Abstract
Cry1Ac toxin‐binding proteins from Helicoverpa armigera brush border membrane vesicles were identified by an improved pull‐down method that involves coupling Cry1Ac to CNBr agarose combined with liquid chromatography–tandem mass spectrometry (LC‐MS/MS). According to the LC‐MS/MS results, Cry1Ac toxin could bind to six classes of aminopeptidase‐N, alkaline phosphatase, cadherin‐like protein, ATP‐binding cassette transporter subfamily C protein (ABCC2), actin, ATPase, polycalin, and some other proteins not previously characterized as Cry toxin‐binding molecules such as dipeptidyl peptidase or carboxyl/choline esterase and some serine proteases. This is the first report that suggests the direct binding of Cry1Ac toxin to ABCC2 in H. armigera.
Keywords: ABCC2, alkaline phosphatase, aminopeptidase‐N, Bacillus thuringiensis, cadherin, Cry1Ac, Helicoverpa armigera.
Introduction
Bacillus thuringiensis is one of the most widely used biopesticides because of its specific insecticidal activity against target insects and its safety for humans and for the environment (Ibrahim et al. 2010; Pardo‐Lopez et al. 2013). Most of the Cry toxins are produced during the sporulation phase of bacterial growth (Bravo et al. 2007). In a complex multistep process including solubilization, activation, binding to different insect gut proteins, membrane insertion, and pore formation, Cry toxins kill the insects by disrupting midgut cells (Bravo et al. 2004; Pigott and Ellar 2007; Pacheco et al. 2009).
Helicoverpa armigera is a serious agricultural pest globally, it is one of the most damaging cotton pests in China and increased frequency of resistance to Cry1Ac cotton has been reported from field populations in northern China (Zhang et al. 2011). Thus, actions to understand the mechanism of action and resistance mechanism of Cry1Ac toxin in this pest are necessary.
Some proteins from insect midgut cells have been shown to bind Cry1A toxins, including cadherin (CAD) (Vadlamudi et al. 1993; Hua et al. 2014), aminopeptidase‐N (APN) (Nakanishi et al. 2002; Rajagopal et al. 2003; Bravo et al. 2004; Chen et al. 2009), alkaline phosphatase (ALP) (Jurat‐Fuentes and Adang 2004; Fernandez et al. 2006; Soberón et al. 2009; Arenas et al. 2010; Upadhyay and Singh 2011; Zuniga‐Navarrete et al. 2013), and actin among others (McNall and Adang 2003; Krishnamoorthy et al. 2007; Bayyareddy et al. 2009). Different strategies were used to identify these proteins as Cry1A‐binding proteins, for instance, several APN molecules have been shown to bind with different Cry toxins by using pull‐down Cry1A‐affinity chromatography assays followed by elution with different buffers: APN1 from Heliothis virescens was eluted with N‐acetyl‐galactosamine (GalNAc) containing buffer as Cry1Ac binding to APN1 relies on GalNac moieties (Luo et al. 1997); in the case of APN2 from Manduca sexta, it was eluted using a high‐pH carbonate buffer (Denolf et al. 1997), while APN4 from H. virescens was eluted with 2.0 mol/L NaSCN buffer (Banks et al. 2001). In this study, we used an improved pull‐down method that in contrast with the previously reported pull‐down assays does not require the elution step. Liquid chromatography–tandem mass spectrometry (LC‐MS/MS) was used for the identification of native proteins from microvilli membrane of midgut cells isolated from H. armigera that bind to Cry1Ac trypsin‐activated toxin. Here we show that the method used to pull down Cry1Ac‐binding proteins allowed the identification of a wide range of insect gut proteins. Some of these proteins had been previously identified by the different methods reported earlier. However, the pull‐down strategy presented here identified several other molecules that had not been characterized as Cry1Ac‐binding molecules. Among these, an ABCC2 transporter, previously showed to be involved in Cry1A action since resistance in different lepidopteran species was linked to ABCC2 mutations (Gahan et al. 2010; Baxter et al. 2011; Atsumi et al. 2012; Xiao et al. 2014), was identified as a Cry1Ac‐binding protein. The role of ABCC2 transporter in the mechanism of action of Cry1A toxin is not known, but it has been proposed that it is a receptor protein involved in facilitating insertion of Cry1A oligomers into the membrane (Heckel 2012). Nevertheless, a direct interaction of Cry1A with ABCC2 from insect gut was not demonstrated until now. Revealing the role of ABCC2 in Cry1Ac mode of action is important for the future improvement of these toxins and for developing tools to counter insect resistance.
Experimental Procedures
Production of Cry1Ac toxin
The HD73 strain producing Cry1Ac toxin was grown in 1/2 LB medium (0.5% NaCl, 0.5% tryptone, and 0.25% yeast extract, w/v) at 220 rpm and 30°C as described previously (Zhou et al. 2015). Cells were grown until complete sporulation observed under optical microscope. The pellet, including crystals, spores, and debris, was collected by centrifugation at 12,000g for 20 min at 4°C, and washed once with 1 mol/L NaCl and then with distilled water. The crystals were dissolved and digested simultaneously in 50 mmol/L Na2CO3, 3% beta‐mercaptoethanol (v/v) pH 10.0, at 37°C for 2 h in the presence of trypsin at 20:1 mass ratio (protein: trypsin). The activated toxin was purified by size‐exclusion chromatography (Superdex 75 10/300 GL, AKTA Avant, GE Healthcare, Uppsala, Sweden) using 20 mmol/L Na2CO3/NaHCO3 (pH 9.5) buffer at 1 mL/min flow rate. The Cry1Ac protoxin, and the purified toxin was analyzed by SDS‐PAGE (8% acrylamide), and gel was stained by Coomassie blue.
Preparation of brush border membrane vesicles (BBMV) from H. armigera larvae
Helicoverpa armigera larvae were kindly provided by Cotton Insect Pests Laboratory (Institute of Plant Protection, Chinese Academy of Agricultural Sciences) and reared with an artificial diet (Liang et al. 1999). The 50% lethal concentration of Cry1Ac against H. armigera was 7.15 μg/g (95% confidence interval: 2.22–17.98 μg/g) as we reported previously (Xue et al. 2008). About 1000 fifth instar larvae were used for BBMV preparation. The larvae were chilled on ice for 10 min before dissection. The midgut tissue was dissected from the larvae, separated from the hindgut and fat body, a longitudinal slit was done in the tissue to remove food bolus, and it was finally washed in phosphate‐buffered saline buffer (PBS buffer: 137 mmol/L NaCl, 2.7 mmol/L KCl, 4.3 mmol/L Na2HPO4, 1.4 mmol/L KH2PO4, pH 7.4). After transfer to Eppendorf tubes, midgut tissue was quickly frozen in liquid nitrogen and stored at −70°C until use. BBMV were prepared according to the method described by Wolfersberger et al. (1987). Purified BBMV (200 μg) were solubilized in 200 μL 1% 3‐[(3‐cholamidopropyl)‐dimethylammonio]‐1‐propanesulfonate (CHAPS)‐containing buffer (150 mmol/L NaCl, 5 mmol/L EGTA, 20 mmol/L Tris, 1% CHAPS, w/v, pH 7.5) on ice for 30 min. After centrifugation at 13,000g for 5 min at 4°C, the soluble BBMV protein was quantified in using the Bradford method with bovine serum albumin (BSA) as a standard.
Pull‐down assay
Purified Cry1Ac toxin was coupled with CNBr agarose (GE Healthcare), according to the manufacturer's instructions. One mg of purified Cry1Ac toxin was incubated with 500 μL CNBr agarose in 0.2 mol/L NaHCO3 buffer (pH 8.3) at room temperature for 6 h. The noncoupled free CNBr was blocked by incubation with 0.1 mol/L glycine in 0.2 mol/L NaHCO3 buffer (pH 8.3) at room temperature for another 6 h. The noncoupled Cry1Ac protein was removed by 10 washes with 500 μL PBS as described by the manufacturer. Finally, the coupled CNBr‐Cry1Ac agarose was stored in 20% ethanol (v/v) at 4°C.
We have previously described the pull‐down assay (Shu et al. 2015), which was improved to identify the Cry8Ea‐binding protein on BBMV from Holotrichia parallela. Briefly, after incubation of 50 μL CNBr‐Cry1Ac agarose with 100 μg solubilized BBMV proteins for 2 h at 4°C, the unbound BBMV proteins were removed by a fast centrifugation at 400g for 30 sec at 4°C. To avoid losing low abundant proteins and proteins with low affinity, the unbound proteins were recovered in the supernatant after first incubation with CNBr‐Cry1Ac and agarose, and were incubated again with additional 50 μL CNBr‐Cry1Ac agarose at 4°C for 2 h. The unbound proteins were removed by centrifugation for 30 sec at 400g at 4°C. The CNBr‐Cry1Ac agarose containing the bound proteins was washed five times with 500 μL PBS supplemented with 1 mol/L NaCl, followed by five washes with 500 μL PBS to remove all unbound proteins. The proteins that remained bound to the CNBr‐Cry1Ac agarose were considered as Cry1Ac‐binding proteins and were dissociated from the agarose by boiling for 10 min in 50 μL SDS‐PAGE loading buffer (100 mmol/L Tris‐Cl, 200 mmol/L DTT, 4% SDS w/v, 0.2% bromophenol blue w/v, 20% glycerol v/v, pH 6.8). The supernatant was separated by SDS‐PAGE (discontinuous acrylamide gradients from 8% to 12% acrylamide). As negative control of this experiment, the activated CNBr agarose was coupled with 0.1 mol/L Tris‐HCl amino methane buffer (pH 8.5) without Cry1Ac protein, blocked as described earlier, and used for incubation with BBMV.
Identification of binding proteins
After stained by Coomassie blue, the SDS‐PAGE gels were divided and cut into seven parts that were sent to Huada Protein Research Center (HPRC) for LC‐MS/MS analysis. MS/MS data were searched by HPRC against ncbi_insecta_201401 201401 (1,786,564 sequences; 642,693,167 residues) database using the Mascot search engine (Matrix Science, London, UK). Search parameters are provided by HPRC as follows: type of search – MS/MS ion search; enzyme – trypsin; fixed modifications – carboxymethyl (C); variable modifications – Gln>pyro‐Glu (N‐term Q) and oxidation (M); mass values – monoisotopic; protein mass – unrestricted; peptide mass tolerance – ±15 ppm; fragment mass tolerance – ±20 mmu; max missed cleavages – 1; instrument type – default; number of queries – 9983. Peptide identifications were accepted if they could be established at greater than 50% probability as specified by the PeptideProphet algorithm (Keller et al. 2002). Protein identifications were accepted if they could be established at greater than 90% probability and contained at least two identified peptides. Protein probabilities were assigned by the ProteinProphet algorithm (Nesvizhskii et al. 2003).
Results
Preparation of Cry1Ac toxin
As shown in Figure 1, Cry1Ac protoxin present in the crystal/spore mixture (lane 1) was solubilized and digested by trypsin to produce the purified activated toxin (lane 2). After purification by Superdex 75, the activated toxin (60 kDa, lane 3) was coupled with CNBr agarose as described in Experimental Procedures.
Figure 1.
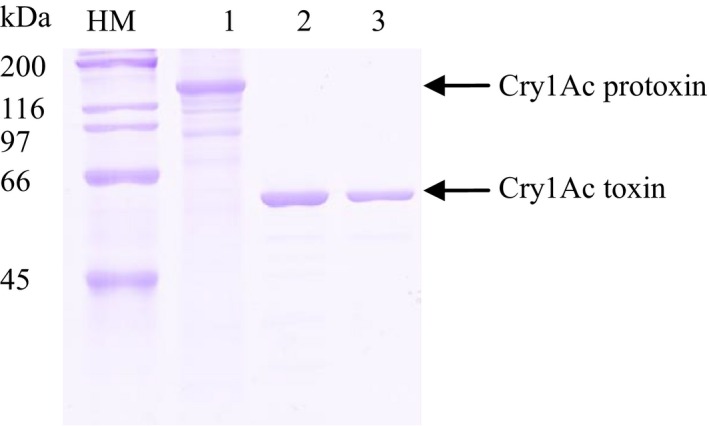
SDS‐PAGE analysis of Cry1Ac (8% acrylamide). HM: high‐range marker; 1: Cry1Ac protoxin from HD73 strain; 2: Cry1Ac toxin activated by trypsin; 3: Cry1Ac toxin purified by gel filtration chromatography.
Detection of Cry1Ac‐binding proteins in H. armigera BBMV
To analyze the Cry1Ac‐binding proteins present in BBMV from H. armigera, BBMV were prepared from fifth instar larvae and solubilized as described in Experimental Procedures. The solubilized BBMV proteins (lane 1 in Fig. 2) were incubated with CNBr‐Cry1Ac‐coupled agarose, and the unbound proteins (lane 2 in Fig. 2) were removed by centrifugation. In order to avoid losing low‐affinity binding proteins or proteins that were less abundant, the unbound proteins (lane 2 in Fig. 2) were incubated again with CNBr‐Cry1Ac‐coupled agarose. The unbound proteins from reincubation with CNBr‐Cry1Ac agarose were removed by centrifugation (lanes 5 in Fig. 2). After exhaustive washing steps, no protein was detected in the final washing buffer according to the analysis of SDS‐PAGE electrophoresis (lanes 3 and 6 in Fig. 2). The Cry1Ac‐binding proteins in the first incubation and in the reincubation step with the CNBr‐Cry1Ac‐coupled agarose were boiled for 10 min in loading buffer and resolved in SDS‐PAGE. The gel was divided into seven regions, respectively, as shown in Figure 2 lanes 4 and 7 (F samples correspond to the first incubation step, while R samples correspond to the reincubation step). These seven regions of the gel were used for LC‐MS/MS analysis. It is important to note that the advantage of the strategy resented here is that no‐elution step was necessary to elute the insect midgut proteins bound to Cry1Ac. The bands of 60 kDa in F‐4 and R‐4 correspond to Cry1Ac toxin that was also dissociated after boiling. Protein bands in R‐1 and R‐2 became weaker compared with F‐1 and F‐2, respectively, although similar protein profiles of Cry1Ac‐binding proteins were obtained in the first incubation and in the reincubation with agarose‐coupled Cry1Ac. However, some protein bands were lost in R‐3 compared with F‐3. In the negative control performed with blocked CNBr agarose without Cry1Ac toxin, no protein bands were observed after SDS‐PAGE analysis (data not shown).
Figure 2.
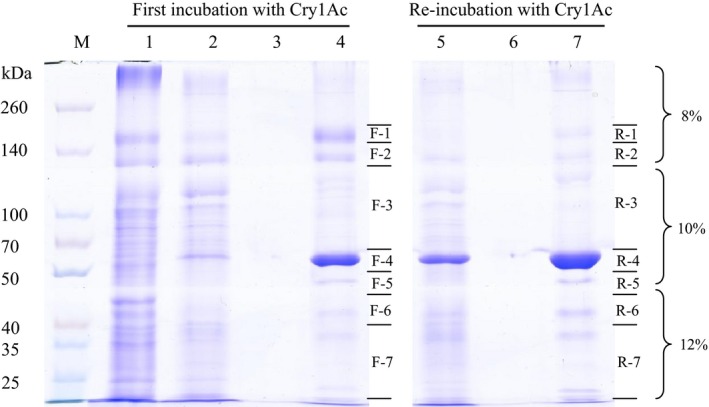
Identification of Cry1Ac‐binding proteins on the BBMV from Helicoverpa armigera using the pull‐down method without elution. Proteins were resolved in PAGE discontinuous acrylamide gradients from 8% to 12%. Lanes 4 and 7 were divided into seven fractions (F‐1 to F‐7 and R‐1 to R‐7) and each fraction was used for LC‐MS/MS analysis. M: marker; 1: proteins from H. armigera BBMV incubated with CNBr‐Cry1Ac‐coupled agarose; 2: unbound proteins after the first incubation with CNBr‐Cry1Ac agarose; 5: unbound proteins after reincubation with CNBr‐Cry1Ac agarose; 3 and 6: proteins found in the elution obtained after last wash with phosphate‐buffered saline (PBS); 4 and 7: proteins bound to CNBr‐Cry1Ac after first and reincubations, respectively.
Identification of Cry1Ac‐binding proteins by LC‐MS/MS
Table 1 shows a list of proteins identified in each fraction of the gel using a significant score higher than 50 as threshold. APN molecules were detected in all samples analyzed. Six different classes of APN as Cry1Ac‐binding proteins were identified. V‐ATPase was detected also in most samples analyzed except for F‐2, R‐1, and R‐2 samples, while polycalin was detected in most samples except for F‐5, F‐7, R‐4, and R‐5 samples. Actin was identified in F‐5 to F‐7, R‐3 and R‐5 to R‐7 samples. Alkaline phosphatase was identified in F‐3 and R‐4. Cadherin‐like protein was identified as Cry1Ac‐binding protein in F‐2 region, while ABCC2 was identified in F‐6. Also, different proteins not previously characterized as Cry1Ac‐binding proteins were identified such as dipeptidyl peptidase, carboxyl/choline esterase, azurocidin‐like serine proteinase, and trypsin‐like protease among others which had a significant score in the LC‐MS/MS analysis (Table 1).
Table 1.
The LC‐MS/MS assay results of binding proteins to Cry1Ac on BBMVs from Helicoverpa armigera
| Fraction | Accession number | Score | Protein description [species]a | Sequence coverage | Fraction | Accession number | Score | Protein description [species] | Sequence coverage |
|---|---|---|---|---|---|---|---|---|---|
| F‐1 | gi|17027158 | 18803 | Aminopeptidase N1 [Ha] | 50% | R‐1 | gi|30961821 | 7938 | Midgut aminopeptidase N 2 [Ha] | 54% |
| gi|30961821 | 15683 | Midgut aminopeptidase N2 [Ha] | 59% | gi|17027158 | 1767 | Aminopeptidase N1 [Ha] | 42% | ||
| gi|514430167 | 1367 | Aminopeptidase N5 [On] | 5% | gi|514430167 | 646 | Aminopeptidase N5 [On] | 4% | ||
| gi|25814968 | 364 | Midgut aminopeptidase APN2 [Ha] | 24% | gi|30961823 | 434 | Midgut aminopeptidase N3 [Ha] | 23% | ||
| gi|512902608 | 214 | PREDICTED: extracellular domains containing protein CG31004‐like isoform X1 [Bm] | 7% | gi|512902608 | 146 | PREDICTED: extracellular domains containing protein CG31004‐like isoform X1[Bm] | 7% | ||
| gi|156968281 | 88 | Multidomain lipocalin [Ha] | 6% | gi|156968281 | 156 | Multidomain lipocalin [Ha] | 11% | ||
| gi|171740913 | 88 | Polycalin [Ha] | 6% | gi|171740913 | 156 | Polycalin [Ha] | 11% | ||
| gi|512926607 | 67 | PREDICTED: V‐type proton ATPase 116 kDa subunit a isoform 1‐like isoform X1 [Bm] | 4% | gi|27818925 | 156 | Aminopeptidase N4 [Ha] | 12% | ||
| gi|498991524 | 58 | PREDICTED: hydroxymethyl pyrimidine/phosphomethyl pyrimidine kinase‐like [Cc] | 10% | ||||||
| F‐2 | gi|17027158 | 7565 | Aminopeptidase N1 [Ha] | 57% | R‐2 | gi|25814968 | 1801 | Midgut aminopeptidase APN2 [Ha] | 37% |
| gi|30961823 | 3593 | Midgut aminopeptidase N3 [Ha] | 41% | gi|27818925 | 2403 | Aminopeptidase N4 [Ha] | 35% | ||
| gi|30961821 | 4265 | Midgut aminopeptidase N2 [Ha] | 56% | gi|30961823 | 1863 | Midgut aminopeptidase N3 [Ha] | 40% | ||
| gi|27818925 | 3902 | Aminopeptidase N4 [Ha] | 33% | gi|171740913 | 270 | Polycalin [Ha] | 11% | ||
| gi|171740923 | 236 | Polycalin [Ha] | 10% | gi|294846780 | 91 | Carboxyl/choline esterase CCE001 g [Ha] | 12% | ||
| gi|336319051 | 82 | Dipeptidyl peptidase [Bb] | 11% | gi|509177421 | 71 | Dipeptidyl peptidase 4, partial [Pa] | 23% | ||
| gi|26051280 | 79 | Cadherin‐like protein [Ha] | 3% | gi|327420450 | 69 | Aminopeptidase 7C [Mc] | 4% | ||
| gi|270001176 | 56 | Hypothetical protein TcasGA2_TC016071 [Tc] | 6% | gi|336319051 | 55 | Dipeptidyl peptidase [Bb] | 10% | ||
| F‐3 | gi|30961821 | 4469 | Midgut aminopeptidase N2 [Ha] | 56% | R‐3 | gi|11062 | 539 | H(+)‐transporting ATPase [Ms] | 27% |
| gi|170791085 | 374 | Aminopeptidase N6 [Ha] | 26% | gi|30961821 | 448 | Midgut aminopeptidase N2 [Ha] | 38% | ||
| gi|17027158 | 4647 | Aminopeptidase N1 [Ha] | 46% | gi|307695440 | 190 | V ATPase A, partial [Ha] | 42% | ||
| gi|194295558 | 57 | Alkaline phosphatase 2 [Ha] | 18% | gi|357629674 | 519 | V‐type proton ATPase catalytic subunit A [Dap] | 28% | ||
| gi|171740893 | 760 | Aminopeptidase N5 [Ha] | 28% | gi|156968281 | 409 | Multidomain lipocalin [Ha] | 15% | ||
| gi|27818925 | 1142 | Aminopeptidase N4 [Ha] | 29% | gi|171740923 | 425 | Polycalin [Ha] | 15% | ||
| gi|307695440 | 517 | V ATPase A, partial [Ha] | 62% | gi|171740893 | 402 | Aminopeptidase N5 [Ha] | 23% | ||
| gi|171740923 | 1155 | Polycalin [Ha] | 21% | gi|27818925 | 399 | Aminopeptidase N4 [Ha] | 21% | ||
| gi|512919199 | 149 | PREDICTED: maltase 1‐like [Bm] | 3% | gi|30961823 | 313 | Midgut aminopeptidase N3 [Ha] | 18% | ||
| gi|30961825 | 1008 | Midgut aminopeptidase N3 [Ha] | 36% | gi|170791085 | 169 | Aminopeptidase N6 [Ha] | 20% | ||
| gi|498991524 | 59 | PREDICTED: hydroxymethyl pyrimidine/phosphomethyl pyrimidinekinase‐like [Cc] | 10% | gi|498991524 | 58 | PREDICTED: hydroxymethyl pyrimidine/phosphomethyl pyrimidinekinase‐like [Cc] | 6% | ||
| gi|17027158 | 124 | aminopeptidase N1 [Ha] | 17% | ||||||
| gi|327420450 | 64 | aminopeptidase 7C [Mc] | 4% | ||||||
| gi|15284015 | 60 | actin [Cy] | 15% | ||||||
| F‐4 | gi|237459 | 1455 | Vacuolar (V‐type) H(+)‐ATPase B subunit [Hv] | 61% | R‐4 | gi|237459 | 725 | Vacuolar (V‐type) H(+)‐ATPase B subunit [Hv] | 46% |
| gi|38455217 | 1225 | Aminopeptidase N1 [Ha] | 32% | gi|25814968 | 118 | Midgut aminopeptidase APN2 [Ha] | 4% | ||
| gi|30961823 | 603 | Midgut aminopeptidase N3[Ha] | 28% | gi|30961823 | 118 | Midgut aminopeptidase N3 [Ha] | 4% | ||
| gi|25814968 | 603 | Midgut aminopeptidase APN2 [Ha] | 28% | gi|112820264 | 74 | Aminopeptidase [Aj] | 2% | ||
| gi|87248463 | 120 | ATP synthase [Bm] | 17% | gi|194295558 | 56 | Alkaline phosphatase 2 [Ha] | 10% | ||
| gi|171740913 | 96 | Polycalin [Ha] | 5% | ||||||
| gi|328709450 | 92 | PREDICTED: hypothetical protein LOC100568862 [Ap] | 5% | ||||||
| gi|456005201 | 53 | Glycosyltransferase 1 [Cs] | 4% | ||||||
| F‐5 | gi|17027158 | 639 | Aminopeptidase N1 [Ha] | 24% | R‐5 | gi|156759 | 144 | Actin [Dm] | 25% |
| gi|87248463 | 375 | ATP synthase [Bm] | 27% | gi|1419687 | 106 | 40‐kDa V‐ATPase subunit [Ms] | 19% | ||
| gi|237459 | 265 | Vacuolar (V‐type) H(+)‐ATPase B subunit [Hv] | 37% | gi|171740901 | 84 | Azurocidin‐like serine proteinase [Ha] | 26% | ||
| gi|7158844 | 165 | Aminopeptidase 3 [Hp] | 5% | gi|87248183 | 84 | Vacuolar ATPase subunit C [Bm] | 20% | ||
| gi|25814968 | 165 | Midgut aminopeptidase APN2 [Ha] | 12% | gi|545919627 | 71 | Putative vacuolar H+‐ATPase V1 sector subunit c [Ca] | 16% | ||
| gi|187942442 | 151 | Cytoplasmic actin [Ai] | 47% | gi|237459 | 81 | Vacuolar (V‐type) H(+)‐ATPase B subunit [Hv] | 12% | ||
| gi|156968281 | 121 | Multidomain lipocalin [Ha] | 9% | gi|189238960 | 66 | PREDICTED: similar to AGAP005845‐PA [Tc] | 16% | ||
| gi|328709450 | 58 | PREDICTED: hypothetical protein LOC100568862 [Ap] | 5% | ||||||
| gi|11178 | 58 | Glyceraldehyde‐3‐phosphate dehydrogenase [Dh] | 4% | ||||||
| gi|7230426 | 57 | Thioredoxinperoxidase 1 [Dm] | 5% | ||||||
| F‐6 | gi|30961821 | 674 | Midgut aminopeptidase N2 [Ha] | 31% | R‐6 | gi|54640128 | 443 | ATPsyn‐beta [Drp] | 36% |
| gi|25814966 | 467 | Midgut aminopeptidase APN1 [Ha] | 25% | gi|509164969 | 255 | ATP synthase [Pa] | 28% | ||
| gi|30961823 | 266 | Midgut aminopeptidase N3 [Ha] | 17% | gi|389609043 | 184 | Vacuolar H[+]‐ATPase SFD subunit [Px] | 33% | ||
| gi|187942442 | 214 | Cytoplasmic actin [Ai] | 41% | gi|509177493 | 175 | Vacuolar ATP synthase subunit H, partial [Pa] | 31% | ||
| gi|299481057 | 203 | Juvenile hormone epoxide hydrolase [Ha] | 20% | gi|194118083 | 119 | GL25088 [Dp] | 7% | ||
| gi|307695438 | 166 | V ATPase C, partial [Ha] | 48% | gi|22725694 | 112 | Aminopeptidase N [Ha] | 5% | ||
| gi|171740901 | 168 | Azurocidin‐like serine proteinase [Ha] | 45% | gi|194165198 | 104 | GK24170 [Dw] | 9% | ||
| gi|296427826 | 118 | ABC transporter family C protein ABCC2 [Hs] | 9% | gi|156759 | 94 | Actin [Dm] | 25% | ||
| gi|156968281 | 72 | Multidomain lipocalin [Ha] | 8% | gi|8810 | 86 | Vacuolar ATPase B subunit [Dm] | 6% | ||
| gi|171740913 | 72 | Polycalin [Ha] | 8% | gi|171740913 | 85 | Polycalin [Ha] | 7% | ||
| gi|328709450 | 52 | PREDICTED: hypothetical protein LOC100568862 [Ap] | 5% | gi|14269425 | 70 | 110 kDa aminopeptidase [Hv] | 2% | ||
| gi|15212555 | 58 | Aminopeptidase N [Ha] | 2% | ||||||
| F‐7 | gi|187942442 | 599 | Cytoplasmic actin [Ai] | 46% | R‐7 | gi|187942442 | 576 | Cytoplasmic actin [Ai] | 56% |
| gi|38455217 | 446 | Aminopeptidase N1 [Ha] | 30% | gi|287945 | 163 | ATP synthase beta subunit [Dm] | 28% | ||
| gi|215434805 | 270 | Trypsin‐like protease [Ha] | 18% | gi|215434805 | 131 | Trypsin‐like protease [Ha] | 18% | ||
| gi|25814968 | 255 | Midgut aminopeptidase APN2 [Ha] | 16% | gi|15212555 | 87 | Aminopeptidase N [Ha] | 6% | ||
| gi|237459 | 234 | Vacuolar (V‐type) H(+)‐ATPase B subunit [Hv] | 47% | gi|389611115 | 85 | Vacuolar H[+]‐ATPase 26kD E subunit [Pp] | 9% | ||
| gi|7158844 | 204 | Aminopeptidase 3 [Hp] | 6% | gi|403487715 | 74 | Glyceraldehyde‐3‐phosphate dehydrogenase, partial [Cl] | 22% | ||
| gi|332025502 | 87 | Myosin‐IB [Ae] | 2% | gi|237459 | 70 | Vacuolar (V‐type) H(+)‐ATPase B subunit [Hv] | 21% | ||
| gi|151384885 | 78 | ADP/ATP translocase [Ha] | 16% | gi|156547293 | 69 | PREDICTED: ADP,ATP carrier protein 2‐like [Nv] | 7% | ||
| gi|40022264 | 76 | Diazepam‐binding inhibitor [Ha] | 17% | gi|194118083 | 65 | GL25088 [Dp] | 5% | ||
| gi|18253049 | 60 | Ribosomal protein L7 [Sf] | 11% | gi|545919639 | 62 | Putative ADP/ATP transporter on adenylatetranslocase [Ca] | 10% | ||
| gi|308055648 | 56 | NADPH cytochrome b5 reductase [Ha] | 20% | gi|171740913 | 55 | Polycalin [Ha] | 3% |
Peptide score distribution. Ions score is −10log(P), where P is the probability that the observed match is a random event. Individual ions scores >38 indicate identity or extensive homology (P < 0.05).
Ae, Acromyrmex echinatior; Ai, Agrotis ipsilon; Aj, Achaea janata; Bb, Biston betularia; Bm, Bombyx mori; Ca, Corethrella appendiculata; Cc, Ceratitis capitata; Cl, Cymothoe lurida; Cs, Chilo suppressalis; Cy, Chironomus yoshimatsui; Da, Drosophila ananassae; Dh, Drosophila hydei; Dm, Drosophila melanogaster; Dp, Drosophila persimilis; Drp, Drosophila pseudoobscura; Dw, Drosophila willistoni; Ha, Helicoverpa armigera; Hp, Helicoverpa punctigera; Hov, Homalodisca vitripennis; Hv, Heliothis virescens; Hs, Heliothis subflexa; Ms, Manduca sexta; Nv, Nasonia vitripennis; On, Ostrinia nubilalis; Pa, Pararge aegeria; Px, Papilio xuthus; Pp, Papilio polytes; Sf, Spodoptera frugiperda; Tc, Tribolium castaneum.
Discussion
The interaction of Cry toxins and their binding proteins in the insect gut highly determines insecticidal activity. Therefore, the identification of binding proteins will facilitate the elucidation of the insecticidal mechanism of Cry toxins. Identification of binding proteins has generally been achieved using affinity chromatography (Denolf et al. 1997; Luo et al. 1997 Banks et al. 2001), ligand blot analysis (Nakanishi et al. 2002; Jurat‐Fuentes and Adang 2004; Arenas et al. 2010), or immunoprecipitation assays (Luo et al. 1996; Bravo et al. 2004). However, some binding proteins may not be identified using these methods due to denaturation of proteins or incompatibility with the elution buffer. In this article, an improved pull‐down method that does not rely on elution of binding proteins after affinity chromatography was used to identify Cry1Ac‐binding proteins on BBMV from H. armigera. Different Cry toxin‐receptors previously described in different insects (e.g., APN, ALP, cadherin) and several novel Cry‐interacting partners were identified, suggesting that this methodology is more comprehensive for identification of Cry toxin‐binding proteins than other methods that require protein elution. However, for many of the proteins identified here as Cry1Ac‐binding proteins, it still remains to be determined if they are involved in Cry1Ac toxicity to H. armigera.
Cry1Ac‐binding proteins were identified in the first incubation and reincubation with CNBr‐Cry1Ac‐coupled agarose. However, no additional Cry1Ac‐binding proteins in the reincubation step with CNBr‐Cry1Ac agarose were identified, compared with the first incubation. Moreover, ABCC2 and cadherin‐like protein, two major molecules involved in Cry1A toxin action, were only detected in the first incubation with CNBr‐Cry1Ac agarose supporting that these two proteins were not highly abundant on BBMV as was previously suggested (Zhang et al. 2012b); it is also possible that their high‐affinity binding could account for their detection only in the first incubation step. Moreover, the size of the identified protein bands corresponding to ABCC2 and cadherin‐like protein was smaller than their predicted native size, and was probably due to degradation that could also explain their identification only in the first incubation step.
Cry1Ac resistance of H. armigera has been shown to be linked to different cadherin allele mutations supporting that cadherin has an important role in Cry1Ac toxicity (Zhang et al. 2012a). Cadherin is well recognized as Cry toxin receptor that after binding to Cry1Ab toxins triggers toxin oligomerization, playing an important role in Cry toxin mechanism of action (Gomez et al. 2014). In the case of H. armigera, it was shown that residues 1217–1461 of cadherin participate in Cry1Ac interaction (Wang et al. 2005).
Among all the receptors of Cry toxins described in different insects, APN is one of the most widely studied. Binding with APN has been shown to be an important step in mediating the toxicity of Cry toxins (Bravo et al. 2013). Thirty‐eight different APN proteins have been reported for 12 different lepidopteran insects and these were classified in five groups (Pigott and Ellar 2007). In H. armigera, at least five different classes of APN were reported (Pigott and Ellar 2007), and APN1 and APN2 from this insect species were expressed in Hi5 insect cells showing that besides being found in the membrane and catalytically active, they were capable to bind Cry1Ac toxin (Rajagopal et al. 2003). In this article, six classes of APNs were identified as Cry1Ac‐binding proteins using the improved pull‐down method. Due to the high abundance on BBMV and degradation, many APN fragments were determined to bind with Cry1Ac, even in gel zones that are much smaller than their predicted native size. APN1 (Rajagopal et al. 2003), APN2 (Rajagopal et al. 2003), APN3 (Gill et al. 1995; Banks et al. 2001), and APN4 (Banks et al. 2001) were reported to bind Cry1Ac. In the case of APN1, it was demonstrated to be functional receptor of Cry1Ac in H. armigera (Sivakumar et al. 2007), and related to insect resistance to Cry1Ac (Zhang et al. 2009; Tiewsiri and Wang 2011). However, it remains to be analyzed if the other APN isoforms such as APN5 and APN6 identified here are involved in Cry1Ac toxicity in H. armigera. In the case of ALP, it was previously shown that ALP is involved in Cry1Ac toxicity in H. armigera since resistance to Cry1Ac correlated with low ALP expression in different resistant colonies (Chen et al. 2015).
Consistent with other studies, actin and V‐type ATPase A were shown to bind Cry toxins (McNall and Adang 2003; Krishnamoorthy et al. 2007; Bayyareddy et al. 2009; Chen et al. 2010). In the mosquito Aedes aegypti, silencing of an actin gene resulted in hypersensitive phenotype to Cry11Aa toxin suggesting that actin is somehow involved in Cry toxicity (Cancino‐Rodezno et al. 2012). Polycalin was reported to bind to Cry toxin in B. mori (Hossain et al. 2004; Pandian et al. 2008) and H. armigera (Angelucci et al. 2008; Ma et al. 2012). However, it was shown that polycalin is not likely to be involved in toxicity in B. mori (Pandian et al. 2010).
We identified several additional proteins with unknown function in Cry toxin mode of action such as dipeptidyl peptidase or carboxyl/choline esterase and serine proteases among others (Table 1). Further studies are required to characterize the binding interactions between Cry1Ac toxin and the proteins identified in this work and to verify the biological functions of these proteins in Cry1Ac toxicity.
The exact role of ABCC2 in the mechanism of action of Cry toxins remains elusive. Mutations in ABCC2 have been shown to be linked to high levels of Cry1Ac resistance in different lepidopteran species including H. armigera (Gahan et al. 2010; Baxter et al. 2011; Atsumi et al. 2012; Xiao et al. 2014). Thus, it was proposed that ABCC2 may be an additional Cry toxin receptor, but no direct evidence of binding between Cry1A toxin and ABCC2 transporter isolated from lepidopteran larvae has been provided until now. Previously, it was shown that the expression of ABCC2 from Bombyx mori in Sf9 insect cells provided binding of Cry1A toxins to cells and increases sensitivity to these toxins, suggesting that interaction between these proteins is important for toxicity (Tanaka et al. 2013). A previous report identified Cry1Ac‐binding proteins in H. armigera after a ligand blot assay performed in two‐dimensional gel electrophoresis (Chen et al. 2010). Similar proteins to those reported here were also identified such as APN, CAD, V‐ATPase, and actin (Chen et al. 2010). Some spots that showed similarities with glutathione ABC transporter from the bacteria Erwinia carotovora was also identified (Chen et al. 2010). However, it is important to mention that this glutathione‐ABC transporter is completely different from the ABCC2 transporter showing only 4% identity in the primary sequence with ABCC2 from Heliothis virescence (accession number GQ332571.1) that is linked with resistance to Cry1Ac toxin. In this work, we were able to identify an ABCC2 protein from H. armigera that has high similarity with an ABCC2 protein from Heliothis subflexa (accession number: ADH16744), which showed 96% identity with the ABCC2 protein from H. virescens. The identification score (−10log(P), where P is the probability that the observed match is a random event) for ABCC2 (score, 118) is in the same magnitude as cadherin (score, 79) that was previously confirmed to be a Cry1Ac‐binding protein (Wang et al. 2005). Still additional experimental evidence is required to show that Cry1Ac directly binds ABCC2 transporter. ABCC2 transporter is a transmembrane protein with a small exposed region outside the membrane (Aller et al. 2009). This structural constraint could explain why ABCC2 was not previously identified as a Cry1A‐binding protein. Pull‐down experiments allows interaction of the bait protein (Cry1Ac) with proteins in their native state. Thus, our data strongly suggests that ABCC2 is a Cry1Ac‐binding protein.
Conclusions
APN, ALP, cadherin, actin, V‐type ATPase A, polycalin, ABCC2, and some other proteins not previously characterized as Cry toxin‐binding molecules (e.g., dipeptidyl peptidase or carboxyl/choline esterase and some serine proteases) were identified as Cry1Ac‐binding protein by the improved pull‐down method. This is the first report that provides evidence of possible direct binding of Cry1Ac toxin to ABCC2 isolated from insect BBMV.
Conflict of Interest
None declared.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant no. 31272115), 863 Project (grant no. 2011AA10A203), and China Postdoctoral Science Foundation (grant no. 2014M560487; 2015T80623).
MicrobiologyOpen 2016; 5(4): 659–669
References
- Aller, S. G. , Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., et al. 2009. Structure of P‐glycoprotein reveals a molecular basis for poly‐specific drug binding. Science 323:1718–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelucci, C. , Barrett‐Wilt G. A., Hunt D. F., Akhurst R. J., East P. D., Gordon K. H., et al. 2008. Diversity of aminopeptidases, derived from four lepidopteran gene duplications, and polycalins expressed in the midgut of Helicoverpa armigera: identification of proteins binding the delta‐endotoxin, Cry1Ac of Bacillus thuringiensis . Insect Biochem. Mol. Biol. 38:685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas, I. , Bravo A., Soberón M., and Gomez I.. 2010. Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin. J. Biol. Chem. 285:12497–12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi, S. , Miyamato K., Yamamoto K., Narukawa J., Kawai S., Sezutsu H., et al. 2012. Single amino acid mutation in an ATP‐binding cassette transporter causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori . Proc. Natl Acad. Sci. USA 109:1591–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks, D. J. , Jurat‐Fuentes J. L., Dean D. H., and Adang M. J.. 2001. Bacillus thuringiensis Cry1Ac and Cry1Fa δ‐endotoxin binding to a novel 110 kDa aminopeptidase in Heliothis virescens is not N‐acetylgalactosamine mediated. Insect Biochem. Mol. Biol. 31:909–918. [DOI] [PubMed] [Google Scholar]
- Baxter, S. W. , Badenes‐Perez F. R., Morrison A., Vogel H., Crickmore N., Kain W., et al. 2011. Parallel evolution of Bt toxin resistance in Lepidoptera. Genetics 189:675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayyareddy, K. , Andacht T. M., Abdullah M. A., and Adang M.. 2009. Proteomic identification of Bacillus thuringiensis subsp. israelensis toxin Cry4Ba binding proteins in midgut membranes from Aedes (Stegomyia) aegypti Linnaeus (Diptera, Culicidae) larvae. Insect Biochem. Mol. Biol. 39:279–286. [DOI] [PubMed] [Google Scholar]
- Bravo, A. , Gomez I., Conde J., Munoz‐Garay C., Sanchez J., Miranda R., et al. 2004. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore‐forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim. Biophys. Acta 1667:38–46. [DOI] [PubMed] [Google Scholar]
- Bravo, A. , Gill S. S., and Soberón M.. 2007. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 49:423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo, A. , Gomez I., Porta H., Garcia‐Gomez B. I., Rodriguez‐Almazan C., Pardo L., et al. 2013. Evolution of Bacillus thuringiensis Cry toxins insecticidal activity. Microb. Biotechnol. 6:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancino‐Rodezno, A. , Lozano L., Oppert C., Castro J. I., Lanz‐Mendoza H., Encarnacion S., et al. 2012. Comparative proteomic analysis of Aedes aegypti larval midgut alter intoxication with Cry11Aa toxin from Bacillus thuringiensis . PLoS ONE 7:e37034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. W. , Aimanova K. G., Pan S., and Gill S. S.. 2009. Identification and characterization of Aedes aegypti aminopeptidase N as a putative receptor of Bacillus thuringiensis Cry11A toxin. Insect Biochem. Mol. Biol. 39:688–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. Z. , Liang G. M., Zhang J., Wu K. M., Guo Y. Y., and Rector B. G.. 2010. Proteomic analysis of novel Cry1Ac binding proteins in Helicoverpa armigera (Hübner). Arch. Insect Biochem. Physiol. 73:61–73. [DOI] [PubMed] [Google Scholar]
- Chen, W. , Liu C., Xiao Y., Zhang D., Zhang Y., Li X., et al. 2015. A toxin‐binding alkaline phosphatase fragment synergizes Bt toxin Cry1Ac against susceptible and resistant H. armigera . PLoS ONE 10:e0126288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denolf, P. , Hendrickx K., Van Damme J., Jansens S., Peferoen M., Degheele D., et al. 1997. Cloning and characterization of Manduca sexta and Plutella xylostella midgut aminopeptidase N enzymes related to Bacillus thuringiensis toxin‐binding proteins. Eur. J. Biochem. 248:748–761. [DOI] [PubMed] [Google Scholar]
- Fernandez, L. E. , Aimanova K. G., Gill S. S., Bravo A., and Soberón M.. 2006. A GPI‐anchored alkaline phosphatase is a functional midgut receptor of Cry11Aa toxin in Aedes aegyptilarvae. Biochem. J. 394:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahan, L. J. , Pauchet Y., Vogel H., and Heckel D. G.. 2010. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 6:e1001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, S. S. , Cowles E. A., and Francis V.. 1995. Identification, isolation, andcloning of a Bacillus thuringiensis CryIAc toxin‐binding protein from the midgut of the lepidopteran insect Heliothis virescens . J. Biol. Chem. 270:27277–27282. [DOI] [PubMed] [Google Scholar]
- Gomez, I. , Sanchez J., Munoz‐Garay C., Matus V., Gill S.S., Soberon M., et al. 2014. Bacillus thuringiensis Cry1A toxins are versatile‐proteins with multiple modes of action: two distinct pre‐pores are involved in toxicity. Biochem. J. 459:383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel, D. G. 2012. Learning the ABCs of Bt: ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pestic. Biochem. Physiol. 104:103–110. [Google Scholar]
- Hossain, D. M. , Shitomi Y., Moriyama K., Higuch M., Hayakawa T., Mitsui T., et al. 2004. Characterization of a novel plasma membrane protein, expressed in the midgut epithelia of Bombyx mori, that binds to Cry1A toxins. Appl. Environ. Microbiol. 70:4604–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, G. , Park Y. J., and Adang M. J.. 2014. Cadherin AdCad1 in Alphitobius diaperinus larvae is a receptor of Cry3Bb toxin from Bacillus thuringiensis . Insect Biochem. Mol. Biol. 45:11–17. [DOI] [PubMed] [Google Scholar]
- Ibrahim, M. A. , Griko N., Junker M., and Bulla L. A.. 2010. Bacillus thuringiensis: a genomics and proteomics perspective. Bioeng. Bugs 1:31–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurat‐Fuentes, J. L. , and Adang M. J.. 2004. Characterization of a Cry1Ac receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur. J. Biochem. 271:3127–3135. [DOI] [PubMed] [Google Scholar]
- Keller, A. , Nesvizhskii A. I., Kolker E., and Aebersold R.. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy, M. , Jurat‐Fuentes J. L., McNall R. J., Andacht T., and Adang M. J.. 2007. Identification of novel Cry1Ac binding proteins in midgut membranes from Heliothis virescens using proteomic analyses. Insect Biochem. Mol. Biol. 37:189–201. [DOI] [PubMed] [Google Scholar]
- Liang, G. M. , Tan W. J., and Guo Y. Y.. 1999. An improvement in the technique of artificial raring cotton bollworm. Plant Prot. 25:15–17. [Google Scholar]
- Luo, K. , Lu Y. J., and Adang M. J.. 1996. A 106 kDa form of aminopeptidase is a receptor for Bacillus thuringiensis CryIC δ‐endotoxin in the brush border membrane of Manduca sexta . Insect Biochem. Mol. Biol. 26:783–791. [Google Scholar]
- Luo, K. , Sangandala S., Masoon L., Mazza A., Brousseau R., and Adang M. J.. 1997. The Heliothis virescens 170 kDa aminopeptidase functions as “receptor A” by mediating specific Bacillus thuringiensis Cry1A δ‐endotoxin binding and pore formation. Insect Biochem. Mol. Biol. 27:735–743. [DOI] [PubMed] [Google Scholar]
- Ma, G. , Rahman M. M., Grant W., Schmidt O., and Asgari S.. 2012. Insect tolerance to the crystal toxins Cry1Ac and Cry2Ab is mediated by the binding of monomeric toxin to lipophoringlycolipids causing oligomerization and sequestration reactions. Dev. Comp. Immunol. 37:184–192. [DOI] [PubMed] [Google Scholar]
- McNall, R. J. , and Adang M. J.. 2003. Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect Biochem. Mol. Biol. 33:999–1010. [DOI] [PubMed] [Google Scholar]
- Nakanishi, K. , Yaoi K., Nagino Y., Hara H., Kitami M., Atsumi S., et al. 2002. Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella their classification and the factors that determine their binding specificity to Bacillus thuringiensis Cry1A toxin. FEBS Lett. 519:215–220. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii, A. I. , Keller A., Kolker E., and Aebersold R.. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658. [DOI] [PubMed] [Google Scholar]
- Pacheco, S. , Gomez I., Gill S. S., Bravo A., and Soberón M.. 2009. Enhancement of insecticidal activity of Bacillus thuringiensis Cry1A toxins by fragments of a toxin‐binding cadherin correlates with oligomer formation. Peptides 30:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandian, G. N. , Ishikawa T., Vaijayanthi T., Hossain D. M., Yamamoto S., Nishiumi T., et al. 2008. Bombyx mori midgut membrane protein P252, which binds to Bacillus thuringiensis Cry1A, is a chlorophyllide‐binding protein, and the resulting complex has antimicrobial activity. Appl. Environ. Microbiol. 74:1324–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandian, G. N. , Ishikawa T., Vaijayanthi T., Hossain D. M., Yamamoto S., Nishiumi T., et al. 2010. Formation of macromolecule complex with Bacillus thuringiensis Cry1A toxins and chlorophyllide binding 252‐kDa lipocalin‐like protein locating on Bombyx mori midgut membrane. J. Membr. Biol. 237:125–136. [DOI] [PubMed] [Google Scholar]
- Pardo‐Lopez, L. , Soberón M., and Bravo A.. 2013. Bacillus thuringiensis insecticidal three‐domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 37:3–22. [DOI] [PubMed] [Google Scholar]
- Pigott, C. R. , and Ellar D. J.. 2007. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 71:255–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal, R. , Agrawal N., Selvapandiyan A., Sivakumar S., Ahmad S., and Bhatnagar R. K.. 2003. Recombinantly expressed isozymic aminopeptidases from Helicoverpa armigera midgut display differential interaction with closely related Cry proteins. Biochem. J. 370:971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, C. , Tan S., Yin J., Soberón M., Bravo A., Liu C., et al. 2015. Assembling of Holotrichia parallela (dark black chafer) midgut tissue transcriptome and identification of midgut proteins that bind to Cry8Ea toxin from Bacillus thuringiensis . Appl. Microbiol. Biotechnol. 99:7209–7218. [DOI] [PubMed] [Google Scholar]
- Sivakumar, S. , Rajagopal R., Venkatesh G. R., Srivastava A., and Bhatnagar R. K.. 2007. Knockdown of aminopeptidase‐N from Helicoverpa armigera larvae and in transfected Sf21 cells by RNA interference reveals its functional interaction with Bacillus thuringiensis insecticidal protein Cry1Ac. J. Biol. Chem. 282:7312–7319. [DOI] [PubMed] [Google Scholar]
- Soberón, M. , Gill S. S., and Bravo A.. 2009. Signaling versus punching hole: how do Bacillus thuringiensis toxins kill insect midgut cells? Cell. Mol. Life Sci. 66:1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, S. , Miyamoto K., Noda H., Jurat‐Fuentes J. L., Yoshizawa Y., Endo H., et al. 2013. The ATP‐binding cassette transporter subfamily C member 2 in Bombyx mori larvae is a functional receptor for Cry toxins from Bacillus thuringiensis . FEBS J. 280:1782–1794. [DOI] [PubMed] [Google Scholar]
- Tiewsiri, K. , and Wang P.. 2011. Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Natl Acad. Sci. USA 108:14037–14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay, S. K. , and Singh P. K.. 2011. Role of alkaline phosphatase in insecticidal action of Cry1Ac against Helicoverpa armigera larvae. Biotechnol. Lett. 33:2027–2036. [DOI] [PubMed] [Google Scholar]
- Vadlamudi, R. K. , Ji T. H., and Bulla L. A. J.. 1993. A specific binding protein from Manduca sexta for the insecticidal toxin of Bacillus thuringiensis subsp. berliner . J. Biol. Chem. 268:12334–12340. [PubMed] [Google Scholar]
- Wang, G. R. , Wu K. M., Liang G. M., and Guo Y. Y.. 2005. Gene cloning and expression of cadherin in midgut of Helicoverpa armigera and its Cry1A binding region. Sci. China Ser. C 48:346–356. [DOI] [PubMed] [Google Scholar]
- Wolfersberger, M. G. , Luthy P., Maurer A., Parenti P., Sacchi F. V., Giordana B., et al. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from larval midgut of the cabbage butterfly (Pierisbrassicae). Comp. Biochem. Physiol. 86:301–308. [Google Scholar]
- Xiao, Y. , Zhang T., Liu C., Heckel D. G., Li X., Tabashnik B. E., et al. 2014. Mis‐splicing of the ABCC2 gene linked with Bt toxin resistance in Helicoverpa armigera . Sci. Rep. 4:6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, J. , Liang G., Crickmore N., Li H., He K., Song F., et al. 2008. Cloning and characterization of a novel Cry1A toxin from Bacillus thuringiensis with high toxicity to the Asian corn borer and other lepidopteran insects. FEMS Microbiol. Lett. 280:95–101. [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Cheng H., Gao Y., Wang G., Liang G., and Wu K.. 2009. Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem. Mol. Biol. 39:421–429. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Yin W., Zhao J., Jin L., Yang Y., Wu S., et al. 2011. Early warning of cotton bollworm resistance associated with intensive planting of Bt cotton in China. PLoS ONE 6:e22874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Tian W., Zhao J., Jin L., Yang J., Liu C., et al. 2012a. Diverse genetic basis of field‐evolved resistance to Bt cotton in cotton bollworm from China. Proc. Natl Acad. Sci. USA 109:10275–10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Tiewsiri K., Kain W., Huang L., and Wang P.. 2012b. Resistance of Trichoplusia ni to Bacillus thuringiensis toxin Cry1Ac is independent of alteration of the cadherin‐like receptor for Cry toxins. PLoS ONE 7:e35991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z. S. , Yang S. J., Shu C. L., Song F. P., Zhou X. P., and Zhang J.. 2015. Comparison and optimization of the method for Cry1Ac protoxin preparation in HD73 strain. J. Integr. Agric. 14:1598–1063. [Google Scholar]
- Zuniga‐Navarrete, F. , Gomez I., Pena G., Bravo A., and Soberón M.. 2013. A Tenebriomolitor GPI‐anchored alkaline phosphatase is involved in binding of Bacillus thuringiensis Cry3Aa to brush border membrane vesicles. Peptides 41:81–86. [DOI] [PubMed] [Google Scholar]


