Abstract
Introduction
Many second-line treatments for advanced non-small-cell lung cancer (NSCLC) have been assessed in randomised controlled trials, but which treatments work the best remains unclear. Novel treatments are being rapidly developed. We need a comprehensive up-to-date evidence synthesis of all these treatments. We present the protocol for a live cumulative network meta-analysis (NMA) to address this need.
Methods and analysis
We will consider trials of second-line treatments in patients with advanced NSCLC with wild-type or unknown epidermal growth factor receptor status. We will consider any single agent of cytotoxic chemotherapy, targeted therapy, combination of cytotoxic chemotherapy and targeted therapy and any combination of targeted therapies. The primary outcomes will be overall survival and progression-free survival. The live cumulative NMA will be initiated with a NMA and then iterations will be repeated at regular intervals to keep the NMA up-to-date over time. We have defined the update frequency as 4 months, based on an assessment of the pace of evidence production on this topic. Each iteration will consist of six methodological steps: adaptive search for treatments and trials, screening of reports and selection of trials, data extraction, assessment of risk of bias, update of the network of trials and synthesis, and dissemination. We will set up a research community in lung cancer, with different groups of contributors of different skills. We will distribute tasks through online crowdsourcing. This proof-of-concept study in second-line treatments of advanced NSCLC will allow one for assessing the feasibility of live cumulative NMA and opening the path for this new form of synthesis.
Ethics and dissemination
Ethical approval is not required because our study will not include confidential participant data and interventions. The description of all the steps and the results of this live cumulative NMA will be available online.
Trial registration number
CRD42015017592.
Keywords: Non-small cell lung cancer, systematic reviews, crowdsourcing
Strengths and limitations of this study.
This proof-of-concept study will be the first live cumulative network meta-analysis (NMA) evaluating second-line treatments in advanced non-small-cell lung cancer (NSCLC) with wild-type or unknown status for epidermal growth factor receptor.
It will address the current gaps by providing a continuously updated panorama of the randomised evidence for all available second-line treatments in advanced NSCLC.
This new approach to evidence synthesis will provide physicians and patients with different levels of information to guide clinical decision-making.
The involvement of a research community in lung cancer will increase the clinical relevance, methodological rigour and practical feasibility of this live cumulative NMA.
A challenge may be the end-user acceptability of this new approach but open access to all data would allow reanalyses for the whole set or subsets of trials.
Introduction
Non-small-cell lung cancer (NSCLC) represents 85% of lung cancer and remains the leading cause of cancer-related deaths worldwide.1 More than 40 second-line treatments have been evaluated over the past decade and their number is continually increasing. As an example, in 2015, the US Food and Drug Administration approved two new treatments: nivolumab, an immune checkpoint inhibitor, and ramucirumab, a vascular endothelial growth factor (VEGF) inhibitor, in combination with docetaxel.2 3 Clinicians and patients who must take medical decisions need to know which treatments work best among all available treatments. They increasingly turn to meta-analyses (MAs) but MAs do not provide an exhaustive up-to-date synthesis of all available treatments and thus prevent from answering easily their questions of interest.
In fact, MAs assess direct comparisons between two treatments and thus focus on specific parts of the existing evidence.4 We have previously shown that, when considered collectively, 29 systematic reviews of second-line treatments in advanced NSCLC published from 2001 to 2015 did not encompass the whole available randomised evidence, with more than 40% of treatments, treatment comparisons and trials missing.5 There are no broad MAs encompassing all available treatments, and which treatments work the best remains unclear.
Moreover, all direct comparisons between available treatments are typically not assessed in randomised controlled trials. A solution could be provided by network meta-analysis (NMA), which allows for comparing all treatments with each other, even if randomised controlled trials are not available for some treatment comparisons.6 To the best of our knowledge, such an NMA is not available for second-line treatments of advanced NSCLC. Two previous NMA have focused on small subsets of treatments (four and six treatments, respectively).7 8
Another potential concern is that when MAs exist, only very few are updated. However, according to the clinical area, a MA may become quickly out-of-date. In a sample of 100 MAs indexed in American College of Physicians (ACP) Journal Club, about one-quarter were out-of-date within 2 years of publication.9 In second-line treatments of advanced NSCLC, clinically important randomised evidence appears much more rapidly.5
Considering the update concern, two types of solutions have been proposed: rapid reviews and living systematic reviews. Rapid reviews streamline traditional systematic review methods in order to synthesise evidence within a shortened timeframe.10 Living systematic reviews are online summaries of health research updated as new research becomes available.11 Rapid updates are particularly needed in therapeutic fields such as advanced NSCLC, in which new second-line treatments emerge quickly.
To account for the need to cover all available evidence, address the lack of some treatment comparisons and to update constantly, we have proposed a new paradigm called ‘live cumulative NMA’. The paradigm consists of a single systematic review and evidence synthesis (including MAs and NMAs) encompassing the whole randomised evidence for all available treatments in a specific condition and continuously updated.5 12
We report the protocol of a live cumulative NMA assessing the relative efficacy and safety of all second-line treatments for advanced NSCLC in patients with wild-type or unknown status for epidermal growth factor receptor (EGFR). We present the different methodological steps and describe the crowdsourcing of a research community in lung cancer for the update process.
Methods and analysis
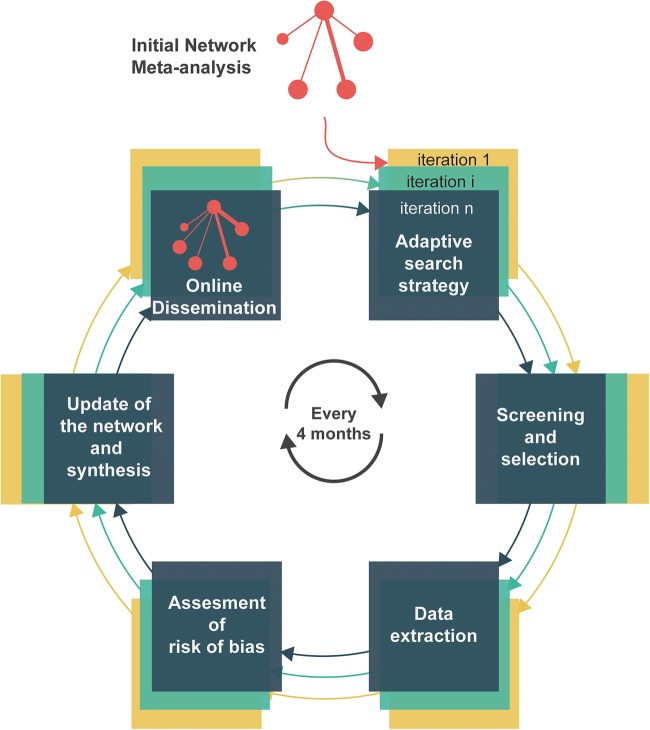
Figure 1 describes the process of a live cumulative NMA of randomised controlled trials. It is initiated with a conventional NMA. Six methodological steps are then repeated at regular intervals to update the NMA over time: adaptive search for treatments and trials, crowd-sourced screening of reports and selection of trials, data extraction, assessment of risk of bias, update of the network of trials and synthesis, and finally dissemination. Here, we present the prespecified methods for the initial NMA and then the methods for iterations. These iterations will be performed by a research community in lung cancer.
Figure 1.
Principles of live cumulative network meta-analysis. Live cumulative network meta-analysis is initiated with an initial network meta-analysis, then six methodological steps are repeated every 4 months. Step 1 is detailed in figure 2. Steps 3–6 are not required if no new trial is available.
Initial NMA
The prespecified methods follow the recommendations of the Cochrane Comparing Multiple Interventions Methods Group13 and were informed by the PRISMA extension statement for systematic reviews incorporating NMAs.14 The initial NMA was registered at PROSPERO (CRD42015017592).
Criteria for considering trials for this review
Types of studies and participants
We will consider randomised trials in patients with advanced NSCLC with wild-type or unknown status for EGFR receiving second-line treatment. We will exclude trials focusing exclusively on patients with EGFR-activating mutation or anaplastic lymphoma kinase rearrangement, because it represents a specific minority subgroup of all advanced NSCLC.15
Types of interventions
We will consider any treatment for second-line of advanced NSCLC with wild-type or unknown status for EGFR. We list eligible treatments that we have identified so far in online supplementary appendix 1; we cannot predict future treatments, we will consider them for inclusion. Treatments can be categorised into: monochemotherapy; targeted therapy (eg, targeting EGFR, VEGF receptor and multitargeted tyrosine kinase inhibitors); combination of a cytotoxic chemotherapy and a targeted therapy and finally combination of two targeted therapies. Of note, targeted therapies include immunotherapies such as treatments targeting programmed death 1 (PD-1) and its ligand PD-L1. We will exclude trials assessing a combination of two cytotoxic drugs because: (1) monochemotherapy is currently the standard treatment for second-line treatment by the American Society of Clinical Oncology (ASCO) Clinical Practice Guideline;16 and (2) in Di Maio MA of individual patient data which analysed six trials (847 patients), doublet chemotherapy increased response rate and progression-free survival (PFS) but was more toxic and did not improve overall survival (OS) compared with single agent.17
Trials assessing second-line treatments versus each other or placebo or best supportive care will be eligible. We will consider trials of second-line therapy and trials including both second-line and third-line therapy, because there is no clinical reason to presume that patients in third-line could not be randomised to any of the treatments (ie, with respect to the transitivity assumption).
bmjopen-2016-011841supp_appendix.pdf (234.4KB, pdf)
Types of outcome measures
The primary outcomes will be OS and PFS.
The secondary outcomes will be:
Objective response defined as a complete response or a partial response, according to the Response Evaluation Criteria in Solid Tumors (RECIST).18
Toxicity evaluated by serious adverse events (SAEs) as defined on ClinicalTrials.gov (https://clinicaltrials.gov/ct2/help/glossary/serious-adverse-event).
And quality of life (QoL), whatever the scale reported in trials.
Search methods for identification of trials
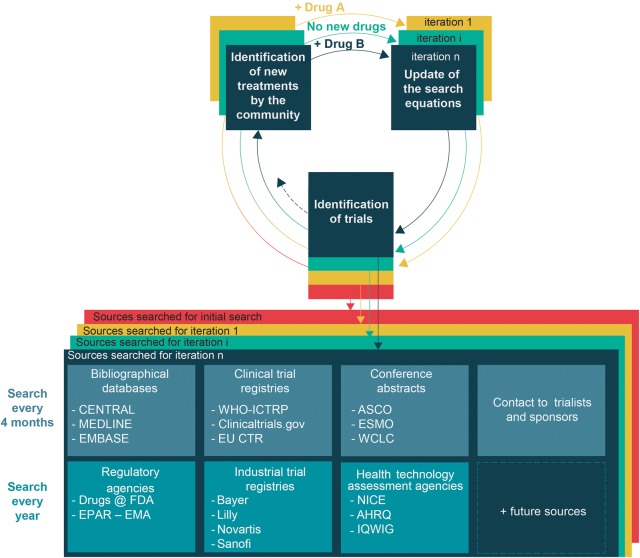
We have designed an exhaustive search strategy according to high standards to identify published and unpublished trials.19 We will search a range of bibliographic databases (CENTRAL, MEDLINE, EMBASE) with no restriction on language, status or year of publication (for search equations see online supplementary appendix 1). We will screen the reference lists of all selected trials and the list of trials selected in previous systematic reviews. We will also search additional sources (figure 2).
Figure 2.
Adaptive search strategy. These different sources will be searched for the initial network meta-analysis and for each iteration. A research community interested in lung cancer will identify new second-line treatments for advanced NSCLC. The search strategy (ie, specific requests for querying the different sources) will be updated over time to identify trials assessing these new treatments. We will also update this adaptive search strategy by querying new sources when they become available (eg, the OpenTrials database50). We will also consider clinical trial data sharing repositories (eg, Clinical Study Data Request or Yale University Open Data Access Project) as potential sources to identify some unpublished trials. AHRQ, Agency for Healthcare Research and Quality; ASCO, American Society of Clinical Oncology; CENTRAL, Cochrane Central Register of Controlled Trials; EU CTR, European Union Clinical Trials Register; EPAR-EMA, European Public Assessment Reports-European Medicines Agency; ESMO, European Society of Medical Oncology; FDA, Food and Drug Administration; IQWIG, Institute for Quality and Efficiency in Health Care; NICE, National Institute for Health and Care Excellence; NSCLC, non-small-cell lung cancer; WCLC, World Conference on Lung Cancer; WHO ICTRP, WHO International Clinical Trials Registry Platform.
Selection of trials
Two reviewers will independently and in duplicate examine titles and abstracts to exclude obvious irrelevant reports. They will then independently examine full-text articles to determine eligibility. Trial authors will be contacted for clarification when necessary. Disagreements will be discussed with a third reviewer. They will document the primary reasons for exclusion.
Data extraction and management
All data will be independently extracted by two reviewers using a standardised form. Disagreements will be discussed with a third reviewer. In case of several reports pertaining to the same trial, they will extract data from the different sources, compare them and in case of discrepancies, will give priority to the first available source among regulatory agency reports, results posted at ClinicalTrials.gov, full-text articles, pharmaceutical reports and conference abstracts. The authors of the trials will be contacted to provide missing data if needed.
For each trial, they will extract the study phase; single-centre or multicentre status; funding source (private, public, both or unclear); number of randomised patients; drugs, dosage, frequency and modality of administration; patient age; stage (IIIB vs IV); performance status (0–1 vs 2); history of smoking (never-smoker vs former or current smoker); proportion of patients with second-line treatment; outcome data: HRs for PFS and OS and their 95% CIs; number of patients with an objective response, number of patients with SAEs and means and SDs for QoL. They will also extract data on trial population characteristics that may act as treatment effect modifiers: gender, histology (non-squamous vs squamous), ethnicity (Asian vs Caucasian) and EGFR mutation status (wild-type vs unknown status).
Geometry of the network
We will produce diagrams to show the amount of evidence in the network of randomised trials for each outcome (OS, PFS, objective response, SAEs and QoL). Each node will be a treatment. An edge will connect two nodes when at least one trial compared the two corresponding treatments. The node size will be proportional to the number of patients randomly allocated to the corresponding treatment and the edge width to the total number of trials between the corresponding treatments.
Assessment of risk of bias in included trials
Two reviewers will assess the risk of bias by using the Cochrane Risk of Bias Tool.20 Disagreements will be discussed with a third reviewer. They will separately assess blinding for objective outcomes (OS) and subjective outcomes (PFS, objective response, SAEs and QoL). Blinding of patients and care providers will be considered at ‘low risk’ if blinding was insured or if the outcome was unlikely to be affected by lack of blinding (OS) and at ‘high risk’ for subjective outcomes if blinding was lacking. Blinding of outcome assessors will be considered at ‘low risk’ if blinding was insured or for an objective outcome; it will be considered at ‘low risk’ if an independent Clinical Endpoint Adjudication Committee assessed subjective outcomes and at ‘high risk’ otherwise.
Measures of treatment effect
For time-to-event end points (OS, PFS), we will use HRs. When HRs are unavailable from trial reports, we will reconstruct individual survival patient data from published Kaplan-Meier curves and will estimate HRs.21 For dichotomous outcomes (objective response, SAEs), we will use ORs. For continuous outcomes (QoL), standardised mean differences will be used.
Data synthesis
We will assess clinical and methodological diversity by comparing summary characteristics of trials and study populations across the different pairwise comparisons between treatments. The assumption of transitivity will be evaluated by comparing the distribution of the potential effect modifiers across different pairwise comparisons.
Two analyses will be performed: first, we will compare individual treatments with each other, and second we will compare the different categories of second-line treatments previously mentioned (monochemotherapy, targeted therapy, combination of a monochemotherapy and a targeted therapy, combination of two targeted therapies and placebo). For this second analysis, trials in which patients in the control arm receive chemotherapy (eg, docetaxel or pemetrexed) at the investigators' discretion will be included.
We will perform pairwise and NMAs by using random-effects models with a Bayesian approach. We will estimate mean relative treatment effects as well as the associated 95% credible intervals. For NMAs, we will calculate the area under the cumulative probability curve (surface under the cumulative ranking curve) and associated credible intervals.22 The statistical homogeneity of results will be assessed by Higgins and Thompson's I2 statistic and then between-trial variance estimates (τ2). For pairwise MA, we will estimate different heterogeneity variances for each pairwise comparison. For NMAs, we will assume a common estimate for the heterogeneity variance across the different comparisons. The magnitude of the heterogeneity parameter will be assessed by comparison with empirical distributions.23–25 We will also assess statistical inconsistency by using state-of-the-art methods based on loop-specific and global network approaches.26
If enough trials are available, we will perform network metaregression or subgroup analyses by using the following effect modifiers as possible sources of heterogeneity and inconsistency between direct and indirect evidence: gender, histology, ethnicity and EGFR status (EGFR wild-type vs unknown).
Successive iterations to keep the NMA up-to-date
In the past 5 years, about two new treatments were analysed in randomised trials of second-line treatments for advanced NSCLC every 4 months.5 Therefore, we must cover the whole randomised evidence for all treatments for this condition but also keep this exhaustive synthesis up-to-date according to the pace of evidence generation.
The six methodological steps will be repeated every 4 months to keep the NMA up-to-date. We have designed these iterations and the update frequency to maintain the high-quality standards of a systematic review and to ensure that they are feasible. Although automated methods are being developed to facilitate systematic reviews, we chose to perform these iterations with manual processes as currently recommended to ensure high quality.27 28 Moreover, we set the update frequency after an assessment of the rhythm of randomised evidence production and the amount of work required to select trials and extract data.5 For instance, we estimated that, at each iteration, the reviewers would have fewer than 400 records to screen and about 4 randomised trials to extract data from, so the anticipated workload is manageable.5
Finally, an innovative aspect will be the creation and involvement of a research community in lung cancer. The involvement of such a community would increase the clinical relevance, methodological rigour and practical feasibility of the NMA. This community will consist of different groups with different skills. In the largest group, people interested in lung cancer will be able to voluntarily report new treatments and new (planned, ongoing, completed) trials of second-line treatments for advanced NSCLC. Second, an open group of international experts in lung cancer (clinicians, triallists or members of cooperative oncology groups) will validate the methodological choices and will be involved in the screening and selection step. For instance, the group would validate selection criteria for population, treatments and outcomes. Finally, a smaller open group of researchers with expertise in systematic review methodology (risk-of-bias assessment and data extraction) and in statistical MA methods, will execute the remaining review steps. The tasks will be distributed to individuals via crowdsourcing to reduce the workload. For instance, records needed to be screened will be allocated at random among experts in lung cancer so that each record is assessed by two independent experts in duplicate.
Adaptive search for new treatments and trials
Contrary to a living systematic review that focuses on a comparison between two treatments, a live cumulative NMA covers the whole evidence for all treatments. Therefore, it needs to continuously identify new evidence for treatments already in the network of trials but also novel treatments. Indeed, several new treatments of the network are already being assessed in planned or ongoing trials, as in the Lung-MAP SWOG S1400 trial.29 Novel treatments can correspond to a new single drug, a combination of a new drug and a previous drug, or a new combination of previous drugs. Thus, an adaptive search strategy will be created to continuously cover all available second-line treatments for advanced NSCLC (figure 2).
We have set up a website at which anyone interested in lung cancer may report new treatments assessed in randomised controlled trials (http://livenetworkmetaanalysis.com/). The community of experts in lung cancer will also monitor various sources to identify novel treatments and will validate treatments notified by the community of people interested in lung cancer.
The search strategy will be then updated by adding relevant keywords so that specific requests of the various sources capture randomised trials of these new treatments (figure 2).
As for the initial NMA, the different sources described in figure 2 will be searched with the last updated search strategy. The search will run from the previous iteration to the current one (covering a 4-month interval) for bibliographical databases, clinical trial registries and conference proceedings. With the search equation being known for each source, a script (html extraction by automated http requests) will be used to automatically and simultaneously query the multiple sources every 4 months.30 For trials identified as completed in clinical trial registries but without posted results or those identified only by a conference proceeding, some trained reviewers will contact triallists to request complete results. A personalised email will be sent with systematic reminders.31
Some experts in lung cancer will search other sources (regulatory agencies, industry trial registries and health technology assessment agencies) once a year. For those sources and some conference proceedings (eg, European Society for Medical Oncology), automated querying cannot be used, and the search will still rely on a manual process.
Screening of reports and selection of trials
We have estimated that there will be about 400 records needed to be screened every 4 months.5 We will apply crowdsourcing of experts in lung cancer: we will distribute these records between experts so that each report will be screened three times. For a group of 12 experts, each will have to screen around 100 records at each iteration. For records not having been twice screened or in case of disagreements, a trained reviewer will be involved and make the final decision.
Since trials may be reported in several articles, abstracts or other reports, some trained reviewers will always check if a new trial report can be linked to a previous report of the same trial. In our previous study, we found a median of two reports for each trial, published about 12 months apart, which corresponds to three iterations.5 Some of the trained reviewers will be asked to link the multiple reports together. A list of all included and excluded trials will be provided with reasons for exclusion. If at least one trial with new results is selected, the subsequent steps will be performed: data extraction, assessment of risk of bias, update of the network of trials and synthesis, and dissemination.
Data extraction
This step will be performed by two of the trained reviewers. The method for extraction will be as previously described except for results posted on ClinicalTrials.gov. In this case, an automatic data extraction process will be used to automatically abstract posted results from ClinicalTrials.gov.32
Assessment of risk of bias
Two of the trained reviewers will assess the risk of bias and discuss disagreements with a third reviewer. Although automated methods such as text mining can assist with risk-of-bias assessments, we opted for a manual approach, as currently recommended.27 28 33 34 Indeed, risk-of-bias assessments may rely on other sources than the published article such as protocols, whereas the automated tools rely on articles only.
Update of the network of trials and synthesis
Every 4 months, each newly identified trials will be incorporated in the network (ie, one network for each outcome (OS, PFS, objective response, SAEs and QoL)). We have estimated that two new treatments will appear every 4 months. Therefore, at each iteration, the NMA will allow for estimating all comparisons between these two new treatments and other treatments already in the network.
A common issue in NMA is the definition of nodes. In fact, treatments assessed in trials may be similar but not identical (eg, different drug administration schedules). The community of experts will be asked, via a group consensus method, if each newly identified treatment will correspond to a new node or to a pre-existing node.35 36 For instance, a drug administered every 3 weeks may be lumped together with the pre-existing node with administration every week. The experts will also be involved in validating changes from the protocol, including decisions about eligibility criteria (eg, Are enriched trials of patients with tumours positive for PI3KCA eligible or not?) or about subgroup analyses.29
The data will be reanalysed every 4 months. New NMAs will be performed by using a Bayesian approach. Since Bayesian inference is not affected by repeated updates, adjustment for multiple testing will not be incorporated to account for the inflated type I error.37 38
Dissemination
In addition to a classical article for dissemination of the live cumulative NMA results, the findings will be disseminated via an open access website so that they can be useful for the community.39 The results will be presented in terms of tables and figures, ensuring that sufficient information is presented to render the paper informative so that the live cumulative NMA becomes a useful tool to help medical decision-making with different levels of information provided. First, the amount of randomised evidence in terms of network graphs for each outcome and at each iteration how the networks of evidence evolve over time will be shown. Second, treatment effects by forest plots, league tables and reporting of treatment rankings will be presented. Third, elements to allow readers to evaluate their level of confidence in the results will be provided, such as assessments of consistency and the risk of bias. To ensure a transparent process, an open access to the protocol (and its amendments), statistical codes, the screening and selection elements (flow diagram, list of included trials, list of excluded trials with reasons for exclusion) and archives of previous iterations will be given. Finally, the characteristics and results of included trials will be detailed to allow for an evaluation of clinical diversity and transitivity. The ability to post comments and discussion will be provided.
Discussion
To the best of our knowledge, we present the first protocol for a live cumulative NMA assessing the relative efficacy and safety of all second-line treatments available for advanced NSCLC in patients with wild-type or unknown EGFR status.
We chose to update the network every 4 months considering the pace of evidence. Once the initial NMA is performed, maintaining the evidence up-to-date over time seems manageable and a reasonable investment over time as compared with the substantial cost of producing the initial synthesis. Indeed, for our previous search up to March 2015, we screened about 8000 records and extracted data for 77 trials, which required a substantial amount of researcher working time; compared with this initial NMA, each iteration would represent about 5% in terms of records needed to screen and trials to extract. Some methodological choices could be adapted during the process, such as the update frequency. One practical issue may be the need to adapt the frequency of updates over time according to the pace of evidence generation, and once some definite conclusions regarding some specific subset of treatments will be found.
The concept of live cumulative NMA allows for moving from a series of MAs (focusing on the comparison between two treatments, at risk of ignoring novel treatments, frequently out-of-date) to a cumulative network of randomised evidence updated shortly after new evidence becomes available. Beyond the question at hand, this new form of synthesis answers the real questions of interest for clinicians, patients and decision-makers. As long as this paradigm of live cumulative NMA is not adopted broadly, there is a substantial risk that randomised evidence is wasted.5
We designed this live cumulative NMA in the field of thoracic oncology as a ‘proof-of-concept’ study. Of note, we will document all practical issues and difficulties encountered to demonstrate that this type of synthesis is feasible. We are aware of many challenges, such as achieving the trade-off between the quality of the evidence synthesis and the time and cost required to perform it. We suggest setting up a research community interested in lung cancer with a partitioning of review tasks via online crowdsourcing to facilitate some steps of live cumulative NMAs. Automated methods are being developed and could further facilitate the conduct of multiple live cumulative NMAs.30 33 34 40–51
We expect this pioneering study will open the path to implement live cumulative NMA. If the concept is proven, it could be applied to other clinical questions. Each topic would be handled by a specific community. It would allow for reducing the number of overlapping MAs and waste in research. Using crowdsourcing and crowdtiming may facilitate the commitment of volunteers and experts. For our clinical question, some participants have been identified. Readers willing to contribute can find information at http://livenetworkmetaanalysis.com/.
Acknowledgments
The authors thank Laura Smales (BioMedEditing, Toronto, Canada) for language revision of the manuscript and Elise Diard for the conception of the figures. The authors also thank the Cochrane Lung Cancer group, Professor Virginie Westeel and Professor Jacques Cadranel for their contributions.
Footnotes
Contributors: PC helped design the study, and wrote the draft protocol. PR conceived and designed the study and helped write the draft protocol. LT conceived and designed the study and wrote the draft protocol. All authors read and approved the final protocol.
Funding: This study was supported by a LEGS POIX 2015 grant (from Chancellerie des Universités de Paris), a grant from the French National Cancer Institute (Institut National du Cancer, INCa) and Cochrane France.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: This article is the protocol of a live cumulative NMA. The authors plan to report transparently the list of all selected trials and provide open access to all extracted data for each trial.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R et al. . Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.US Food and Drug Administration. Highlights of prescribing information: CYRAMZA (ramucirumab). http://www.accessdata.fda.gov/drugsatfda_docs/label/2014/125477s007lbl.pdf
- 3.US Food and Drug Administration. Highlights of prescribing information: OPDIVO™ (nivolumab). http://www.accessdata.fda.gov/drugsatfda_docs/label/2015/125527s000lbl.pdf
- 4.Haidich AB, Pilalas D, Contopoulos-Ioannidis DG et al. . Most meta-analyses of drug interventions have narrow scopes and many focus on specific agents. J Clin Epidemiol 2013;66:371–8. 10.1016/j.jclinepi.2012.10.014 [DOI] [PubMed] [Google Scholar]
- 5.Créquit P, Trinquart L, Yavchitz A et al. . Wasted research when systematic reviews fail to provide a complete and up-to-date evidence synthesis: the example of lung cancer. BMC Med 2016;14:8 10.1186/s12916-016-0555-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cipriani A, Higgins JP, Geddes JR et al. . Conceptual and technical challenges in network meta-analysis. Ann Intern Med 2013;159:130–7. 10.7326/0003-4819-159-2-201307160-00008 [DOI] [PubMed] [Google Scholar]
- 7.Hawkins N, Scott DA, Woods BS et al. . No study left behind: a network meta-analysis in non-small-cell lung cancer demonstrating the importance of considering all relevant data. Value Health 2009;12:996–1003. 10.1111/j.1524-4733.2009.00541.x [DOI] [PubMed] [Google Scholar]
- 8.Popat S, Mellemgaard A, Fahrbach K et al. . Nintedanib plus docetaxel as second-line therapy in patients with non-small-cell lung cancer: a network meta-analysis. Future Oncol 2015;11:409–20. 10.2217/fon.14.290 [DOI] [PubMed] [Google Scholar]
- 9.Shojania KG, Sampson M, Ansari MT et al. . How quickly do systematic reviews go out of date? A survival analysis. Ann Intern Med 2007;147:224–33. 10.7326/0003-4819-147-4-200708210-00179 [DOI] [PubMed] [Google Scholar]
- 10.Polisena J, Garritty C, Kamel C et al. . Rapid review programs to support health care and policy decision making: a descriptive analysis of processes and methods. Syst Rev 2015;4:26 10.1186/s13643-015-0022-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott JH, Turner T, Clavisi O et al. . Living systematic reviews: an emerging opportunity to narrow the evidence-practice gap. PLoS Med 2014;11:e1001603 10.1371/journal.pmed.1001603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vandvik PO, Brignardello-Petersen R, Guyatt GH. Living cumulative network meta-analysis to reduce waste in research: a paradigmatic shift for systematic reviews? BMC Med 2016;14:59 10.1186/s12916-016-0596-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comparing Multiple Interventions Methods Group. Protocol template for a Cochrane intervention review that compares multiple interventions 2014. http://methods.cochrane.org/cmi/sites/methods.cochrane.org.cmi/files/uploads/Protocol%20for%20Cochrane%20Reviews%20with%20Multiple%20Interventions.pdf
- 14.Hutton B, Salanti G, Caldwell DM et al. . The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: checklist and explanations. Ann Intern Med 2015;162:777–84. 10.7326/M14-2385 [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Chavez A, Thomas A, Rajan A et al. . Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology phase II basket trial. J Clin Oncol 2015;33:1000–7. 10.1200/JCO.2014.58.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masters GA, Temin S, Azzoli CG et al. . Systemic therapy for stage IV non-small-cell lung cancer: American Society of Clinical Oncology clinical practice guideline update. J Clin Oncol 2015;33:3488–515. 10.1200/JCO.2015.62.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Maio M, Chiodini P, Georgoulias V et al. . Meta-analysis of single-agent chemotherapy compared with combination chemotherapy as second-line treatment of advanced non-small-cell lung cancer. J Clin Oncol 2009;27:1836–43. 10.1200/JCO.2008.17.5844 [DOI] [PubMed] [Google Scholar]
- 18.Therasse P, Arbuck SG, Eisenhauer EA et al. . New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000;92:205–16. 10.1093/jnci/92.3.205 [DOI] [PubMed] [Google Scholar]
- 19.Chan AW. Out of sight but not out of mind: how to search for unpublished clinical trial evidence. BMJ 2012;344:d8013 10.1136/bmj.d8013 [DOI] [PubMed] [Google Scholar]
- 20.Higgins JP, Altman DG, Gøtzsche PC et al. . The Cochrane Collaboration's tool for assessing risk of bias in randomised trials. BMJ 2011;343:d5928 10.1136/bmj.d5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guyot P, Ades AE, Ouwens MJ et al. . Enhanced secondary analysis of survival data: reconstructing the data from published Kaplan-Meier survival curves. BMC Med Res Methodol 2012;12:9 10.1186/1471-2288-12-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salanti G, Ades AE, Ioannidis JPA. Graphical methods and numerical summaries for presenting results from multiple-treatment meta-analysis: an overview and tutorial. J Clin Epidemiol 2011;64:163–71. 10.1016/j.jclinepi.2010.03.016 [DOI] [PubMed] [Google Scholar]
- 23.Turner RM, Davey J, Clarke MJ et al. . Predicting the extent of heterogeneity in meta-analysis, using empirical data from the Cochrane Database of Systematic Reviews. Int J Epidemiol 2012;41:818–27. 10.1093/ije/dys041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner RM, Jackson D, Wei Y et al. . Predictive distributions for between-study heterogeneity and simple methods for their application in Bayesian meta-analysis. Stat Med 2015;34:984–98. 10.1002/sim.6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhodes KM, Turner RM, Higgins JP. Predictive distributions were developed for the extent of heterogeneity in meta-analyses of continuous outcome data. J Clin Epidemiol 2015;68:52–60. 10.1016/j.jclinepi.2014.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dias S, Welton NJ, Sutton AJ et al. . Evidence synthesis for decision making 4: inconsistency in networks of evidence based on randomized controlled trials. Med Decis Making 2013;33:641–56. 10.1177/0272989X12455847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moher D, Liberati A, Tetzlaff J et al. . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 2009;339:b2535 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins JP, Green S. Cochrane handbook for systematic reviews of interventions. Updated March 2011. http://handbook.cochrane.org [Google Scholar]
- 29.Decatris MP, Farrugia D, O'Byrne KJ. Clinician perspective on molecular profiling of non-small-cell lung cancer. J Clin Oncol 2016;34:884–6. 10.1200/JCO.2015.65.2040 [DOI] [PubMed] [Google Scholar]
- 30.Smalheiser NR, Lin C, Jia L et al. . Design and implementation of Metta, a metasearch engine for biomedical literature retrieval intended for systematic reviewers. Health Inf Sci Syst 2014;2:1 10.1186/2047-2501-2-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maruani A, Boutron I, Baron G et al. . Impact of sending email reminders of the legal requirement for posting results on ClinicalTrials.gov: cohort embedded pragmatic randomized controlled trial. BMJ 2014;349:g5579 10.1136/bmj.g5579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tasneem A, Aberle L, Ananth H et al. . The database for aggregate analysis of ClinicalTrials.gov (AACT) and subsequent regrouping by clinical specialty. PLoS ONE 2012;7:e33677 10.1371/journal.pone.0033677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall IJ, Kuiper J, Wallace BC. Automating risk of bias assessment for clinical trials. IEEE J Biomed Health Inform 2015;19:1406–12. 10.1109/JBHI.2015.2431314 [DOI] [PubMed] [Google Scholar]
- 34.Millard LA, Flach PA, Higgins JP. Machine learning to assist risk-of-bias assessments in systematic reviews. Int J Epidemiol 2016;45:266–77. 10.1093/ije/dyv306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Giovane C, Vacchi L, Mavridis D et al. . Network meta-analysis models to account for variability in treatment definitions: application to dose effects. Stat Med 2013;32:25–39. 10.1002/sim.5512 [DOI] [PubMed] [Google Scholar]
- 36.Mosseri J, Trinquart L, Nizard R et al. . Meta-analysis of a complex network of non-pharmacological interventions: the example of femoral neck fracture. PLoS ONE 2016;11:e0146336 10.1371/journal.pone.0146336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higgins JP, Whitehead A, Simmonds M. Sequential methods for random-effects meta-analysis. Stat Med 2011;30:903–21. 10.1002/sim.4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolakopoulou A, Mavridis D, Salanti G. Prospective use of network meta-analysis using formal statistical monitoring. 36th Annual Conference of the International Society for Clinical Biostatistics; 23–27 August 2015 Utrecht, The Netherlands. [Google Scholar]
- 39.Pulverer B. EMBO Press—a new way to publish. EMBO J 2014;33:1–2. 10.1002/embj.201387566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rathbone J, Hoffmann T, Glasziou P. Faster title and abstract screening? Evaluating Abstrackr, a semi-automated online screening program for systematic reviewers. Syst Rev 2015;4:80 10.1186/s13643-015-0067-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsafnat G, Dunn A, Glasziou P et al. . The automation of systematic reviews. BMJ 2013;346:f139 10.1136/bmj.f139 [DOI] [PubMed] [Google Scholar]
- 42.Tsafnat G, Glasziou P, Choong MK et al. . Systematic review automation technologies. Syst Rev 2014;3:74 10.1186/2046-4053-3-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miwa M, Thomas J, O'Mara-Eves A et al. . Reducing systematic review workload through certainty-based screening. J Biomed Inform 2014;51:242–53. 10.1016/j.jbi.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Mara-Eves A, Thomas J, McNaught J et al. . Using text mining for study identification in systematic reviews: a systematic review of current approaches. Syst Rev 2015;4:5 10.1186/2046-4053-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace BC, Trikalinos TA, Lau J et al. . Semi-automated screening of biomedical citations for systematic reviews. BMC Bioinformatics 2010;11:55 10.1186/1471-2105-11-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llewellyn A, Whittington C, Stewart G et al. . The use of Bayesian networks to assess the quality of evidence from research synthesis: 2. Inter-rater reliability and comparison with standard GRADE assessment. PLoS ONE 2015;10:e0123511 10.1371/journal.pone.0123511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart GB, Higgins JP, Schünemann H et al. . The use of Bayesian networks to assess the quality of evidence from research synthesis: 1. PLoS ONE 2015;10:e0114497 10.1371/journal.pone.0114497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shanahan DR. A living document: reincarnating the research article. Trials 2015;16:151 10.1186/s13063-015-0666-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Open Meta Analysis. http://openmetaanalysis.github.io.
- 50. OpenTrials | All the Data oAtT, Linked. http://opentrials.net.
- 51.Mo Y, Kontonatsios G, Ananiadou S. Supporting systematic reviews using LDA-based document representations. Syst Rev 2015;4:172 10.1186/s13643-015-0117-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2016-011841supp_appendix.pdf (234.4KB, pdf)