Abstract
Lactobacillus plantarum produces a number of antimicrobial peptides (bacteriocins) that mostly target closely related bacteria. Although bacteriocins are important for the ecology of these bacteria, very little is known about how the peptides target sensitive cells. In this work, a putative membrane protein receptor of the two‐peptide bacteriocin plantaricin JK was identified by comparing Illumina sequence reads from plantaricin JK‐resistant mutants to a crude assembly of the sensitive wild‐type Weissella viridescens genome using the polymorphism discovery tool VAAL. Ten resistant mutants harbored altogether seven independent mutations in a gene encoding an APC superfamily protein with 12 transmembrane helices. The APC superfamily transporter thus is likely to serve as a target for plantaricin JK on sensitive cells.
Keywords: Antibacterial activity, bacteriocins, membrane proteins, mode of action
Introduction
Many species of lactic acid bacteria produce nonmodified, ribosomally synthesized antibacterial peptides (bacteriocins) that kill target bacteria by membrane permeabilization (Nissen‐Meyer et al. 2009; Cotter et al. 2013). In many cases, the bacteriocins do not act by directly perturbing the membrane lipids, but rather by binding to a specific membrane protein (the “bacteriocin receptor”), where the interaction between peptide and receptor protein leads to membrane leakage and cell death (Ramnath et al. 2000; Héchard and Sahl 2002; Diep et al. 2007; Uzelac et al. 2013; Kjos et al. 2014). Although the events that lead to leakage are not well characterized, there are indications that the mechanism involves conformational changes in the receptor protein upon binding of the bacteriocin (Diep et al. 2007; Kjos et al. 2010a). This receptor‐mediated mechanism may explain both the extreme potency of many bacteriocins (antimicrobial activity in the pico‐ to nanomolar range) and their often narrow inhibitory spectra. Also involved in the mode of action of bacteriocins are the immunity proteins that bacteriocin‐producing cells synthesize to protect themselves against the action of their own bacteriocins. In the receptor‐mediated model of bacteriocin action, the immunity protein acts by interacting with the bacteriocin‐bound receptor and thereby prevents membrane leakage (Diep et al. 2007).
Bacteriocins are of interest both due to their importance for interactions between bacterial species in natural habitats, and as alternatives to classical antibacterials to combat pathogens and food spoilage bacteria. However, we still lack knowledge about the nature of the interactions between bacteriocins, receptors, and immunity proteins, and in many cases we do not even know the identities of these three components. Generally, the immunity protein is encoded in the same operon as the bacteriocin (van Belkum et al. 1991; Cotter et al. 2013) and thus easy to identify. The identification of the receptor protein of a given bacteriocin is less trivial, and the receptors of many bacteriocins are consequently still unknown. The IIC and IID subunits of the mannose phosphotransferase system were, by a combination of genetic and biochemical analyses, shown to act as receptors for the pediocin‐like (class IIa) bacteriocins (Ramnath et al. 2000; Gravesen et al. 2002; Héchard and Sahl 2002; Diep et al. 2007). The same transport system also functions as a receptor for lactococcin A and microcin E492, bacteriocins that structurally are quite different from each other and from the pediocin‐like bacteriocins (Bieler et al. 2006; Diep et al. 2007). A glucose‐specific PTS system is also involved in sensitivity to the glycosylated bacteriocin sublancin. However, in this case the mode of action does not seem to involve compromised membrane integrity or pore formation (Garcia De Gonzalo et al. 2015). Other identified bacteriocin receptor proteins are the maltose ABC transporter (garvicin ML, a class IIc bacteriocin) (Gabrielsen et al. 2012) and the metallopeptidase YvjB (the class IId bacteriocin LsbB) (Uzelac et al. 2013).
Recently we identified the receptor protein of lactococcin G, a two‐peptide bacteriocin belonging to class IIb (Kjos et al. 2014). Bacteriocins in this class consist of two different peptides that must be present in equimolar amounts for full antibacterial activity. By whole‐genome sequencing of lactococcin G‐resistant mutants of a sensitive strain, we could identify the lactococcin G receptor as the membrane‐embedded enzyme UppP, an undecaprenyl pyrophosphate phosphatase that is involved in cell‐wall synthesis. When compared with the annotated Genbank genome of the target strain, the resistant mutants all had point mutations in or close to the uppP gene leading to production of a truncated protein or to reduced protein expression. In the present work, we have used a similar approach to identify a protein involved in the mode of action of plantaricin JK, a two‐peptide bacteriocin produced by strains of Lactobacillus plantarum (Anderssen et al. 1998). Like lactococcin G, plantaricin JK is most active when the two peptides are present in equimolar amounts, although the peptides also show a much reduced activity on their own. Plantaricin JK is encoded in the plantaricin locus of L. plantarum by the genes plnJ and plnK, which are cotranscribed with the immunity genes plnLR (Diep et al. 1995, 2009; Kjos, Snipen, et al. 2010b). The two genes encode peptides of 25 (PlnJ) and 32 (PlnK) amino acids that act together to permeabilize the membrane of susceptible cells, resulting in a drop in electric potential and pH gradient, and eventually cell death (Moll et al. 1999). The peptides form amphiphilic alpha‐helices and interact via so‐called GxxxG motifs (Hauge et al. 1999; Rogne et al. 2009). Plantaricin JK is known to only target strains that are closely related to the producer (Anderssen et al. 1998), but the underlying reasons for this have remained unknown. Here, we shed light on the mode of action of plantaricin JK, information that will be important for the further understanding of how plantaricin genes contribute to the population ecology of lactobacilli (Riley and Wertz 2002).
Experimental Procedures
Production and purification of plantaricin JK
The PlnJ and PlnK peptides of plantaricin JK were obtained by solid‐phase synthesis as previously described (Hauge et al. 1999). Synthetic plantaricin EF peptides were obtained from Genscript. The concentrations of the peptides in solution were estimated by measuring the absorbance at 280 nm and using an absorbance coefficient calculated from the content of aromatic amino acids.
Bacteriocin activity assays
The antimicrobial activity of plantaricin JK and plantaricin EF against sensitive and resistant target cells was tested using a 96‐well microtiter plate‐based activity assay similarly as previously described (Oppegård et al. 2007, 2008, 2010). When measuring the antimicrobial activity of plantaricin JK and EF against wild‐type and mutant Weissella viridescens, overnight (stationary phase) cultures of these cells were diluted about 1:50 in MRS medium, and 200 μL of this cell suspension was added to each well together with twofold dilutions of the bacteriocin. The microtiter plates were then incubated for 6–8 h at 30°C before the growth inhibition was measured spectrophotometrically at 600 nm. The minimum inhibitory concentration (MIC) is defined as the peptide concentration (the sum of both peptides [in a 1: 1 ratio]) that inhibited growth by 50%.
Generation of plantaricin JK‐resistant mutants
The plantaricin JK‐sensitive strain W. viridescens NCDO 1655 (MIC about 0.5 nmol/L) was used as a source of spontaneous mutants with increased resistance to the bacteriocin. This strain was used since it is also sensitive to plantaricin EF. The strain was routinely cultured in MRS media without additions. A culture of W. viridescens from a single colony was diluted about 1:100 with MRS media containing twofold dilutions of plantaricin JK (at concentrations ranging from about 1–20 nmol/L). The cells were cultured about 24 h at 30°C in 6‐mm wells of a 96‐well microtiter plate. Cultures from wells in which cell growth was obtained were diluted 1:10 with MRS medium and stored at either 30°C or 4°C for about 100 h, and then diluted about 1:100 with MRS media containing twofold dilutions of plantaricin JK (concentrations ranging from about 5–50 nmol/L). The cells were then cultured between 70 to 100 h at 30°C in 6‐mm wells of a 96‐well microtiter plate. Cells from wells in which cell growth was obtained were plated on MRS agar plates and individual colonies were picked. Cells from each colony were tested for resistance to plantaricin JK using the bacteriocin activity assay. This resulted in 10 plantaricin JK‐resistant strains (JK1‐6, JK8‐11). To assess the stability of the resistance phenotype, the resistant cells were grown for several generations in MRS medium without added bacteriocin, and then again tested for resistance. We observed no decrease in resistance after prolonged growth in the absence of bacteriocin, indicating that resistance was due to mutation rather than to adaptation.
Isolation of genomic DNA and whole‐genome sequencing
DNA was isolated from 1.5 mL overnight cultures of the sensitive wild‐type strain and the 10 resistant mutants using the Qiagen Blood and Tissue Kit (Qiagen NV, The Netherlands) according to the producer's recommendations. DNA samples were submitted to The Norwegian Sequencing Centre (sequencing.uio.no). The samples were sequenced by 16× multiplexing in an Illumina MiSeq instrument, giving 1.5–2.5 million 250 nt long paired‐end reads per strain.
Assembly of the reference genome
Since no reference genome of W. viridescens NCDO 1655 is available in the public databases, we made assemblies of the wild‐type paired‐end sequence reads (in some instances after merging overlapping paired‐end reads with FLASH (Magoc and Salzberg 2011)) using publicly available software for de novo assembly (ABySS (Simpson et al. 2009), SPAdes (Bankevich et al. 2012), Velvet (Zerbino and Birney 2008), and the assembly program in Geneious (Geneious version 7 created by Biomatters, available from http://www.geneious.com)). The raw assemblies were then used as reference genomes in the polymorphism detection software VAAL (Nusbaum et al. 2008).
Comparison of Illumina reads from the mutated strains with the assembled reference genome
Illumina reads were compared with the wild‐type genome contigs using the polymorphism discovery tool VAAL.
Contigs harboring differences reported by VAAL were annotated for probable gene content using a combination of Glimmer 3 and Blast searches. VAAL hits were evaluated and verified by aligning relevant reads to individual contigs using the Align/Assemble tool and the Find Variations/SNPs tool in the Geneious suite. This showed that all mutations detected by VAAL were located in position with a high coverage of mutant reads (154–484×), that both strands were equally represented in the reads, and that the mutations were present in 90–100% of the reads (detailed results not shown).
Results
Increased plantaricin JK resistance of mutated W. viridescens strains
Strains that are sensitive to plantaricin JK include L. plantarum, L. sakei, and W. viridescens (previously known as L. viridescens (Collins et al. 1993)), all close relatives of the producer. Sensitive W. viridescens NCDO 1665 was exposed to increasing concentrations of plantaricin JK in liquid culture prior to plating. Using this method, we collected 10 W. viridescens colonies with increased resistance against plantaricin JK. As shown in Table 1, the MIC values for the strains derived from the 10 isolated colonies were 100–600 times higher than for the parental strain (0.5 nmol/L). Genomic DNA was isolated from each of the 10 strains with increased resistance, as well as the parental strain, and sequenced.
Table 1.
Increased resistance of W. viridescens mutants toward plantaricin JK and plantaricin EF. The mutants were isolated and minimum inhibitory concentration (MIC) values determined as detailed in the Experimental procedures section
| Mutant strain | Fold increase of MIC value | |
|---|---|---|
| Plantaricin JK | Plantaricin EF | |
| JK1 | 100 ± 20 | 1 |
| JK2 | 300 ± 100 | 1 |
| JK3 | 300 ± 100 | 1 |
| JK4 | 600 ± 200 | 2 ± 1 |
| JK5 | 600 ± 200 | 1 |
| JK6 | 600 ± 200 | 1 |
| JK8 | 300 ± 100 | 1 |
| JK9 | 300 ± 100 | 1 |
| JK10 | 100 ± 50 | 1 |
| JK11 | 400 ± 100 | 7 ± 3 |
Strains that produce plantaricin JK, for example, L. plantarum C11, also produce another two‐peptide bacteriocin, plantaricin EF (Anderssen et al. 1998; Diep et al. 2009). W. viridescens NCDO 1655 is even more sensitive to this bacteriocin than to plantaricin JK (MIC = 0.05 nmol/L). To investigate the specificity of the mutants, we also tested how the plantaricin EF sensitivity was affected. As shown in Table 1, there was no significant reduction in the sensitivity of the mutants, compared to the wild‐type strain, except for mutant JK11, which was marginally more resistant to plantaricin EF than the wild‐type. This indicates that the increased resistance of the mutants to plantaricin JK is not a general effect on the bacteriocin resistance, but an effect that is specific for plantaricin JK.
Assembly of a reference genome
As described in the Experimental procedures section, we made several assemblies of the wild‐type genome, using a selection of assembly tools. The Geneious assembly program used on FLASH‐merged reads gave the lowest number of contigs (243), and this assembly was the only one that gave a reasonably low number of polymorphic sites in the subsequent VAAL analysis. Thus, this 243‐contig assembly was used for further analyses. The contigs added up to a total of 1.59 Mbp. A 34 contig assembly of the genome of a different W. viridescens strain, DMS 20410, was recently added to the NCBI database (NZ_JQBM01000000, published 12‐11‐2015). The genome representation is described as full, and the genome size is given as 1,537,173 bp. Assuming that the genome sizes of the two strains are similar, this would indicate that the 243 contigs represent close to the full genome.
Localization of mutations
Whole‐genome sequence reads from the 10 highly plantaricin JK‐resistant colonies as well as wild‐type reads were compared to the reference genome using VAAL. In total, 58 differences between the sequence reads from the analyzed strains and the reference genome contigs were detected (Table S1). Of these, 11 differences were present in the bacteriocin‐sensitive wild‐type, as well as in several or all of the resistant strains. All these differences were localized to regions of the assembly with a very low coverage (1–5× rather than the average 50×), that is, regions in the assembly where errors in the consensus sequence are most likely to occur. Consequently, these 11 differences were regarded as assembly errors and were not considered in the further analysis. Likewise, another 11 differences reported by VAAL as present in several of the mutant genomes were localized to low‐coverage regions with unreliable consensus sequences; these were also excluded from further consideration (the excluded differences are labeled in red in Table S1). The remaining 36 differences reported by VAAL between resistant and wild‐type genomes are given in Table 2.
Table 2.
Differences in genome sequence between the sensitive wild‐type and resistant mutants of W. viridescens. The Table is based on Supplementary Table 1, but false positives have been removed as detailed in the Methods section
| Contig no. | Position in contig | Coverage in assembly | Genetic location of difference | Mutant strains | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VT1 (wild‐type) | JK1 | JK2 | JK3 | JK4 | JK5 | JK6 | JK8 | JK9 | JK10 | JK11 | ||||
| Contig 3 | 15,305 | 5 | Silent mutation in D‐Ala‐D‐Ala carboxypeptidase | X | ||||||||||
| Contig 7 | 17,428 | 16 | ACG‐ATG = 40T‐M in S4 RNA‐binding domain protein | X | X | |||||||||
| Contig 10 | 2518 | 44 | CGA‐TGA = 104R‐stop in APC family amino acid‐polyamine‐organocation transporter, 610 aa | X | ||||||||||
| Contig 10 | 2739 | 30 | TTC‐TTTC = 177F‐frameshift in APC family amino acid‐polyamine‐organocation transporter | X | X | X | ||||||||
| Contig 10 | 3337 | 18 | GCG‐ACG = 377A‐T in APC family amino acid‐polyamine‐organocation transporter | X | ||||||||||
| Contig 10 | 3367 | 19 | TCT‐CCT = 387S‐P in APC family amino acid‐polyamine‐organocation transporter | X | ||||||||||
| Contig 10 | 3393 | 23 | TGG‐TGA = 395W‐stop in APC family amino acid‐polyamine‐organocation transporter | X | ||||||||||
| Contig 10 | 3482 | 21 | ATG‐AAG = 425M‐K in APC family amino acid‐polyamine‐organocation transporter | X | ||||||||||
| Contig 10 | 3491 | 22 | CAT‐CGT = 428H‐R in APC family amino acid‐polyamine‐organocation transporter | X | X | |||||||||
| Contig 10 | 12,129 | 6 | GGT‐AGT = G‐S in nucleic acid‐binding protein | X | ||||||||||
| Contig 11 | 2629 | 6 | GTA‐ATA = V‐I in predicted metal‐dependent hydrolase | X | ||||||||||
| Contig 12 | 3366 | 15 | GTG‐GCG = V‐A in Met‐tRNA formyl transferase | X | ||||||||||
| Contig 15 | 5486 | 18 | Intergenic region according to Glimmer, no BlastX hits | X | ||||||||||
| Contig 17 | 10,149 | 45 | Just downstream of Glimmer orf 10 branched chain amino acid aminotransferase | X | ||||||||||
| Contig 23 | 2907 | 10 | ATT‐GTT = I‐V in UTP–glucose‐1‐P uridylyltransferase | X | ||||||||||
| Contig 23 | 13,717 | 32 | GTG‐GGTG = frameshift in glutamine ABC transporter, permease/substrate‐binding protein | X | X | |||||||||
| Contig 26 | 9532 | 60 | ATT‐GTT = I‐V in purH, bifunctional phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase | X | X | |||||||||
| Contig 28 | 11,172 | 14 | GGA‐GGGA = frameshift 55 aa from end of integral membrane protein | X | ||||||||||
| Contig 36 | 7084 | 57 | GCT‐GTT = A‐V in oxoacyl‐ACP synthase | X | ||||||||||
| Contig 37 | 167 | GAA‐GAG = E‐E silent mutation in aspartate kinase | X | |||||||||||
| Contig 42 | 4874 | 6 | CTA‐CCA = L‐P in putative dienelactone hydrolase (no gene predicted by Glimmer) | X | X | X | ||||||||
| Contig 47 | 2473 | 7 | Deletion of T in Intergenic region according to Glimmer | |||||||||||
| Contig 64 | 3734 | 4 | AGT‐AGC = silent mutation in nucleotide‐binding protein | X | ||||||||||
| Contig 69 | 1283 | 14 | GGT‐AGT = G‐S in gene with similarity to HTH AraC regulatory protein | X | ||||||||||
| Contig 73 | 2467 | 19 | Frameshift after amino acid 187 in PTS system mannose family transporter subunit IID protein. | X | ||||||||||
| Contig 77 | 233 | 28 | CAC‐CAT = Silent mutation in pyruvate carboxylase | X | X | X | ||||||||
| Contig 79 | 2182 | 15 | GAC‐AAC = D‐N in putative uncharacterized protein. Verified to be a mutation | X | ||||||||||
| Contig 84 | 4263 | 8 | BLAST and Glimmer: not coding region, downstream of GTP‐binding protein TypA gene | X | ||||||||||
| Contig 85 | 8117 | 5 | AAA‐AAAA = frameshift toward end of Glimmer prediction, no Blast similarity to anything | X | ||||||||||
| Contig 103 | 928 | 81 | ATT‐ATC = silent mutation in glutathione reductase | X | ||||||||||
| Contig 111 | 1948 | 30 | AAT‐AAC = silent mutation in oxidoreductase | X | ||||||||||
| Contig 114 | 6645 | 2 | GGA‐GAA = G‐E in MccC family protein – putative peptidase | X | ||||||||||
| Contig 132 | 1556 | 6 | Intergenic region | X | ||||||||||
| Contig 146 | 971 | 23 | CAG‐CGG = Q‐R in acetylornithine deacetylase | X | ||||||||||
| Contig 153 | 1131 | 2 | 6 nt downstream of penicillin‐binding protein/beta‐lactamase | X | ||||||||||
| Contig 160 | 2010 | 5 | 6 nt downstream of conserved hypothetical protein, putative receptor | X | ||||||||||
Remarkably, all 10 plantaricin JK‐resistant strains harbored mutations in a region in contig 10 between positions 2518 and 3491. Contig 10 is 31 kb long, and the coverage in this part of the contig was 13–45 fold. The gene prediction software Glimmer 3 (Delcher et al. 2007) predicted a gene in contig 10 from position 2209 to 4041. A comparison of the encoded amino acid sequence (610 aa) with the Genbank nonredundant protein database using Blast revealed that the predicted gene encoded a protein with 87–88% similarity to proteins from various Weissella, Leuconostoc, and Lactobacillus species. These proteins are annotated as APC family amino acid‐polyamine‐organocation transporters, amino acid permeases, and amino acid transporters. A comparison of the sequence to the Pfam database of protein domains revealed the presence of a Pfam family AA_permease_2 domain (PF13520). The amino acid sequence of the protein, and the positions of predicted transmembrane helices (predicted using Geneious) are shown in Fig. 1, along with the positions of the mutations in the resistant isolates.
Figure 1.
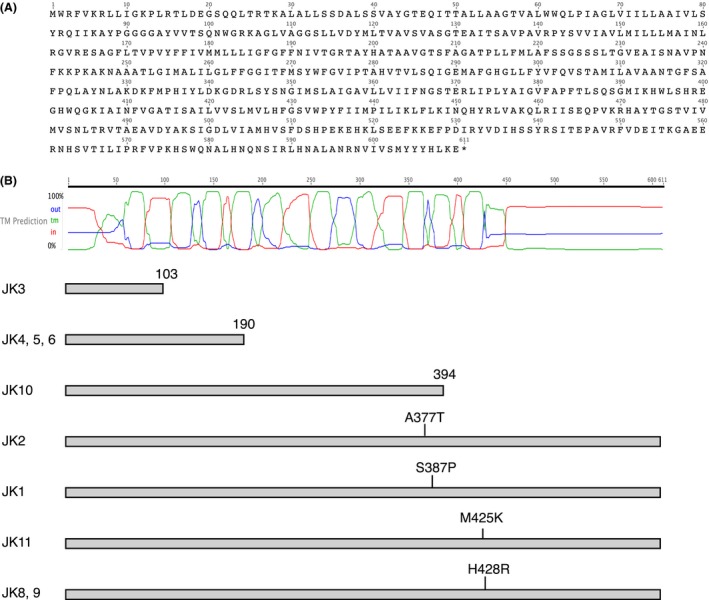
The APC superfamily protein encoded by the gene that was mutated in the 10 plantaricin JK‐resistant strains. (A) The amino acid sequence of the protein. (B) The predicted membrane topology of the protein. The green curve shows predicted transmembrane regions, the red curve cytoplasmic regions, and the blue curve extracellular regions. Below these curves, the protein products expected from the gene mutations are shown schematically for the 10 mutants.
The transmembrane (TM) prediction tool in the Geneious suite predicted 12 TM helices in the encoded protein (Fig. 1A). The predicted positions of the 12 TM helices correspond to their positions in other members of this protein superfamily (Reig et al. 2007). The resistant strains JK4, 5, and 6 contain an identical nucleotide change in the gene and may be results of a single mutational event. The same is the case for JK8 and 9. Three of the identified mutations lead to truncation of the protein: in JK3 after amino acid 103 (between TM helix 2 and 3), in JK4, 5, and 6 after amino acid 190 (in the middle of TM helix 5, due to a frameshift mutation) and in JK10 after amino acid 394 (after TM helix 10). The remaining four independent mutations are single amino acid changes occurring between position 377 and 428 (TM helix 10 and the predicted extracellular region between TM helix 11 and 12).
Nine of the 10 resistant strains contained one or more additional mutations located outside the APC family transporter gene (Table 2). However, these mutations appear to be randomly occurring in different genes, and we could not find any patterns that could explain the resistant phenotype. Importantly, in one strain (JK3 with 300‐fold increased resistance), the mutation in the APC superfamily gene was the only difference to the wild‐type genome that was detected.
We have made several efforts to verify the essential role of the APC family protein in the function of plantaricin JK, both by reintroducing the wild‐type gene in resistant W. viridescens mutants and by introducing the gene in resistant Lactobacillus strains using different cloning strategies. However, we did not succeed in obtaining viable cells carrying the intact transgene of this large membrane protein.
Discussion
Identification of mutations leading to resistance toward plantaricin JK
In our previous identification of the receptor of the two‐peptide bacteriocin lactococcin G (Kjos et al. 2014), we identified mutations in the sensitive target strains L. lactis IL1403 and MG1363, for which extensively annotated, fully assembled genomes were available in the sequence databases. In the present work, no reference genome was available for any plantaricin JK‐sensitive strains. As an alternative we used a crude assembly of the W. viridescens genome. Typically, crude assemblies of microbial genomes by short reads will consist of a large number of contigs (in our case 243) that cannot easily be assembled further into a single genome, due to errors in the assembly process. These errors are mainly due to repetitive regions in the genome (e.g., genes for ribosomal RNA). Assembly software will frequently misplace reads from such regions, leading to deterioration of consensus sequences and consequent problems in the further assembly. When compared with mutant sequence reads, errors in the consensus sequences may erroneously be reported as polymorphisms. We found that most of such false positives could be removed by including a comparison of the wild‐type sequence reads and the assembly constructed from them in the polymorphism analysis, and by disregarding differences reported in low‐coverage regions of the assembly. Long‐read sequencing such as Pacbio SMRT sequencing might provide a better alternative for contig assembly but is at the moment still significantly more expensive than short read Illumina sequencing.
The APC family protein is a likely plantaricin JK receptor
The results from the sequence comparisons revealed a clear correlation between plantaricin JK resistance and mutations in a contig 10 gene encoding an APC family protein, and thus strongly suggest that the APC family protein is involved in the mode of action of the bacteriocin that eventually leads to membrane leakage (Moll et al. 1999). The most likely role of the APC protein would be as a receptor protein. The APC protein has several of the properties expected for a bacteriocin receptor: similar to other identified bacteriocin receptors (Diep et al. 2007; Gabrielsen et al. 2012; Uzelac et al. 2013; Kjos et al. 2014) it is a transmembrane protein, with regions of the polypeptide chain exposed on the outer surface of the membrane. Like several of the other identified bacteriocin receptors (mannose PTS permease, glucose PTS permease, and maltose ABC transporter) it is a transport protein, which means that it possesses some kind of pore‐opening mechanism that could be hijacked and forced to open due to bacteriocin binding. Furthermore, the APC family protein sequence of W. viridescens is only 76.4% identical to the closest relative in the public databases and thus considerably different from its homologs in other sequenced Weissella and Lactobacillus strains. Assuming that plantaricin JK uses this protein as a sequence‐specific target, this may explain the narrow inhibitory spectrum of this bacteriocin, which only targets a small subset of strains within these genera.
Also relevant here is the fact that except for JK11, which was marginally (fivefold) more resistant than the wild‐type, the mutants with increased resistance toward plantaricin JK are as sensitive as the wild‐type toward the two‐peptide bacteriocin plantaricin EF. This indicates that the resistance was specific against plantaricin JK, and that plantaricin JK and EF have different receptors, consistent with an earlier study that showed that plantaricin EF causes leakage of small monovalent cations, but not anions, whereas plantaricin JK causes leakage of some anions, but not cations (Moll et al. 1999).
Position of the mutations in the APC family protein
The APC superfamily of transporters specific for amino acids, polyamines, and organocations is one of the largest protein superfamilies and has members in all kingdoms of life (Jack et al. 2000; Reig et al. 2007). In the motif database Pfam the APC superfamily has been subdivided into 20 families, and based on sequence comparisons, the putative plantaricin JK receptor should belong to the AA_permease_2 family.
As shown in Figure 1B, three of the seven identified mutations lead to truncation of the APC family protein and removal of two, seven, and ten TM helices, respectively. Most likely, these truncations all result in an inactive protein. Four of the mutations lead to amino acid substitutions. Two of these are present in TM helix 10, which according to the prediction tool MEMSAT‐SVM ((Nugent and Jones 2012), http://bioinf.cs.ucl.ac.uk/psipred/?memsatsvm=1) may be a pore‐lining helix, whereas the remaining two are localized to the extracellular (according to predictions) region between TM helix 11 and 12. The positioning of these mutations to the vicinity of a membrane pore and to an extracellular loop, respectively, is fully consistent with their effect on the action of the bacteriocin.
Generally, there is a good correlation between the nature of the mutations and the degree of resistance they lead to. Thus, two of the truncations (JK3 and JK4, 5, 6) lead to 3–600 fold increased resistance, whereas the point mutations seem to have a somewhat smaller effect. It is, however, noteworthy that the mutation in JK10, leading to truncation after amino acid 394, gives a lower increase in the resistance than the point mutations in position 425 and 428 in JK8, 9, and 11.
Our identification of an APC family protein as a putative membrane protein receptor of plantaricin JK is fully based on the observed accumulation of point mutations in the encoding gene, where the mutations in some cases lead to the introduction of premature stop codons and in other cases to amino acid residue changes in regions that presumably are important for the bacteriocin/protein interaction. We will continue our efforts to produce the protein by heterologous expression or in vitro expression, both to further verify its function as a plantaricin JK receptor and to study the molecular interactions between the protein, the bacteriocin, and the immunity protein.
Conflict of Interest
None declared.
Supporting information
Table S1. Each row represents a polymorphism reported by VAAL, with the position of the polymorphism detailed in the leftmost column, followed by information about the strain distribution (wild‐type and 10 mutants) of the polymorphism.
MicrobiologyOpen 2016; 5(4): 700–708
References
- Anderssen, E. L. , Diep D. B., Nes I. F., Eijsink V. G. H., and Nissen‐Meyer J.. 1998. Antagonistic activity of Lactobacillus plantarum C11: two new two‐peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Appl. Environ. Microbiol. 64:2269–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich, A. , Nurk S., Antipov D., Gurevich A. A., M. Dvorkin , Kulikov A. S., et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single‐cell sequencing. J. Comput. Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum, M. J. , Hayema B. J., Jeeninga R. E., Kok J., and Venema G.. 1991. Organization and nucleotide sequences of two lactococcal bacteriocin operons. Appl. Environ. Microbiol. 57:492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieler, S. , Silva F., Soto C., and Belin D.. 2006. Bactericidal activity of both secreted and nonsecreted microcin E492 requires the mannose permease. J. Bacteriol. 188:7049–7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, M. D. , Samelis J., and Metaxopoulos J.. 1993. Taxonomic studies on some leuconostoc‐like organisms from fermented sausages: description of a new genus Weissella for the Leuconostoc paramesenteroides . J. Appl. Bacteriol. 75:595–603. [DOI] [PubMed] [Google Scholar]
- Cotter, P. D. , Ross R. P., and Hill C.. 2013. Bacteriocins ‐ a viable alternative to antibiotics? Nat. Rev. Microbiol. 11:95–105. [DOI] [PubMed] [Google Scholar]
- Delcher, A. L. , Bratke K. A., Powers E. C., and Salzberg S. L.. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics(Oxford, England) 23:673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep, D. B. , Håvarstein L. S., and Nes I. F.. 1995. A bacteriocin‐like peptide induces bacteriocin synthesis in Lactobacillus plantarum C11. Mol. Microbiol. 18:631–639. [DOI] [PubMed] [Google Scholar]
- Diep, D. B. , Skaugen M., Salehian Z., Holo H., and Nes I. F.. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl Acad. Sci. USA 104:2384–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep, D. B. , Skaugen M., Salehian Z., Holo H., and Nes I. F.. 2009. An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum . Peptides 30:1562–1574. [DOI] [PubMed] [Google Scholar]
- Gabrielsen, C. , Brede D. A., Hernández P. E., Nes I. F., and Diep D. B.. 2012. The maltose ABC transporter in Lactococcus lactis facilitates high‐level sensitivity to the circular bacteriocin garvicin ML. Antimicrob. Agents Chemother. 56:2908–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia De Gonzalo, C. V. , Denham E. L., Mars R. A. T., J. Stülke , van der Donk W. A., and van Dijl J. M.. 2015. The phosphoenolpyruvate: sugar phosphotransferase system is involved in sensitivity to the glucosylated bacteriocin sublancin. Antimicrob. Agents Chemother. 59:6844–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravesen, A. , Ramnath M., Rechinger K. B., Andersen N., Jänsch L., Héchard Y.. 2002. High‐level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes . Microbiology(Reading) 148:2361–2369. [DOI] [PubMed] [Google Scholar]
- Hauge, H. H. , Mantzilas D., Eijsink V. G. H., and Nissen‐Meyer J.. 1999. Membrane‐mimicking entities induce structuring of the two‐peptide bacteriocins plantaricin E/F and plantaricin J/K. J. Bacteriol. 181:740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Héchard, Y. , and Sahl H.‐G.. 2002. Mode of action of modified and unmodified bacteriocins from Gram‐positive bacteria. Biochimie 84(5–6):545–557. [DOI] [PubMed] [Google Scholar]
- Jack, D. L. , Paulsen I. T., and Saier M. H. Jr. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology(Reading) 146:1797–1814. [DOI] [PubMed] [Google Scholar]
- Kjos, M. , Salehian Z., Nes I. F., and Diep D. B.. 2010a. An extracellular loop of the mannose phosphotransferase system component IIC is responsible for specific targeting by class IIa bacteriocins. J. Bacteriol. 192:5906–5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos, M. , Snipen L., Salehian Z., Nes I. F., and Diep D. B.. 2010b. The abi proteins and their involvement in bacteriocin self‐immunity. J. Bacteriol. 192:2068–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos, M. , Oppegård C., Diep D. B., Nes I. F., Nissen‐Meyer J., and Kristensen T.. 2014. Sensitivity to the two‐peptide bacteriocin lactococcin G is dependent on UppP, an enzyme involved in cell‐wall synthesis. Mol. Microbiol. 92:1177–1187. [DOI] [PubMed] [Google Scholar]
- Magoc, T. , and Salzberg S. L.. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics(Oxford, England) 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, G. N. , van den Akker E., Hauge H. H., Nissen‐Meyer J., Nes I. F., and Konings W. N.. 1999. Complementary and overlapping selectivity of the two‐peptide bacteriocins plantaricin EF and JK. J. Bacteriol. 181:4848–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen‐Meyer, J. , Rogne P., Oppegård C., Haugen H. S., and Kristiansen P. E.. 2009. Structure‐function relationships of the non‐lanthionine‐containing peptide (class II) bacteriocins produced by Gram‐positive bacteria. Curr. Pharm. Biotechnol. 10:19–37. [DOI] [PubMed] [Google Scholar]
- Nugent, T. , and Jones D. T.. 2012. Detecting pore‐lining regions in transmembrane protein sequences. BMC Bioinformatics 13:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum, C. , Ohsumi T. K., Gomez J., Aquadro J., Victor T. C., Warren R. M., et al. 2008. Sensitive, specific polymorphism discovery in bacteria using massively parallel sequencing. Nat. Methods 6:67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppegård, C. , Fimland G., Thorbaek L., and Nissen‐Meyer J.. 2007. Analysis of the two‐peptide bacteriocins lactococcin G and enterocin 1071 by site‐directed mutagenesis. Appl. Environ. Microbiol. 73:2931–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppegård, C. , Schmidt J., Kristiansen P. E., and Nissen‐Meyer J.. 2008. Mutational analysis of putative helix‐helix interacting GxxxG‐motifs and tryptophan residues in the two‐peptide bacteriocin lactococcin G. Biochemistry 47:5242–5249. [DOI] [PubMed] [Google Scholar]
- Oppegård, C. , Emanuelsen L., Thorbek L., Fimland G., and Nissen‐Meyer J.. 2010. The lactococcin G immunity protein recognizes specific regions in both peptides constituting the two‐peptide bacteriocin lactococcin G. Appl. Environ. Microbiol. 76:1267–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramnath, M. , Beukes M., Tamura K., and Hastings J. W.. 2000. Absence of a putative mannose‐specific phosphotransferase system enzyme IIAB component in a leucocin A‐resistant strain of Listeria monocytogenes, as shown by two‐dimensional sodium dodecyl sulfate‐polyacrylamide gel electrophoresis. Appl. Environ. Microbiol. 66:3098–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reig, N. , del Rio C., Casagrande F., Ratera M., Gelpí J. L., and Torrents D.. 2007. Functional and structural characterization of the first prokaryotic member of the L‐amino acid transporter (LAT) family: a model for APC transporters. J. Biol. Chem. 282:13270–13281. [DOI] [PubMed] [Google Scholar]
- Riley, M. A. , and Wertz J. E.. 2002. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56:117–137. [DOI] [PubMed] [Google Scholar]
- Rogne, P. , Haugen C., Fimland G., Nissen‐Meyer J., and Kristiansen P. E.. 2009. Three‐dimensional structure of the two‐peptide bacteriocin plantaricin JK. Peptides 30:1613–1621. [DOI] [PubMed] [Google Scholar]
- Simpson, J. T. , Wong K., Jackman S. D., Schein J. E., Jones S. J. M., and Birol I.. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19:1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzelac, G. , Kojic M., Lozo J., Aleksandrzak‐Piekarczyk T., Gabrielsen C., Kristensen T.. 2013. A Zn‐dependent metallopeptidase is responsible for sensitivity to LsbB, a class II leaderless bacteriocin of Lactococcus lactis subsp. lactis BGMN1‐5. J. Bacteriol. 195:5614–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino, D. R. , and Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Each row represents a polymorphism reported by VAAL, with the position of the polymorphism detailed in the leftmost column, followed by information about the strain distribution (wild‐type and 10 mutants) of the polymorphism.


