Abstract
In this work, we explore the refactoring of the circuitry of λ phage by engineering a new‐to‐nature regulator that responds to an ad hoc input signal that behaves orthogonal with respect to the host cell. We tailored a chimeric regulator, termed Qλ, between the CI protein of the λ phage and the BzdR repressor from Azoarcus sp. strain CIB that responds to benzoyl‐CoA. When the Qλ was expressed in the appropriate Escherichia coli cells, it was able to reprogram the lytic/lysogenic λ phage decision according to the intracellular production of benzoyl‐CoA. Our results are also an example of how generating new artificial regulators that respond to effectors of choice may be useful to control different cellular processes.
Keywords: Azoarcus, protein engineering, synthetic biology, transcriptional regulation
Introduction
Gene switches and protein devices have wide utility in synthetic biology and multiple elements are needed to construct advanced circuitry or to control organism physiology. The λ phage lytic/lysogenic decision circuit represents a network that has been extensively used for circuit reprogramming (Vohradsky 2001; Atsumi and Little 2004; Little 2010). Most of the studies involved the two antagonistic repressors, CI and Cro (Astromoff and Ptashne 1995; Little 2010). The λCI repressor encoded by the cI gene of λ phage represses the P L and P R promoter, and therefore the λ phage lytic cycle (Stayrook et al. 2008; Hochschild and Lewis 2009). The N‐terminal domain of CI (λNCI; residues 1–92) mediates in DNA‐binding, and is connected by a 37‐residue linker to the C‐terminal domain (λCCI; residues 93–101) that participates in the dimerization of the repressor and also in the interaction between dimers through a protease‐sensitive subdomain (Pabo and Lewis 1982; Astromoff and Ptashne 1995; Bell et al. 2000). The CI repressor is expressed from the P RM promoter and binds cooperatively to two operators, O R1 and O R2, to repress the P R lytic promoter and thus blocking transcription of the cro gene (Reichardt and Kaiser 1971; Hawley and McClure 1982; Michalowski et al. 2004). When Escherichia coli senses an environmental stress signal, such as cell and/or phage DNA damage, for example by ultraviolet irradiation, thymine starvation, or other environmental stress signals, the λ prophage is induced to the lytic cycle by the RecA‐mediated proteolytic cleavage of the CI repressor (Herskowitz 1973; Craig and Roberts 1981). Cro protein, that is essential for λ phage lytic development (Schubert et al. 2013), is expressed from P R and prevents CI expression, presumably by virtue of its high affinity for the O R3 operator, where it can bind to repress P RM by direct CI positive feedback.
The CI repressor can be ascribed to the HTH‐XRE‐family of proteins (SMART release, available on the Web Wide Web at http://smart.embl-heidelberg.de/smart/do_annotation.pl?BLAST=DUMMY&DOMAIN=HTH_XRE). The XRE (xenobiotic response element) family of transcriptional regulators is the second most extended family of regulators in bacteria with an helix‐turn‐helix (HTH) DNA‐binding motif similar to that of the well‐characterized Cro protein of λ phage (Roberts et al. 1977; Sauer et al. 1982; Barragán et al. 2005). Environmental signals that activate XRE‐family proteins are widely variable, and they range from small molecular effectors to large proteins. Although the XRE‐family regulators share a characteristic N‐terminal HTH DNA‐binding domain (Pfam01381), their C‐terminal effector‐binding region is highly variable. It is well known that the N‐terminal and linker domains of CI (λNLCI) lack the motifs involved in dimerization (Di Lallo et al. 1999). Therefore, λNLCI has been used as a genetic tool to study the ability of certain proteins, or protein domains, to dimerize after their fusion with the λNLCI domain, by monitoring the repression of the target P R promoter (Di Lallo et al. 1999). However, as far as we know, none of these chimeric CI regulators was tested for its ability to reprogram the lifestyle of the λ phage.
In this work, we explore the refactoring of the circuitry of λ phage by engineering a new‐to‐nature regulator that responds to an ad hoc input signal that does not interfere with the normal metabolic network of the host cell, that is, it is to some extent orthogonal, and hence avoid unexpected changes in the behavior of the circuit. To design such regulatory protein, we have engineered a chimera between the CI protein and the BzdR protein from bacterium Azoarcus sp. strain CIB. BzdR is a transcriptional regulator involved in the control of the anaerobic catabolism of benzoate that recognizes benzoyl‐CoA as inducer molecule (Barragán et al. 2005). The multidomain organization of BzdR consists of an N‐terminal domain, harboring a HTH fold, connected through a linker sequence to a C‐terminal domain (LCBzdR; residues 91–298) that shows similarity to the E. coli shikimate kinase I (SKI) and recognizes the inducer molecule benzoyl‐CoA alleviating BzdR repression on the target promoter (Barragán et al. 2005; Durante‐Rodríguez et al. 2010, 2013). Here, we tailored an interkingdom chimeric regulator, termed Qλ, factored by fusion of two functional modules, a DNA‐binding domain from the CI repressor of λ phage and an effector‐recognition domain from the bacterial BzdR protein (Fig. 1). The Qλ, when expressed in the appropriate E. coli cells was able to reprogram the lytic/lysogenic λ phage decision according to the intracellular production of the orthogonal input signal benzoyl‐CoA. Our results are also an example of how generating artificial regulators that respond to selected input signals to control different cellular processes.
Figure 1.
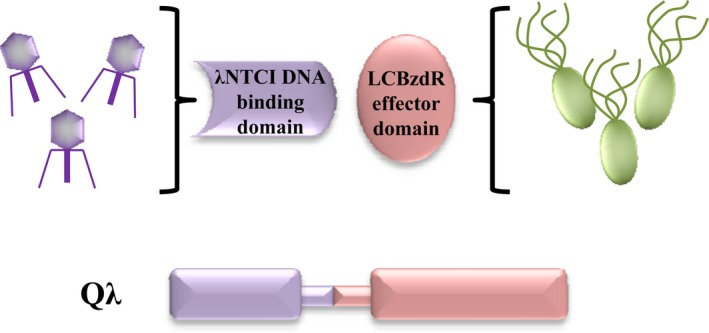
Rationale of the construction of the interkingdom Qλ chimera. Qλ is a chimera factored by fusion of two functional modules, a DNA‐binding domain from the CI repressor of λ phage (λ NLCI) and an effector‐recognition domain from the bacterial BzdR protein (LCBzdR). λ NLCI, N‐terminal and linker domains of CI.
Material and Methods
Bacterial strains, bacteriophages, plasmids, and growth conditions
The E. coli strains, phages, as well as the plasmids used in this work, are listed in Table 1. Escherichia coli cells were grown at 37°C on lysogeny broth (LB) medium (Sambrook and Rusell 2001). Suspension Medium (SM) medium for bacteria and λ phage dilutions were prepared as described by Miller (1972). Where appropriate, antibiotics were added at the following concentrations: ampicillin, 100 μg mL−1; kanamycin 50 μg mL−1; chloramphenicol, 30 μg mL−1.
Table 1.
Bacterial strains, bacteriophages, and plasmids used in this work
| Strain or plasmid | Relevant phenotype and/or genotype | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | endA1 hsdR17 supE44 thi‐1 recA1 gyrA(Nalr) | Sambrook and Rusell (2001) |
| relA1Δ(argF‐lac) U169 depR ø80dlacΔ(lacZ)M15 | ||
| MV1190 | Δ(lac‐pro), thi, supE, Δ(srl‐recA)306::Tn10, (F' traD36) | Bio‐Rad |
| lacI q ZΔM15, proAB | ||
| Bacteriophages | ||
| λ phage | EMBL4 | Frischauf et al. (1983) |
| Plasmids | ||
| pCI‐CAT | Apr, pC132 derivative harboring the cat gene of Tn906 in‐frame with the 5′ portion of λCI | Di Lallo et al. (1999) |
| pNLCI | Apr, pCI‐CAT derivate that express the λNLCI module | This work |
| pQλ | Apr, pNLCI derivative that express the Qλ chimera under control of the lac operator | This work |
| pCK01BzdA | Cmr, pCK01 derivative that express the bzdA gene under the control of the Plac promoter | Barragán et al. (2005) |
| pSJ6 | Kmr, broad‐host‐range plasmid that includes a 4.0 kb | Jaenecke et al. (1996) |
| NotI cassette harboring the fusion P R :tnp'::'lacZ | ||
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Kmr, kanamycin resistant.
Molecular biology techniques
Recombinant DNA techniques were carried out by published methods (Sambrook and Rusell 2001). Plasmid DNA was prepared with High Pure plasmid isolation kit (Roche Applied Science, Penzberg, Germany). DNA fragments were purified with Gene‐Clean Turbo (Qbiogene, Inc., Carlsbad, CA, USA). Oligonucleotides were supplied by Sigma‐Aldrich (Carlsbad, CA, USA). All cloned inserts and DNA fragments were confirmed by DNA sequencing through an ABI Prism 377 automated DNA sequencer (Applied Biosystems Inc., Waltham, MA, USA). Transformation of E. coli cells was carried out by using the RbCl method (Sambrook and Rusell 2001) or by electroporation (Gene Pulser; Bio‐Rad, Hercules, CA, USA).
Plasmids construction
Plasmid pNLCI expressing the λNLCI protein was constructed by deleting the cat gene, encoding chloramphenicol acetyl transferase, from the pCI‐CAT plasmid (Di Lallo et al. 1999). To this end, plasmid pCI‐CAT was HincII/SmaI double digested and further religated. Plasmid pQλ was constructed by cloning into HincII/BamHI double‐digested pNLCI plasmid a 642‐bp StuI/BamHI LCBzdR gene fragment that codes for the linker and the C‐terminal domain of BzdR. LCBzdR was PCR‐amplified from the Azoarcus sp. CIB genome by using oligonucleotides 3‐newlynk (5′‐GAAGGCCTGCGCGAGGAGGCGGAGCAGTC‐3′; an engineered StuI site is underlined) and 5‐C2 (5′‐CGGGATCCTCAGCGTGCCAGGACTTCGAGG‐3′; an engineered BamHI site is underlined).
Enzymatic assays
β‐galactosidase activities were measured using permeabilized cells as described by Miller (1972).
Bacteriophage infection assays
We tested the immunity to λ infection as described by Di Lallo et al. (1999). Bacterial strains were grown in LB to about 1 × 108 cells mL−1, centrifuged and resuspended in SM supplemented with 10 mmol/L MgSO4 and 5 mmol/L CaCl2, and poured with soft agar on LB plates. Spots of several dilutions of λ stock phage were then added, the plates were incubated for 18 h at 37°C, and the number of lysis plaques was determined.
Results and Discussion
Engineering a new‐to‐nature Qλ regulator
With the aim of repurpose an existing regulatory module to make it responsive to metabolic effectors of choice and refactoring the λ lytic/lysogenic decision circuit, we accomplished the construction of a CI‐derived chimera, termed Qλ, made of two functional modules, that is, λNLCI for binding to the P R promoter and LCBzdR for binding to the effector signal (Fig. 2A). As indicated in the Introduction, BzdR is the transcriptional regulator that controls the P N promoter responsible for the anaerobic catabolism of benzoate in Azoarcus sp. CIB. LCBzdR consists of the C‐terminal domain of the BzdR repressor that recognizes specifically benzoyl‐CoA as its effector molecule, and the linker region that was shown to be essential to transfer the conformational changes induced by benzoyl‐CoA to the N‐terminal DNA‐binding domain, thus leading to the release of the repressor from the P N target promoter (Durante‐Rodríguez et al. 2010, 2013). Accordingly, a functional Qλ chimera might be able to repress the λ P R promoter in the absence of benzoyl‐CoA, but should be able to de‐repress this promoter in the presence of benzoyl‐CoA. Thus, the rationale of our strategy was to express a functional Qλ chimera in a recombinant E. coli strain able to synthesize benzoyl‐CoA, and then confirm that after λ infection, the lytic cycle was responding to the new artificial input signal, benzoyl‐CoA. Moreover, since benzoyl‐CoA is not present in natural E. coli strains, it has a minimal impact in the general metabolism of the host cell which, in turn, should not interfere with the effector of the new regulatory circuit, that is, benzoyl‐CoA should behave to some extent orthogonal in respect to the host cell.
Figure 2.
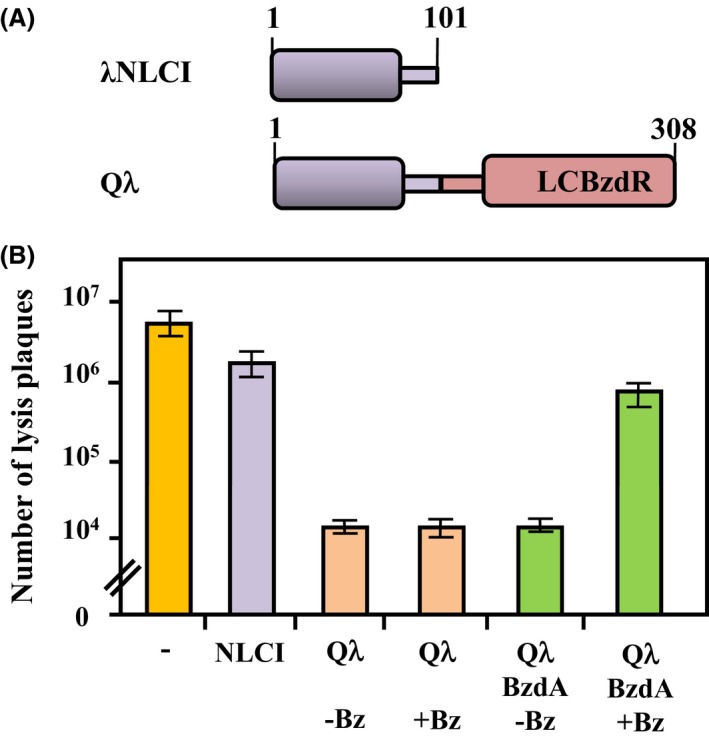
Modular architecture and functionality of the Qλ chimera. (A) Scheme of the modular architecture of the λ NLCI protein and Qλ chimera. The N‐terminal domain and linker region of CI (residues 1–101 from CI protein) are shown in violet. The C‐terminal domain and linker region of BzdR (residues 91–298 from BzdR protein) are shown in red. (B) Escherichia coli MV1190 parental strain (−) or MV1190 strain carrying plasmids pNLCI, expresess λ NLCI (NLCI), pQ λ, expresess Qλ (Qλ), and pQ λ plus pCK01BzdA, expresses BzdA (BzdA), were grown in LB medium in the absence (−Bz) or presence (+Bz) of 3 mmol/L benzoate, and then infected with λ phage as detailed in Material and Methods. Bars represent the average number of lysis plaques observed from three independent infection experiments ±SD. λ NLCI, N‐terminal and linker domains of CI.
The λNLCI protein and the Qλ chimera were constructed and expressed under control of the Plac promoter in plasmids pNLCI and pQλ (Table 1), respectively, as indicated in Material and Methods. To validate the functionality of these proteins, we accomplished an immunity test to λ infection. To this end, we infected the parental E. coli MV1190 strain with the λ phage, obtaining an average of 6.4 × 106 lysis plaques (Fig. 2B). When the recipient bacteria contained plasmid pNLCI expressing the λNLCI protein, no significant variation in the number of the phage plaques was observed (Fig. 2B), indicating that, as expected, λNLCI behaved as a nonfunctional CI repressor. However, when the recipient cells expressed the Qλ chimera, a decrease in the number of lysis plaques of about three orders of magnitude was observed (Fig. 2B). This result indicates that the Qλ chimera confers immunity to λ infection and therefore, suggests that this synthetic regulator is able to efficiently repress the lytic cycle of the λ phage. To check now whether the production of benzoyl‐CoA in the recipient cells could modulate the activity of the Qλ chimera and hence, the level of immunity to λ infection, we constructed an E. coli MV1190 strain harboring plasmid pCK01BzdA (Table 1) that expresses the bzdA gene (Barragán et al. 2005). bzdA encodes the benzoate‐CoA ligase that converts benzoate into benzoyl‐CoA as the first step in the anaerobic benzoate degradation pathway from Azoarcus sp. CIB, and it confers the ability to generate intracellular benzoyl‐CoA when cloned in E. coli cells grown in the presence of benzoate (López‐Barragán et al. 2004). Although the E. coli MV1190 cells expressing the Qλ and BzdA proteins were significantly immune to λ infection when grown in the absence of benzoate, they increased significantly their susceptibility to the lytic effect of λ phage when grown in the presence of benzoate (Fig. 2B). As expected, in the absence of the BzdA protein, benzoate does not cause any effect on the E. coli immunity to λ phage (Fig. 2B). Thus, these results suggest that the Qλ chimera is able to recognize benzoyl‐CoA as effector molecule, leading to the alleviation of the repression on the P R promoter that controls the lytic cycle of λ phage. In summary, all these results have validated that the new‐to‐nature Qλ regulator represents a successful strategy to reprogram the life cycle of λ phage.
The benzoyl‐CoA‐dependent Qλ/P R regulatory system
The tight repression of the P R promoter of λ phage by the CI repressor and its activation in the presence of CI inducers is a very useful trait that has been extensively used to develop conditional gene expression systems that respond to different environmental signals (Meyer et al. 1980). However, most of the CI/P R derived regulatory couples that respond to a chemical signal rely on the transcriptional regulation of the cI gene, or on the native regulatory cascade that leads to the inactivation of the CI repressor. Therefore, it becomes interesting to explore new chemical signals that could be used to modulate directly the activity of the CI regulator within the target host cell. Since benzoyl‐CoA appears to be a metabolite that is not present in most microorganisms, it becomes a nice input signal to develop conditional expression systems that do not interfere, that is, orthogonal, with the general metabolism of the host cell. The results presented above suggested that the Qλ chimera was able to repress the P R promoter involved in the lytic/lysogenic decision of the λ phage, and that the presence of benzoyl‐CoA within the cell alleviated such repression mechanism. To confirm the control of the P R promoter by the Qλ repressor and the benzoyl‐CoA inducer, we monitored the expression of a P R ::lacZ reporter fusion. To this end, E. coli MV1190 strain harboring plasmid pSJ6, which expresses the P R ::lacZ fusion (Table 1), was transformed with plasmids pNLCI or pQλ (Table 1), and the β‐galactosidase production was monitored. Figure 3 shows that cells expressing the λNTCI protein have a P R promoter activity (around 400 Miller units) similar to that of control cells lacking the CI repressor, revealing that, as expected, the λNTCI protein is unable to repress the P R promoter. On the contrary, cells expressing the Qλ chimera exhibited a 10‐fold decrease in the β‐galactosidase activity (Fig. 3), indicating that Qλ was repressing efficiently the P R promoter. When the E. coli cells expressing the Qλ chimera and the BzdA ligase were grown in the absence of benzoate, the P R promoter activity was inhibited (Fig. 3), but a significant activation of P R was observed when increasing concentrations of benzoate was added to the culture medium, reaching β‐galactosidase levels similar to those of the control cells lacking CI at benzoate concentration above 0.5 mmol/L (Fig. 3).
Figure 3.
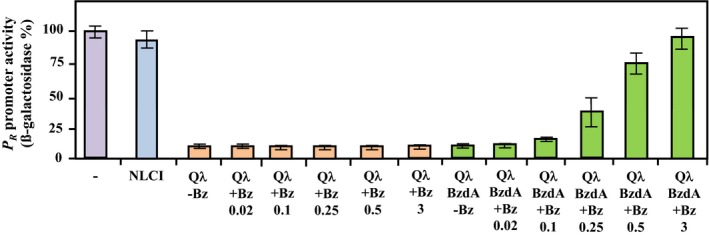
The Qλ chimera controls the PR promoter in a benzoyl‐CoA‐dependent manner. Escherichia coli MV1190 cells carrying plasmid pSJ6, expresses PR ::lacZ, (−), or MV1190 cells carrying plasmid pSJ6 and plasmids pNLCI (NLCI), pQ λ (Qλ), or pQ λ plus pCK01BzdA (BzdA), were grown in LB medium in the absence (−Bz) or presence (+Bz) of 0.02, 0.1, 0.25, 0.5, or 3 mmol/L benzoate. β‐galactosidase activities were measured as described in Material and Methods, and they are represented as a percentage of the activity from E. coli MV1190 (pSJ6) cells (400 Miller units). Graphed values are the average from three independent experiments ±SD. NLCI, N‐terminal, and linker domains of CI.
Analytical centrifugation and electron microscopy studies demonstrated that BzdR is a dimeric protein in solution (Durante‐Rodríguez et al. 2010), but C‐BzdR behaves as a globular monomer (Durante‐Rodríguez et al. 2013). Moreover, we have shown previously that chimeras that include the linker region from BzdR fused to monomeric domains or complete proteins (e.g., shikimate kinase) render functional dimeric proteins (Durante‐Rodríguez et al. 2013). Since we have demonstrated here that Qλ is able to repress the activity of the P R promoter (Fig. 3), and it is known that λNLCI lacks the motifs involved in dimerization and therefore, does not bind to P R (Di Lallo et al. 1999), the linker sequence from LCBzR might be involved in the generation of a functional dimeric Qλ chimera. It was suggested that the LCBzdR domain has a LID‐like region that suffers a significant conformational change in the presence of the benzoyl‐CoA inducer molecule, which would be then transmitted to the NBzdR domain through the linker region. This benzoyl‐CoA‐induced conformational change triggers a structural modification at the DNA‐binding surfaces of the protein without affecting its dimeric state, which would end with the release of the regulator from the target promoter (Durante‐Rodríguez et al. 2010); a similar mechanism of action for the Qλ chimera controlling the P R promoter can be predicted (Fig. 4).
Figure 4.

Model describing the Qλ chimera as a functional device. Comparison of the native and the artificial control of the λ PR promoter by the CI repressor and the Qλ chimera, respectively. The CI repressor binds and represses the PR promoter, and it releases from the DNA after certain environmental stress signals, such as UV radiation, leading to the de‐repression of PR. The Qλ chimera is also able to repress the PR promoter, but de‐repression is triggered by the presence of benzoyl‐CoA, an inducer molecule that behaves orthogonal with respect to the host cell.
All these results allowed us to conclude that the Qλ/P R regulatory couple becomes a new artificial regulatory system that responds to an unusual inducer molecule, benzoyl‐CoA, and it constitutes an interesting alternative to current conditional gene expression systems based on the CI repressor and the P R promoter.
Conclusions
This work shows that the interkingdom chimera Qλ is a functional protein that retains the activities predicted for its two constituent domains, namely recognition and binding to the target P R promoter by the N‐terminal λNLCI domain, and to the inducer molecule benzoyl‐CoA by the C‐terminal LCBzdR domain (Fig. 4). The Qλ chimera was used to refactor the lytic/lysogenic decision circuit of the λ phage using benzoyl‐CoA as the input signal of the engineered circuitry (Fig. 4). In this sense, Qλ device constitutes a molecular gate able to execute an ORN logic operation (Silva‐Rocha and de Lorenzo 2011). This ORN logic gate shows three elements, that is, the transcriptional regulator Qλ, the inducer benzoyl‐CoA and the target promoter P R, and the output is the induction of the lytic cycle except in the case of the presence of Qλ without the inducer benzoyl‐CoA (Fig. 5).
Figure 5.
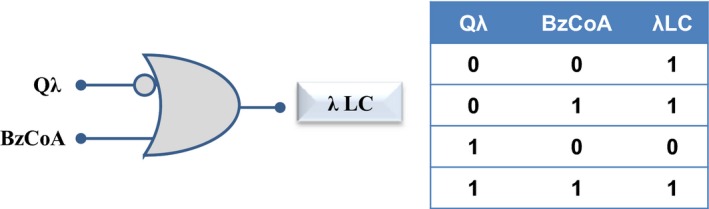
Logic gate and layout of Qλ device activity. At the left is presented the standard graphic annotation of the ORN gate representing the Qλ device activity. At the right, is shown the associated truth table of the ORN gate. BzCoA, benzoyl‐CoA; λ LC, λ lytic cycle.
Modular experimental systems are needed to artificially control biological networks. As demonstrated here with the Qλ chimera, LCBzdR becomes a novel functional module that might be useful in the field of synthetic biology for engineering regulators à la carte that respond to benzoyl‐CoA, an inducer molecule that behaves orthogonal with respect to most microbial cells. Although this work has shown the value of the LCBzdR module by addressing the control of the λ P R promoter, one can foresee the fusion of this module to the DNA‐binding domain of a wide variety of transcriptional regulators that control the activity of promoters driving the expression of genes involved in many different cellular processes. These novel devices designed à la carte would be useful to reprogram crucial cell functions, but also for engineering novel conditional gene expression systems of biotechnological interest that respond to benzoyl‐CoA as the effector molecule.
Conflict of Interest
None declared.
Acknowledgments
This work was supported by the Ministry of Economy and Competitiveness of Spain (grants number BIO2009‐10438, BIO2012‐39501), and the European Union FP7 (grant agreement no. 311815 [SYNPOL project]). The authors also thank to V. de Lorenzo and J. L. García for critical reading of the manuscript and inspiring discussions, A. Valencia for the technical assistance, and Secugen S. L. for help with DNA sequencing.
MicrobiologyOpen 2016; 5(4): 575–581
References
- Astromoff, A. , and Ptashne M.. 1995. A variant of λ repressor with an altered pattern of cooperative binding to DNA sites. Proc. Natl. Acad. Sci. USA 92:8110–8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi, S. , and Little J. W.. 2004. Regulatory circuit design and evolution using phage λ . Genes Dev. 18:2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragán, M. J. , Blazquez B., Zamarro M. T., Mancheño J. M., García J. L., Díaz E., et al. 2005. BzdR, a repressor that controls the anaerobic catabolism of benzoate in Azoarcus sp. CIB, is the first member of a new subfamily of transcriptional regulators. J. Biol. Chem. 280:10683–10694. [DOI] [PubMed] [Google Scholar]
- Bell, C. E. , Frescura P., Hochschild A., and Lewis M.. 2000. Crystal structure of the λ repressor C‐terminal domain provides a model for cooperative operator binding. Cell 101:801–811. [DOI] [PubMed] [Google Scholar]
- Craig, N. L. , and Roberts J. W.. 1981. Function of nucleoside triphosphate and polynucleotide in Escherichia coli recA protein‐directed cleavage of phage lambda repressor. J. Biol. Chem. 256:8039–8044. [PubMed] [Google Scholar]
- Di Lallo, G. , Ghelardini P., and Paolozzi L.. 1999. Two‐hybrid assay: construction of an Escherichia coli system to quantify homodimerization ability in vivo. Microbiology 145:1485–1490. [DOI] [PubMed] [Google Scholar]
- Durante‐Rodríguez, G. , Valderrama J. A., Mancheño J. M., et al. 2010. Biochemical characterization of the transcriptional regulator BzdR from Azoarcus sp. CIB. J. Biol. Chem. 285:35694–35705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante‐Rodríguez, G. , Mancheño J. M., Rivas G., et al. 2013. Identification of a missing link in the evolution of an enzyme into a transcriptional regulator. PLoS One 8:e57518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf, A. M. , Lehrach H., Poustka A., and Murray N.. 1983. λ replacement vectors carrying polylinker sequences. J. Mol. Biol. 170:827–842. [DOI] [PubMed] [Google Scholar]
- Hawley, D. K. , and McClure W. R.. 1982. Mechanism of activation of transcription initiation from the lambda P RM promoter. J. Mol. Biol. 157:493–525. [DOI] [PubMed] [Google Scholar]
- Herskowitz, I. 1973. Control of gene expression in bacteriophage lambda. Annu. Rev. Genet. 7:289–324. [DOI] [PubMed] [Google Scholar]
- Hochschild, A. , and Lewis M.. 2009. The bacteriophage lambda CI protein finds an asymmetric solution. Curr. Opin. Struct. Biol. 19:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenecke, S. , de Lorenzo V., Timmis K. N., and Díaz E.. 1996. A stringently controlled expression system for analysing lateral gene transfer between bacteria. Mol. Microbiol. 21:293–300. [DOI] [PubMed] [Google Scholar]
- Little, J. W. 2010. Evolution of complex gene regulatory circuits by addition of refinements. Curr. Biol. 20:R724–R734. [DOI] [PubMed] [Google Scholar]
- López‐Barragán, M. J. , Carmona M., Zamarro M. T., et al. 2004. The bzd gene cluster, coding for anaerobic benzoate catabolism, in Azoarcus sp. strain CIB. J. Bacteriol. 186:5762–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, B. J. , Maurer R., and Ptashne M.. 1980. Gene regulation at the right operator (O R) of bacteriophage lambda. II. O R1, O R2, and O R3: their roles in mediating the effects of repressor and cro. J. Mol. Biol. 139:163–194. [DOI] [PubMed] [Google Scholar]
- Michalowski, C. B. , Short M. D., and Little J. W.. 2004. Sequence tolerance of the phage lambda P RM promoter: implications for evolution of gene regulatory circuitry. J. Bacteriol. 186:7988–7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- Pabo, C. O. , and Lewis M.. 1982. The operator‐binding domain of λ repressor: structure and DNA recognition. Nature 298:443–447. [DOI] [PubMed] [Google Scholar]
- Reichardt, L. , and Kaiser A. D.. 1971. Control of lambda repressor synthesis. Proc. Natl. Acad. Sci. USA 68:2185–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, T. M. , Shimatake H., Brady C., and Rosenberg M.. 1977. Sequence of Cro gene of bacteriophage lambda. Nature 270:274–275. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , and Rusell D. W.. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- Sauer, R. T. , Yocum R. R., Doolittle R. F., Lewis M., and Pabo C. O.. 1982. Homology among DNA‐binding proteins suggests use of a conserved super‐secondary structure. Nature 298:447–451. [DOI] [PubMed] [Google Scholar]
- Schubert, R. A. , Dodd I. B., Egan J. B., and Shearwin K. E.. 2013. Cro's role in the CI‐Cro bistable switch is critical for λ's transition from lyogeny to lytic development. Genes Dev. 21:2461–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva‐Rocha, R. , and de Lorenzo V.. 2011. Implementing an OR–NOT (ORN) logic gate with components of the SOS regulatory network of Escherichia coli . Mol. Biosyst. 7:2389–2396. [DOI] [PubMed] [Google Scholar]
- Stayrook, S. , Jaru‐Ampornpan P., Ni J., Hochschild A., and Lewis M.. 2008. Crystal structure of the lambda repressor and a model for pairwise cooperative operator binding. Nature 452:1022–1025. [DOI] [PubMed] [Google Scholar]
- Vohradsky, J. 2001. Neural model of the genetic network. J. Biol. Chem. 276:36168–36173. [DOI] [PubMed] [Google Scholar]


