Abstract
Background
Vaginal lactobacilli offer protection against recurrent urinary infections, bacterial vaginosis, and vaginal candidiasis.
Objective
To characterise the isolated vaginal lactobacilli strains for their probiotic properties and to compare their probiotic potential.
Methods
The Lactobacillus strains were isolated from vaginal samples by conventional culturing and identified by sequencing of the 16S rDNA fragment. Several functional properties were detected (production of hydrogen peroxide and lactic acid; antagonistic activity against Escherichia coli, Candida albicans, Candida glabrata, and Gardnerella vaginalis; auto-aggregation and adhesiveness) as well as safety (haemolytic activity, antibiotic susceptibility, presence of transferrable resistance genes).
Results
A total of 135 vaginal lactobacilli strains of three species, Lactobacillus crispatus (56%), Lactobacillus jensenii (26%), and Lactobacillus gasseri (18%) were characterised using several functional and safety tests. Most of L. crispatus (89%) and L. jensenii (86%) strains produced H2O2. The best lactic acid producers were L. gasseri (18.2±2.2 mg/ml) compared to L. crispatus (15.6±2.8 mg/ml) and L. jensenii (11.6±2.6 mg/ml) (p<0.0001; p<0.0001, respectively). L. crispatus strains showed significantly higher anti-E. coli activity compared to L. jensenii. L. gasseri strains expressed significantly lower anticandidal activity compared to L. crispatus and L. jensenii (p<0.0001). There was no significant difference between the species in antagonistic activity against G. vaginalis. Nearly a third of the strains were able to auto-aggregate while all the tested strains showed a good ability to adhere to HeLa cells. None of the tested lactobacilli caused haemolysis. Although phenotypical resistance was not found to ampicillin, chloramphenicol, erythromycin, gentamycin, tetracycline, and vancomycin, the erm(B), tet(M), and tet(K) were detected in some strains. All strains were resistant to metronidazole, trimethoprim/sulfamethoxazole, and kanamycin.
Conclusions
Our study revealed that the production of different antimicrobial metabolites is highly strain-specific and that the metabolites are not correlated with each other. L. crispatus displays better antagonistic activity against E. coli and Candida spp. than L. gasseri and L. jensenii; therefore; a potential probiotic candidate could be found among L. crispatus strains.
Keywords: vaginal lactobacilli, probiotic, Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus jensenii
Lactobacilli are gram-positive, strict or aerotolerant anaerobic, rod-shaped bacteria that belong to microbiota of human gastrointestinal and genitourinary tract. It has been suggested that lactobacilli in vaginal microbiota of a healthy premenopausal woman offer protection against different diseases including recurrent urinary infections, bacterial vaginosis, and vaginal candidiasis. Thus, vaginal lactobacillar probiotics may prevent and/or treat these conditions. Moreover, it has been shown recently that also the orally administered vaginal strains of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 may restore vaginal microbiota (1). There are lots of probiotics for gastrointestinal tract application but only a few probiotics for urogenital tract currently on the market. Former studies have shown that probiotic properties are highly species- and strain-specific (2). Therefore, several thorough in vitro tests as well as in vivo animal studies and clinical trials are needed for developing new probiotics. The probiotic candidate must be assessed for the presence of functional properties including the production of antimicrobial compounds (e.g. hydrogen peroxide, organic acids, and bacteriocins) and direct antimicrobial activity against particular pathogens.
Though the genus Lactobacillus has a recognised safety status, as well as having a long history of safe use for food fermentation, several criteria must be taken into consideration when selecting a safe probiotic Lactobacillus strain (3–5). For safety reasons, the in vitro tests should include correct identification of a putative probiotic strain to the species level, determination of antibiotic resistance pattern, and testing for haemolytic activity.
The main objective of this study was to characterise previously isolated vaginal lactobacilli for their probiotic properties and to compare the probiotic potential between different Lactobacillus species.
Materials and methods
Participants and sample collection
Vaginal samples were obtained from volunteer women participating in the study of reproductive tract microbiome. The study was approved by the Ethics Review Committee on Human Research of the University of Tartu, Estonia (protocol No. 193/T-16, 31 May 2010). A total of 70 women from infertile couples (48 from partners of healthy men, 22 from partners of men with inflammatory prostatitis) and 64 healthy fertile women were enrolled in this study.
Bacterial and yeast strains
Lactobacillus strains
All vaginal specimens were inoculated onto MRS agar (de Man-Rogosa Sharpe agar, Oxoid Ltd., Basingstoke, UK) and the plates were incubated both microaerobically and anaerobically at 37°C for 48 h. Discrete colonies were picked and subcultured to obtain pure cultures. Lactobacilli were preliminarily identified to the genus level by Gram stain, colony morphology, and negative catalase test. Furthermore, lactobacilli were identified by sequencing of the 16S rDNA fragment as described earlier (6).
Opportunistic microorganisms
Staphylococcus aureus ATCC 25923 and Streptococcus pyogenes ATCC 19615 were used as positive controls for testing haemolytic activity of vaginal lactobacilli.
The antagonistic activity of putative probiotic strains was assessed against the following target bacteria: facultative anaerobic pyelonephritic strain of Escherichia coli ATCC 700336, cystitic strain of E. coli ATCC 700414, three clinical isolates of E. coli (isolated from women with bacterial vaginosis), Candida albicans ATCC 32032, Candida albicans RTN 071, Candida glabrata RTN 009, Gardnerella vaginalis ATCC 14018 (DSM 4944), and one clinical isolate of G. vaginalis.
Detection of hydrogen peroxide production
All lactobacilli were tested for the production of H2O2 on a tetramethylbenzidine (TMB, Sigma Chemical Co., St. Louis, Mo, USA) agar plate (7). The lactobacilli strains were plated on TMB agar containing TMB, horseradish peroxidase, Brucella agar base, hemin, starch, and vitamin K, and incubated for 48 h at 37°C in anaerobic conditions. After exposure to ambient air, hydrogen peroxide–producing colonies turned blue. Change of colour was assessed semi-quantitatively after 30 min. Colour intensity was designated as follows: − (negative),+(weak positive), ++ (moderate positive), and +++ (strong positive).
Detection of lactic acid production
The production of lactic acid was estimated by gas chromatography as described by Holdeman et al. (8). Analysis was performed after cultivation of lactobacilli at 37°C in modified MRS broth for 48 h in an anaerobic glove chamber (Concept 400, Biotrace, Bridgend, UK) with a gas mixture of CO2/H2/N2:5/5/90%. The samples were then centrifuged at 6,000 g for 10 min.
Lactic acid methylate samples were prepared by adding 0.4 g sodium chloride, 0.2 ml 50% sulphuric acid, and 2 ml methanol in 1 ml prepared culture broth. Tubes were heated at 60°C for 1.5 h; 1 ml water and 0.5 ml chloroform were added. The emulsion form was centrifuged at 3,000 rpm for 3 min. One microliter of the chloroform sample was injected into the GC column. The HP Chemical Station for GC system (A.06 revision) was used. The gas chromatograph (Hewlett Packard, Palo Alto, CA, USA) was equipped with a hydrogen flame ionisation detector, an autosampler (model 7683), and a capillary GC column HP-INNOWax (Hewlett-Packard) of 15 m×0.25 mm (i.d.), coated with cross-linked polyethylene glycol (film thickness 0.15 mm). The detector temperature was 250°C and the injector temperature was 200°C, respectively. The oven temperature programme was a gradient system; the initial temperature was 60°C for 1 min that was increased to 120°C at a rate of 10°C/min and held there for 10 min. Lactic acid (Sigma-Aldrich, St. Louis, Mo, USA) was used as a standard. Methylated stock solutions were a different concentration of each acid, namely 0.15, 0.3, 0.6, 1.2, 2.4, 3.6, and 4.8 mg/ml. Each standard solution was injected in duplicate to obtain its retention time and area under the curve. The chromatographic peak areas were integrated with a Hewlett-Packard networking integrator.
Detection of haemolytic activity
A single line of lactobacilli culture (grown in MRS broth for 48 h) was streaked onto blood agar plates containing human, sheep, or horse blood. The haemolytic activity of putative probiotics was verified visually after 24 h and 48 h of incubation in anaerobic (90% N2, 5% CO2, 5% H2) environments. Blood agar plates were examined for signs of β-haemolysis (clear zones around colonies), α-haemolysis (green halo around colonies), or γ-haemolysis (no zones around colonies) (9). The Staphylococcus aureus strain (ATCC 25923) and Streptococcus pyogenes strains (ATCC 19615) were used as positive controls.
Detection of auto-aggregation
Auto-aggregation was tested according to the modified method described previously (10). The strains were incubated in MRS broth (de Man Rogosa Sharp, Oxoid Ltd., Basingstoke, UK) for 18 h at 37°C in anaerobic conditions (CO2/H2/N2: 5/5/90%). Thereafter, lactobacilli were washed twice in PBS buffer and re-suspended in PBS to the concentration of 108 CFU/ml (McFarland 3.0). The suspension was vortexed for 10 s. Next, 0.2 ml of the upper layer was pipetted into the wells of the microtiter plate, and optical density (OD) at 600 nm was measured (A0). Second OD detection (At) was performed after the suspension was stored at room temperature for 2 h. Auto-aggregation was detected with the following calculation:
Auto-aggregation was also estimated visually – positive score was recorded when clearly visible sand-like particles were formed by the aggregated cells and settled in the bottom of the tubes, leaving a clear supernatant. The results were scored on a 3-point scale, depending on the time of the auto-aggregation. If the auto-aggregation was detected within 15 min, the reaction was scored at 3; within 15–30 min, the score was considered 2, and within 30–60 min, the reaction was scored as 1 (11). All measurements were performed three times and the average of separate detections was calculated.
Detection of adhesion
Altogether, 10 different L. crispatus strains with auto-aggregation properties and L. rhamnosus GG ATCC 53103 as a positive control were tested for their ability to adhere to HeLa cells. For adhesion testing, 2 ml of HeLa cell suspension was added to each well in six-well tissue culture plate at a concentration of 2×105 cells/ml and incubated in a 5% CO2 atmosphere at 37°C for 48 h until the cells were grown to approximately 60% confluence (12, 13). After 48 h, growing cells were twice washed with PBS.
Lactobacilli were incubated for 48 h at 37°C in liquid MRS (0.1 ml of seedbank in 4 ml of MRS), centrifuged at 3,000 rpm for 4 min and twice washed with PBS. Lactobacilli suspension was prepared in PBS according to McFarland 3.0. Lactobacilli suspension (0.1 ml) was added to 0.9 ml DMEM/Ham's F-12 with stable glutamine (GE Healthcare, Logan, Utah) and incubated in a 1 ml well at 37°C for 1 h in 10% CO2. Next, cells were washed in 1 ml of PBS five times and 1 ml of 0.05% Triton X100 was added for 10 min (14). Applying a 10-fold dilution method, lactobacilli were seeded onto MRS agar; the colonies were counted after incubation and expressed in% of adhesion (15).
N1 – the amount of adhered bacteria (log10CFU/ml); N0 – the amount of applied bacteria (log10CFU/ml).
All lactobacilli strains were tested in two repeats three times.
Antibiotic susceptibility testing
Susceptibility to ampicillin, chloramphenicol, clindamycin, erythromycin, gentamycin, kanamycin, metronidazole, nitrofurantoin, norfloxacin, streptomycin, tetracycline, trimethoprim/sulfamethoxazole, and vancomycin was assessed according to the European Union Commission recommendations for probiotic safety (16, 17).
The minimum inhibitory concentrations (MICs) were determined by the Etest® method (BioMérieux, Marcy l'Etoile, France). The assay was performed by suspending individual colonies picked up from fresh cultures on LSM agar (90% Iso-Sensitest (Oxoid Ltd., Basingstoke, UK) and 10% MRS (Oxoid)) plates incubated for 24 h at 37°C in an anaerobic glove chamber (Concept 400, Biotrace, Bridgend, UK) to McFarland standard 1 in sterile 0.85% NaCl solution. The suspension was swabbed in three directions on LSM agar and allowed to dry before applying the Etest strips (18, 19). Plates were incubated at 37°C for 48 h under anaerobic conditions before final results were recorded. MICs were read directly from the test strip according to the instructions of the manufacturer. Strains showing MICs less than EFSA's breakpoints were considered sensitive; otherwise, they were considered resistant (16, 17).
Detection of antimicrobial resistance genes
Total DNA of Lactobacillus strains was extracted using a QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany) following the manufacturer's protocol. The PCR assay mix (total volume, 50 µl) contained 20 pmol of each primer (Table 1), 1XPCR buffer (Fermentas, Lithuania), each deoxynucleoside triphosphate at a concentration of 200 µM, and 2U Taq DNA polymerase. A 50 ng portion of purified total DNA was used as a template. In a first PCR assay, tet genes encoding ribosomal protection proteins (RPP) were detected with degenerated primers DI and DII (20). If positive, additional PCR assays were performed with primers specific for the tetM, tetO, tetS, tetW, and tetQ genes. All Lactobacillus strains were tested for the presence of the tetracycline efflux genes tetK and tetL. Polymerase chain reaction amplicons for tet genes were performed in an Eppendorf PCR System (Eppendorf AG, Hamburg, Germany) with the following programmes: initial denaturation at 94°C for 5 min; 35 cycles of 94°C for 1 min, corresponding annealing temperature (Table 1) for 1 min, and 72°C for 2 min; and a final extension step at 72°C for 10 min. Amplification specifications for integrase Int1 were as follows: 94°C for 5 min, followed by 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 7 min. Amplicons were electrophoresed on 1.5% agarose gel, and a 1-kb or 100 bp ladders (Fermentas) were used as a molecular size marker.
Table 1.
PCR primers and amplification conditions
| Primer pair | Target gene | Sequence 5′–3′ | Annealing temp (°C) | Amplicon size (bp) | Reference for primers |
|---|---|---|---|---|---|
| DI DII |
RPP | GAYACNCCNGGNCAYRTNGAYTT GCCCARWANGGRTTNGGNGGNACYTC |
50 | 1,083 | Clermont et al., 1997 (20) |
| DI TetM-R |
tet(M) | GAYACNCCNGGNCAYRTNGAYTT CACCGAGCAGGGATTTCTCCAC |
55 | 1,513 | Clermont et al., 1997 (20) |
| TetS-F TetS-R |
tet(S) | ATCAAGATATTAAGGAC TTCTCTATGTGGTAATC |
55 | 573 | Charpentier et al., 1993 (21) |
| TetO-F TetO-R |
tet(O) | AATGAAGATTCCGACAATTT CTCATGCGTTGTAGTATTCCA |
55 | 781 | Sougakoff et al., 1987 (22) |
| TetK-F TetK-R |
tet(K) | TTATGGTGGTTGTAGCTAGAAA AAAGGGTTAGAAACTCTTGAAA |
55 | 348 | Gevers et al., 2003 (23) |
| TetL-F TetL-R |
tet(L) | GTMGTTGCGCGCTATATTCC GTGAAMGRWAGCCCACCTAA |
55 | 696 | Gevers et al., 2003 (23) |
| TetQ-F TetQ-R |
tet(Q) | AGAATCTGCTGTTTGCCAGTG CGGAGTGTCAATGATATTGCA |
56 | 169 | Ge et al., 2007 (24) |
| TetW-F TetW-R |
tet(W) | GAGAGCCTGCTATATGCCAGC GGGCGTATCCACAATGTTAAC |
64 | 168 | Kastner et al., 2006 (25) |
| ErmB-F ErmB-R |
erm(B) | CATTTAACGACGAAACTGGC GGAACATCTGTGGTATGGCG |
56 | 405 | Jensen et al., 1999 (26) |
| ermA-F ermA-R |
erm (A) | AAGCGGTAAACCCCTCTGA TTCGCAAATCCCTTCTCAAC |
55 | 190 | Klare et al., 2007 (27) |
| Int | int | GGCATCCAAGCAGCAAG AAGCAGACTTGACCTGA |
55 | – | Levesque et al., 1995 (28) |
Detection of antagonistic activity
Antagonistic activity of lactobacilli against urinary pathogens
The following target bacteria were used for detection of antimicrobial activity of lactobacilli: E. coli ATCC 700414, E. coli ATCC 700336, and three E. coli strains that were previously isolated from women with bacterial vaginosis.
The inhibitory effect of vaginal strains on E. coli strains was determined by a modified agar spot method as described previously (29, 30). Briefly, the tested lactobacilli cultures were spotted (1 µl) on the surface of MRS agar (20 ml) and incubated anaerobically for 24 h at 37°C. An overnight culture of E. coli (approximately 8×108 cells) was mixed with soft agar (0.7%) nutrient agar (20 ml) (Oxoid Ltd., Basingstoke, UK) and poured over the plate in which lactobacilli were grown. After anaerobic incubation for 24 h at 37°C, the plates were checked for inhibition zones around Lactobacillus spots and their diameters were recorded. All tests concerning the detection of antimicrobial activity were repeated three times in three independent experiments.
Antagonistic activity of lactobacilli against Candida spp.
The following target yeasts were used for detection of anticandidal activity of lactobacilli: Candida albicans ATCC 32032, Candida albicans RTN 071, and Candida glabrata RTN 009.
Anticandidal activity was detected by a modified agar overlay technique described by Strus et al. (31). Briefly, Lactobacillus isolate was cultured in MRS broth for 24 h at 37°C in anaerobic conditions. Afterwards, lactobacilli were cultured in the middle of the MRS agar plate as a 2-cm wide stripe using a sterile cotton swab. The plates were incubated in anaerobic conditions at 37°C for 48 h. The Lactobacillus spp. growth was then overlaid with melted MRS agar. After solidification of the agar overlay, a suspension of C. albicans or C. glabrata (obtained from overnight culture of Sabouraud agar (Oxoid)), in sterile saline with 0.5 MacFarland's score turbidity was streaked over the agar surface with a cotton swab. The plates were preincubated at 4°C for 4 h in a refrigerator, then kept at 37°C for 24 h in aerobic conditions, and left for 24 h at room temperature before reading the results. After adequate growth, the zones of inhibition of Candida spp. were evaluated using a measure of Lactobacillus spp. streak line with inhibition zone and divided by 2.
Antagonistic activity of lactobacilli against Gardnerella vaginalis strains
The reference strain of Gardnerella vaginalis ATCC 14018 (DSM 4944) and one clinical isolate of G. vaginalis were used as target bacteria.
Culture supernatants of selected lactobacilli strains were prepared by cultivation of lactobacilli in MRS broth anaerobically at 37°C for 48 h. Thereafter, removal of the cells by centrifugation at 6,000 g for 15 min was done. The pH of each supernatant was measured (Hanna Instruments HI 9024C, Singapore) and supernatants were sterilised using 0.20 µm millipore filters (Sarstedt, Nümbrecht, Germany). Natural cell-free culture supernatants of isolated vaginal lactobacilli were examined for their antibacterial activity on 96-well microtiter plates.
G. vaginalis strains were grown on BD Gardnerella Selective Agar with 5% human blood in an anaerobic environment for 48 h. G. vaginalis colonies were suspended in sterile saline to achieve a turbidity of about 0.5 McFarland.
Furthermore, G. vaginalis suspensions (20 µl), 5% human serum (Sigma-Aldrich, St. Louis, Mo, USA) in Schaedler broth (Oxoid) (150 µl), and natural cell-free supernatants (30 µl) were transferred to a 96-well microplate (served as test well). G. vaginalis suspensions (20 µl) and 5% human serum in Schaedler broth (180 µl) transferred into microplate served as the positive control (control well). Growth and growth inhibition of G. vaginalis was determined by monitoring the changes in OD at 600 nm as measured with a absorbance microplate reader (Sunrise, Tecan, Austria) after 0 and 48 h incubation. Readings from the 0 h incubation time were used to determine the absorbance due to media and inoculum.
The percentage of reduction in growth of target bacteria due to cell-free supernatant was calculated as per the following formula:
Experiments were repeated three times with two replicates. The inhibition of G. vaginalis growth in percent was ranked as high (>78.0%), intermediate (67.3–77.0%), or low (<67.2%).
Statistical analysis
Statistical analysis was performed by using R-2.14.0 (A Language and Environment, www.r-project.org). Data of lactic acid production and antagonistic activity of putative probiotic lactobacilli strains were analysed using a Student's t-test or Mann-Whitney rank sum test with the Bonferroni correction for multiple groups. Data were expressed as mean±standard deviation. Comparison of Lactobacillus counts of different subject groups was performed using the χ2-test. Spearman rank order correlation was used to compare hydroxide peroxide and lactic acid productions. Differences were considered significant when p-value was <0.05.
Results
A total of 135 vaginal lactobacilli were isolated from reproductive-aged women. All isolated vaginal Lactobacillus strains were finally identified by sequencing of the 16S rDNA fragment and belonged to three species: Lactobacillus crispatus (56%), Lactobacillus jensenii (26%), and Lactobacillus gasseri (18%). There was no significant difference in the distribution of these three species among healthy and infertile couple groups (data not shown).
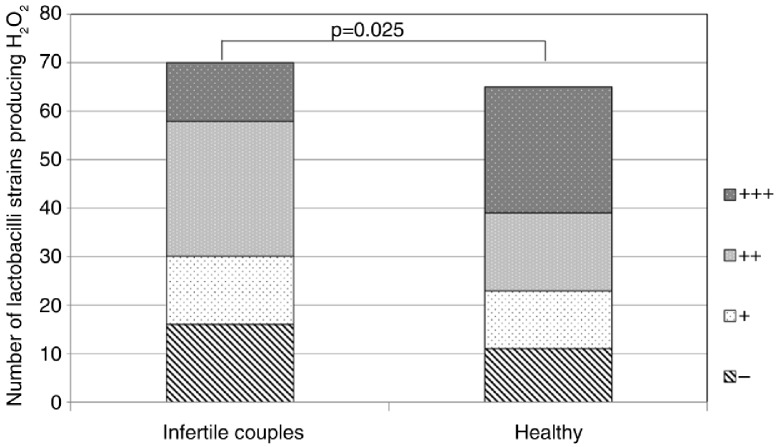
Production of hydrogen peroxide was tested in all 135 vaginal lactobacilli strains by the semi-qualitative TMB-peroxidase assay. In total, 108 Lactobacillus strains out of 135 produced some amount of hydrogen peroxide. Most of L. crispatus (89%) and L. jensenii (86%) strains and only 42% of L. gasseri strains produced hydrogen peroxide (in comparison with L. crispatus (p<0.0001) and in comparison with L. jensenii (p=0.0003)). Moreover, lactobacilli strains of healthy women expressed higher production of hydrogen peroxide compared to strains from infertile couples (p=0.025) (Fig. 1).
Fig. 1.
H2O2 production in lactobacilli strains of different study groups (135 Lactobacillus strains). −, negative; +, weak positive; ++, moderate positive; + + +, strong positive.
Lactic acid production was measured among hydrogen peroxide–producing Lactobacillus strains. Among the tested Lactobacillus strains, the best lactic acid producers were L. gasseri (18.2±2.2 mg/ml) followed by L. crispatus (15.6±2.8 mg/ml) and L. jensenii (11.6±2.6 mg/ml) (p<0.0001; p<0.0001, respectively). Lactic acid production did not correlate with hydrogen peroxide production and no differences were found between the donors’ groups.
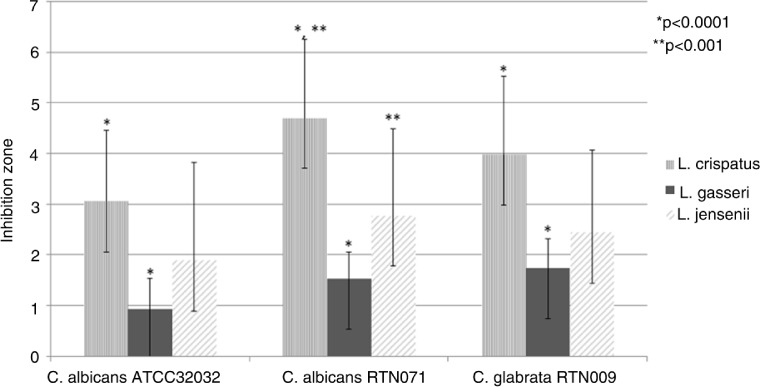
Furthermore, 56 lactobacilli strains were selected according to their production of hydrogen peroxide and lactic acid for testing antagonistic activity toward E. coli and Candida spp. strains using modified agar spot method. L. crispatus strains showed the highest anti-E. coli activity (Table 2) and the highest antagonistic activity against both Candida albicans and Candida glabrata (Fig. 2).
Table 2.
Antimicrobial activity of vaginal lactobacilli isolates against urogenital E. coli strains determined by a modified agar-spot method (inhibition zones in mm, expressed as mean±SD)
| Species | E. coli RTN038a | E. coli RTN047a | E. coli RTN100a | E. coli ATCC 700414b | E. coli ATCC 700336b |
|---|---|---|---|---|---|
| L. crispatus | 8.62±1.25* | 10.70±1.21 | 9.36±1.67** | 10.14±1.00*** | 10.42±1.28 |
| L. gasseri | 7.89±1.10 | 10.07±1.22 | 8.74±2.74 | 9.92±1.52 | 9.63±1.24 |
| L. jensenii | 6.47±2.23* | 9.67±1.75 | 6.90±3.18** | 8.58±2.66*** | 8.82±3.15 |
Three E. coli strains were previously isolated from women with bacterial vaginosis
reference strains: pyelonephritic strain of E. coli ATCC 700336 and cystitic strain of E. coli ATCC 700414. *p=0.0002; **p=0.003; ***p=0.006.
Fig. 2.
Anticandidal activity of vaginal lactobacilli (56 Lactobacillus strains). Significant differences in inhibition of Candida spp. between Lactobacillus spp.: *p<0.0001, *p<0.001.
Thereafter, 31 most promising lactobacilli strains were selected out of 56 for further experiments. We found that the strains of all three lactobacilli species had quite similar antagonistic activity against two different G. vaginalis strains (data not shown). Only 12 lactobacilli strains out of 31 were able to auto-aggregate and there was no statistical difference in auto-aggregation among different Lactobacillus species (data not shown). None of the 31 lactobacilli showed auto-aggregation within the 15 min detection period. All tested L. crispatus strains adhered to HeLa cells; the adhesion percentage was between 92.1 and 78.5%.
Safety assessment tests showed that none of the tested lactobacilli caused the lysis of erythrocytes on human, sheep, or horse blood agar while complete lysis (β-haemolysis) was registered in case of positive controls (Streptococcus pyogenes and Staphylococcus aureus). No resistance was detected to ampicillin, chloramphenicol, erythromycin, gentamycin, tetracycline, and vancomycin. All the tested 31 lactobacilli strains were resistant to metronidazole, trimethoprim/sulfamethoxazole, and kanamycin, and 21 L. crispatus strains out of 26 as well as all L. gasseri and L. jensenii were resistant to norfloxacin (Table 3). Resistance genes detected in the vaginal Lactobacillus strains are listed in Table 4. Although all tested strains were phenotypically susceptible to tetracycline and erythromycin, we detected the following resistance genes in some strains: erm(B), tet(M), and tet(K).
Table 3.
Antibiotic susceptibility of the 31 Lactobacillus strains measured by E-test method
| AM | CL | CM | EM | GM | KM | MZ | NI | NX | SM | TC | TS | VA | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. crispatus (n=26) | S | 26 | 26 | 24 | 26 | 26 | 0 | 0 | 26 | 5 | 23 | 26 | 0 | 26 |
| R | 0 | 0 | 2 | 0 | 0 | 26 | 26 | 0 | 21 | 3 | 0 | 26 | 0 | |
| L. gasseri (n=3) | S | 3 | 3 | 0 | 3 | 3 | 0 | 0 | 2 | 0 | 2 | 3 | 0 | 3 |
| R | 0 | 0 | 3 | 0 | 0 | 3 | 3 | 1 | 3 | 1 | 0 | 3 | 0 | |
| L. jensenii (n=2) | S | 2 | 2 | 2 | 2 | 2 | 0 | 0 | 2 | 0 | 1 | 2 | 0 | 2 |
| R | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 2 | 1 | 0 | 2 | 0 |
AM, ampicillin; CL, chloramphenicol; CM, clindamycin; EM, erythromycin; GM, gentamycin; KM, kanamycin; MZ, metronidazole; NI, nitrofurantoin; NX, norfloxacin; SM, streptomycin; TC, tetracycline; TS, trimethoprim/sulfamethoxazole; VA, vancomycin; S, susceptible; R, resistant according to EFSA 2008.
Table 4.
Antimicrobial resistance genes in 31 selected Lactobacillus strains
| erm(B) | tet(M) | tet(S) | tet(O) | tet(K) | tet(L) | RPP | ||
|---|---|---|---|---|---|---|---|---|
| L. crispatus (n=26) | Present | 14 | 0 | 0 | 0 | 6 | 0 | 6 |
| Absent | 12 | 26 | 26 | 26 | 20 | 26 | 20 | |
| L. gasseri (n=3) | Present | 3 | 0 | 0 | 0 | 2 | 0 | 2 |
| Absent | 0 | 3 | 3 | 3 | 1 | 3 | 1 | |
| L. jensenii (n=2) | Present | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| Absent | 1 | 1 | 2 | 2 | 2 | 2 | 1 |
RPP, tet genes encoding ribosomal protection proteins.
Discussion
Lactobacillus crispatus was the most frequent cultured species in vaginal samples followed by L. gasseri and L. jensenii that corresponds to previous studies (32, 33). The presence of L. crispatus is considered a major determinant to the stability of the normal vaginal microbiota in women of reproductive age (34). In general, vaginal lactobacilli offer protection from different diseases including recurrent urinary infections, bacterial vaginosis, and vaginal candidiasis through production of different antimicrobial compounds (e.g. hydrogen peroxide, lactic acid) and capacity to adhere and compete for adhesion sites in the vaginal epithelium (35). Since these beneficial properties are highly strain-specific, several species and strains must be screened when new probiotics are developed.
In our study, the lactobacilli strains isolated from healthy women produced significantly higher amounts of hydrogen peroxide compared to strains of infertile couple's women. It has been shown earlier that H2O2-producing lactobacilli are present in the vagina of most healthy women but are absent in most women with bacterial vaginosis (36). In our study, 108 Lactobacillus strains out of 135 produced hydrogen peroxide. The majority of L. crispatus and L. jensenii, and less than half of L. gasseri strains produced hydrogen peroxide. This corresponds to the previous data from literature indicating that L. crispatus and L. jensenii produce hydrogen peroxide more frequently and in larger quantities than L. gasseri (7).
Lactic acid is another powerful antimicrobial compound produced by lactic acid bacteria. The production of lactic acid by lactobacilli results in lowering pH that is important for preventing the colonisation and proliferation of non-indigenous pathogenic organisms in the vagina (35). Lactic acid production has been detected in earlier studies among vaginal lactobacilli isolates. Gil et al. (37) reported that L. salivarius was the highest lactic acid producer. In our study, the best lactic acid producers among tested Lactobacillus species were L. gasseri, followed by L. crispatus and L. jensenii.
We subsequently tested antagonistic activity of lactobacilli towards selected pathogens using three different modified methods. L. crispatus strains showed significantly higher anti-E. coli activity compared to L. jensenii, while L. gasseri strains expressed significantly lower anti-candidal activity compared to L. crispatus and L. jensenii. Inhibition of E. coli and Candida spp. by vaginal lactobacilli has been reported in several studies (37, 38). Although Aroutcheva et al. (39) have previously showed an inhibitory effect of bacteriocin obtained by vaginal lactobacilli against G. vaginalis, we did not find significant differences among our lactobacilli strains in antagonistic activity against two different G. vaginalis strains.
The colonisation and adhesion of bacteria is probably influenced by auto-aggregation. Testing auto-aggregative ability is an indicator of adhesion property as it favours colonisation through the formation of a bacterial film and also contributes to the exclusion of pathogens (40). We found that auto-aggregation was expressed in one third of lactobacilli strains only. At the same time, the strains expressed good adhesion ability to HeLa cells.
Although lactobacilli have a long history of safe use, they may cause clinical conditions like bacteraemia and endocarditis in rare cases (41). Therefore, several safety tests should be carried out. Safety assessment of the lactobacilli strains in our study included detection of haemolysis, antibiotic susceptibility, and transferable antibiotic resistance genes. Our tested strains appeared to be safe, since none of the tested lactobacilli strains caused the lysis of erythrocytes of human, sheep, or horse blood. As for antibiotic resistance, several studies provide the evidence of resistance to erythromycin and/or tetracycline among human faecal Lactobacillus isolates (42). In the last decade, some concerns have been expressed that the microorganisms used in food can be vehicles for transmission of antibiotic resistance genes (16, 17). Therefore, safety assessment of Lactobacillus strains should include the detection of their antibiotic resistance profile (43). In our study, no resistance among the tested lactobacilli was found to ampicillin, chloramphenicol, erythromycin, gentamycin, tetracycline, and vancomycin. Nevertheless, high level of resistance to norfloxacin, metronidazole, and trimethoprim–sulfamethoxazole is similar to some other reports (42, 44) that could be interpreted as high natural resistance to these antibiotics. Although all the tested strains were phenotypically susceptible to tetracycline and erythromycin, we detected the following resistance genes in some strains: erm(B), tet(M), and tet(K). Co-occurrence of several genes is necessary to display phenotypic resistance. Some studies have indicated a higher risk of antibiotic resistance transfer than previously believed, especially during treatment with antibiotics (45–47). Therefore, it is important to avoid Lactobacillus strains carrying resistance genes during the novel probiotic development.
In conclusion, our study revealed that the production of different antimicrobial metabolites is highly strain-specific and that the metabolites are not correlated with each other. L. crispatus displays better antagonistic activity against E. coli and Candida spp. than L. gasseri and L. jensenii; therefore, a potential probiotic candidate could be found among L. crispatus strains.
Acknowledgements
This study was supported by Estonian Research Council (grant No. IUT34-19), Estonian Ministry of Education and Research (grant No. KOGU-HUMB), and Enterprise Estonia (grants No. EU30020; EU48695).
Conflict of interest and funding
No conflict of interest declared. The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
References
- 1.Macklaim JM, Clemente JC, Knight R, Gloor GB, Reid G. Changes in vaginal microbiota following antimicrobial and probiotic therapy. Microb Ecol Health Dis. 2015;26 doi: 10.3402/mehd.v26.27799. 27799, doi: http://dx.doi.org/10.3402/mehd.v26.27799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanders ME. Probiotics: definition, sources, selection, and uses. Clin Infect Dis. 2008;46(Suppl 2):S58–61; discussion S144–51. doi: 10.1086/523341. [DOI] [PubMed] [Google Scholar]
- 3.EFSA. Guidance on the scientific requirements for health claims related to antioxidants, oxidative damage and cardiovascular health. EFSA J. 2011;9:2474. doi: 10.2903/j.efsa.2018.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.FAO/WHO. Report of a joint FAO/WHO working group on drafting guidelines for the evaluation of probiotics in food. London, Ontario, Canada: 2002. Guidelines for the evaluation of probiotics in food. [Google Scholar]
- 5.Reid G, Jass J, Sebulsky MT, McCormick JK. Potential uses of probiotics in clinical practice. Clin Microbiol Rev. 2003;16:658–72. doi: 10.1128/CMR.16.4.658-672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smidt I, Kiiker R, Oopkaup H, Lapp E, Rööp T, Truusalu K, et al. Comparison of detection methods for vaginal lactobacilli. Benef Microbes. 2015;6:747–51. doi: 10.3920/BM2014.0154. [DOI] [PubMed] [Google Scholar]
- 7.Eschenbach DA, Davick PR, Williams BL, Klebanoff SJ, Young-Smith K, Critchlow CM, et al. Prevalence of hydrogen peroxide-producing Lactobacillus species in normal women and women with bacterial vaginosis. J Clin Microbiol. 1989;27:251–6. doi: 10.1128/jcm.27.2.251-256.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holdeman LV, Cato EP, Moore WEC. Anaerobe laboratory manual. Blacksburg, VA: Virginia Polytechnic Institute and State Laboratory; 1977. [Google Scholar]
- 9.Ruoff KL. Algorithm for Identification of Aerobic Gram-Positive Cocci. In: Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, editors. Manual of Clinical Microbiology. 9th ed. Washington, DC: ASM Press; 2006. pp. 365–367. (2007) [Google Scholar]
- 10.Kos B, Suskovic J, Vukovic S, Simpraga M, Frece J, Matosic S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol. 2003;94:981–7. doi: 10.1046/j.1365-2672.2003.01915.x. [DOI] [PubMed] [Google Scholar]
- 11.Bujnakova D, Kmet V. Aggregation of animal lactobacilli with O157 enterohemorrhagic Escherichia coli . J Vet Med B Infect Dis Vet Public Health. 2002;49:152–4. doi: 10.1046/j.1439-0450.2002.00526.x. [DOI] [PubMed] [Google Scholar]
- 12.Kaewnopparat S, Dangmanee N, Kaewnopparat N, Srichana T, Chulasiri M, Settharaksa S. In vitro probiotic properties of Lactobacillus fermentum SK5 isolated from vagina of a healthy woman. Anaerobe. 2013;22:6–13. doi: 10.1016/j.anaerobe.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Verdenelli MC, Coman MM, Cecchini C, Silvi S, Orpianesi C, Cresci A. Evaluation of antipathogenic activity and adherence properties of human Lactobacillus strains for vaginal formulations. J Appl Microbiol. 2014;116:1297–307. doi: 10.1111/jam.12459. [DOI] [PubMed] [Google Scholar]
- 14.Goh YJ, Azcarate-Peril MA, O'Flaherty S, Durmaz E, Valence F, Jardin J, et al. Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2009;75:3093–105. doi: 10.1128/AEM.02502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik S, Petrova MI, Claes IJ, Verhoeven TL, Busschaert P, Vaneechoutte M, et al. The highly autoaggregative and adhesive phenotype of the vaginal Lactobacillus plantarum strain CMPG5300 is sortase dependent. Appl Environ Microbiol. 2013;79:4576–85. doi: 10.1128/AEM.00926-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.EFSA. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012;10:2740. [Google Scholar]
- 17.EFSA. Technical guidance. Update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. Prepared by the Panel on Additives and Products or Substances used in Animal Feed (Question No. EFSA-Q-2008-004) EFSA J. 2008;732:1–15. doi: 10.2903/j.efsa.2008.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayrhofer S, Domig KJ, Mair C, Zitz U, Huys G, Kneifel W. Comparison of broth microdilution, Etest, and agar disk diffusion methods for antimicrobial susceptibility testing of Lactobacillus acidophilus group members. Appl Environ Microbiol. 2008;74:3745–8. doi: 10.1128/AEM.02849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubio R, Jofre A, Martin B, Aymerich T, Garriga M. Characterization of lactic acid bacteria isolated from infant faeces as potential probiotic starter cultures for fermented sausages. Food Microbiol. 2014;38:303–11. doi: 10.1016/j.fm.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 20.Clermont D, Chesneau O, De Cespedes G, Horaud T. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob Agents Chemother. 1997;41:112–6. doi: 10.1128/aac.41.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charpentier E, Gerbaud G, Courvalin P. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene. 1993;131:27–34. doi: 10.1016/0378-1119(93)90665-p. [DOI] [PubMed] [Google Scholar]
- 22.Sougakoff W, Papadopoulou B, Nordmann P, Courvalin P. Nucleotide-sequence and distribution of gene tet(O) encoding tetracycline resistance in Campylobacter coli . FEMS Microbiol Lett. 1987;44:153–9. [Google Scholar]
- 23.Gevers D, Danielsen M, Huys G, Swings J. Molecular characterization of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl Environ Microbiol. 2003;69:1270–5. doi: 10.1128/AEM.69.2.1270-1275.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge B, Jiang P, Han F, Saleh NK, Dhiman N, Fedorko DP, et al. Identification and antimicrobial susceptibility of lactic acid bacteria from retail fermented foods. J Food Protect. 2007;70:2606–12. doi: 10.4315/0362-028x-70.11.2606. [DOI] [PubMed] [Google Scholar]
- 25.Kastner S, Perreten V, Bleuler H, Hugenschmidt G, Lacroix C, Meile L. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst Appl Microbiol. 2006;29:145–55. doi: 10.1016/j.syapm.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Jensen LB, Frimodt-Moller N, Aarestrup FM. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol Lett. 1999;170:151–8. doi: 10.1111/j.1574-6968.1999.tb13368.x. [DOI] [PubMed] [Google Scholar]
- 27.Klare I, Konstabel C, Werner G, Huys G, Vankerckhoven V, Kahlmeter G, et al. Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J Antimicrob Chemother. 2007;59:900–12. doi: 10.1093/jac/dkm035. [DOI] [PubMed] [Google Scholar]
- 28.Levesque C, Piche L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–91. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobsen CN, Rosenfeldt Nielsen V, Hayford AE, Moller PL, Michaelsen KF, Paerregaard A, et al. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;65:4949–56. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schillinger U, Lucke FK. Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol. 1989;55:1901–6. doi: 10.1128/aem.55.8.1901-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strus M, Kucharska A, Kukla G, Brzychczy-Wloch M, Maresz K, Heczko PB. The in vitro activity of vaginal Lactobacillus with probiotic properties against Candida . Infect Dis Obstet Gynecol. 2005;13(2):69–75. doi: 10.1080/10647440400028136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Pena MD, Castro-Escarpulli G, Aguilera-Arreola MG. Lactobacillus species isolated from vaginal secretions of healthy and bacterial vaginosis-intermediate Mexican women: a prospective study. BMC Infect Dis. 2013;13:189. doi: 10.1186/1471-2334-13-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pendharkar S, Magopane T, Larsson PG, de Bruyn G, Gray GE, Hammarstrom L, et al. Identification and characterisation of vaginal lactobacilli from South African women. BMC Infect Dis. 2013;13:43. doi: 10.1186/1471-2334-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verstraelen H, Verhelst R, Claeys G, De Backer E, Temmerman M, Vaneechoutte M. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol. 2009;9:116. doi: 10.1186/1471-2180-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borges S, Silva J, Teixeira P. The role of lactobacilli and probiotics in maintaining vaginal health. Arch Gynecol Obstet. 2014;289:479–89. doi: 10.1007/s00404-013-3064-9. [DOI] [PubMed] [Google Scholar]
- 36.Klebanoff SJ, Hillier SL, Eschenbach DA, Waltersdorph AM. Control of the microbial flora of the vagina by H2O2-generating lactobacilli. J Infect Dis. 1991;164:94–100. doi: 10.1093/infdis/164.1.94. [DOI] [PubMed] [Google Scholar]
- 37.Gil NF, Martinez RC, Gomes BC, Nomizo A, De Martinis EC. Vaginal lactobacilli as potential probiotics against Candida spp. Braz J Microbiol. 2010;41:6–14. doi: 10.1590/S1517-83822010000100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoyancheva G, Marzotto M, Dellaglio F, Torriani S. Bacteriocin production and gene sequencing analysis from vaginal Lactobacillus strains. Arch Microbiol. 2014;196:645–53. doi: 10.1007/s00203-014-1003-1. [DOI] [PubMed] [Google Scholar]
- 39.Aroutcheva A, Gariti D, Simon M, Shott S, Faro J, Simoes JA, et al. Defense factors of vaginal lactobacilli. Am J Obstet Gynecol. 2001;185:375–9. doi: 10.1067/mob.2001.115867. [DOI] [PubMed] [Google Scholar]
- 40.Collado MC, Surono I, Meriluoto J, Salminen S. Indigenous dadih lactic acid bacteria: cell-surface properties and interactions with pathogens. J Food Sci. 2007;72:M89–93. doi: 10.1111/j.1750-3841.2007.00294.x. [DOI] [PubMed] [Google Scholar]
- 41.Snydman DR. The safety of probiotics. Clin Infect Dis. 2008;46(Suppl 2):S104–11; discussion S144–51. doi: 10.1086/523331. [DOI] [PubMed] [Google Scholar]
- 42.Delgado S, Florez AB, Mayo B. Antibiotic susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr Microbiol. 2005;50:202–7. doi: 10.1007/s00284-004-4431-3. [DOI] [PubMed] [Google Scholar]
- 43.Bernardeau M, Vernoux JP, Henri-Dubernet S, Gueguen M. Safety assessment of dairy microorganisms: the Lactobacillus genus. Int J Food Microbiol. 2008;126:278–85. doi: 10.1016/j.ijfoodmicro.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 44.Danielsen M, Wind A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int J Food Microbiol. 2003;82:1–11. doi: 10.1016/s0168-1605(02)00254-4. [DOI] [PubMed] [Google Scholar]
- 45.Jacobsen L, Wilcks A, Hammer K, Huys G, Gevers D, Andersen SR. Horizontal transfer of tet(M) and erm(B) resistance plasmids from food strains of Lactobacillus plantarum to Enterococcus faecalis JH2–2 in the gastrointestinal tract of gnotobiotic rats. FEMS Microbiol Ecol. 2007;59:158–66. doi: 10.1111/j.1574-6941.2006.00212.x. [DOI] [PubMed] [Google Scholar]
- 46.Toomey N, Monaghan A, Fanning S, Bolton D. Transfer of antibiotic resistance marker genes between lactic acid bacteria in model rumen and plant environments. Appl Environ Microbiol. 2009;75:3146–52. doi: 10.1128/AEM.02471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JH, Novak JT, Knocke WR, Pruden A. survival of antibiotic resistant bacteria and horizontal gene transfer control antibiotic resistance gene content in anaerobic digesters. Front Microbiol. 2016;7:263. doi: 10.3389/fmicb.2016.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]