3043. doi: 10.1200/JCO.2010.33.8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blackford A, Serrano OK, Wolfgang CL, et al. SMAD4 gene mutations are associated with poor prognosis in pancreatic cancer. Clin Cancer Res. 2009;15(14):4674–4679. doi: 10.1158/1078-0432.CCR-09-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winter JM, Tang LH, Klimstra DS, et al. Failure patterns in resected pancreas adenocarcinoma: lack of predicted benefit to SMAD4 expression. Ann Surg. 2013;258(2):331–335. doi: 10.1097/SLA.0b013e31827fe9ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Biankin AV, Morey AL, Lee CS, et al. DPC4/Smad4 expression and outcome in pancreatic ductal adenocarcinoma. J Clin Oncol. 2002;20(23):4531–4542. doi: 10.1200/JCO.2002.12.063. [DOI] [PubMed] [Google Scholar]
- 80.Penafuerte C, Bautista-Lopez N, Bouchentouf M, et al. Novel TGF-beta antagonist inhibits tumor growth and angiogenesis by inducing IL-2 receptor-driven STAT1 activation. J Immunol. 2011;186(12):6933–6944. doi: 10.4049/jimmunol.1003816. [DOI] [PubMed] [Google Scholar]
- 81.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366(6456):704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 82.Liggett W, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J Clin Oncol. 1998;16(3):1197–1206. doi: 10.1200/JCO.1998.16.3.1197. [DOI] [PubMed] [Google Scholar]
- 83.Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17(17):5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caldas C, Hahn SA, da Costa LT, et al. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nature Genet. 1994;8(1):27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 85.Schutte M, Hruban RH, Geradts J, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57(15):3126–3130. [PubMed] [Google Scholar]
- 86.Goldstein AM, Fraser MC, Struewing JP, et al. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16 INK4 mutations. N Engl J Med. 1995;333(15):970–975. doi: 10.1056/NEJM199510123331504. [DOI] [PubMed] [Google Scholar]
- 87.Aguirre AJ, Bardeesy N, Sinha M, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heilmann AM, Perera RM, Ecker V, et al. CDK4/6 and IGF1 receptor inhibitors synergize to suppress the growth of p16INK4A–deficient pancreatic cancers. Cancer Res. 2014;74(14):3947–3958. doi: 10.1158/0008-5472.CAN-13-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu F, Korc M. Cdk4/6 Inhibition induces epithelial–mesenchymal transition and enhances invasiveness in pancreatic cancer cells. Mol Cancer Ther. 2012;11(10):2138–2148. doi: 10.1158/1535-7163.MCT-12-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Finn RS, Crown JP, Lang I, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16(1):25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 91.Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thompson D, Easton DF. Cancer incidence in BRCA1 mutation carriers. J Natl Cancer Inst. 2002;94(18):1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 93.Consortium BCL. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst. 1999;91(15):1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 94.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 95.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nature Rev Cancer. 2003;3(3):155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 96.Falck J, Mailand N, Syljuåsen RG, et al. The ATM–Chk2–Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410(6830):842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 97.Xu B, O’Donnell AH, Kim S-T, Kastan MB. Phosphorylation of serine 1387 in Brca1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer Res. 2002;62(16):4588–4591. [PubMed] [Google Scholar]
- 98.Banin S, Moyal L, Shieh SY, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281(5383):1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 99.Vaughn JP, Cirisano FD, Huper G, et al. Cell cycle control of BRCA2. Cancer Res. 1996;56(20):4590–4594. [PubMed] [Google Scholar]
- 100.Goggins M, Hruban RH, Kern SE. BRCA2 is inactivated late in the development of pancreatic intraepithelial neoplasia: evidence and implications. Am J Pathol. 2000;156(5):1767–1771. doi: 10.1016/S0002-9440(10)65047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nature Rev Cancer. 2012;12(9):587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- 102.Polyak K, Garber J. Targeting the missing links for cancer therapy. Nature Med. 2011;17(3):283–284. doi: 10.1038/nm0311-283. [DOI] [PubMed] [Google Scholar]
- 103.Golan T, Kanji Z, Epelbaum R, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer. 2014;111(6):1132–1138. doi: 10.1038/bjc.2014.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lundgren K, Walworth N, Booher R, et al. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell. 1991;64(6):1111–1122. doi: 10.1016/0092-8674(91)90266-2. [DOI] [PubMed] [Google Scholar]
- 105.Rajeshkumar N, De Oliveira E, Ottenhof N, et al. MK-1775, a potent Wee1 inhibitor, synergizes with gemcitabine to achieve tumor regressions, selectively in p53-deficient pancreatic cancer xenografts. Clin Cancer Res. 2011;17(9):2799–2806. doi: 10.1158/1078-0432.CCR-10-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Daud AI, Ashworth MT, Strosberg J, et al. Phase I dose-escalation trial of checkpoint kinase 1 inhibitor MK-8776 as monotherapy and in combination with gemcitabine in patients with advanced solid tumors. J Clin Oncol. 2015;57:5027. doi: 10.1200/JCO.2014.57.5027. [DOI] [PubMed] [Google Scholar]
- 107.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nature Rev Genet. 2007;8(4):286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 108.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jones S, Li M, Parsons DW, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutation. 2012;33(1):100–103. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends Mol Med. 2007;13(9):373–380. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guan B, Wang TL, Shih IM. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71(21):6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Helming KC, Wang X, Wilson BG, et al. ARID1B is a specific vulnerability in ARID1A–mutant cancers. Nature Med. 2014;20(3):251–254. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Levy P, Baraitser M. Coffin-Siris syndrome. J Med Genet. 1991;28(5):338. doi: 10.1136/jmg.28.5.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ruault M, Brun ME, Ventura M, et al. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene. 2002;284(1):73–81. doi: 10.1016/s0378-1119(02)00392-x. [DOI] [PubMed] [Google Scholar]
- 115.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature Rev Genet. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee J, Kim DH, Lee S, et al. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl Acad Sci. 2009;106(21):8513–8518. doi: 10.1073/pnas.0902873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29(5):258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gómez-Gaviro M, Domínguez-Luis M, Canchado J, et al. Expression and regulation of the metalloproteinase ADAM-8 during human neutrophil pathophysiological activation and its catalytic activity on L-selectin shedding. J Immunol. 2007;178(12):8053–8063. doi: 10.4049/jimmunol.178.12.8053. [DOI] [PubMed] [Google Scholar]
- 119.Valkovskaya N, Kayed H, Felix K, et al. ADAM8 expression is associated with increased invasiveness and reduced patient survival in pancreatic cancer. J Cell Mol Med. 2007;11(5):1162–1174. doi: 10.1111/j.1582-4934.2007.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grützmann R, Lüttges J, Sipos B, et al. ADAM9 expression in pancreatic cancer is associated with tumour type and is a prognostic factor in ductal adenocarcinoma. Br J Cancer. 2004;90(5):1053–1058. doi: 10.1038/sj.bjc.6601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Puolakkainen P, Koski A, Vainionpää S, et al. Anti-inflammatory macrophages activate invasion in pancreatic adenocarcinoma by increasing the MMP9 and ADAM8 expression. Med Oncol. 2014;31(3):1–9. doi: 10.1007/s12032-014-0884-9. [DOI] [PubMed] [Google Scholar]
- 122.Ishikawa N, Daigo Y, Yasui W, et al. ADAM8 as a novel serological and histochemical marker for lung cancer. Clin Cancer Res. 2004;10(24):8363–8370. doi: 10.1158/1078-0432.CCR-04-1436. [DOI] [PubMed] [Google Scholar]
- 123.Ellenrieder V, Alber B, Lacher U, et al. Role of MT-MMPs and MMP-2 in pancreatic cancer progression. Int J Cancer. 2000;85(1):14–20. doi: 10.1002/(sici)1097-0215(20000101)85:1<14::aid-ijc3>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 124.Rasmussen HS, McCann PP. Matrix metalloproteinase inhibition as a novel anticancer strategy: a review with special focus on batimastat and marimastat. Pharmacol Ther. 1997;75(1):69–75. doi: 10.1016/s0163-7258(97)00023-5. [DOI] [PubMed] [Google Scholar]
- 125.Bramhall S, Schulz J, Nemunaitis J, et al. A double-blind placebo-controlled, randomised study comparing gemcitabine and marimastat with gemcitabine and placebo as first line therapy in patients with advanced pancreatic cancer. Br J Cancer. 2002;87(2):161–167. doi: 10.1038/sj.bjc.6600446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moore M, Hamm J, Dancey J, et al. Comparison of gemcitabine versus the matrix metalloproteinase inhibitor BAY 12-9566 in patients with advanced or metastatic adenocarcinoma of the pancreas: a phase III trial of the National cancer institute of Canada clinical trials group. J Clin Oncol. 2003;21(17):3296–3302. doi: 10.1200/JCO.2003.02.098. [DOI] [PubMed] [Google Scholar]
- 127.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti–PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kazim S, Malafa MP, Coppola D, et al. Selective nuclear export inhibitor KPT-330 enhances the antitumor activity of gemcitabine in human pancreatic cancer. Mol Cancer Ther. 2015;14(7):1570–1581. doi: 10.1158/1535-7163.MCT-15-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 130.Zeng G, Germinaro M, Micsenyi A, et al. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. 2006;8(4):279–289. doi: 10.1593/neo.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Al-Aynati MM, Radulovich N, Riddell RH, Tsao MS. Epithelial-cadherin and beta-catenin expression changes in pancreatic intraepithelial neoplasia. Clin Cancer Res. 2004;10(4):1235–1240. doi: 10.1158/1078-0432.ccr-03-0087. [DOI] [PubMed] [Google Scholar]
- 132.Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. J Clin Oncol. 2008;26(17):2806–2812. doi: 10.1200/JCO.2008.16.6702. [DOI] [PubMed] [Google Scholar]
- 133.Nusse R, Varmus HE. Wnt genes. Cell. 1992;69(7):1073–1087. doi: 10.1016/0092-8674(92)90630-u. [DOI] [PubMed] [Google Scholar]
- 134.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li YJ, Wei ZM, Meng YX, Ji XR. Beta-Catenin up-regulates the expression of cyclinD1, c-myc and MMP-7 in human pancreatic cancer: relationships with carcinogenesis and metastasis. World J Gastroenterol. 2005;11:2117–2123. doi: 10.3748/wjg.v11.i14.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Taniguchi H, Yamamoto H, Hirata T, et al. Frequent epigenetic inactivation of Wnt inhibitory factor-1 in human gastrointestinal cancers. Oncogene. 2005;24(53):7946–7952. doi: 10.1038/sj.onc.1208910. [DOI] [PubMed] [Google Scholar]
- 137.Tanaka Y, Kato K, Notohara K, et al. Frequent beta-catenin mutation and cytoplasmic/nuclear accumulation in pancreatic solid-pseudopapillary neoplasm. Cancer Res. 2001;61(23):8401–8404. [PubMed] [Google Scholar]
- 138.Yu M, Ting DT, Stott SL, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487(7408):510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Marson A, Foreman R, Chevalier B, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3(2):132. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434(7035):843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 141.Cui J, Jiang W, Wang S, et al. Role of Wnt/beta-catenin signaling in drug resistance of pancreatic cancer. Curr Pharm Des. 2012;18(17):2464–2471. doi: 10.2174/13816128112092464. [DOI] [PubMed] [Google Scholar]
- 142.Gurney A, Axelrod F, Bond CJ, et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci U S A. 2012;109(29):11717–11722. doi: 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 144.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of Hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 145.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15(6):801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med. 2009;9(7):873–886. doi: 10.2174/156652409789105570. [DOI] [PubMed] [Google Scholar]
- 147.Thayer SP, di Magliano MP, Heiser PW, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425(6960):851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhao C, Chen A, Jamieson CH, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458(7239):776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bailey JM, Swanson BJ, Hamada T, et al. Sonic hedgehog promotes desmoplasia in pancreatic cancer. Clin Cancer Res. 2008;14(19):5995–6004. doi: 10.1158/1078-0432.CCR-08-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rudin CM, Hann CL, Laterra J, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361(12):1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rudin C, Jimeno A, Miller W. A phase I study of IPI-926, a novel hedgehog pathway inhibitor, in patients (pts) with advanced or metastatic solid tumors. J Clin Oncol. 2011;29(1):3014. [Google Scholar]
- 153.Williams R. Discontinued drugs in 2012: oncology drugs. Expert Opin Investig Drugs. 2013;22(12):1627–1644. doi: 10.1517/13543784.2013.847088. [DOI] [PubMed] [Google Scholar]
- 154.Kim EJ, Sahai V, Abel EV, et al. Pilot clinical trial of hedgehog pathway inhibitor GDC-0449 (Vismodegib) in combination with gemcitabine in patients with metastatic pancreatic adenocarcinoma. Clin Cancer Res. 2014;20(23):5937–5945. doi: 10.1158/1078-0432.CCR-14-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rhim AD, Oberstein PE, Thomas DH, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25(6):735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kidd T, Brose K, Mitchell KJ, et al. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 1998;92(2):205–215. doi: 10.1016/s0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- 157.Wang B, Xiao Y, Ding BB, et al. Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell. 2003;4(1):19–29. doi: 10.1016/s1535-6108(03)00164-8. [DOI] [PubMed] [Google Scholar]
- 158.Dallol A, Da Silva NF, Viacava P, et al. SLIT2, a human homologue of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res. 2002;62(20):5874–5880. [PubMed] [Google Scholar]
- 159.Mehlen P, Delloye-Bourgeois C, Chédotal A. Novel roles for Slits and netrins: axon guidance cues as anticancer targets? Nature Rev Cancer. 2011;11(3):188–197. doi: 10.1038/nrc3005. [DOI] [PubMed] [Google Scholar]
- 160.Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649–6461. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 161.Allenspach EJ, Maillard I, Aster JC, Pear W. Notch signaling in cancer. Cancer Biol Ther. 2002;1(5):466–476. doi: 10.4161/cbt.1.5.159. [DOI] [PubMed] [Google Scholar]
- 162.Miyamoto Y, Maitra A, Ghosh B, et al. Notch mediates TGFalpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3(6):565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 163.Bailey JM, Singh PK, Hollingsworth MA. Cancer metastasis facilitated by developmental pathways: sonic hedgehog, notch, and bone morphogenic proteins. J Cell Biochem. 2007;102(4):829–839. doi: 10.1002/jcb.21509. [DOI] [PubMed] [Google Scholar]
- 164.Bao B, Wang Z, Ali S, et al. Notch-1 induces epithelial-mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells. Cancer Lett. 2011;307(1):26–36. doi: 10.1016/j.canlet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 165.Wang Z, Li Y, Kong D, et al. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009;69(6):2400–2407. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang Z, Banerjee S, Li Y, et al. Down-regulation of Notch-1 inhibits invasion by inactivation of nuclear factor-kappaB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res. 2006;66(5):2778–2784. doi: 10.1158/0008-5472.CAN-05-4281. [DOI] [PubMed] [Google Scholar]
- 167.Lee JY, Song SY, Park JY. Notch pathway activation is associated with pancreatic cancer treatment failure. Pancreatology. 2014;14(1):48–53. doi: 10.1016/j.pan.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 168.O’Reilly EM, Smith LS, Bendell JC, et al. Phase Ib of anticancer stem cell antibody OMP-59R5 (anti-Notch2/3) in combination with nab-paclitaxel and gemcitabine (Nab-P plus Gem) in patients (pts) with untreated metastatic pancreatic cancer (mPC) J Clin Oncol. 2014 [Google Scholar]
- 169.Waddell N, Pajic M, Patch A-M, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]